Abstract
Many biochemical processes, and the proteins and cofactors involved, have been defined for the eukaryote Saccharomyces cerevisiae. This understanding has been largely derived through the awesome power of yeast genetics. The proteins responsible for the reactions that build complex molecules and generate energy for the cell have been integrated into web-based tools that provide classical views of pathways. The Yeast Pathways in the Saccharomyces Genome Database (SGD) is, however, the only database created from manually curated literature annotations. In this protocol, gene function is explored using phenotype annotations to enable hypotheses to be formulated about a gene’s action. A common use of the SGD is to understand more about a gene that was identified via a phenotypic screen or found to interact with a gene/protein of interest. There are still many genes that do not yet have an experimentally defined function and so the information currently available can be used to speculate about their potential function. Typically, computational annotations based on sequence similarity are used to predict gene function. In addition, annotations are sometimes available for phenotypes of mutations in the gene of interest. Integrated results for a few example genes will be explored in this protocol. This will be instructive for the exploration of details that aid the analysis of experimental results and the establishment of connections within the yeast literature.
MATERIALS
Equipment
Internet-connected computer with web browser
METHOD
At any step in this protocol, support is available via the comprehensive Help documents maintained by SGD. These can be accessed via the help button at the top of each page or, for specific features, a small red button with a question mark in its center is provided and will link to the help pages specific for that feature.
The SGD is continually updated; therefore, specific items presented in this protocol may not appear on the SGD website exactly as described.
Exploring Biochemical Pathway Annotation
This series of steps explores the gene CAT2, describing the pathway tools available from SGD and focusing on the Locus pages. Locus pages are at the core of SGD, where information about each chromosomal feature is provided. The collected information is divided into several tabs by topic. The default starting tab is the Summary page, which provides an overview of a gene and its product.
-
1
Open the URL www.yeastgenome.org in any modern web browser (e.g., Chrome, Firefox, or Safari).
-
2
In the upper right Search box slowly enter “acetylcarnitine,” and observe the list of displayed matches that become more specific as additional letters are added. (By the time you have entered “acetylc” you are down to two choices, one representing our entry of choice.) Press return at any time to select the top string to see a full list of all entries available in the database matching that string, or mouse click to select one of the alternative items of interest. To continue with this exercise, select “acetylcarnitine.”
The “Search” box in the upper right of all SGD pages is the starting point to locate things of interest. Most of the textural information included at SGD has been indexed and is provided as choices from the Search box. Often, as text is entered, you will find additional items of interest that you can immediately explore using this search feature. -
3
From the search results page click on “CAT2” to display the CAT2 Locus Summary page. To go directly to the CAT2 Locus Summary page enter “CAT2” in the Search box and press return.
When the search text matches a gene name, you are directly taken to the matching Locus Summary page. The Locus Summary page includes basic information about a chromosomal feature, in this case the CAT2 gene. The Description states that Cat2p is carnitine acetyl-CoA transferase and that it is located in both mitochondria and peroxisomes. The Description includes the citation for this information. There are several subsections to the page that provide detailed annotations using ontologies—for example, the Gene Ontology subsection leads to Molecular Function, Biological Process, and Cellular Component ontologies. -
4
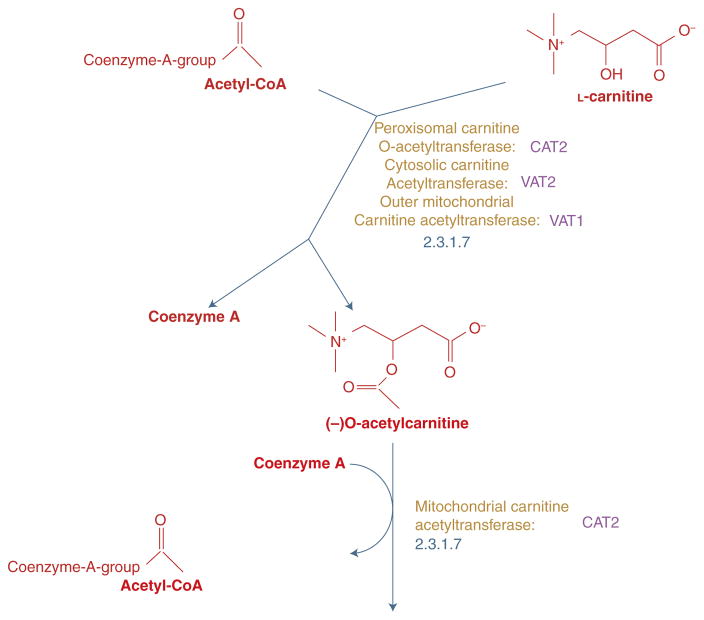
To explore the “Pathways” in which this enzyme participates, select “carnitine shuttle” under the Pathways subcategory to be taken to the biochemical pathway tool called Yeast Pathways. View the graphical representation for the carnitine shuttle, including a summary of the products and intermediates, that is provided. At the top of the page, select the button “More Detail” once or twice to zoom into the pathway illustration and view the chemical structures of the molecules involved in this reaction (see Fig. 1).
Cat2p is one of the three enzymes that catalyzes L-carnitine to O-acetylcarnitine using the cofactor acetyl-CoA to donate an acetate group resulting in Coenzyme A. Here you will also learn that Cat2p is the peroxisomal carnitine O-acetyltransferase.Yeast Pathways was created using MetaCyc software (Caspi et al. 2014). -
5
Explore the pathway by moving the mouse cursor over gene names, compounds, cofactors, enzyme names, and reactions to bring up more information. Also click on the highlighted text. For example, click on Coenzyme A to see a detailed molecular structure, other reactions that it is involved in, and hyperlinks to other web resources. Click on the hyperlink to PubChem, for example, and you are taken to the chemical component database at the NCBI (pubchem.ncbi.nlm.nih.gov).
