Abstract
Neurodegenerative diseases pose an extraordinary threat to the world’s aging population, yet no disease-modifying therapies are available. Although genome-wide association studies (GWASs) have identified hundreds of risk loci for neurodegeneration, the mechanisms by which these loci influence disease risk are largely unknown. Here, we investigated the association between common genetic variants at the 7p21 locus and risk of the neurodegenerative disease frontotemporal lobar degeneration. We showed that variants associated with disease risk correlate with increased expression of the 7p21 gene TMEM106B and no other genes; co-localization analyses implicated a common causal variant underlying both association with disease and association with TMEM106B expression in lymphoblastoid cell lines and human brain. Furthermore, increases in the amount of TMEM106B resulted in increases in abnormal lysosomal phenotypes and cell toxicity in both immortalized cell lines and neurons. We then combined fine-mapping, bioinformatics, and bench-based approaches to functionally characterize all candidate causal variants at this locus. This approach identified a noncoding variant, rs1990620, that differentially recruits CTCF in lymphoblastoid cell lines and human brain to influence CTCF-mediated long-range chromatin-looping interactions between multiple cis-regulatory elements, including the TMEM106B promoter. Our findings thus provide an in-depth analysis of the 7p21 locus linked by GWASs to frontotemporal lobar degeneration, nominating a causal variant and causal mechanism for allele-specific expression and disease association at this locus. Finally, we show that genetic variants associated with risk of neurodegenerative diseases beyond frontotemporal lobar degeneration are enriched in CTCF-binding sites found in brain-relevant tissues, implicating CTCF-mediated gene regulation in risk of neurodegeneration more generally.
Keywords: frontotemporal dementia, frontotemporal lobar degeneration, TMEM106B, GWAS, eQTL, causal variant, functional variant, CTCF, chromatin architecture, Capture-C
Introduction
Neurodegenerative diseases are a leading cause of disability and death in the developed world, and the number of individuals affected by these diseases is poised to increase as the world population ages. There are still no disease-modifying therapies for the major late-onset neurodegenerative diseases, such as Alzheimer disease (AD), Parkinson disease (PD), frontotemporal lobar degeneration (FTLD), and amyotrophic lateral sclerosis (ALS).1 To generate novel leads for tackling this growing problem, researchers have performed many genome-wide association studies (GWASs) involving >100,000 individuals affected by the various neurodegenerative diseases and have identified >200 genetic risk loci.2 Although genetic risk loci have been utilized, singly or in aggregate, for refining predictions of disease risk,3, 4 the greatest potential for these GWAS-identified loci could lie in the identification of novel disease mechanisms.5
However, the interpretation of disease-associated risk loci is complicated. The “sentinel” variant, usually a single-nucleotide polymorphism (SNP) identified by a GWAS, is rarely the specific change in DNA sequence—or “causal” variant—that results at the molecular level in a mechanistic change. Instead, in most cases, tens or hundreds of genetic variants at each locus are in strong linkage disequilibrium (LD) with the sentinel variant, constituting a set of co-inherited variants—or haplotype—any of which could be the underlying cause of increased disease risk.6 Indeed, the risk-associated haplotype can span multiple genes, making even the gene to which a GWAS signal belongs unclear. Given these complexities, it is perhaps unsurprising that none of the GWAS-identified neurodegenerative-disease risk loci, with the exception of common variants near SNCA (MIM: 163890), which had already been implicated before the GWAS era in the development of PD,7 have been characterized in molecular detail. Yet, such a molecularly precise understanding of a GWAS-identified genetic risk locus is a likely prerequisite for downstream therapeutic development.
FTLD is a neurodegenerative dementia affecting ∼10–20 per 100,000 persons between the ages of 45 and 64 years, making FTLD the second-most-common early-onset dementia.8, 9 FTLD is a fatal, untreatable disease, such that death typically occurs within ∼8 years after diagnosis.8 Noncoding SNPs in chromosomal region 7p21 have been associated with risk of the major neuropathological FTLD subtype (FTLD-TDP), characterized by pathological inclusions of TDP-43 (transactive response element DNA-binding protein 43 kDa).10 The association between this locus and FTLD-TDP has been replicated,11, 12, 13 and the major T allele of the sentinel SNP, rs1990622, yielded an odds ratio of ∼1.6 for disease development.10 The genotype at rs1990622 also associates with penetrance and age at disease onset in Mendelian forms of FTLD-TDP,12, 14, 15, 16 risk of development of cognitive impairment in the related disorder ALS,17 and interestingly, cognitive reserve in older adults without neurodegenerative diseases.18, 19 Genotypes at this locus have not been associated with non-brain-related traits or diseases.2
We and others have implicated a gene in this region, TMEM106B (MIM: 613413), as the causal gene.20, 21, 22 Although some studies have suggested a potential functional role for a nonsynonymous SNP in TMEM106B exon 6,23, 24 proposed mechanisms differ between these studies. Moreover, other groups have failed to replicate any molecular or cellular effects of the exon 6 SNP,21, 25 a result that agrees with our own observations. Thus, studies to date have not explained how genetic variation at the 7p21 locus affects the function of TMEM106B or another gene and thereby contributes to the pathogenesis of FTLD-TDP.
In this study, we were able to demonstrate that (1) common GWAS-implicated variants associated with FTLD-TDP are correlated with expression levels of TMEM106B, whereby increased expression correlates with the risk haplotype; (2) incremental increases in TMEM106B expression are associated with incremental increases in cell toxicity; (3) the risk allele of a candidate causal variant (rs1990620) in complete LD with rs1990622, the GWAS sentinel SNP, increases recruitment of the chromatin-organizing protein CCCTC-binding factor (CTCF) downstream of TMEM106B; and (4) long-range chromatin-looping interactions involving the CTCF site and other distal regulatory elements at the TMEM106B locus are stronger on the risk haplotype. Together, these data provide a molecularly detailed mechanism for the effect of common genetic variation at this locus on risk of neurodegenerative disease.
Material and Methods
eQTL Analyses
The GWAS sentinel SNP, rs1990622,10 was queried for association with all transcripts genome-wide with the Genotype-Tissue Expression (GTEx) database of expression quantitative trait loci (eQTLs),26 consisting of 7,051 samples and representing 44 different tissues from 449 healthy donors. GTEx eQTL plots were generated with SNiPA.27 Conditional analyses and fine-mapping were performed with HapMap3-imputed genotypes from a published multi-ethnic eQTL study of lymphoblastoid cell lines (LCLs)28 as previously described.29 In brief, gene expression data were normalized to the empirical average quantiles across all samples. Subsequently, the distribution of each gene expression trait was transformed to the quantiles of the standard normal distribution separately within each population. The effects of known and unknown covariates were controlled for by principal-component analysis. A cis-eQTL scan was performed by regression of the additive effect of each SNP within 1 Mb of TMEM106B on gene expression by Bayesian regression, as implemented in SNPTEST.30
Colocalization Analyses
In order to assess the probability that the TMEM106B cis-eQTL and the FTLD-TDP GWAS signal share the same causal variant, we applied the COLOC R package.31 We assessed evidence of co-localization by using all SNPs that were within 1 Mb of the lead GWAS variant and were in common between the GWAS and GTEx eQTL study. GTEx eQTL data26 were downloaded from the GTEx Portal (see Web Resources). We ran COLOC with default parameters and assessed evidence of the posterior probability of a shared causal variant (“PP.H4”).
Analysis of LD Structure at the TMEM106B Locus
We visualized the combined CEU (Utah residents with ancestry from northern and western Europe) genotype data from HapMap phases I, II, and III32 in HaploView to assign LD blocks33 after filtering out SNPs with a minor allele frequency [MAF] < 0.001 and requiring that 90% of informative pairwise LD values within a block represented strong LD. Pairwise LD between variants at the TMEM106B locus was determined with both HapMap and 1000 Genomes data34 visualized either in HaploView or on HaploReg v.4.1.35
Cell Culture
HeLa cells were cultured in DMEM with 10% fetal bovine serum (FBS), 1% l-glutamine, and 1% penicillin-streptomycin. LCLs were obtained from the Coriell Institute for Medical Research and were cultured in RPMI with 15% FBS, 1% l-glutamine, and 1% penicillin-streptomycin. Jurkat cells were cultured in RPMI with 10% FBS, 1% l-glutamine, and 1% penicillin-streptomycin.
Neuronal Culture, Transfection, and Immunofluorescence Microscopy
Primary hippocampal mouse neurons were isolated and cultured as previously described.22 Genetic constructs for TMEM106B-GFP overexpression were delivered by nucleofection (Lonza Amaxa Nucleofector 2b) as previously described.36 Immunofluorescence labeling experiments with anti-LAMP1 antibodies (at 1 μg/mL; 1D4B, Developmental Studies Hybridoma Bank) were performed and cells were imaged as previously described.36
TMEM106B Overexpression Experiments
TMEM106B expression constructs designed to increase the amount of TMEM106B by 2-fold, 5-fold, and 20-fold, as previously described,22 were transfected into HeLa cells with Lipofectamine 2000 according to the manufacturer protocols. Cells transfected with a 5/TO construct were used as the baseline (1×) condition. 48 hr after transfection, ten bright-field images were taken across three biological replicates for each condition at 100× on a Life Technologies EVOS FL microscope. Image files were assigned random identifiers, and for each image, a blinded individual counted the number of cells that displayed the vacuolar phenotype, defined by having at least one clear punctate vacuolar structure. This experiment was repeated three times, and the results were pooled. For assessment of cytotoxicity, the same transfection protocol was carried out; however, 48 hr after transfection, cells were spun down and resuspended in trypan-blue-containing culture DMEM, and the proportion of trypan-blue-positive cells was determined with a hemocytometer. This experiment was also carried out three times. Western blots were performed for all six experiments as described previously,22 and the data were pooled together for the quantification shown in Figure 3B. The effect of increased amounts of TMEM106B on the vacuolar phenotype and cell death was assessed with a one-way ANOVA.
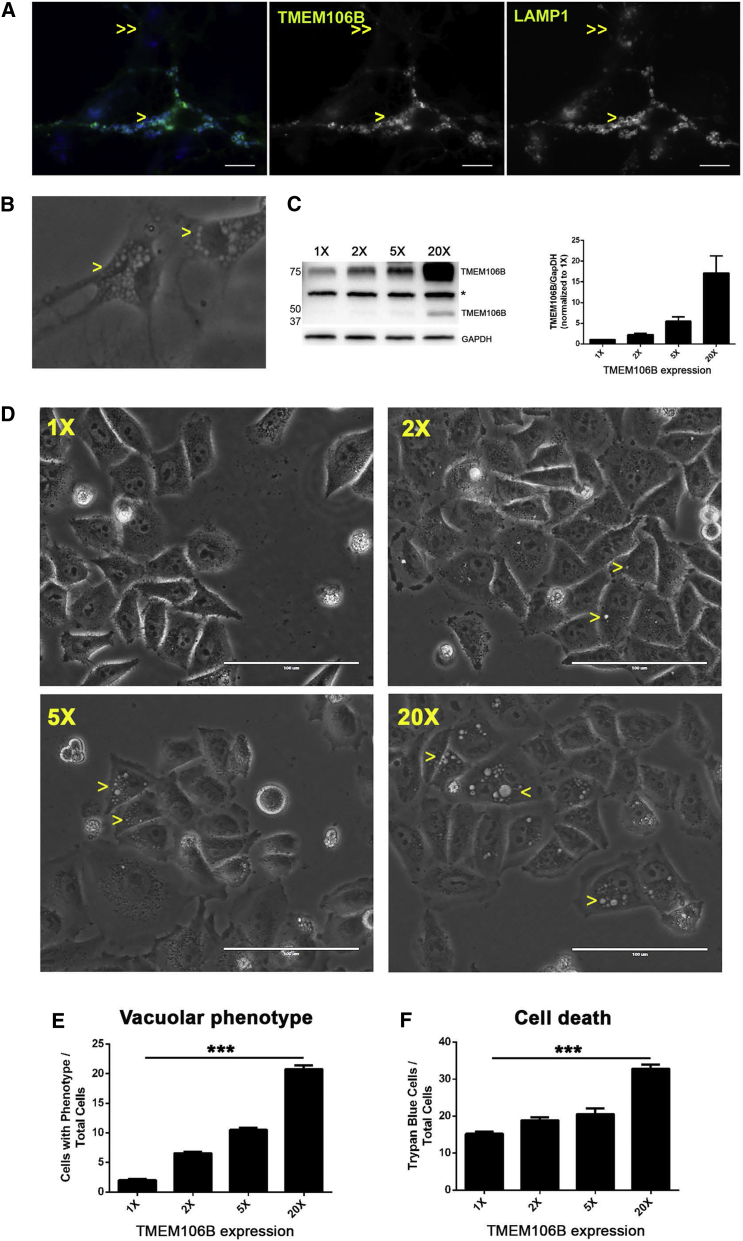
Figure 3.
