Abstract
DHX30 is a member of the family of DExH-box helicases, which use ATP hydrolysis to unwind RNA secondary structures. Here we identified six different de novo missense mutations in DHX30 in twelve unrelated individuals affected by global developmental delay (GDD), intellectual disability (ID), severe speech impairment and gait abnormalities. While four mutations are recurrent, two are unique with one affecting the codon of one recurrent mutation. All amino acid changes are located within highly conserved helicase motifs and were found to either impair ATPase activity or RNA recognition in different in vitro assays. Moreover, protein variants exhibit an increased propensity to trigger stress granule (SG) formation resulting in global translation inhibition. Thus, our findings highlight the prominent role of translation control in development and function of the central nervous system and also provide molecular insight into how DHX30 dysfunction might cause a neurodevelopmental disorder.
Introduction
ATP-dependent unwinding of RNA secondary structures by RNA helicases (RHs) is required for most aspects of RNA metabolism, including synthesis, nuclear processing and export, translation, and storage and decay of RNA, as well as ribonucleoprotein (RNP) assembly.1, 2, 3 Six superfamilies of RHs are known,2 with superfamily 2 containing more than 50 human members characterized by a DExD or DExH signature in their Walker B motif, thus termed DDX and DHX proteins, respectively.4 Several RHs have been assigned to multiple cellular functions, often operating in large RNP complexes, while others appear to be restricted to a particular cellular process.2 Whereas a large body of structural and functional data has been accumulated for RHs from model organisms, such as the yeast DExH protein Prp43, for most human RHs the exact function remains unknown.
Human genetic studies have recently begun to address the pathological relevance of altered RH function; thus, somatic mutations in DDX3X (MIM: 300160) observed in various tumors were found to disrupt global translation.5 Moreover, germline mutations in the same gene are associated with intellectual disability (MIM: 300958),6 pointing to a role of translational control in proper development and function of the nervous system.
Here we describe disease-causing de novo missense mutations in DHX30 (MIM: 616423, RefSeq NM_138615.2) in individuals affected by intellectual disability and global developmental delay. DHX30 belongs to the DExH family of RNA helicases, and has until now mostly escaped scientific attention. We show that DHX30 is indeed an RNA-dependent ATPase, and that all mutations interfere with either RNA binding or ATPase activity. Importantly, protein variants of DHX30 interfere also with global translation by inducing the formation of stress granules.
Material and Methods
Research Subjects
Written informed consent for all subjects was obtained in accordance with protocols approved by the respective ethics committees of the institutions involved in this study.
Genetic Analysis
Some of the investigators presenting affected individuals in this study were connected through GeneMatcher, a web-based tool for researchers and clinicians working on identical genes.7 Trio whole-exome sequencing (trio-WES) experiments, data annotation, and interpretation were performed in nine different centers with slightly different procedures using methods that were described previously. Briefly, trio-WES in families A, C, D, E, J, and L were performed with a SureSelect Human All Exon (Agilent, Santa Clara, CA, USA), and sequenced on a HiSeq2000, HiSeq2500 or HiSeq4000 platform (Illumina, San Diego, CA, USA), as described before.8, 9, 10, 11, 12, 13 Trio-WES in families B, H, and K was performed with a SOLiD-Optimized SureSelect Human Exome Kit (Agilent version 2, 50 Mb), followed by SOLiD 4 System sequencing (Life Technologies) as previously described.14, 15 Trio-WES in family I was performed using the SeqCapEZ VCR 2.0 (Roche NimbleGen) and sequenced on the HiSeq 2000 Sequencer (Illumina, San Diego, CA, USA) as previously described.16 Genetic analyses in families F and G were described before.17 All putative de novo variants were validated and confirmed by Sanger sequencing, by standard procedure.
Expression Constructs
cDNA coding for transcript variant 1 of human DHX30 was obtained from Origene; the DHX30 coding sequence was subcloned into EcoRI/SmaI sites of pEGFP-C3 (Clontech), allowing for expression of DHX30-WT carrying an N-terminal GFP-tag (GFP-DHX30). In parallel, several constructs were generated in pEGFP-N2, leading to expression of DHX30 variants carrying a C-terminal GFP-tag (DHX30-GFP); in particular, we generated a construct corresponding to transcript variant 3; the protein product of this cDNA carries a putative N-terminal mitochondrial targeting sequence. Missense mutations found in affected individuals were introduced into the pEGFP-C3 based vector, and into the mitochondrial construct using Quick-Change II site directed mutagenesis kit (Agilent, Waldbronn, Germany), with mutagenic oligonucleotides designed based on the Quick-Change instruction manual. All constructs were verified by Sanger sequencing.
Cell Culture, Transfection, and Immunocytochemistry
Human embryonic kidney 293T (HEK293T) and human bone osteosarcoma epithelial (U2OS) cells were grown on cell culture dishes and coverslips, respectively, utilizing Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Cells were transfected with TurboFect transfection reagent (ThermoFisher Scientific) according to the manufacturer’s recommendations. Fixation of U2OS cells and immuno-cytochemical analysis was performed as previously described18 employing the following antibodies at manufacturers’ recommended dilutions: anti-DHX30 rabbit polyclonal (Bethyl, #A302-218A), anti-DDX3X mouse monoclonal (BioLegend, #658602), anti-Mitochondria mouse monoclonal (Abcam, #ab3298), anti-puromycin mouse monoclonal (Millipore, #MABE343), and goat anti-mouse, anti-rat, and anti-rabbit IgG coupled to either Alexa Fluor 488, Alexa Fluor 546, or Alexa Fluor 635, respectively (ThermoFisher Scientific). A custom made anti-ATXN2 rat monoclonal antibody was used at a 1:10 dilution. Recombinant DHX30 fusion proteins were directly visualized via their GFP-tag. Coverslips were mounted with ProLong Diamond Antifade Mountant with DAPI (ThermoFisher Scientific). Images were acquired utilizing a confocal microscope (Leica TCS SP8, 63x/1.25 objective) and processed using ImageJ, Corel Paint Shop Pro X, macromedia FreeHand MX and PowerPoint software.
