Abstract
The Saccharomyces Genome Database (SGD) is the main community repository of information for the budding yeast, Saccharomyces cerevisiae. The SGD has collected published results on chromosomal features, including genes and their products, and has become an encyclopedia of information on the biology of the yeast cell. This information includes gene and gene product function, phenotype, interactions, regulation, complexes, and pathways. All information has been integrated into a unique web resource, accessible via http://yeastgenome.org. The website also provides custom tools to allow useful searches and visualization of data. The experimentally defined functions of genes, mutant phenotypes, and sequence homologies archived in the SGD provide a platform for understanding many fields of biological research. The mission of SGD is to provide public access to all published experimental results on yeast to aid life science students, educators, and researchers. As such, the SGD has become an essential tool for the design of experiments and for the analysis of experimental results.
INTRODUCTION
The wisdom of the budding yeast research community is represented by the body of experimental work published in the last five decades. Access to these results is available to anyone who has the opportunity to explore this literature and a lot of time. Reviews on specific areas of yeast biology are also available; however, for the student of yeast biology, it is a challenge to digest the literature. What if a researcher is interested in only a small portion of what is known on a particular topic or gene, the relevant information being spread across many papers? What about bioinformaticists or computer scientists who wish to use computational analyses to explore results that are in the supplemental pages of many papers? The answer to accessing relevant information is an encyclopedic database of integrated experimental results that is annotated by expert biocurators. This database is the Saccharomyces Genome Database (SGD; www.yeastgenome.org; Cherry et al. 2012).
Historically, the core of the SGD comprises chromosomal features—defined regions of the chromosome that are associated with a function or product. Information on protein-coding genes and non-protein-coding RNA genes, such as tRNA and rRNA genes, is typically what is available. The basic entry point is a gene name that leads directly to summary information about the locus. A keyword describing a function, phenotype, selective condition, or text from an abstract will also provide entry into SGD. Gene or chromosomal regions can be identified for exploration using a DNA or protein sequence with an integrated BLAST sequence search tool. Unique identifiers from many sources also provide valid entry points, such as protein and DNA sequence accession numbers, PubMed and NCBI identifiers, author names, and Gene Ontology (GO) function terms. The information provided by SGD has been gathered and is maintained by a group of scientists working as biocurators and software developers. This team is devoted to providing users with up-to-date information and connections to all the major research resources and tools that allow previously published data to be explored and the user’s own results to be interpreted. The goal is to aid experimental design and the analysis of results as well as to facilitate education and further study of budding yeast.
SGD MAINTAINS THE S. cerevisiae REFERENCE GENOME SEQUENCE
The SGD is the keeper of S. cerevisiae nomenclature and reference genome sequence (Engel et al. 2014). It has been responsible for maintaining this important community documentation since 1996. The reference genome has been updated several times since its first release in 1996, typically for a single region only after specific rules were met; for example, both strands from a region in the S288C strain were sequenced to confirm that a change was necessary. As described by Engel et al. (2014), a major update of the reference genome sequence occurred in 2010 to incorporate data generated by next-generation sequencing from a single colony. The sequence of this reference is deemed to be of such high quality that now only gross changes to the sequence will be considered. This is because any single colony is likely to have at least one nucleotide difference compared with its parent, excluding any sequence differences that arise because of technical errors. Thus, it is appropriate to assume that any minor differences observed are the result of allelic variation compared to the reference.
ANNOTATIONS AND ONTOLOGIES IN THE SGD
The SGD provides thousands of annotations for yeast genes and their products. Experimental results are captured as annotations using detailed precise vocabulary, typically in the form of an ontology, and include details of an experiment’s evidence and methods and the appropriate literature citation. An ontology is a highly structured form of controlled vocabulary. Each entry in the ontology is commonly called a term. The name of the term is a short descriptive phrase, such as “carnitine dehydratase activity.” However, each term also has a definition, similar to the definition of an English word found within a dictionary, which provides the complete usage and detailed explanation of the word/term. It is critical to consult the term’s definition because the distinction between terms can be subtle. One important difference between the definition of an ontology term and that of an English word is that the term has only one meaning; however, there can be synonyms. The creation of ontology terms and their definitions often involves debate but the result has been a descriptive language. The use of ontologies has been successful in unifying communication between scientific communities and in providing a standard dictionary for topics such as molecular functions, biological processes, mutant phenotypes, chemical properties, and structures. In addition to terms and definitions, ontologies require a relationship between terms to define the type of connection (relationship). In an ontology, a term can have more than one parent term, the term above it in the ontology, as well as more than one child, terms below it in the ontology. GO is a system used to describe gene function and is used extensively in SGD annotations. Further details on the GO project are available from several sources (Gene Ontology Consortium 2013). GO annotations are often used to illustrate the structure of an ontology, but many other ontologies are also used to construct annotations in SGD, as is the case in all modern biological databases.