Dose-Dependent Effects on Cell Toxicity Are Seen with Different Amounts of TMEM106B
(A) Mouse hippocampal neurons were nucleofected with TMEM106B-GFP, resulting in transient overexpression of TMEM106B in some cells (single arrowheads) and endogenous amounts of TMEM106B in neighboring cells (double arrowheads). Neurons were then visualized for TMEM106B (middle panel) or the lysosomal marker LAMP1 (right panel). Neurons with increased amounts of TMEM106B (single arrowheads) formed enlarged vacuoles in which TMEM106B (green) and LAMP1 (blue) co-localized (left panel shows merged color images of middle and right panels). In contrast, vacuoles were absent in neurons with endogenous amounts of TMEM106 (double arrowheads), which showed punctate LAMP1 staining. Scale bar, 10 μm.
(B) The vacuolar phenotype (single arrowheads) was readily observed in two neighboring neurons by bright-field imaging.
(C) Western blot of TMEM106B levels in the absence (1×) and presence (2×, 5×, and 20×) of various TMEM106B expression constructs transfected into HeLa cells. The bands at ∼75 and ∼40 kDa represent dimeric and monomeric forms of TMEM106B, respectively. A non-specific band is indicated by the asterisk. Quantification was performed for blots from six independent experiments (±SEM), demonstrating reliable expression levels of each construct.
(D) Representative bright-field images demonstrate a dose-dependent vacuolar phenotype in cells. Yellow arrowheads indicate cells exhibiting the phenotype.
(E and F) Quantification of the number of cells exhibiting (E) the vacuolar phenotype and (F) cell death is shown for each expression paradigm across three independent experiments.
Asterisks denote statistical significance (p < 0.001 by ANOVA).
Cell-Line Haplotype Phasing
In order to confirm all cell lines used for mRNA stability and chromosome conformation capture (Capture-C) experiments as TMEM106B haplotype heterozygotes, we first performed TaqMan SNP genotyping assays, as previously described,15 to confirm heterozygosity of rs1990622 and marker SNPs in strong or complete LD with rs1990622 (rs3807865, r2 = 0.9; rs6966915, r2 = 1; rs3173615, r2 = 1; and rs1468803, r2 = 1). For the cell lines used for the Capture-C experiments, we also analyzed the CTCF-binding region by Sanger sequencing to confirm heterozygosity of the three completely linked candidate causal variants: rs1990622, rs1990621, and rs1990620. Heterozygosity of the promoter SNP rs4721056 (r2 = 0.5 with rs1990622) was confirmed by genotyping as well, but because of the lower LD between this SNP and rs1990622, we also PCR amplified the region containing rs4721056 and three SNPs in strong LD with rs1990622 (rs7781670, r2 = 0.9; rs1019309, r2 = 0.9; and rs1019307, r2 = 0.89) in the three Capture-C cell lines. This amplicon was cloned into the multiple cloning site (MCS) of the pGL3-Promoter vector, and Sanger sequencing of individual clones confirmed that no cell lines had mixed haplotypes comprising these SNPs, thus linking the risk allele of rs4721056 to the risk haplotype in all cell lines.
Experiments of mRNA Stability
Three LCLs homozygous for the TMEM106B risk haplotype and three LCLs homozygous for the protective haplotype were treated with 1 μg/μL actinomycin D, and RNA was extracted 0, 1, 2, 4, 8, and 24 hr after treatment. RT-qPCR was performed as previously described22 for quantifying TMEM106B expression (normalized to 18S RNA, which decayed by only ∼14% in 24 hr) at each time point. Decay curves for mRNA stability were compared by two-way ANOVAs. This experiment was performed on each of the six cell lines twice for a total of six biological replicates for each haplotype.
Epigenomic Prioritization of Candidate-Variant-Containing cis-Regulatory Elements
We used the UCSC Genome Browser (hg19)37 to visualize ENCODE DNase hypersensitivity (DHS)38 and transcription factor (TF) chromatin immunoprecipitation sequencing (ChIP-seq)39 tracks for all cell types tested in the ENCODE Project, as well as the chromatin-state segmentation40 track for the GM12878 LCL line. We also used the WashU EpiGenome Browser41 to visualize NIH Roadmap EpiGenome Project42 data. Specifically, we analyzed the H3K4me1, H3K4me3, and H3K27ac histone marks, as well as chromatin-state segmentation, in LCLs, primary leukocytes, and all human brain samples. We determined which of the top eQTL SNPs from the fine-mapping overlapped a cis-regulatory element (CRE) predicted to be active in LCLs on the basis of the presence of DHS, TF binding, or an active chromatin state and determined whether any of these regions had epigenetic evidence of activity in primary leukocytes and/or brain from Roadmap EpiGenome data. Putative CREs were then tested for allele-specific activity in downstream assays.
In situations where more than one SNP was found in a candidate CRE (e.g., for the CTCF-binding-site CRE), we used the vertebrate JASPAR Database43 and RegulomeDB44 to perform in silico analyses to predict which individual SNPs might disrupt known TF motifs.
Reporter Assays
To test putative LCL CREs with epigenetic evidence of potential enhancer activity, we cloned each region with either the risk- or protective-haplotype SNP alleles, by using 1000 Genomes phase I45 haplotype information, into the upstream MCS of the pGL3-Promoter luciferase reporter vector (catalog no. E1761, Promega) with restriction enzymes KpnI and NheI (catalog nos. R0142 and R0131, respectively, New England BioLabs). We tested two regions of 481 and 444 bp, each of which encompassed the DHS, TF binding, and candidate causal variants. We transfected 2.5 × 106 LCLs by using program Y-001 and nucleofection solution V on the Lonza Nucleofector 2b and using the pGL3-Basic vector (with no enhancer; catalog no. E1751, Promega) as a negative control and the pGL3-Control vector (with an SV40 enhancer; catalog no. E1741, Promega) as a positive control (Figure S3A). Three or four biological replicates were included for each construct in each experiment, and four independent experiments were performed for each candidate CRE. 24 hr after transfection, cell lysates were isolated with the Promega Dual-Luciferase Reporter Assay System (catalog no. E1910, Promega), as described previously,22 for luciferase readout. We used two-tailed t tests to test for statistically significant differences in reporter activity.
ENCODE Data Mining for CTCF Binding and DHS
Using the ENCODE portal (see Web Resources and Table S3), we downloaded the BAM read alignment files for all CTCF ChIP-seq experiments that showed a CTCF peak at the region containing rs1990620, as well as for the DNase digital genomic footprinting (DGF) experiments performed in cell lines that showed a DHS site sequencing peak at this region. We analyzed raw sequencing reads containing rs1990620 to identify cell lines heterozygous at the TMEM106B FTLD-TDP risk haplotype. We summed reads for risk and protective alleles across all heterozygous cell types and across all three SNPs in the CTCF-binding region and assessed deviation from a 50:50 proportion by using a two-tailed binomial sign test.
Electrophoretic Mobility Shift Assays
Nuclear extract was obtained from LCLs and human occipital cortex with the Thermo Fisher Scientific Nuclear and Cytoplasmic Extraction Reagents Kit (catalog no. 78833). A 61 bp 5′ biotinylated DNA probe containing the risk or protective allele of rs1990620 at position 31, with 30 bp of genomic sequence on either side, was incubated with extract and competed with excess amounts of unlabeled oligonucleotide containing either the risk or protective allele of rs1990620 in competition electrophoretic mobility shift assays (EMSAs). As a negative control, an unlabeled oligonucleotide derived from the intronic candidate CRE was also investigated in competition EMSAs. We performed supershift assays with probes biotinylated on either the 5′ or 3′ side, as designated in the text, to determine the presence or absence of specific proteins in the shifts observed on EMSA; supershift assays used 2 μL of anti-NFYA (catalog no. sc-10779X, Santa Cruz Biotechnology), anti-PU.1 (catalog no. sc-352X, Santa Cruz Biotechnology), or anti-CTCF (catalog no. 07-729, EMD Millipore) antibody. Standard EMSA protocols from the Thermo Fisher Scientific LightShift Chemiluminescent EMSA Kit (catalog no. 20148) were used.
Hi-C Data Visualization
To examine the chromatin architecture at and around the TMEM106B locus, we visualized in situ Hi-C heatmaps generated from LCLs46 by using the cloud-based software Juicebox.47
Capture-C
Capture-C was performed similarly to the methods described in previous reports.48, 49 In brief, we generated 3C libraries by fixing 10 × 106 cells with formaldehyde and then digesting and ligating them with DpnII. We sonicated phenol-chloroform-extracted DNA to produce 200–300 bp fragments and prepared sequencing libraries with the NEBNext DNA Library Prep Master Mix Set (catalog no. E6040, Illumina). 10 μg of each capture library underwent multiplexed PCR with unique index oligonucleotides. Hybridization with 60 bp biotinylated capture probes (Table S5) was performed with the SeqCap EZ Hybridization and Wash Kit (catalog no. 05634261001, Roche). In brief, 3C libraries were dried with heat in a thermocycler and resuspended with hybridization reagents. 2 μL (3 pmoles) of pooled capture probes for each bait region was added to the resuspended libraries and incubated for 72 hr in a thermocycler at 47°C. After the captured material was isolated with streptavidin beads and PCR, an additional 24 hr of capture was performed. The capture probes, ordered from Integrated DNA Technologies, flank DpnII cut sites that are proximal to marker SNPs in the TMEM106B promoter region and CTCF site and are designed to contain marker SNPs that distinguish between haplotypes for captured ligation products. Promoter capture experiments used rs4721056 to distinguish haplotypes, and CTCF-site capture experiments used rs1990620 and rs1990621 to distinguish haplotypes (see Cell-Line Haplotype Phasing). Samples were pooled and sequenced on one lane of an Illumina HiSeq 2500 with 125 bp paired-end reads, yielding ∼230 million read pairs. Two LCLs and Jurkat cells were captured at both regions, with two technical replicates for each capture, by different individuals in tandem.
Capture-C Data Analyses
Quality control was performed with FastQC (see Web Resources), and pre-processing and read alignment were performed as previously described.50 In brief, the paired-end reads were reconstructed into single reads with FLASH, digested in silico with the DpnII2E.pl script, and mapped with Bowtie1.51 The aligned reads were then analyzed with the CCanalyser3.pl script. Interactions were tested for significance with fourSig52 (default parameters and a window size of five Dpn II restriction enzyme fragments). First, we tested interactions for significance by using all reads mapping to chromosome 7. Then, we restricted our analyses to interactions within the topologically associated domain (TAD) and sub-TAD to test for significant interactions within these regions.