ATPase Assay
Transfected HEK293T cells were lysed in 1 mL of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl pH 8.0; 150 mM NaCl; 0.1% SDS; 0.5% sodium deoxycholate; 1% NP-40; 5 mM EDTA) and lysates were cleared by centrifugation at 20,000 x g for 20 min at 4°C. GFP-containing proteins were purified from the supernatant by immunoprecipitation using 20 μl of GFP-Trap_A matrix (Chromotek, Munich, Germany). Precipitates were washed twice in RIPA buffer, and twice in phosphate free ATPase assay buffer (40 mM KCl; 35 mM HEPES pH 7.5; 5 mM MgCl2; prepared in plastic ware to avoid phosphate contamination). Precipitates were then incubated in 50 μl phosphate free buffer supplemented with 2 mM ATP and 2 mM DTT at 30°C for 30 min (for assaying ATPase activity in the absence of exogenous RNA). After brief centrifugation (1 min, 1000 x g), the supernatant was removed and precipitate samples were incubated in phosphate free buffer containing 2 mM ATP; 2 mM DTT, and 100 μg/ml yeast RNA for 30 min at 30°C (for assaying ATPase activity in the presence of exogenous RNA). The amount of free phosphate released by ATP hydrolysis was determined photometrically using Biomol Green reagent (Enzo Life Sciences, Lörrach, Germany). Subsequently, the amount of bead-attached DHX30 protein was determined by western blotting using anti-GFP (Covance). In each case, ATPase activity was normalized to the amount of GFP-tagged DHX30 protein attached to the GFP-trap matrix.
RNA Immunoprecipitation
Immunoprecipitation of recombinant proteins from lysates of transfected HEK293T cells via GFP-Trap_A (Chromotek), RNA purification from precipitates and real-time PCR with TaqMan probes were essentially performed as previously described.18 TaqMan Gene Expression Assays (ThermoFisher Scientific) for the following human genes were used: AES (Assay ID Hs01081012_m1), B4GAT1 (Hs04194311_s1) and MRPL11 (Hs00601653_g1).
Puromycin Incorporation Assay
Transfected U2OS cells were pulse labeled with puromycin (Invitrogen; 1 μg/ml) for 30 min. Immuno-cytochemistry was performed as described above.
Statistics
Statistical evaluation was performed depending on the experiment by either two-tailed unpaired Student’s t test or ANOVA followed by Dunnett’s multiple comparisons test using Prism 6 (GraphPad), as specifically indicated for each experiment in figure legends. P values expressed as ∗(p < 0.05), ∗∗(p < 0.01), and ∗∗∗(p < 0.001) were considered significant. ns, indicates no significant difference between the groups.
Results
Using trio whole-exome sequencing (WES) or in a single case using singleton-WES followed by targeted Sanger-resequencing, we identified six different de novo mutations in DHX30 in 12 unrelated individuals affected by GDD and ID. Individuals A and B spoke 20 and 4 words, respectively, whereas the others remained non-verbal. All individuals had delayed milestones of motor development. Six have never learned to walk without support, whereas the others have an ataxic gait and are only able to walk short distances. Further, subtle and only partially overlapping facial dysmorphisms were observed. The clinical phenotype further included muscular hypotonia in all affected individuals, feeding difficulties and brain anomalies on MRI in nine, autistic features and sleep disturbances in seven, and strabismus and joint hypermobility in six individuals (Figure S1, Table 1 and Supplemental Note).
Table 1.