The references listed in the References subcategory on the CAT2 Locus Summary page provide the experimental results for these annotations. The Yeast Pathways page presents the gene name as CAT2 (gene nomenclature), but, here, the protein name, Cat2p (protein nomenclature) is used, because the function of the protein is known.
FIGURE 1.
Reaction catalyzed by Cat2p. Yeast Pathways provides a detailed illustration (redrawn for clarity) of the reaction, including molecular structures of reactants and products.
Exploring Mutant Phenotype Annotation
-
6
Search for AIM17 via the Search box and go to its summary page.
The summary page description indicates that Aim17p has been purified from mitochondria and that mutants have an impact on mitochondrial genome loss. The GO annotations subsection states that the Molecular Function is unknown. Thus no experimental results have been published that describe this protein’s function in the cell. -
7
Select the Phenotype tab, then select the “Phenotype details” button at top right.
A bar graph indicates there are several annotations of AIM17 from knockouts, that is null mutations, and that more were obtained from large-scale survey experiments than from classical genetic experiments. -
8
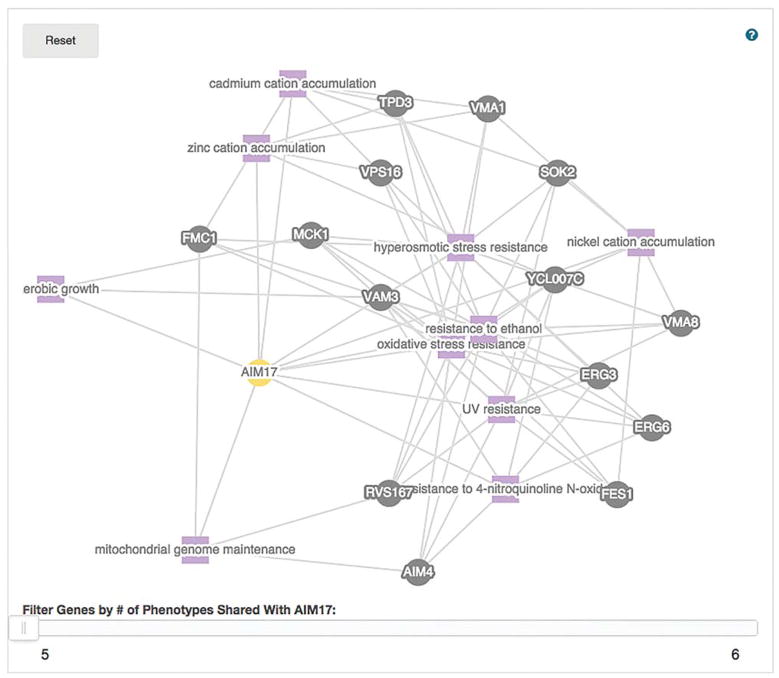
Scroll to the bottom of the page to view a network visualization of the phenotype observables and the other genes that are annotated to them.
Controlled vocabularies are used for mutant phenotypes, thus allowing all genes that have the same mutant phenotype to have a consistent annotation. This consistency is important as it guarantees your search for a particular phenotype will find all genes with that specific mutant phenotype. The use of free text would not provide such a guarantee. You will find this type of network visualization for different types of information on other tabs. -
9
Experiment by selecting the yellow AIM17 ball and dragging it around.
In this way the display changes and you can explore the relationship of AIM17 to the observables and the other genes that have been annotated to the same observables (see Fig. 2). -
10
Move the slider at the bottom of the network to change the number of phenotypes shared between AIM17 and other genes.
Notice that the dark balls represent other genes that share eight phenotype observables. We know that this gene product is localized to the mitochondria and that the null mutant has similar phenotypes to a variety of characterized genes. This set of phenotypes and genes may lead you to a hypothesis about the function of AIM7. -
11
From the Phenotype Annotations table click the phenotype “nickel cation accumulation: increased” button to view all the genes annotated to the same observable phenotype (nickel cation accumulation) and qualifier (increased). Toward the top of the page, select the text “nickel cation accumulation” to see all annotations to this observable phenotype and a diagram of the relationship of this term within the Yeast Phenotype Ontology (Harris et al. 2013).
FIGURE 2.
Network of AIM17 with other genes that share five phenotype annotations. AIM17, yellow ball, is linked with other genes, dark balls, via phenotype annotations, purple squares. The user can dissect regions that have a complex structure by dragging the balls or squares. The network can also be simplified by increasing the number of shared annotations required for the linkage to be shown by moving the slider at the bottom of the graph.
Acknowledgments
I am grateful to all the present and past staff of the Saccharomyces Genome Database project for their dedication to accuracy and service to life science educators and researchers. I also want to thank the yeast research community for their support and suggestions. This work was supported by the National Human Genome Research Institute (grant number U41 HG001315) and funding for open access charge was provided by the National Institutes of Health. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Human Genome Research Institute or the National Institutes of Health.
References
- Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014;42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Lock A, Bähler J, Oliver SG, Wood V. FYPO: The fission yeast phenotype ontology. Bioinformatics. 2013;29:1671–1678. doi: 10.1093/bioinformatics/btt266. [DOI] [PMC free article] [PubMed] [Google Scholar]