To determine whether the long-range interactions captured by the hybridization probes show allelic bias, we first estimated each SNP’s technical (non-biological) bias, which could reflect capture bias, mapping bias, or other sources of technical bias. We estimated technical bias by using two independent approaches. For the first approach, we enumerated the risk- and protective-allele reads containing each marker SNP from ligated fragments that mapped directly adjacent to the probe sequences (i.e., mapping to the 5 kb window containing the probe sequences or the immediately adjacent 5 kb windows at each capture site) for each experiment. In the second approach, we enumerated the risk- and protective-allele reads containing each marker SNP from ligated fragments mapping at a great distance from TMEM106B (i.e., on chromosome 7 but outside of the 1Mb TMEM106B TAD, as defined by the LCL Hi-C heatmap).46 In each case, we assumed that the interactions too close or too far from TMEM106B do not represent functional interactions. For the adjacent probe-proximal interactions, ligations can occur artifactually as a result of chromosomal proximity.53 For the interactions outside the TMEM106B TAD, low read count per interaction suggests that these are not true interactions, consistent with the fourSig analyses described in Figure S8. Technical bias estimated by these two approaches is likely to be conservative—some true biological interactions could be “discounted” in this way, whereby false negatives are created to most reliably account for false-positive interactions. Despite different underlying assumptions, technical-bias estimates derived from the two methods, which are based on thousands of reads for each marker SNP, agree with each other to within <1% for all SNPs.
In each case, we compared read counts for interactions originating from each haplotype (1) with an expected proportion of 0.5 (in analyses without normalization for technical bias) or (2) with the proportions observed from the technical-bias estimates described above (in analyses normalized for technical bias) by using a two-tailed binomial sign test.
CTCF-Site SNP-Enrichment Analysis
We identified all SNPs that have been associated with risk of adult-onset neurodegenerative diseases (FTLD, AD, PD, ALS, and related conditions) at a genome-wide-significant level (p ≤ 5 × 10−8) by using the NHGRI-EBI GWAS Catalog.2 This list contains 200 SNPs associated with 29 traits; nine of the SNPs were not identifiable by the Genomic Regulatory Elements and GWAS Overlap (GREGOR) algorithm,54 resulting in a final list of 191 neurodegeneration risk SNPs (Table S7). As a comparator group, we identified all SNPs associated with risk of leukemia and lymphoma from the same GWAS Catalog, which yielded 177 SNPs; three of these SNPs were not identifiable by GREGOR, resulting in a final list of 174 leukemia and lymphoma risk SNPs (Table S8). We then identified CTCF-binding sites in seven brain-relevant cell and tissue types (hippocampal astrocytes, cerebellar astrocytes, BE2_C neuroblastoma, retinoic acid (RA)-treated SK-N-SH neuroblastoma, choroid plexus epithelial cells, H54 glioblastoma, and brain microvascular endothelial cells) and two leukocyte-relevant cell types (lymphoblastic GM12878 and leukocytic K562 cell lines) by using all available optimal irreproducible-discovery-rate-thresholded peak BED files from ENCODE CTCF ChIP-seq experiments.39, 55 We used the GREGOR pipeline to determine whether the disease-associated SNPs or their LD proxies (at an r2 value ≥ 0.8) were more abundant in CTCF-binding sites in both disease-matched and non-disease-matched cell types than control SNPs that were matched for MAF, number of LD proxies, and distance to the nearest gene.54
To separate CTCF-binding sites common to both brain-relevant and leukocyte-relevant cell and tissue types from CTCF-binding sites unique to each group, we removed any peaks that overlapped by at least one base pair between brain-relevant and leukocyte-relevant cell and tissue types from the CTCF ChIP-seq datasets (nine datasets total: seven in brain-relevant and two in leukocyte-relevant cell and tissue types).
Results
Genetic Variation at the 7p21 Locus Associates with TMEM106B Expression
It is increasingly recognized that many GWAS-implicated variants associated with disease risk can confer their effects by altering the expression levels of nearby genes.7, 56, 57, 58, 59, 60, 61, 62, 63 In the case of the 7p21 FTLD-TDP risk locus, several studies have demonstrated such an eQTL effect for TMEM106B in multiple human tissue types, including brain and Epstein-Barr virus (EBV)-immortalized B LCLs.28, 64, 65, 66 We therefore systematically investigated the 7p21 locus for all eQTL effects in order to confirm the TMEM106B eQTL effect and to exclude other potential causal genes at this locus.
Analysis of data from 44 tissue types represented in the GTEx project26 demonstrated a robust association between the genotype at rs1990622 (the GWAS sentinel SNP) and TMEM106B expression in several cell types from healthy individuals. Specifically, multiple brain regions showed an association between the FTLD-TDP risk allele at rs1990622 and increased expression of TMEM106B, and the tissue types with the strongest eQTL effects in this direction across the GTEx data were LCLs (n = 114, p = 1.9 × 10−6), transformed fibroblasts (n = 272, p = 3.0 × 10−7) (Figure 1A), spleen (n = 89, p = 4.4 × 10−3), and the hippocampal brain region (n = 81, p = 1.3 × 10−4) and the nucleus accumbens brain region (n = 93, p = 0.01; Figure 1B). No other transcript was significantly associated genome-wide with the rs1990622 genotype, consistent with other published large-scale eQTL studies.64, 65 The association observed between rs1990622 and TMEM106B mRNA expression in human brain data corroborates previous reports of other samples.66
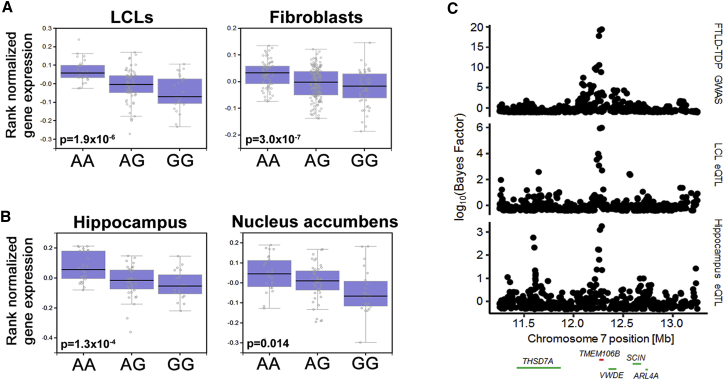
Figure 1.
Analysis of eQTL Effects at TMEM106B
(A and B) Boxplots from the GTEx data demonstrate the association between TMEM106B expression and the rs1990622 genotype (A = risk allele) in peripheral cell types (A) and human brain regions (B). Data from LCLs (n = 114), fibroblasts (n = 272), hippocampus (n = 81), and nucleus accumbens (n = 93) are shown. Black lines indicate median expression levels, lower and upper bounds of boxes indicate 25th and 75th percentile expression levels, respectively, and circles outside whiskers denote outliers. Each circle represents an individual sample.
(C) Association plots of the 2 Mb region centered on rs1990622 indicate the association between SNPs genotyped in the FTLD-TDP GWAS and FTLD-TDP (top), TMEM106B expression in GTEx LCLs (middle), and TMEM106B expression in GTEx hippocampal samples (bottom). Genomic coordinates are from the UCSC Genome Browser hg19 reference assembly, and RefSeq genes (TMEM106B highlighted in red) are indicated below the plots.
To determine whether these eQTL signals are likely to result from the same functional genetic variant(s) underlying risk of FTLD-TDP, we performed colocalization analyses over a 2 Mb region centered on TMEM106B. Both the LCL and hippocampal eQTL association signals overlapped the association signal for FTLD-TDP risk (Figure 1C). Specifically, the LCL eQTL signal had a 97% posterior probability of representing the same signal as the association with FTLD-TDP risk, thus making LCLs an attractive cellular model for investigating the molecular underpinnings of disease association at this locus.
The TMEM106B locus on 7p21 is harbored within a 36 kb LD block in samples from individuals of European ancestry, the population in which the original FTLD-TDP GWAS was performed (Figure 2A). This LD block encompasses the TMEM106B promoter, the entirety of TMEM106B, and extends ∼10 kb downstream of the gene. According to 1000 Genomes data,67 the block contains 104 genetic variants that are in strong, but not perfect, LD with rs1990622 (r2 ≥ 0.8; Figure 2A). Indeed, in-depth examination of the eQTL effect in human LCL (Figure 2B) and hippocampal (Figure 2C) samples from GTEx revealed that dozens of variants in strong LD with rs1990622 are associated with TMEM106B expression to a similar degree. We thus asked whether more than one eQTL signal occurs in this region and what the candidate causal variant(s) underlying association with disease and TMEM106B expression might be.
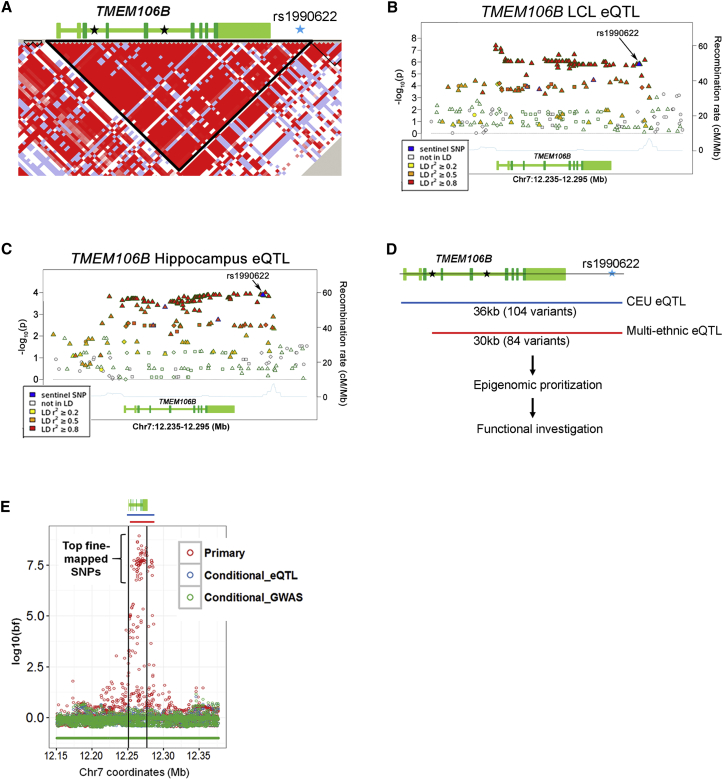
Figure 2.
LD Structure and Candidate Causal Variants at the TMEM106B Locus
(A) TMEM106B is located within a ∼36 kb LD block (inverted black triangle) in populations of European ancestry (CEU [Utah residents with ancestry from northern and western Europe]). The gene structure is indicated above the LD plot; coding exons are in dark green, UTRs and introns are in light green, and SNPs associated with FTLD-TDP by GWASs (including the sentinel SNP, rs1990622, in blue) are indicated with stars.
(B and C) The TMEM106B eQTL effect extends across the 36 kb LD block in LCLs (B) and hippocampal samples (C) from GTEx; SNPs in strong LD with rs1990622 (indicated in blue and with an arrow) show the strongest association with TMEM106B expression.
(D and E) Analysis of a multi-ethnic LCL eQTL study truncates the region of association on the 5′ end, and the remaining candidate causal variants span a ∼30 kb region (compare red lines to blue lines in D and E). Conditional analyses performed on the TMEM106B eQTL effect with the data from individuals of multiple ethnicities are shown in (E). Each circle represents a SNP; genomic positions are on the x axis, and associations with TMEM106B expression are on the y axis (log10-transformed Bayes factor). TMEM106B and regions of eQTL association are indicated above the plot and are color coded as in (D). The primary multi-ethnic eQTL analysis (red) demonstrates a strong association between a SNP cluster and TMEM106B expression. Conditioning this analysis on either the top eQTL SNP (blue) or the sentinel GWAS SNP (green) resulted in loss of an association signal at this locus (i.e., no highly associated SNPs are shown in blue or green). Genomic coordinates are from the UCSC Genome Browser hg19 reference assembly.