Clinical Characteristics of Affected Individuals with DHX30 Alterations
| Clinical findings | Proband A | Proband B | Proband C | Proband D | Proband E | Proband F | Proband G | Proband H | Proband I | Proband J | Proband K | Proband L |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Female | Male | Female | Female | Male | Female | Female | Female | Female | Male | Female | Male |
| Age at last examination (years) | 3 9/12 | 13 | 6 3/12 | 8 | 17 | 14 | 14 2/12 | 8 | 6 1/12 | 4 8/12 | 6 5/12 | 4 8/12 |
| Intellectual disability | + | + | + | + | + | + | + | + | + | + | + | + |
| Age of first words (years) | 1 8/12 | 4 | – | – | – | – | – | – | – | – | – | – |
| Speech ability | 20 words | 4 words | non-verbal | non-verbal | non-verbal | non-verbal | non-verbal | non-verbal | non-verbal | non-verbal | non-verbal | non-verbal |
| Motor development delay | + | + | + | + | + | + | + | + | + | + | + | + |
| Muscular hypotonia | + | + | + | + | + | + | + | + | + | + | + | + |
| Age of walking (years) | 2 8/12 | 6 | – | – | 6 | – | 3 | – | – | – | 5 | 8 |
| Gait abnormalities | ataxic | ataxic | no independent walking | no independent walking | ataxic; only short distances independent | no independent walking | ataxic | no independent walking | no independent walking | no independent walking | ataxic | ataxic |
| Autistic features | + | – | + | – | + | + | + | – | + | – | + | – |
| Sleep disturbance | + | + | + | + | – | – | + | + | + | – | – | – |
| Seizures | + | – | – | + | – | – | – | – | + | – | – | – |
| Feeding difficulties | + | + | – | – | + | + | + | + | + | + | – | + |
| Strabismus | + | – | + | – | – | – | – | – | + | + | + | + |
| Joint hypermobility | + | – | + | – | + | + | – | – | + | – | + | – |
| Cerebral MRI anomalies | – | – | – | delayed myelination, cerebellar atrophy, enlarged ventricles | cortical atrophy, dilated ventricles | mild cerebral atrophy | delayed myelination | cerebral atrophy, delayed myelination, dilated ventricles | cerebral atrophy | delayed myelination, dilated ventricles, corpus callosal abnormalities | delayed myelination, cerebellar atrophy, dilated ventricles | delayed myelination, cerebellar atrophy, dilated ventricles |
| DHX30 alteration | c.1478G>A p.Arg493His | c.1478G>A p.Arg493His | c.1685A>G p.His562Arg | c.2342G>A p.Gly781Asp | c.2342G>A p.Gly781Asp | c.2344C>T p.Arg782Trp | c.2344C>T p.Arg782Trp | c.2344C>T p.Arg782Trp | c.2353C>T p.Arg785Cys | c.2353C>T p.Arg785Cys | c.2353C>T p.Arg785Cys | c.2354G>A p.Arg785His |
+, present; –, absent;
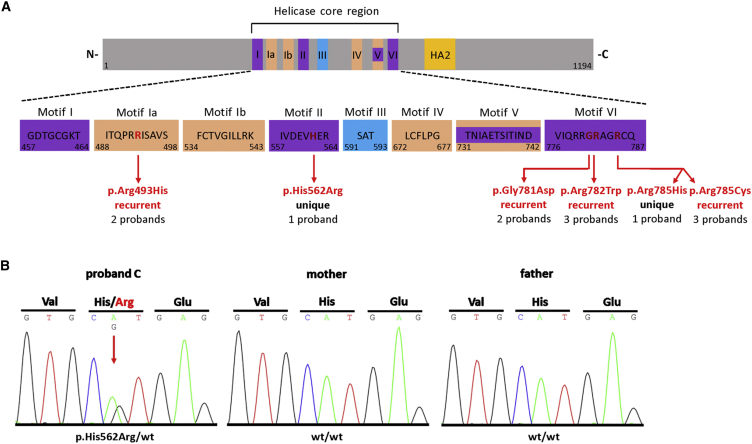
DHX30 belongs to the DExH family of RHs. Within its helicase core region, there are eight highly conserved motifs which are, based on homology to other superfamily 2 helicases,19 predicted to mediate either ATP binding/hydrolysis or RNA recognition (Figure 1 and Figure S2). Interestingly, each mutation reported herein leads to a substitution of a conserved residue within one of these motifs. We predicted the functional relevance of these residues based on homology to other superfamily 2 helicases19 and on published structures of the RH Prp43.20 In more detail, c.1478G>A, (p.Arg493His) identified in two individuals affects motif Ia, and Arg493 is likely a key residue mediating RNA binding.1 All other amino acid substitutions identified lie within motifs which we predicted to be responsible for ATP binding and/or hydrolysis. Namely, c.1685A>G, (p.His562Arg) identified in a single individual affects motif II, also referred to as Walker B motif, which binds β and γ phosphate and coordinates ATP hydrolysis.19 Further, c.2342G>A, (p.Gly781Asp), identified in two individuals, c.2344C>T, (p.Arg782Trp), identified in three individuals, c.2353C>T, (p.Arg785Cys), also identified in three individuals, and c.2354G>A, (p.Arg785His), unique, all result in amino acid alterations residing in motif VI that binds γ phosphate and coordinates, together with motifs I and II, ATP binding and hydrolysis in other DExH family members19, 21 (Figure 1). Indeed, both Arg782 and Arg785 correspond to arginine residues that directly contact the γ phosphate.1 Further evidence for the functional relevance of these highly conserved residues comes from in vivo analyses of the RH Prp43 in Saccharomyces cerevisiae. Substitutions of Arg150 and His218, the corresponding residues in the yeast protein to Arg493 and His562 in human DHX30 (Figure S2B), results in a cold sensitive growth retardation.22, 23 Similarly, the separate exchange of either Gly426, Arg427, and Arg430, corresponding to Gly781, Arg782, and Arg785 in DHX30, respectively (Figure S2B), is lethal.22
Figure 1.
Identified Variants Are Localized within Conserved Helicase Motifs of DHX30
(A) Top: Schematic protein structure of DHX30 showing conserved motifs of the helicase core region and the helicase associated domain (HA2). Nucleotide-interacting motifs (I, II, and VI) are shown in purple, nucleic acid-binding motifs (Ia, Ib, and IV) in orange, motif V, which binds nucleic acid and interacts with nucleotides, in purple and orange, and motif III, which couples ATP hydrolysis to RNA unwinding, in blue. (N- N terminus; C- C terminus). Bottom: Amino acids within conserved motifs of the helicase core region. The position of the first and last amino acid within each motif is denoted below left and right, respectively. The position of the de novo mutations identified in this study are marked with vertical red arrows and shown in red.
(B) Sanger sequence electropherograms of parts of DHX30 after PCR amplification of genomic DNA of the affected individual C and his parents, exemplifying de novo status of the mutations identified here. The amino acid translation is shown in the three-letter code above the DNA sequence. The red arrow indicates the heterozygous mutation at c.1685A>G, (p.His562Arg), present only in the DNA sample of the affected individual.