SGD USER INTERFACE
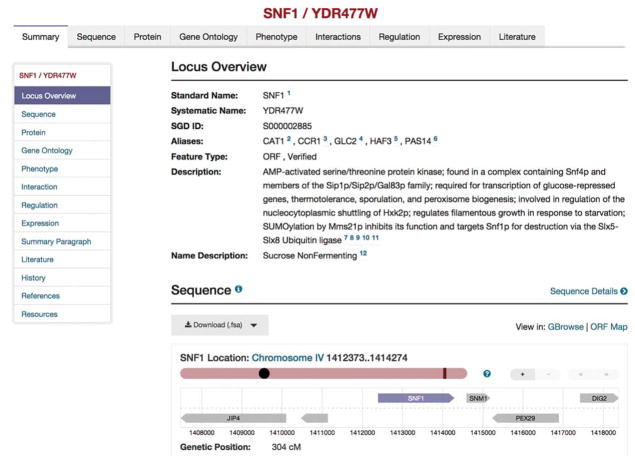
The main entry point into SGD is the home page, www.yeastgenome.org. Across the top of all SGD pages is a purple bar with several features collected into groups of similar purpose. For example, the drop-down menus, “Analyze” for computational tools or “Function” for collections of experimental results such as microarray expression or mutant phenotype data, can be selected. Also at the upper right corner of every page is located the “Search” box. Here text can be entered to search the contents of the database. For example, if the word “actin” is slowly typed into the search box a list appears and changes with the addition of each letter. Entry of “actin” should bring down many phrases that contain “actin.” The results of a search include all the entries for all types of information found within SGD that matches the input string. Selecting a search result takes the user to the relevant page; for example, selecting the “actin” search result, ACT1, brings up the Locus Overview page, which presents the collected information on ACT1 and its product. The locus overview page for another gene, SNF1 (AMP-activated serine/threonine protein kinase), is illustrated in Figure 1. All of the information provided by SGD via its web pages or search tools can be downloaded from its dedicated file retrieval site by selecting the “Download” button at the top of every SGD page. This takes the user to http://downloads.yeastgenome.org.
FIGURE 1.
Locus summary page for SNF1. The locus summary page provides a summary of the information that has been compiled for a gene. Complete information about the gene is available within the topic pages, listed as tabs across the top of the window. Each page also provides quick navigation within the page via the left-hand menu.
The accompanying protocols provide guidance on navigating SGD. See Protocol: The Saccharomyces Genome Database: Exploring Biochemical Pathways and Mutant Phenotypes (Cherry 2015a), Protocol: The Saccharomyces Genome Database: Advanced Searching Methods and Data Mining (Cherry 2015b), Protocol: The Saccharomyces Genome Database: Gene Product Annotation of Function, Process, and Component (Cherry 2015c), and Protocol: The Saccharomyces Genome Database: Exploring Genome Features and Their Annotations (Cherry 2015d).
Acknowledgments
I am grateful to all the present and past staff of the Saccharomyces Genome Database project for their dedication to accuracy and their service to life science educators and researchers. I also want to thank the yeast research community for their support and suggestions. This work was supported by the National Human Genome Research Institute (grant number U41 HG001315) and Funding for open access charge was provided by the National Institutes of Health. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Human Genome Research Institute or the National Institutes of Health.
References
- Cherry JM. The Saccharomyces Genome Database: Exploring biochemical pathways and mutant phenotypes. Cold Spring Harb Protoc. 2015a doi: 10.1101/pdb.prot088898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM. The Saccharomyces Genome Database: Advanced searching methods and data mining. Cold Spring Harb Protoc. 2015b doi: 10.1101/pdb.prot088906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM. The Saccharomyces Genome Database: Gene product annotation of function, process, and component. Cold Spring Harb Protoc. 2015c doi: 10.1101/pdb.prot088914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM. The Saccharomyces Genome Database: Exploring genome features and their annotations. Cold Spring Harb Protoc. 2015d doi: 10.1101/pdb.prot088922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: The genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel SR, Dietrich FS, Fisk DG, Binkley G, Balakrishnan R, Costanzo MC, Dwight SS, Hitz BC, Karra K, Nash RS, et al. The reference genome sequence of Saccharomyces cerevisiae: Then and now. G3 (Bethesda) 2014;4:389–398. doi: 10.1534/g3.113.008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. Gene Ontology annotations and resources. Nucleic Acids Res. 2013;41:D530–D535. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]