We began by honing the region of eQTL association. To do this, we performed a second eQTL analysis of LCLs from eight ethnic populations28 by reasoning that the different haplotype structures seen in disparate populations might refine the 36 kb LD block of association seen in individuals of European ancestry.68 We found that the addition of these populations could truncate the region of association with TMEM106B expression on the 5′ end, effectively removing the promoter and first two exons of the gene and reducing the number of potential causal variants to 84 (75 SNPs and 9 indels; Figures 2D and 2E).
We then performed conditional analyses by using the refined region of association from the multi-ethnic analysis.28 Conditioning on either the GWAS sentinel SNP, rs1990622, or the most significant eQTL SNP, rs6948844 (r2 = 0.95 with rs1990622), yielded no variants that demonstrated any residual association with TMEM106B expression within a 2 Mb region (Figure 2E). These results suggest that there is only one eQTL signal at this locus. Moreover, they corroborate the colocalization analyses suggesting that the same causal variant underlies both association with LCL TMEM106B expression and neurodegenerative disease risk.
Increased Levels of TMEM106B Expression Correlate with Increased Cellular Toxicity
If the causal variant responsible for association with TMEM106B expression levels confers risk of neurodegeneration, one would expect incremental changes in TMEM106B expression to lead to incremental effects on cellular health. We and others have previously shown that increased amounts of TMEM106B mRNA and protein result in the development of enlarged LAMP1+ late endosomes and lysosomes appearing as vacuolar structures in multiple cell types, including neurons, in association with impairment in lysosomal degradative function.21, 22, 25, 36 Indeed, lysosomal defects in general have been heavily implicated in multiple neurodegenerative diseases, including FTLD.69, 70 However, the magnitudes of reported eQTL effects in human tissues are often modest,26, 71 and so we sought to understand the effects of incremental increases in the amount of TMEM106B on disease-relevant measures such as (1) the development of the previously reported vacuolar phenotype of abnormal lysosomes and (2) cell toxicity.
To do this, we first confirmed the previously reported vacuolar phenotype, readily demonstrated by both immunofluorescence (Figure 3A) and bright-field (Figure 3B) imaging, in neurons and HeLa cells. We then employed three different TMEM106B constructs that reliably produced a spectrum of increased amounts of TMEM106B ranging from ∼2× to ∼20× (Figures 3C). In HeLa cells, in which protein amounts can be well controlled and in which the lysosomal phenotype has been well described22, 36 and is readily quantifiable, we found that with each incremental increase in the amount of TMEM106B over baseline, the percentage of cells exhibiting the vacuolar phenotype of enlarged lysosomes (Figures 3D and 3E), as well as the percentage of cell death (Figure 3F), increased. Indeed, even at modest (2×) increases in protein amounts, the percentage of cells exhibiting the vacuolar phenotype tripled, and cell death increased by 20% at 48 hr. Together, these findings suggest that genetic variation at the 7p21 locus might influence risk of neurodegeneration by altering TMEM106B-expression-dependent effects on lysosomal function and cellular health.
A Candidate Causal Regulatory Region
We next sought to identify the causal variant or variants responsible for allele-specific regulation of TMEM106B expression and, by extension, risk of FTLD-TDP. Steady-state levels of TMEM106B transcript depend on both the production of mRNA and its stability. We first considered the possibility of differential mRNA stability. In multiple LCLs homozygous at the TMEM106B locus, we found that mRNA stability did not differ between risk-haplotype homozygotes and protective-haplotype homozygotes (Figure S1), suggesting that differences in the production of mRNA account for the observed eQTL effect.
To identify variants that might have transcriptional regulatory effects, we examined the 84 candidate variants (75 SNPs and 9 indels) located within the refined region of eQTL association (Figures 4A and S2). We used data from ENCODE72 and the NIH Roadmap EpiGenome Project42 to determine (1) whether each variant was located in a predicted LCL CRE, as determined by DHS, TF binding, or an active chromatin state; and (2) whether any such CREs were also predicted to be active in primary leukocytes, brain, or brain-relevant cell lines (Material and Methods).
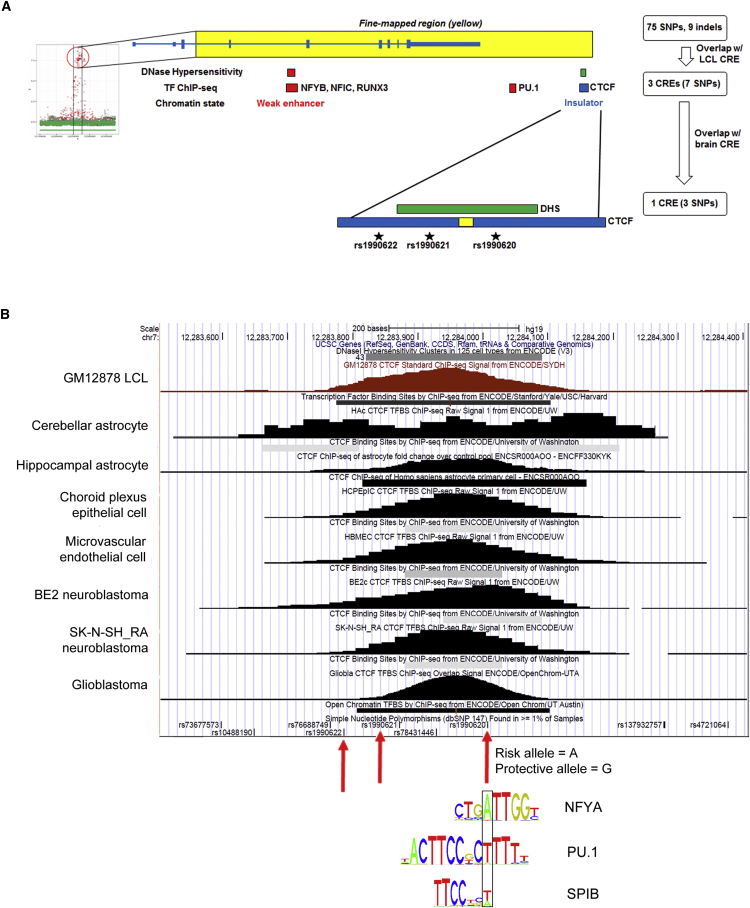
Figure 4.
Prioritization of Putative CREs Harboring Candidate Functional Variants
(A) The 84 variants from the eQTL fine-mapping (left) were prioritized on the basis of overlap with predicted CREs in LCLs (red boxes and text), neuronal and glial cell lines (green), or all three (blue) according to ENCODE and Roadmap EpiGenome data (see flow chart on the right). This analysis yielded seven SNPs in three potential CREs as candidate causal variants. Only one CRE—an intergenic CTCF-binding site (CTCF motif represented as a yellow rectangle)—was predicted to be active in brain-relevant cell lines; this CTCF-binding CRE contains three SNPs in complete LD, including the GWAS sentinel SNP, rs1990622.
(B) A UCSC Genome Browser snapshot of the CTCF-binding region shows the ENCODE DHS track and CTCF ChIP-seq peaks and signals in LCLs and all brain-relevant cell lines. The three candidate causal variants are indicated with red arrows, and the location of rs1990620 in motifs for the TFs NFYA, PU.1, and SPIB are indicated by the black box. In each case, the protective (G) allele disrupts the motifs.
Surprisingly, only seven SNPs spanning three candidate regulatory regions displayed evidence of cis-regulatory activity in LCLs (Figures 4A and S2 and Tables S1 and S2). Three SNPs located in intron 4 of TMEM106B overlapped a predicted LCL enhancer that displayed DHS and binding of the TFs NFIC, RUNX3, and NFYB (Figures 4A and S2B). A fourth SNP downstream of TMEM106B overlapped a binding site for the TF SPI1 (PU.1) in LCLs (Figures 4A and S2C). Although both the intron 4 and downstream intergenic candidate CREs also demonstrated chromatin states suggestive of enhancer activity in multiple leukocyte cell types, neither region appeared to be active in brain tissue, according to Roadmap EpiGenome data. Moreover, in all cell types, neither region bore the H3K27ac histone mark, which has been reported to distinguish active enhancers.73, 74, 75 Indeed, empirical testing of the intron 4 and downstream intergenic CREs in LCL luciferase reporter assays revealed little or no enhancer activity for these CREs (Figure S3). Furthermore, the risk and protective haplotype versions of these regions did not differ in reporter activity, suggesting that none of the overlapping SNPs affect any potential regulatory activity (Figures S3B and S3C).
The remaining candidate regulatory region contained the remaining three completely linked SNPs, one of which was rs1990622, the GWAS sentinel SNP. In virtually all ENCODE-tested cell types, including LCLs and neuronal and glial cell lines, this putative CRE displayed binding for the mammalian chromatin organizing protein CTCF (Figures 4B and S2D). Furthermore, this region lacked activity-associated histone marks in LCLs, primary leukocytes, and brain and was thus assigned an “insulator” chromatin state in LCLs (Figures 4B and S2D). We investigated this region for potential allele-specific effects in the context of CTCF and CTCF-mediated chromatin interactions.
Common Variation at rs1990620 Affects Binding of CTCF at the TMEM106B Locus
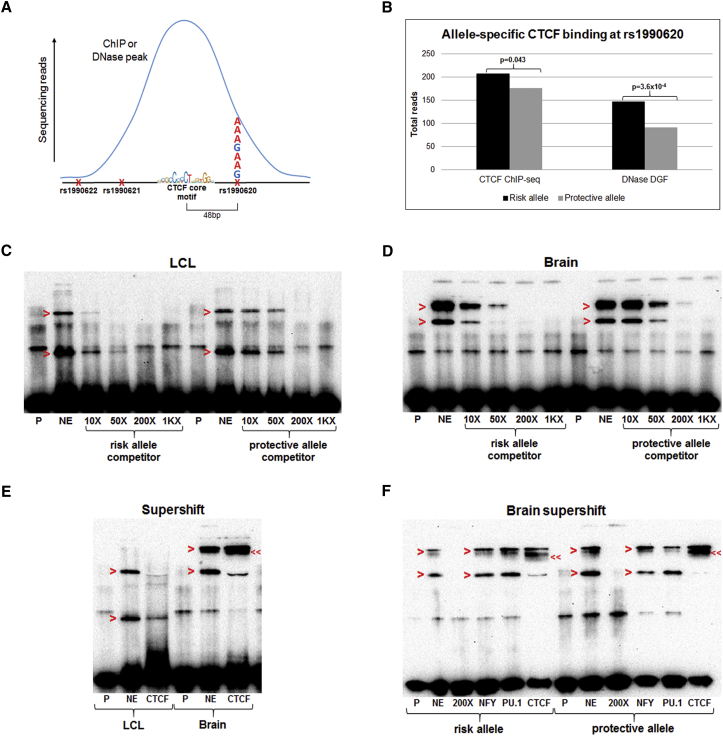
We first sought evidence for allele-specific binding of CTCF to our candidate regulatory region. To do this, we analyzed ENCODE CTCF ChIP-seq and DNase DGF experiments. CTCF ChIP-seq was performed on 99 ENCODE cell lines, 95 of which showed a CTCF peak at the putative CRE. We were able to confirm 20 of these cell lines as heterozygous for the TMEM106B haplotype by examining reads containing rs1990620, the SNP closest to (48 bp from) the CTCF core motif and covered by the most reads (Figure 5A and Tables S3 and S4). By analyzing the reads covering rs1990620, we found significant enrichment of CTCF binding to the risk-associated A allele (p = 0.043; Figure 5B and Table S4). The other two completely linked SNPs were covered by significantly fewer reads but showed similar enrichment of risk alleles, suggesting that this result was not due to technical bias in read mapping at rs1990620 (Table S4). Moreover, in the BE2 neuroblastoma line, the only cell line (of the 20 analyzed in aggregate) with adequate read numbers for individually detecting a difference in allelic proportions of the magnitude seen, we also found a significant enrichment of risk allele reads (64 risk versus 40 protective allele reads, p = 0.024). Finally, we identified six cell lines that were heterozygous at rs1990620 and interrogated by DNase DGF (Table S3). In these lines, we found that the chromosome bearing the risk A allele was significantly more sensitive to DNase cleavage (p < 0.001; Figure 5B), consistent with an open chromatin state and potential regulatory activity,76 as well as a previously reported large-scale DHS QTL study.77
Figure 5.