In line with the functional importance of the six residues affected in our allelic series, it is worth noting that all DHX30 mutations identified herein are exceedingly rare; none of them is present in dbSNP, 1000 Genomes, or the ExAC or the gnomAD browser, indicating that they represent rare variants. In addition, based on the ExAC sequencing data, DHX30 is predicted to be very intolerant to missense mutations, ranked 31 out of 18.000 analyzed genes by its missense Z score of 6.82,24 which is even higher than the average Z score for genes involved in developmental disorders.25 Taken together, the genetic data, especially the recurrence of mutations, supported by published structural and functional data on other RHs provide strong evidence for the pathogenicity of the identified mutations.
During murine embryogenesis, Dhx30/HelG is strongly expressed in neural cells, and its biallelic loss leads to perinatal lethality in mice exhibiting early development defects in the central nervous system.26 Therefore, DHX30 constitutes an excellent candidate gene for human neurodevelopmental disorders. In an international large-scale sequencing study, we recently reported four of the individuals presented herein and suggested DHX30 to be a plausible candidate gene for developmental disorders.17 However, given the large size of the previous study, the corresponding clinical picture remained unclear, and none of the reported alterations was scrutinized by an experimental approach.
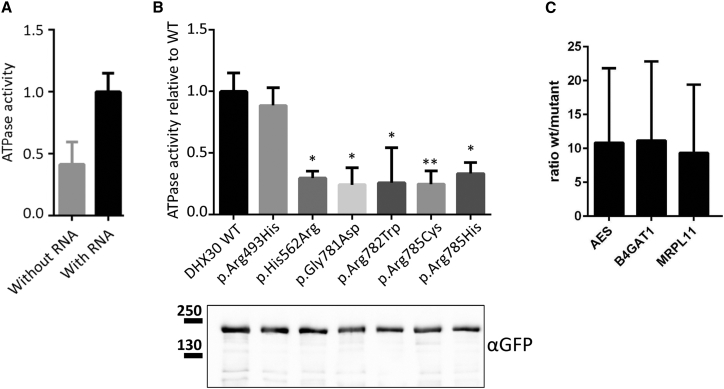
Therefore, to corroborate our findings, we developed an ATPase assay using transiently transfected HEK293T cells. To assess whether wild-type DHX30 (DHX30-WT) acts as an RNA-dependent ATPase, we immunoprecipitated DHX30-GFP and incubated it with ATP in the presence or absence of RNA. Indeed, ATPase activity, determined by a progressive increase of free phosphate concentration in vitro, was significantly stimulated by adding RNA, thus showing that DHX30 hydrolyses ATP in an RNA-dependent fashion (Figure 2A). When compared to DHX30-WT, all protein variants harboring amino acid substitutions in ATP binding motifs II and VI (p.His562Arg, p.Gly781Asp, p.Arg782Trp, p.Arg785Cys, and p.Arg785His) exhibit markedly reduced ATPase activities (Figure 2B). These data confirm our structural predictions and provide strong evidence for the pathogenicity of the respective mutations. Noteworthy, the p.Arg493His amino acid exchange affecting the putative RNA-binding motif Ia did not alter RNA-dependent ATPase activity of DHX30.
Figure 2.
Recombinant Protein Variants of DHX30 Affect Either ATPase Activity or RNA-Binding
(A) GFP-tagged DHX30-WT was immunoprecipitated from HEK293T cell lysates using GFP-Trap_A matrix and assayed for ATPase activity first in the absence and then in the presence of exogenous RNA. Values are normalized on ATPase activity obtained with RNA.
(B) ATPase assays were repeated for WT and protein variants of GFP-DHX30 in the presence of RNA. In each case, ATPase activity was normalized to the amount of DHX30 protein, as determined by western blotting using anti-GFP. ∗,∗∗: significantly different from DHX30-WT (∗p < 0.05; ∗∗p < 0.01; n = 4; ANOVA, followed by Dunnett’s multiple comparisons test).
(C) RNAs extracted from GFP-Trap_A precipitates of DHX30-WT, p.Arg493His, and GFP-mCherry fusion proteins obtained from transfected HEK293T cells were subjected to gene-expression analysis using TaqMan probes for specific human mRNAs. The bar graph displays the fold enrichment of AES, B4GAT1, and MRPL11 transcripts, respectively, in DHX30-WT compared to p.Arg493His precipitates. Vertical lines indicate SD.
To analyze the impact of p.Arg493His on RNA binding, we performed an RNA immunoprecipitation analysis with transfected HEK293T cells.18 For this, RNP complexes containing recombinant DHX30-WT or p.Arg493His, respectively, were affinity purified and subjected to quantitative real-time RT-PCR to examine the presence of three distinct putative target mRNAs. Targets were randomly selected from a publicly available eCLIP (enhanced version of the crosslinking and immunoprecipitation) dataset obtained in HepG2 cells (accession ENCSR565DGW), generated by the ENCODE project.27, 28, 29 A detailed view of the respective DHX30 eCLIP hits is shown in Figure S3. We determined that the p.Arg493His amino acid substitution leads to a 9- to 11-fold decrease in the amount of AES (MIM: 600188), B4GAT1 (MIM: 605517), and MRPL11 (MIM: 611826) mRNAs associated with DHX30. Importantly, only negligible amounts of the three investigated transcripts co-purified with the GFP-mCherry control protein (Figure 2C and Figure S4). These data indicate that the p.Arg493His exchange in DHX30 strongly interferes with its in vivo binding capacity to certain target RNAs, yet does not completely abolish RNA recognition.