The Risk Allele of rs1990620 Preferentially Recruits CTCF in LCLs and Brain
(A) Schematic of the approach to determining allelic bias in CTCF ChIP-seq and DNase DGF experiments. The rs1990620 SNP (48 bp from the CTCF core motif) was analyzed for the number of reads containing the risk or protective allele in heterozygous samples showing a CTCF ChIP-seq or DNase DGF peak at this region.
(B) The risk allele of rs1990620 increased CTCF binding and DHS at this region, according to data from 20 and 6 cell types heterozygous at this locus, respectively (Tables S3 and S4). In the DGF paradigm, higher read counts correspond to higher density of DNase cleavage sites.
(C and D) A 5′ biotinylated probe (P) containing the rs1990620 risk allele was incubated with nuclear extract (NE) from LCLs (C) and human brain (D). In both extracts, the shifted probe-protein complexes (red arrowheads) were more efficiently competed with an unlabeled competitor oligonucleotide (at 10×, 50×, 200×, or 1,000× [1K×] the concentration of the labeled probe) containing the risk allele instead of the protective allele, indicating preferential binding of a nuclear factor or complex to the risk allele.
(E) The addition of an anti-CTCF antibody (lane labeled “CTCF”) diminished both LCL shifts and one of the two brain shifts (red arrowheads), corresponding in molecular weight to the higher LCL shift. Moreover, in brain extracts, an even-higher-molecular-weight supershift (double arrowheads) appeared after the addition of anti-CTCF antibody.
(F) The addition of anti-CTCF antibody, but not anti-NFY or anti-PU.1 antibody (indicated below lane), affected the EMSA shifts (red arrowheads) produced with both the risk and protective allele probes in brain extract. As seen in (E), the addition of the CTCF antibody also produced a supershift to a higher molecular weight (double arrowheads), indicating the presence of CTCF in the shifted complex.
We corroborated these data with in vitro investigations of CTCF binding by EMSAs. In addition to being closest to the CTCF core motif, rs1990620 is the only SNP in this region that is located in a DNase footprint, according to RegulomeDB.44 Furthermore, JASPAR43 and RegulomeDB predict disruption of a completely conserved nucleotide in the nuclear factor YA (NFYA) consensus motif, as well as a less conserved nucleotide in the consensus motifs for the E-twenty-six (ETS) TFs SPI1 (PU.1) and SPIB, as determined by allelic differences in rs1990620 (Figure 4B). We thus tested for rs1990620 allelic differences in the ability to produce shifts in electrophoretic mobility for brain and LCL extracts. We also tested for the presence of CTCF, NFYA, and PU.1 in the shifted complexes.
Utilizing a competition EMSA approach, we found that the risk allele of rs1990620 was more effective at shifting a protein complex in nuclear extracts from LCLs (Figure 5C) and human brain (Figures 5D and S4), whereas a negative control competitor could not outcompete the probe even at 200-fold excess concentration (Figure S5). Moreover, the addition of an anti-CTCF antibody resulted in the disappearance or diminishing of shifts in both LCL and brain extract, as well as the appearance of a supershift in the brain extract (Figure 5E), demonstrating the presence of CTCF in the shifted complex. To confirm specificity for CTCF in the shifted brain-extract complex, we compared supershift assays by using anti-CTCF, anti-NFY, and anti-PU.1 antibodies, and we performed supershift assays by using probes with both the risk and protective alleles at rs1990620. Probes for both risk and protective alleles shifted in electrophoretic mobility with the addition of brain extracts, and in both cases, the addition of anti-CTCF antibody produced a supershift, as well as the disappearance of one of the shifted bands. However, similar EMSA changes were not seen with the addition of either anti-NFY or anti-PU.1 antibody (Figure 5F). Moreover, the same supershift was seen after the addition of anti-CTCF antibody to probes biotinylated at either the 5′ end (Figure 5F) or 3′ end (Figure S6). Finally, similar EMSA investigations with probes spanning the rs1990621 or rs1990622 SNP did not demonstrate consistent allele-specific effects (data not shown).
In summary, gel shift assays and ChIP-seq-based investigation of differential CTCF binding per allele indicate that common variation at a single SNP, rs1990620, might underlie haplotype-specific effects on CTCF recruitment.
Long-Range Interactions Involving TMEM106B Demonstrate Haplotype-Specific Effects
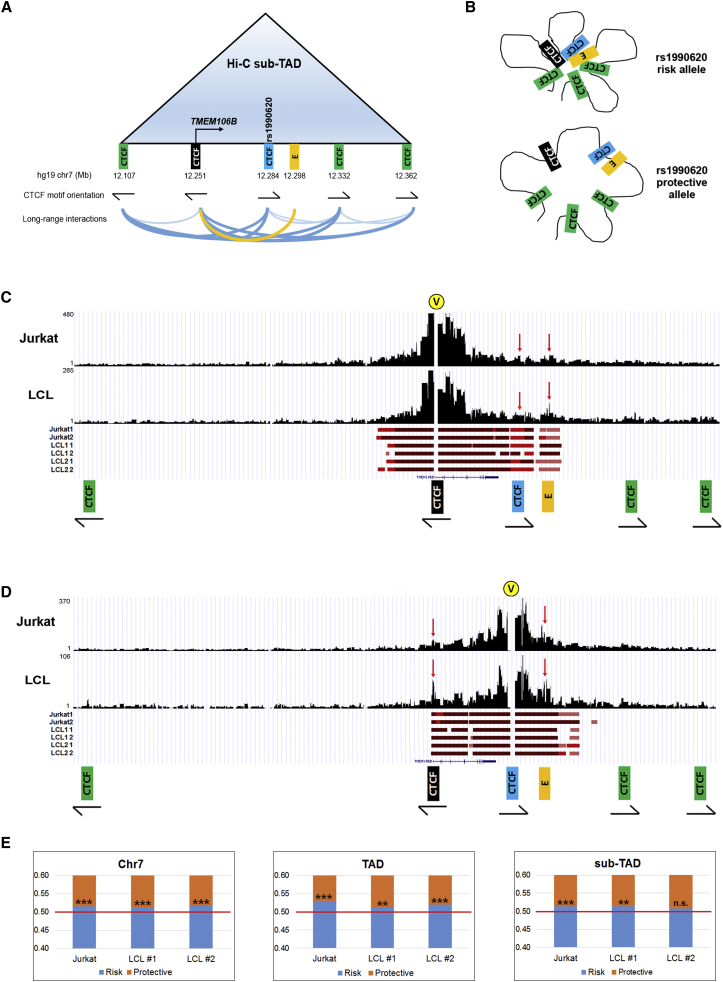
Rapidly emerging evidence suggests that CTCF plays a major role in shaping the three-dimensional architecture of the mammalian genome.78, 79 In particular, CTCF has been reported to contribute to the formation of TADs, which could be central to enhancer-promoter interactions and insulator function.80, 81, 82, 83 Because our candidate rs1990620-containing CTCF-binding CRE lacks activity-associated histone marks, we hypothesized that this CRE might function as an architectural CTCF site by contributing to the formation of a TAD involving the TMEM106B locus.
To investigate this possibility, we analyzed published high-resolution in situ LCL Hi-C heatmaps46 at the TMEM106B locus. Interestingly, these heatmaps indicate that the rs1990620-containing CRE is located within a small ∼250 kb TAD (sub-TAD) that is part of a larger ∼1 Mb TAD (Figure S7) and is involved in multiple long-range chromatin-looping interactions (Figure 6A). Moreover, one of the major interactions occurs between this region and the TMEM106B promoter, which is also located within the sub-TAD, implicating a role for this CTCF site in TMEM106B regulation (Figure 6A). Notably, within this sub-TAD, there are also Hi-C interactions between the promoter and several other CTCF sites, as well as a predicted TMEM106B enhancer84 located ∼13 kb downstream of the CTCF site (Figure 6A). This enhancer harbors no disease- or expression-associated variants and is predicted to be active in multiple primary cell types, including leukocytes and neurons, on the basis of the production of short bidirectional (enhancer RNA) transcripts.84 Thus, analysis of existing LCL Hi-C data confirmed the involvement of our candidate CRE in a TAD at the TMEM106B locus.
Figure 6.
Haplotype-Specific Long-Range Chromatin Interactions at the TMEM106B Locus
(A) Schematic representation of the TMEM106B sub-TAD and interactions among distal regulatory elements according to published LCL Hi-C data.46 The black CTCF site is located at the TMEM106B promoter, the blue CTCF site contains rs1990620, and the gold rectangle labeled “E” represents a transcriptionally active enhancer.84 Note that the CTCF motifs present at the sub-TAD boundaries (12.107 and 12.362) follow the convergent orientation (arrows indicate direction and strand) most commonly reported for interacting CTCF sites.46, 85 Blue lines at the bottom indicate Hi-C interactions between CTCF sites; darker blue lines indicate interactions between CTCF sites in convergent orientation.
(B) Model illustrating how allele-specific CTCF binding at rs1990620 might affect sub-TAD structure and long-range interactions at this locus. More contact among distal regulatory elements occurs on the risk-associated haplotype.
(C and D) Capture-C experimental data for representative Jurkat and LCL samples; raw read coverage is shown on the y axis for interactions captured by probes for (C) the TMEM106B promoter and (D) the rs1990620-containing CTCF site. Significant interactions within the sub-TAD for each cell line and replicate (three cell lines with two technical replicates each) are indicated with red bars below the coverage plots; darker shades of red indicate higher-confidence interactions. Yellow circles marked “V” indicate viewpoints (capture sites). Red arrows indicate interactions between the promoter, the rs1990620 CTCF site, and enhancer.
(E) Allelic bias in long-range interactions involving the TMEM106B promoter across all of chromosome 7 (left), the 1 Mb TAD (middle), and the 250 kb sub-TAD (right) containing TMEM106B. Read-count proportions from capture experiments containing either the risk (blue) or protective (orange) allele of a marker SNP are shown; in each case, more interactions with the TMEM106B promoter involve the risk haplotype. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; n.s. = non-significant.
Given the observed allele-specific effects on CTCF recruitment, we further hypothesized that rs1990620 might influence TMEM106B expression and, by extension, neurodegenerative disease risk through differential effects on CTCF-mediated interactions between distal regulatory elements within this TAD. Specifically, carriers of the rs1990620 risk allele might more efficiently recruit CTCF at this locus, resulting in increased long-range CTCF-mediated chromatin interactions (Figure 6B).