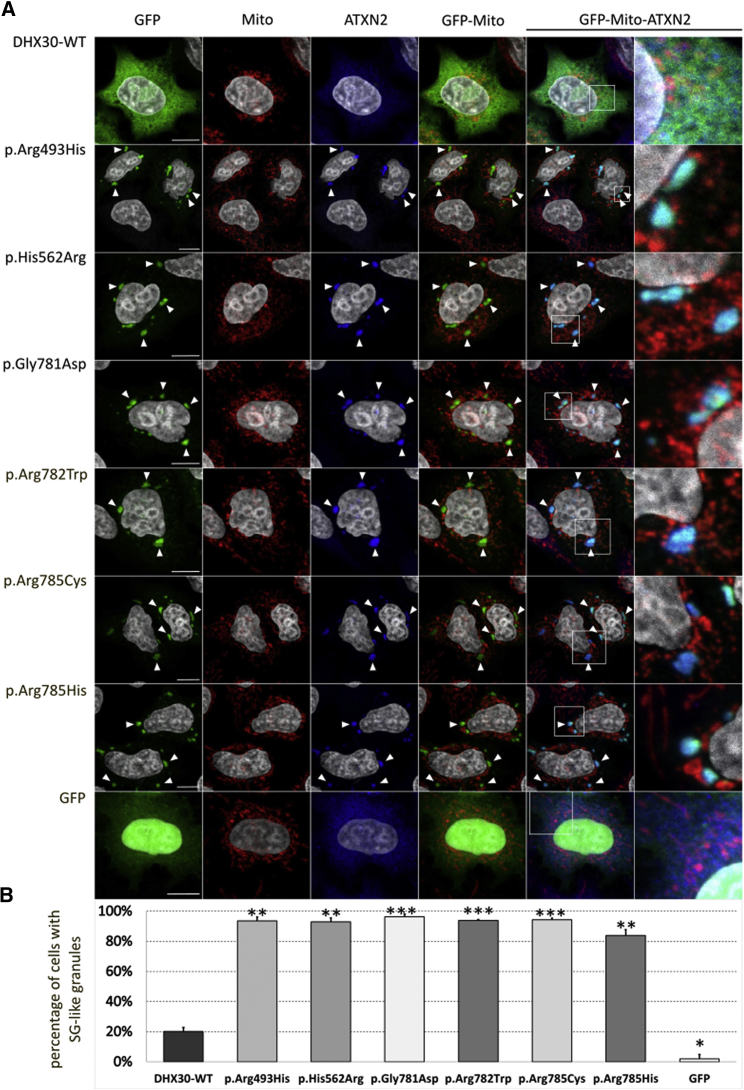
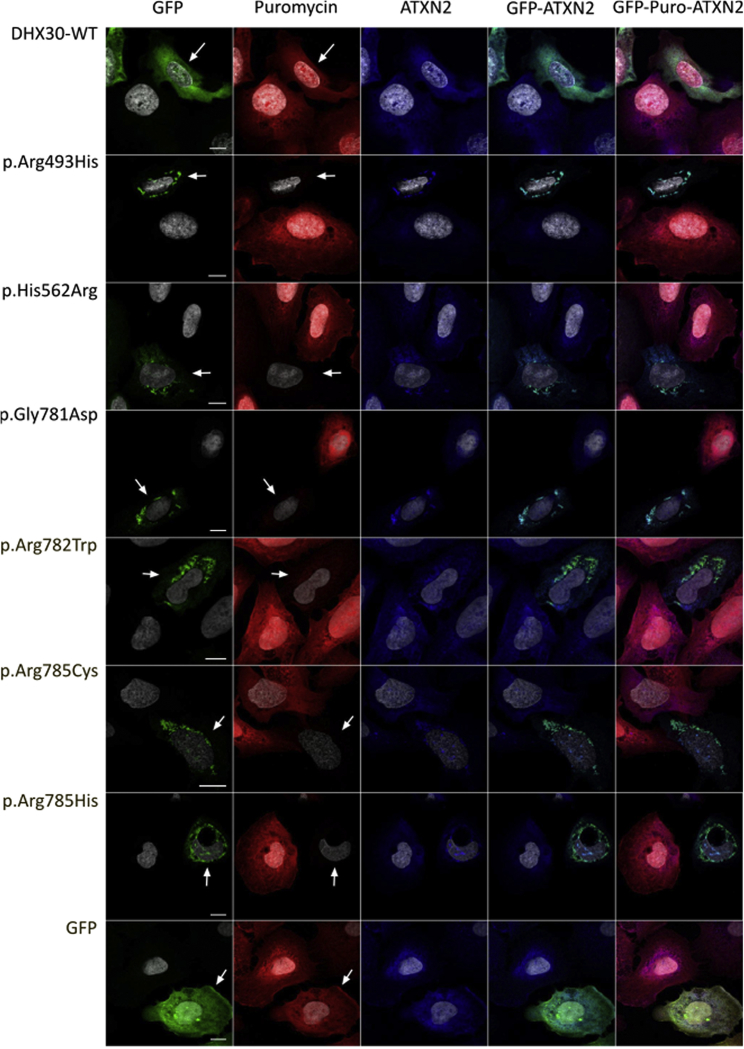
Next, we investigated the subcellular localization of WT and protein variants of DHX30 in U2OS cells. GFP-tagged DHX30-WT was mostly diffusely localized throughout the cytoplasm (Figure 3) with a slight accumulation in mitochondria, consistent with the distribution of endogenous DHX30 (Figure S5A) and previous findings.30 In contrast, in the majority of transfected cells all protein variants strongly accumulated in discrete cytoplasmic foci (> 80%; Figure 3), which were identified as SGs via co-labeling with Ataxin-2 (ATXN2, MIM: 601517), a well-established SG marker.31 SGs are cytoplasmic RNPs that form in cells during stress responses when translation initiation rates of most mRNAs are decreased.32 Remarkably, in response to heat stress endogenous DHX30 also alters its diffuse cytoplasmic localization to accumulate in SGs (Figure S5B). These findings show that independent from exogenous stressors, protein variants of DHX30 exhibit a strongly increased propensity to induce SG assembly compared to DHX30-WT. To further determine if SG formation induced by DHX30 protein variants alters protein synthesis, we monitored global translation rates in transfected versus untransfected U2OS cells. In this assay, puromycin incorporation into newly synthesized peptides directly reflects the translation rate and can be visualized via immunocytochemistry.33 While expression of all tested recombinant DHX30 protein variants dramatically decreased puromycin incorporation compared to neighboring untransfected cells, puromycin labeling of cells expressing DHX30-WT was similar to that of untransfected cells (Figure 4). Taken together, the above data suggest that protein variants of DHX30 significantly increased the propensity of stress granule formation and thus lead to a global decrease in protein synthesis.
Figure 3.
Recombinant Protein Variants of DHX30 Initiate the Formation of SG-like Cytoplasmic Aggregates
(A) Immunocytochemical detection of DHX30-GFP fusion proteins (GFP, green), mitochondria (Mito, red), and endogenous ATXN2 (blue) in transfected U2OS cells. Regions shown at high magnification in the rightmost panels are indicated by boxes. Whereas wild-type DHX30-GFP preferentially resides throughout the cytoplasm and GFP accumulates in nuclei, recombinant protein variants of DHX30 induce the genesis of cytoplasmic foci containing endogenous SG-marker ATXN2 (arrowheads). Nuclei are identified via DAPI staining (gray).
(B) Bar graph indicating the percentage of transfected cells, in which recombinant GFP proteins induce the emergence of SG-like structures. Vertical bars indicate SD. Statistical analysis was performed using unpaired Student’s t test to individually compare each protein variant to wild-type DHX30-GFP (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; n > 300 from two independent transfections).
Figure 4.
SG Formation Initiated by Protein Variants of DHX30 Selectively Inhibits Global Translation in Transfected U2OS Cells
Puromycin incorporation assay in U2OS cells expressing DHX30-GFP fusion proteins (GFP, green). Translation is monitored by staining against puromycin (Puro, red), SGs are detected by ATXN2 (blue) and nuclei via DAPI staining (gray). While cells expressing wild-type DHX30-GFP or GFP display puromycin labeling comparable to neighboring untransfected cells, puromycin incorporation is strongly diminished in cells expressing recombinant protein variants of DHX30. Arrows indicate transfected cells. Note the correlation between SG assembly and lack of puromycin staining.
Discussion
Most RHs are involved in several, non-mutually exclusive aspects of RNA metabolism. Similarly, DHX30 was suggested to control different phases of the RNA life cycle as well as ribosome assembly in mitochondria.30, 34, 35 However, none of the affected individuals presented here displays clear clinical signs of mitochondriopathies. Thus, it is rather unlikely that the mutations identified here primarily affect mitochondrial function. Actually, our in vitro characterization of these mutations has uncovered an additional role for DHX30 related to SG assembly and global translation control. Notably, aberrant SG assembly and clearance with concomitant global translation impairment have been observed in a broad range of neurodegenerative and neurodevelopmental diseases. Examples include amyotrophic lateral sclerosis and frontotemporal dementia (MIM: 612069),36 spinocerebellar ataxia type 2 (MIM: 183090),31 Fragile X syndrome (MIM: 300624)37, 38 and Renpenning syndrome (MIM: 309500).39 Indeed, accurate translation throughout development has recently emerged as a key factor for proper formation and maintenance of complex neural circuits,40 thereby regulating learning, memory, and behavior. Notably, SGs temporarily arrest mRNA translation upon both exogenous and endogenous stressors.32 Because the human organism is, even in utero, under constant exposure to both endogenous and exogenous stressors, we hypothesize that the mutations identified here generate a chronic condition whereby pervasive and pronounced SG hyper-assembly induces impairments in the local regulation of translation.
Whereas we show a distinct molecular defect for each DHX30 variant with respect to either RNA binding or ATPase activity, one might also speculate that disease-associated sequence alterations in DHX30 could affect protein folding or stability. Indeed, protein levels of GFP-tagged variants are somewhat reduced when compared to WT DHX30, as evident from both fluorescence microscopy (Figures 3 and 4) and western blots of lysates of transfected cells (Figure S6). However, this reduced accumulation level might actually be caused by the induction of SGs which is observed upon expression of DHX30 variants. As SG formation leads to reduced general translation, DHX30 variants (but not the WT protein) might indeed limit their own production. Given that DHX30-WT is readily incorporated into SGs e.g., upon heat stress, we assume that also the endogenous DHX30 protein is present in SGs upon expression of variant forms of the protein. These findings might point to a possible dominant negative effect of the mutations identified here.
Interestingly, homozygous deficiency of HelG/DHX30 in mice results in early embryonic lethality, whereas heterozygous mice are apparently normal and fertile.26 However, no in-depth phenotypic analysis of heterozygous mice, including behavioral tests for learning and memory formation, has been performed up to date. Thus, the so-far published data on DHX30-deficient mice do not allow for a direct comparison of phenotypes with the individuals described here. Nevertheless, they might indicate that the missense mutations described in our study might have a more severe effect than the loss of one copy of the gene.
In conclusion, we report that heterozygous missense mutations in DHX30 cause a syndrome characterized by GDD, ID, severe speech impairment and gait abnormalities, and highlight the role of proper translation in neurodevelopment.