To test this, we adapted a recently developed variation of Capture-C48 to agnostically capture all interactions involving our candidate CRE region, as well as the TMEM106B promoter. Specifically, we coupled 3C library preparation with a probe-capture step to enrich for interactions involving our two regions of interest (Table S5). Importantly, we designed our capture probes not to overlap SNPs (thus giving probes equal opportunity to bind to either allele) while also localizing to regions within 60 bp of one or more marker SNPs (thus allowing for analysis of captured interactions in a haplotype-specific manner). We performed our Capture-C experiments in three different cell lines—two different LCLs and the T-cell-leukemia-derived Jurkat cell line, to represent both B and T cell lineages. We included Jurkat cells because cis-regulatory landscapes might be shared across leukocyte lineages, and the TMEM106B eQTL effect has also been reported in primary T cells.86, 87 All three of these cell lines are heterozygous for the TMEM106B haplotype.
When analyzing all long-range (≥2 kb) interactions mapping to chromosome 7, we found that statistically significant interactions for all three cell lines (based on an FDR threshold of 1% from fourSig)52 were largely centered on the ∼1 Mb TAD containing TMEM106B (Figure S8). When restricting the application of fourSig to regions within the TAD (thus increasing the background model), we found that virtually all statistically significant interactions mapped precisely to the sub-TAD (Figure S9). Thus, our data confirm the hierarchical nature of the chromatin architecture previously reported at this locus in LCLs by in situ Hi-C and further suggest that both B- and T-cell-derived cell lines share common TAD boundaries at the 7p21 locus. Importantly, all interactions between the five sub-TAD CTCF sites (including the TMEM106B promoter) and the enhancer were statistically significant in every sample regardless of whether we captured interactions at the TMEM106B promoter or our candidate CTCF-binding CRE (Figure S9). Moreover, the outermost CTCF sites followed a convergent orientation, consistent with sites delineating boundaries of a topological domain.
To obtain a finer-grained understanding of the most meaningful interactions at our locus, we further restricted the fourSig analysis to the ∼250 kb sub-TAD, which further increased the background threshold for significance. Under these conditions, statistically significant interactions among the TMEM106B promoter, the rs1990620-containing CTCF site, and the enhancer emerged (Figures 6C and 6D). Although these interactions were significant for both LCLs and Jurkat cells, they were qualitatively more apparent in LCLs, suggesting that these interactions might be more functionally important in LCLs. Together, these results implicate the CTCF site and the enhancer as potential key regulators of TMEM106B.
Recent studies have suggested that genes involved in CTCF-associated long-range interactions tend to be more transcriptionally active than genes not involved in such interactions.46, 85 Therefore, we asked whether the number of long-range chromatin-looping interactions involving the risk haplotype, which preferentially binds CTCF and expresses TMEM106B at higher levels, is significantly higher than the number of interactions involving the protective haplotype in these heterozygous cell lines (model depicted in Figure 6B). In all three cell lines, we observed significantly more interactions captured with the promoter probes occurring on the risk haplotype, and the effects were consistent regardless of whether we analyzed all interactions mapping to chromosome 7 or restricted the analysis to interactions within the TAD or sub-TAD (Figure 6E and Table S6). When interactions were captured with probes targeting the rs1990620-containing CTCF-binding site, fewer reads were obtained, and no apparent difference was detected between raw reads involving the risk haplotype and those involving the protective haplotype.
Given that various sources of technical bias (e.g., capture bias, alignment bias, and bias in duplicate removal) can influence allele-specific high-throughput sequencing analyses, we next compared the number of long-range interactions involving each haplotype (“true” interactions) against the number of reads aligning to regions directly adjacent to the bait regions (“false” interactions). We assumed that regions directly adjacent to the bait regions would be subject to artifactual ligations simply because of chromosomal proximity, as has been previously suggested.53 Although some true functional interactions could be lost in this way (creating false negatives), this approach let us test for false-positive differences in interactions between the two haplotypes by capturing biases in read alignment or other technical steps of the Capture-C protocol.
After adjustment for technical bias, we still observed significant enrichment of promoter-captured interactions on the risk haplotype in two of three cell lines (p = 1.98 × 10−2 for Jurkat cells and p = 5.78 × 10−3 for LCL 2 for significant deviation from expected proportions). Moreover, the same two cell lines demonstrated a significant enrichment of CTCF-site-captured interactions on the risk haplotype regardless of whether probes captured interactions adjacent to the rs1990620 SNP (p = 3.28 × 10−3 for Jurkat cells and p = 4.37 × 10−3 for LCL 2) or the rs1990621 SNP (p < 1.00 × 10−6 for Jurkat cells and p = 2.38 × 10−2 for LCL 2). Thus, even after bias corrections that were most likely conservative (Material and Methods), two of three cell lines exhibited stronger long-range interactions on the chromosome bearing the FTLD-TDP risk haplotype. Moreover, the preferential involvement of the FTLD-TDP risk haplotype in these long-range interactions was invariant to the “viewpoint” used (i.e., whether captured at the TMEM106B promoter or at the distal CTCF-binding site).
Together, these data suggest that SNP-specific effects on CTCF recruitment might alter the genomic architecture at the TMEM106B locus and thus lead to alterations in gene expression.
Common Genetic Variants Associated with Neurodegenerative Diseases Are Enriched in Brain CTCF-Binding Sites
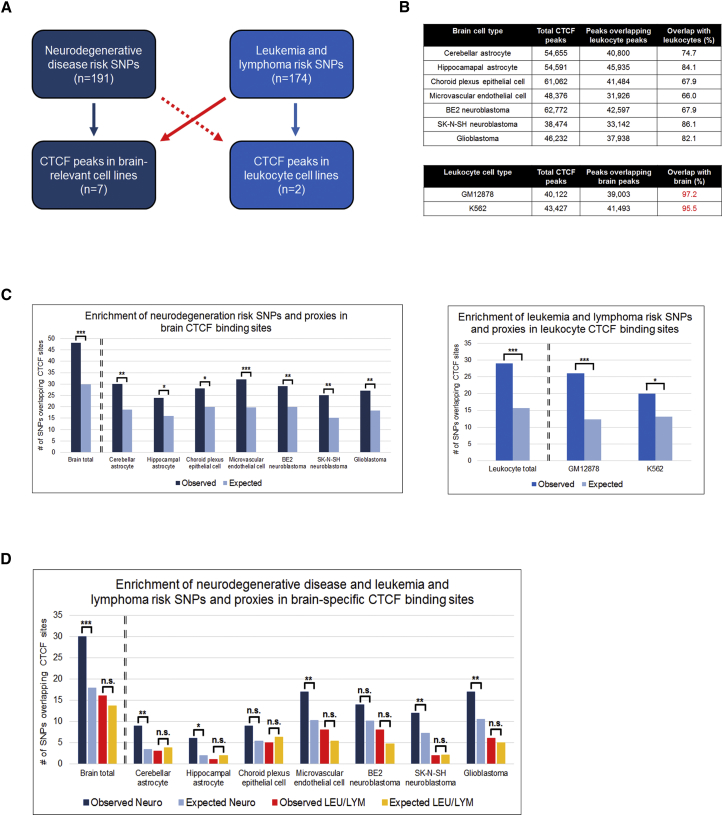
CTCF is emerging as a master regulator of mammalian gene expression through its widespread influences on genomic architecture.78, 79, 88 Having uncovered an allele-specific effect on the expression of TMEM106B, a frontotemporal dementia-associated genetic risk factor most likely mediated by CTCF, we hypothesized that CTCF-mediated effects might play a more general role in conferring risk of neurodegeneration.
To test this, we identified all published SNPs associated with risk of four major neurodegenerative diseases (FTLD, AD, PD, and ALS) at a genome-wide statistical significance level. 200 risk SNPs for neurodegenerative disease were identified from the NHGRI-EBI GWAS Catalog,2 and 191/200 SNPs (nine SNPs could not be matched with the algorithm used; Table S7) and their LD proxies (r2 ≥ 0.8) were investigated for the extent of overlap with brain CTCF-binding sites identified by ChIP-seq72 (Figure 7A, dark-blue boxes).
Figure 7.
Risk SNPs for Neurodegenerative Disease Are Enriched in Brain-Specific CTCF-Binding Sites
(A) Using the GWAS Catalog, we identified 191 risk SNPs for neurodegenerative disease and 174 risk SNPs for lymphoma and leukemia. We then determined the overlap between disease risk SNPs, as well as their LD proxies, and CTCF-binding sites either in disease-relevant cell lines (“matched” analyses, indicated by blue arrows) or in disease-irrelevant cell lines (“unmatched” analyses, indicated by red arrows).
(B) To perform the “unmatched” analyses, we identified a set of CTCF-binding sites that were brain specific (i.e., found in brain-relevant cell types but absent in leukocyte-relevant cell types) and a set of CTCF-binding sites that were leukocyte specific (i.e., found in leukocyte-relevant cell types but absent in brain-relevant cell types). Whereas brain-specific CTCF-binding sites represented 14%–34% of total brain CTCF-binding sites, only 2%–4% of total leukocyte CTCF-binding sites were specific to leukocytes.
(C) Neurodegenerative risk SNPs were significantly enriched in CTCF-binding sites in all seven brain-relevant cell lines (left), and lymphoma and leukemia risk SNPs were significantly enriched in CTCF-binding sites in the leukocytic GM12878 and K562 cell lines (right).
(D) When we constrained our analysis to only the brain-specific CTCF-binding sites, risk SNPs for neurodegenerative disease (Neuro; blue bars) remained significantly enriched in CTCF-binding sites in five of seven brain-relevant cell lines. However, leukemia and lymphoma risk SNPs (LEU/LYM; red and orange bars) were not significantly enriched in brain-specific CTCF-binding sites.
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; n.s., non-significant.
Seven human-brain-relevant CTCF ChIP-seq datasets (Figure 7B, top) were identified from ENCODE data.72 Across the combined set of all seven datasets, using the GREGOR algorithm,54 we found a highly significant ∼1.6-fold enrichment of neurodegenerative-disease SNPs and their LD proxies overlapping CTCF-binding sites (p < 0.0001; Figure 7C). Moreover, the risk SNPs for neurodegenerative disease were significantly enriched in CTCF-binding sites in each of the seven brain-relevant tissue and cell types individually; the most significant enrichments were seen in brain-derived microvascular endothelial cells (1.6-fold enrichment, p < 0.001), cerebellar astrocytes (1.6-fold enrichment, p < 0.01), and RA-treated SK-N-SH neuroblastoma cells (1.6-fold enrichment, p < 0.01; Figure 7C).
We next asked whether the overlap between disease risk SNPs and CTCF-binding sites was specific to (1) neurodegenerative diseases or (2) brain-relevant tissue and cell types. To answer these questions, we analyzed 174 SNPs implicated in risk of leukemia or lymphoma (Table S8), as well as CTCF-binding sites in leukocyte-relevant cell types. In particular, we asked whether results for “matched” analyses (Figure 7A, blue arrows: SNPs implicated in brain-relevant diseases and paired with CTCF-binding sites in brain-relevant tissues; SNPs implicated in leukocyte-relevant diseases and paired with CTCF-binding sites in leukocyte-relevant cells) would differ from results for “unmatched” analyses (Figure 7A, red arrows: SNPs implicated in brain-relevant diseases and paired with CTCF-binding sites in leukocyte-relevant cells; SNPs implicated in leukocyte-relevant diseases and paired with CTCF-binding sites in brain-relevant tissues).
As seen for the “matched” analysis of neurodegenerative diseases, leukemia and lymphoma risk SNPs were significantly enriched in CTCF-binding sites for leukocyte-relevant cell types (1.8-fold enrichment, p < 0.001; Figure 7C). Furthermore, a “matched” analysis of neurodegenerative-disease risk SNPs in CTCF peaks that were found only in brain-relevant cell lines showed significant enrichment (Figure 7D, blue bars, 1.7-fold enrichment, p < 0.001). However, an “unmatched” analysis of leukemia and lymphoma risk SNPs in these brain-specific CTCF sites showed no enrichment (Figure 7D, orange bars). We were unable to perform an “unmatched” analysis of neurodegenerative-disease risk SNPs in CTCF-binding sites for leukocyte-relevant cell types because >95% of CTCF-binding sites found in brain-relevant tissues were also found in leukocyte-relevant cells, precluding our ability to identify CTCF-binding sites unique to leukocytes (Figure 7B, bottom).
Together, these results suggest that allele-specific effects on CTCF-mediated gene-regulation programs might underlie additional risk loci for other diseases. Moreover, despite some evidence of cell-type-specific regulatory mechanisms, the extensive overlap between leukocyte CTCF-binding sites and brain CTCF-binding sites provides further evidence that the CTCF-mediated chromatin interactions described here in LCLs are applicable to the brain as well.
Discussion
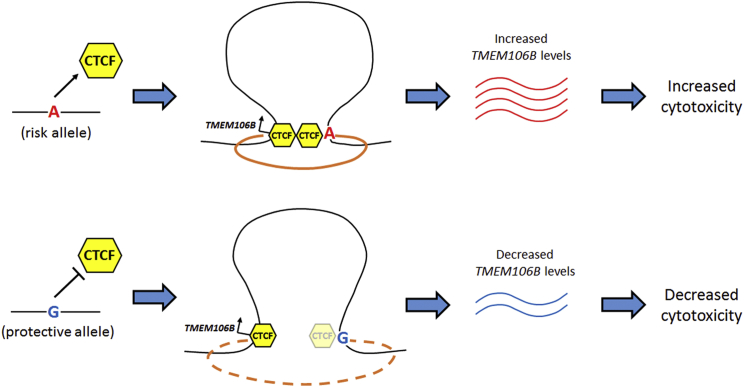
Here, we functionally dissect a locus first linked to the human neurodegenerative disease FTLD by a GWAS. Through a combination of data mining and bench-based experimental studies of human-derived tissues, we have demonstrated that common variants linked to FTLD by a GWAS associate with haplotype-specific expression of TMEM106B in multiple tissues (including the human brain), that this effect might depend on haplotype-specific effects on recruitment of CTCF and corresponding haplotype-specific effects on long-range chromatin interactions, and that incremental increases in the amount of TMEM106B have effects on cell toxicity (Figure 8). Thus, we provide functional characterization of the mechanism by which a causal variant at a GWAS-implicated locus might exert downstream effects on target gene expression and FTLD risk.
Figure 8.
Working Model of the Molecular Mechanism Underlying the 7p21 Association with Neurodegeneration
The risk-associated allele of the causal variant (rs1990620) preferentially recruited CTCF, resulting in haplotype-specific effects on long-range chromatin interactions with downstream effects of increased TMEM106B expression. Increased TMEM106B expression led to increased cytotoxicity and corresponding risk of neurodegeneration.
We note that our model makes certain assumptions; future work in these areas will be a valuable addition to the findings of the current study. First, we presume that allele-based effects on long-range chromatin interactions found in leukocyte cell lines can be extended to tissues and cell types relevant to neurodegeneration. Justification for this assumption comes from our EMSA data demonstrating the preferential recruitment of CTCF by the rs1990620 risk allele in both LCLs and brain, the significant overlap (>95%) between CTCF-binding sites found in leukocytes and CTCF-binding sites found in brain, and our colocalization analysis suggesting a 97% posterior probability of a shared causal variant for both the TMEM106B LCL eQTL effect and association with FTLD. Second, we presume that the CTCF-mediated long-range interactions preferentially involving the FTLD risk haplotype result in increased rather than decreased expression of TMEM106B. These results are consistent with published studies implicating a role for TADs in facilitating transcriptional activation of genes within the TADs through long-range regulatory effects.46, 78, 85 In addition, TADs with allele-specific CTCF-mediated long-range interactions have been associated with allele-specific expression of the genes within these TADs, such that stronger interactions correlate with increased gene transcription.85 Furthermore, our model is consistent with emerging literature on the role of CTCF in the delineation of TADs, insofar as the rs1990620-containing CTCF site investigated in our study is located within a sub-TAD between CTCF-binding sites delineating TAD boundaries (see Figure 6A). As such, binding to the rs1990620-containing CTCF site would not be expected to have the insulating properties of a boundary site and might instead strengthen intra-TAD interactions between the TMEM106B promoter and the downstream enhancer. Indeed, in the data shown in Figure 6, the promoter-enhancer interaction is qualitatively the strongest interaction seen in our Capture-C experiments. Third, we presume that incremental (versus all-or-none) allele-based effects on recruitment of CTCF, long-range chromatin interactions, or TMEM106B expression can nevertheless explain disease association. In this regard, we note that similar modest, incremental effects have been reported for many cases of allele-specific expression,7, 26, 63, 71, 89, 90, 91 including allele-specific expression differences associated with neuropsychiatric disease variants.7, 92 Indeed, conceptually, such an incremental effect is not surprising for common genetic variants that confer only slightly increased odds of developing a disease, yet they could still shed light on important disease mechanisms.
Aspects of the work presented here could be more broadly applicable to common-variant effects on risk of many neurodegenerative diseases or, even more broadly, to many common-variant-trait associations. For example, here, the genotype at the causal noncoding variant rs1990620 alters FTLD risk through an effect on TMEM106B expression. The enrichment of disease-associated variants in predicted cis-regulatory regions56, 57 and the overlap between these variants and variants associated with gene expression levels (eQTLs)57, 93, 94, 95 suggest that many common variants identified by GWASs might act by modulating gene expression.
Moreover, we found that even beyond the example of rs1990620 and FTLD, SNPs associated with risk of other neurodegenerative diseases by GWASs and SNPs associated with risk of leukemia and lymphoma by GWASs are both enriched in CTCF-binding sites. These results suggest that allele-specific modulation of gene-expression programs influenced by CTCF in particular could underlie additional risk factors for other human diseases. Indeed, our results agree with previous studies demonstrating enrichment of trait-associated variants in CTCF-binding sites,57 although no such enrichment has been seen when only CTCF sites that lack histone modifications are considered.96 It is possible, of course, that our reported enrichment of disease SNPs found in CTCF-binding sites is somewhat artifactual. However, the fact that the leukemia and lymphoma risk SNPs did not overlap CTCF-binding sites found in brain tissues in our “mismatched” analysis argues against this possibility.
Key aspects of our study are worth emphasizing. First, in the (few) post-GWAS studies that have mechanistically elucidated causal variants affecting disease risk through eQTL effects,7, 58, 59, 60, 62, 63, 97, 98 most of the proposed causal variants lie in regions with enhancer- or promoter-associated features. In contrast, our results characterize a GWAS causal variant located within an architectural cis-regulatory element. That is, although some GWAS causal variants have been reportedly involved in allele-specific long-range chromatin interactions, such as enhancer-promoter interactions, our results suggest a direct effect of a GWAS causal variant on higher-order chromatin architecture. Second, the degree of molecular precision provided here is largely absent in the characterization of neurodegenerative-disease loci first discovered by GWASs. However, this level of mechanistic detail illuminating the genetic regulation and biological function of GWAS-derived loci is certainly needed if we are to translate the thousands of “leads” obtained in this way into potential avenues for therapeutic interventions. In this context, the strategy illustrated here of prioritizing variants on the basis of the wealth of newly available genomic data and subsequently targeting investigation to cell-culture systems could be more broadly applicable to the study of common variants associated with other human diseases.
Acknowledgments
We would like to thank Dr. Jonathan Schug of the University of Pennsylvania for conceptual and technical expertise in high-throughput sequencing experiments and Nathaniel D. Berkowitz for computational assistance. Funding sources for this work include the NIH (RO1 NS082265, UO1 HL129998, RO1 MH101822, and F31 NS090892) and the Burroughs Wellcome Fund.
Published: October 19, 2017
Footnotes
Supplemental Data include nine figures and eight tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.09.004.
Accession Numbers
The Capture-C data from lymphoblastoid and Jurkat cell lines has been deposited in the Sequence Read Archive under BioProject accession number PRJNA413093 (study SRP119373).
Web Resources
ENCODE, https://www.encodeproject.org/
FastQC, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
GTEx Portal, https://www.gtexportal.org/home/
HaploReg, http://archive.broadinstitute.org/mammals/haploreg/haploreg.php
JASPAR Database, http://jaspar.genereg.net/
NHGRI-EBI GWAS Catalog, https://www.ebi.ac.uk/gwas/
OMIM, http://omim.org/
RegulomeDB, http://www.regulomedb.org/
Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra
UCSC Genome Browser, https://genome.ucsc.edu/
WashU EpiGenome Browser, http://epigenomegateway.wustl.edu/browser/
Supplemental Data
References
- 1.Chen S., Zheng J.C. Translational Neurodegeneration, a platform to share knowledge and experience in translational study of neurodegenerative diseases. Transl. Neurodegener. 2012;1:1. doi: 10.1186/2047-9158-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abraham G., Inouye M. Genomic risk prediction of complex human disease and its clinical application. Curr. Opin. Genet. Dev. 2015;33:10–16. doi: 10.1016/j.gde.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M., DeStefano A.L., Kara E., Bras J., Sharma M., International Parkinson’s Disease Genomics Consortium (IPDGC) Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized GENetics Initiative (PROGENI) 23andMe. GenePD. NeuroGenetics Research Consortium (NGRC) Hussman Institute of Human Genomics (HIHG) Ashkenazi Jewish Dataset Investigator. Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE) North American Brain Expression Consortium (NABEC) United Kingdom Brain Expression Consortium (UKBEC) Greek Parkinson’s Disease Consortium. Alzheimer Genetic Analysis Group Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramanan V.K., Saykin A.J. Pathways to neurodegeneration: mechanistic insights from GWAS in Alzheimer’s disease, Parkinson’s disease, and related disorders. Am. J. Neurodegener. Dis. 2013;2:145–175. [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards S.L., Beesley J., French J.D., Dunning A.M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soldner F., Stelzer Y., Shivalila C.S., Abraham B.J., Latourelle J.C., Barrasa M.I., Goldmann J., Myers R.H., Young R.A., Jaenisch R. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533:95–99. doi: 10.1038/nature17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bang J., Spina S., Miller B.L. Frontotemporal dementia. Lancet. 2015;386:1672–1682. doi: 10.1016/S0140-6736(15)00461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seelaar H., Rohrer J.D., Pijnenburg Y.A., Fox N.C., van Swieten J.C. Clinical, genetic and pathological heterogeneity of frontotemporal dementia: a review. J. Neurol. Neurosurg. Psychiatry. 2011;82:476–486. doi: 10.1136/jnnp.2010.212225. [DOI] [PubMed] [Google Scholar]
- 10.Van Deerlin V.M., Sleiman P.M., Martinez-Lage M., Chen-Plotkin A., Wang L.S., Graff-Radford N.R., Dickson D.W., Rademakers R., Boeve B.F., Grossman M. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat. Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Zee J., Van Langenhove T., Kleinberger G., Sleegers K., Engelborghs S., Vandenberghe R., Santens P., Van den Broeck M., Joris G., Brys J. TMEM106B is associated with frontotemporal lobar degeneration in a clinically diagnosed patient cohort. Brain. 2011;134:808–815. doi: 10.1093/brain/awr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finch N., Carrasquillo M.M., Baker M., Rutherford N.J., Coppola G., Dejesus-Hernandez M., Crook R., Hunter T., Ghidoni R., Benussi L. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology. 2011;76:467–474. doi: 10.1212/WNL.0b013e31820a0e3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernández I., Rosende-Roca M., Alegret M., Mauleón A., Espinosa A., Vargas L., Sotolongo-Grau O., Tárraga L., Boada M., Ruiz A. Association of TMEM106B rs1990622 marker and frontotemporal dementia: evidence for a recessive effect and meta-analysis. J. Alzheimers Dis. 2015;43:325–334. doi: 10.3233/JAD-132432. [DOI] [PubMed] [Google Scholar]