Data Availability
The identified DHX30 variants have been deposited to the Leiden Open (source) Variation Database (LOVD) (https://databases.lovd.nl/shared/variants/DHX30/unique) under following accession numbers: 0000195646, 0000195647, 0000195648, 0000195649, 0000195650, 0000195651, 0000195652, and 0000195653. The raw whole-exome sequencing data that support the findings in affected individual cannot be made publicly available for reasons of affected individuals’ confidentiality. Qualified researchers may apply for access to these data, pending institutional review board approval. All other data generated or analyzed during this study are included in this published article (and its Supplemental Data files).
Acknowledgments
We thank the family members for their participation and collaboration, Hans-Hinrich Hönck (Institute for Human Genetics, UKE Hamburg) for excellent technical assistance and Tim Kreienkamp (Hamburg) for help with extracting eCLIP data. This work was funded in part by local funding (Forschungsförderungsfonds der Medizinischen Fakultät des Universitätsklinikums Hamburg-Eppendorf [FFM], to D.L.), Deutsche Forschungsgemeinschaft through SPP1935 “Deciphering the RNP Code” (to H.-J.K. and S. Kindler), the US National Human Genome Research Institute (NHGRI)/National Heart Lung and Blood Institute (NHLBI; grant number UM1HG006542 to the Baylor-Hopkins Center for Mendelian Genomics), NHGRI grant to Baylor College of Medicine Human Genome Sequencing Center (U54HG003273) and J.E.P. (K08 HG008986), National Institute of Neurological Disorders and Stroke (NINDS) (R01NS05829 to J.R.L.), the French Ministry of Health and the Health Regional Agency from Poitou-Charentes (HUGODIMS, 2013, RC14_0107). Confocal microscopes were provided by UKE microscopic imaging facility (umif). eCLIP data were produced by the ENCODE consortium (Lab: Gene Yeo at UCSD). Acknowledgments of the DDD Study and a list of C4RCD Research Group members appear in the Supplemental Note. Baylor College of Medicine (BCM) and Miraca Holdings Inc. have formed a joint venture with shared ownership and governance of the Baylor Genetics (BG), which performs clinical exome sequencing. J.R.L. has stock ownership in 23andMe and Lasergen, is a paid consultant for Regeneron Pharmaceuticals, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. K.L.H. is a full time employee of Ambry Genetics.
Published: November 2, 2017
Footnotes
Supplemental Data include five figures and Supplemental Experimental Procedures and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.09.014.
Contributor Information
Davor Lessel, Email: d.lessel@uke.de.
Hans-Jürgen Kreienkamp, Email: kreienkamp@uke.de.
Web Resources
1000 Genomes, http://browser.1000genomes.org/index.html
ENCODE, https://www.encodeproject.org/
Ensembl Genome Browser, http://www.ensembl.org/index.html
ExAC Browser, http://exac.broadinstitute.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Linder P., Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 2.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laggerbauer B., Achsel T., Lührmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl. Acad. Sci. USA. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Umate P., Tuteja N., Tuteja R. Genome-wide comprehensive analysis of human helicases. Commun. Integr. Biol. 2011;4:118–137. doi: 10.4161/cib.4.1.13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valentin-Vega Y.A., Wang Y.D., Parker M., Patmore D.M., Kanagaraj A., Moore J., Rusch M., Finkelstein D., Ellison D.W., Gilbertson R.J. Cancer-associated DDX3X mutations drive stress granule assembly and impair global translation. Sci. Rep. 2016;6:25996. doi: 10.1038/srep25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snijders Blok L., Madsen E., Juusola J., Gilissen C., Baralle D., Reijnders M.R., Venselaar H., Helsmoortel C., Cho M.T., Hoischen A., DDD Study Mutations in DDX3X Are a Common Cause of Unexplained Intellectual Disability with Gender-Specific Effects on Wnt Signaling. Am. J. Hum. Genet. 2015;97:343–352. doi: 10.1016/j.ajhg.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobreira N., Schiettecatte F., Boehm C., Valle D., Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum. Mutat. 2015;36:425–431. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hempel M., Cremer K., Ockeloen C.W., Lichtenbelt K.D., Herkert J.C., Denecke J., Haack T.B., Zink A.M., Becker J., Wohlleber E. De Novo Mutations in CHAMP1 Cause Intellectual Disability with Severe Speech Impairment. Am. J. Hum. Genet. 2015;97:493–500. doi: 10.1016/j.ajhg.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Küry S., Besnard T., Ebstein F., Khan T.N., Gambin T., Douglas J., Bacino C.A., Craigen W.J., Sanders S.J., Lehmann A. De Novo Disruption of the Proteasome Regulatory Subunit PSMD12 Causes a Syndromic Neurodevelopmental Disorder. Am. J. Hum. Genet. 2017;100:352–363. doi: 10.1016/j.ajhg.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ta-Shma A., Zhang K., Salimova E., Zernecke A., Sieiro-Mosti D., Stegner D., Furtado M., Shaag A., Perles Z., Nieswandt B. Congenital valvular defects associated with deleterious mutations in the PLD1 gene. J. Med. Genet. 2017;54:278–286. doi: 10.1136/jmedgenet-2016-104259. [DOI] [PubMed] [Google Scholar]
- 11.Isidor B., Küry S., Rosenfeld J.A., Besnard T., Schmitt S., Joss S., Davies S.J., Lebel R.R., Henderson A., Schaaf C.P. De Novo Truncating Mutations in the Kinetochore-Microtubules Attachment Gene CHAMP1 Cause Syndromic Intellectual Disability. Hum. Mutat. 2016;37:354–358. doi: 10.1002/humu.22952. [DOI] [PubMed] [Google Scholar]
- 12.Deciphering Developmental Disorders S., Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banuelos E., Ramsey K., Belnap N., Krishnan M., Balak C., Szelinger S., Siniard A.L., Russell M., Richholt R., De Both M. Case Report: Novel mutations in TBC1D24 are associated with autosomal dominant tonic-clonic and myoclonic epilepsy and recessive Parkinsonism, psychosis, and intellectual disability. F1000Res. 2017;6:553. doi: 10.12688/f1000research.10588.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleefstra T., Kramer J.M., Neveling K., Willemsen M.H., Koemans T.S., Vissers L.E., Wissink-Lindhout W., Fenckova M., van den Akker W.M., Kasri N.N. Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am. J. Hum. Genet. 2012;91:73–82. doi: 10.1016/j.ajhg.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Ligt J., Willemsen M.H., van Bon B.W., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 16.Farwell K.D., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Tippin Davis B., Baxter R.M., Zeng W., Mroske C., Parra M.C. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 17.Eldomery M.K., Coban-Akdemir Z., Harel T., Rosenfeld J.A., Gambin T., Stray-Pedersen A., Küry S., Mercier S., Lessel D., Denecke J. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med. 2017;9:26. doi: 10.1186/s13073-017-0412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miroci H., Schob C., Kindler S., Ölschläger-Schütt J., Fehr S., Jungenitz T., Schwarzacher S.W., Bagni C., Mohr E. Makorin ring zinc finger protein 1 (MKRN1), a novel poly(A)-binding protein-interacting protein, stimulates translation in nerve cells. J. Biol. Chem. 2012;287:1322–1334. doi: 10.1074/jbc.M111.315291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanner N.K., Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 20.Tauchert M.J., Fourmann J.B., Lührmann R., Ficner R. Structural insights into the mechanism of the DEAH-box RNA helicase Prp43. eLife. 2017;6:6. doi: 10.7554/eLife.21510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caruthers J.M., McKay D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 22.Martin A., Schneider S., Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J. Biol. Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka N., Schwer B. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry. 2006;45:6510–6521. doi: 10.1021/bi052656g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng H.J., Tsukahara M., Liu E., Ye L., Xiong H., Noguchi S., Suzuki K., Ji Z.S. The novel helicase helG (DHX30) is expressed during gastrulation in mice and has a structure similar to a human DExH box helicase. Stem Cells Dev. 2015;24:372–383. doi: 10.1089/scd.2014.0077. [DOI] [PubMed] [Google Scholar]
- 27.Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat. Methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium E.P., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C., Gabdank I., Narayanan A.K., Ho M., Lee B.T. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016;44(D1):D726–D732. doi: 10.1093/nar/gkv1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., Bogenhagen D.F. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J. Biol. Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- 31.Nonhoff U., Ralser M., Welzel F., Piccini I., Balzereit D., Yaspo M.L., Lehrach H., Krobitsch S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahboubi H., Stochaj U. Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochim. Biophys. Acta. 2017;1863:884–895. doi: 10.1016/j.bbadis.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt E.K., Clavarino G., Ceppi M., Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 34.Minczuk M., He J., Duch A.M., Ettema T.J., Chlebowski A., Dzionek K., Nijtmans L.G., Huynen M.A., Holt I.J. TEFM (c17orf42) is necessary for transcription of human mtDNA. Nucleic Acids Res. 2011;39:4284–4299. doi: 10.1093/nar/gkq1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antonicka H., Shoubridge E.A. Mitochondrial RNA Granules Are Centers for Posttranscriptional RNA Processing and Ribosome Biogenesis. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.030. February 12, 2015. https://doi.org/10.1016/j.celrep.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 36.Li Y.R., King O.D., Shorter J., Gitler A.D. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanderklish P.W., Edelman G.M. Differential translation and fragile X syndrome. Genes Brain Behav. 2005;4:360–384. doi: 10.1111/j.1601-183X.2005.00134.x. [DOI] [PubMed] [Google Scholar]
- 38.Gareau C., Houssin E., Martel D., Coudert L., Mellaoui S., Huot M.E., Laprise P., Mazroui R. Characterization of fragile X mental retardation protein recruitment and dynamics in Drosophila stress granules. PLoS ONE. 2013;8:e55342. doi: 10.1371/journal.pone.0055342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunde S.A., Musante L., Grimme A., Fischer U., Müller E., Wanker E.E., Kalscheuer V.M. The X-chromosome-linked intellectual disability protein PQBP1 is a component of neuronal RNA granules and regulates the appearance of stress granules. Hum. Mol. Genet. 2011;20:4916–4931. doi: 10.1093/hmg/ddr430. [DOI] [PubMed] [Google Scholar]
- 40.Darnell J.C., Richter J.D. Cytoplasmic RNA-binding proteins and the control of complex brain function. Cold Spring Harb. Perspect. Biol. 2012;4:a012344. doi: 10.1101/cshperspect.a012344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The identified DHX30 variants have been deposited to the Leiden Open (source) Variation Database (LOVD) (https://databases.lovd.nl/shared/variants/DHX30/unique) under following accession numbers: 0000195646, 0000195647, 0000195648, 0000195649, 0000195650, 0000195651, 0000195652, and 0000195653. The raw whole-exome sequencing data that support the findings in affected individual cannot be made publicly available for reasons of affected individuals’ confidentiality. Qualified researchers may apply for access to these data, pending institutional review board approval. All other data generated or analyzed during this study are included in this published article (and its Supplemental Data files).