- 14.Cruchaga C., Graff C., Chiang H.H., Wang J., Hinrichs A.L., Spiegel N., Bertelsen S., Mayo K., Norton J.B., Morris J.C., Goate A. Association of TMEM106B gene polymorphism with age at onset in granulin mutation carriers and plasma granulin protein levels. Arch. Neurol. 2011;68:581–586. doi: 10.1001/archneurol.2010.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher M.D., Suh E., Grossman M., Elman L., McCluskey L., Van Swieten J.C., Al-Sarraj S., Neumann M., Gelpi E., Ghetti B. TMEM106B is a genetic modifier of frontotemporal lobar degeneration with C9orf72 hexanucleotide repeat expansions. Acta Neuropathol. 2014;127:407–418. doi: 10.1007/s00401-013-1239-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Blitterswijk M., Mullen B., Nicholson A.M., Bieniek K.F., Heckman M.G., Baker M.C., DeJesus-Hernandez M., Finch N.A., Brown P.H., Murray M.E. TMEM106B protects C9ORF72 expansion carriers against frontotemporal dementia. Acta Neuropathol. 2014;127:397–406. doi: 10.1007/s00401-013-1240-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vass R., Ashbridge E., Geser F., Hu W.T., Grossman M., Clay-Falcone D., Elman L., McCluskey L., Lee V.M., Van Deerlin V.M. Risk genotypes at TMEM106B are associated with cognitive impairment in amyotrophic lateral sclerosis. Acta Neuropathol. 2011;121:373–380. doi: 10.1007/s00401-010-0782-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhinn H., Abeliovich A. Differential Aging Analysis in Human Cerebral Cortex Identifies Variants in TMEM106B and GRN that Regulate Aging Phenotypes. Cell Syst. 2017;4:404–415.e5. doi: 10.1016/j.cels.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 19.White C.C., Yang H.S., Yu L., Chibnik L.B., Dawe R.J., Yang J., Klein H.U., Felsky D., Ramos-Miguel A., Arfanakis K. Identification of genes associated with dissociation of cognitive performance and neuropathological burden: Multistep analysis of genetic, epigenetic, and transcriptional data. PLoS Med. 2017;14:e1002287. doi: 10.1371/journal.pmed.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang C.M., Fellerer K., Schwenk B.M., Kuhn P.H., Kremmer E., Edbauer D., Capell A., Haass C. Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J. Biol. Chem. 2012;287:19355–19365. doi: 10.1074/jbc.M112.365098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brady O.A., Zheng Y., Murphy K., Huang M., Hu F. The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum. Mol. Genet. 2013;22:685–695. doi: 10.1093/hmg/dds475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen-Plotkin A.S., Unger T.L., Gallagher M.D., Bill E., Kwong L.K., Volpicelli-Daley L., Busch J.I., Akle S., Grossman M., Van Deerlin V. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J. Neurosci. 2012;32:11213–11227. doi: 10.1523/JNEUROSCI.0521-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jun M.H., Han J.H., Lee Y.K., Jang D.J., Kaang B.K., Lee J.A. TMEM106B, a frontotemporal lobar dementia (FTLD) modifier, associates with FTD-3-linked CHMP2B, a complex of ESCRT-III. Mol. Brain. 2015;8:85. doi: 10.1186/s13041-015-0177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholson A.M., Finch N.A., Wojtas A., Baker M.C., Perkerson R.B., 3rd, Castanedes-Casey M., Rousseau L., Benussi L., Binetti G., Ghidoni R. TMEM106B p.T185S regulates TMEM106B protein levels: implications for frontotemporal dementia. J. Neurochem. 2013;126:781–791. doi: 10.1111/jnc.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stagi M., Klein Z.A., Gould T.J., Bewersdorf J., Strittmatter S.M. Lysosome size, motility and stress response regulated by fronto-temporal dementia modifier TMEM106B. Mol. Cell. Neurosci. 2014;61:226–240. doi: 10.1016/j.mcn.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold M., Raffler J., Pfeufer A., Suhre K., Kastenmüller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics. 2015;31:1334–1336. doi: 10.1093/bioinformatics/btu779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stranger B.E., Montgomery S.B., Dimas A.S., Parts L., Stegle O., Ingle C.E., Sekowska M., Smith G.D., Evans D., Gutierrez-Arcelus M. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown C.D., Mangravite L.M., Engelhardt B.E. Integrative modeling of eQTLs and cis-regulatory elements suggests mechanisms underlying cell type specificity of eQTLs. PLoS Genet. 2013;9:e1003649. doi: 10.1371/journal.pgen.1003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 34.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Busch J.I., Unger T.L., Jain N., Tyler Skrinak R., Charan R.A., Chen-Plotkin A.S. Increased expression of the frontotemporal dementia risk factor TMEM106B causes C9orf72-dependent alterations in lysosomes. Hum. Mol. Genet. 2016;25:2681–2697. doi: 10.1093/hmg/ddw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinrichs A.S., Raney B.J., Speir M.L., Rhead B., Casper J., Karolchik D., Kuhn R.M., Rosenbloom K.R., Zweig A.S., Haussler D., Kent W.J. UCSC Data Integrator and Variant Annotation Integrator. Bioinformatics. 2016;32:1430–1432. doi: 10.1093/bioinformatics/btv766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.K., Cheng C., Mu X.J., Khurana E., Rozowsky J., Alexander R. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X., Wang T. Using the Wash U Epigenome Browser to examine genome-wide sequencing data. Curr. Protoc. Bioinformatics. 2012;Chapter 10:10. doi: 10.1002/0471250953.bi1010s40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.Y., Chou A., Ienasescu H. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durand N.C., Robinson J.T., Shamim M.S., Machol I., Mesirov J.P., Lander E.S., Aiden E.L. Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst. 2016;3:99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hughes J.R., Roberts N., McGowan S., Hay D., Giannoulatou E., Lynch M., De Gobbi M., Taylor S., Gibbons R., Higgs D.R. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 2014;46:205–212. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- 49.Hakim O., Misteli T. SnapShot: Chromosome confirmation capture. Cell. 2012;148:1068.e1–1068.e2. doi: 10.1016/j.cell.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davies J.O., Telenius J.M., McGowan S.J., Roberts N.A., Taylor S., Higgs D.R., Hughes J.R. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat. Methods. 2016;13:74–80. doi: 10.1038/nmeth.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams R.L., Jr., Starmer J., Mugford J.W., Calabrese J.M., Mieczkowski P., Yee D., Magnuson T. fourSig: a method for determining chromosomal interactions in 4C-Seq data. Nucleic Acids Res. 2014;42:e68. doi: 10.1093/nar/gku156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cairns J., Freire-Pritchett P., Wingett S.W., Várnai C., Dimond A., Plagnol V., Zerbino D., Schoenfelder S., Javierre B.M., Osborne C. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 2016;17:127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt E.M., Zhang J., Zhou W., Chen J., Mohlke K.L., Chen Y.E., Willer C.J. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics. 2015;31:2601–2606. doi: 10.1093/bioinformatics/btv201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schaub M.A., Boyle A.P., Kundaje A., Batzoglou S., Snyder M. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang Q., Whitington T., Gao P., Lindberg J.F., Yang Y., Sun J., Väisänen M.R., Szulkin R., Annala M., Yan J. A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nat. Genet. 2014;46:126–135. doi: 10.1038/ng.2862. [DOI] [PubMed] [Google Scholar]
- 60.Bauer D.E., Kamran S.C., Lessard S., Xu J., Fujiwara Y., Lin C., Shao Z., Canver M.C., Smith E.C., Pinello L. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dunning A.M., Michailidou K., Kuchenbaecker K.B., Thompson D., French J.D., Beesley J., Healey C.S., Kar S., Pooley K.A., Lopez-Knowles E., EMBRACE. GEMO Study Collaborators. HEBON. kConFab Investigators Breast cancer risk variants at 6q25 display different phenotype associations and regulate ESR1, RMND1 and CCDC170. Nat. Genet. 2016;48:374–386. doi: 10.1038/ng.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smemo S., Tena J.J., Kim K.H., Gamazon E.R., Sakabe N.J., Gómez-Marín C., Aneas I., Credidio F.L., Sobreira D.R., Wasserman N.F. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Claussnitzer M., Dankel S.N., Kim K.H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 65.Liang L., Morar N., Dixon A.L., Lathrop G.M., Abecasis G.R., Moffatt M.F., Cookson W.O. A cross-platform analysis of 14,177 expression quantitative trait loci derived from lymphoblastoid cell lines. Genome Res. 2013;23:716–726. doi: 10.1101/gr.142521.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu L., De Jager P.L., Yang J., Trojanowski J.Q., Bennett D.A., Schneider J.A. The TMEM106B locus and TDP-43 pathology in older persons without FTLD. Neurology. 2015;84:927–934. doi: 10.1212/WNL.0000000000001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dunning A.M., Durocher F., Healey C.S., Teare M.D., McBride S.E., Carlomagno F., Xu C.F., Dawson E., Rhodes S., Ueda S. The extent of linkage disequilibrium in four populations with distinct demographic histories. Am. J. Hum. Genet. 2000;67:1544–1554. doi: 10.1086/316906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fraldi A., Klein A.D., Medina D.L., Settembre C. Brain Disorders Due to Lysosomal Dysfunction. Annu. Rev. Neurosci. 2016;39:277–295. doi: 10.1146/annurev-neuro-070815-014031. [DOI] [PubMed] [Google Scholar]
- 70.Nixon R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 71.Dimas A.S., Deutsch S., Stranger B.E., Montgomery S.B., Borel C., Attar-Cohen H., Ingle C., Beazley C., Gutierrez Arcelus M., Sekowska M. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zentner G.E., Tesar P.J., Scacheri P.C. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crawford G.E., Holt I.E., Whittle J., Webb B.D., Tai D., Davis S., Margulies E.H., Chen Y., Bernat J.A., Ginsburg D. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maurano M.T., Haugen E., Sandstrom R., Vierstra J., Shafer A., Kaul R., Stamatoyannopoulos J.A. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat. Genet. 2015;47:1393–1401. doi: 10.1038/ng.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Merkenschlager M., Nora E.P. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu. Rev. Genomics Hum. Genet. 2016;17:17–43. doi: 10.1146/annurev-genom-083115-022339. [DOI] [PubMed] [Google Scholar]
- 79.Ong C.T., Corces V.G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guo Y., Xu Q., Canzio D., Shou J., Li J., Gorkin D.U., Jung I., Wu H., Zhai Y., Tang Y. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lupiáñez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang Z., Luo O.J., Li X., Zheng M., Zhu J.J., Szalaj P., Trzaskoma P., Magalska A., Wlodarczyk J., Ruszczycki B. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peters J.E., Lyons P.A., Lee J.C., Richard A.C., Fortune M.D., Newcombe P.J., Richardson S., Smith K.G. Insight into Genotype-Phenotype Associations through eQTL Mapping in Multiple Cell Types in Health and Immune-Mediated Disease. PLoS Genet. 2016;12:e1005908. doi: 10.1371/journal.pgen.1005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Raj T., Rothamel K., Mostafavi S., Ye C., Lee M.N., Replogle J.M., Feng T., Lee M., Asinovski N., Frohlich I. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Won H., de la Torre-Ubieta L., Stein J.L., Parikshak N.N., Huang J., Opland C.K., Gandal M.J., Sutton G.J., Hormozdiari F., Lu D. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tewhey R., Kotliar D., Park D.S., Liu B., Winnicki S., Reilly S.K., Andersen K.G., Mikkelsen T.S., Lander E.S., Schaffner S.F., Sabeti P.C. Direct Identification of Hundreds of Expression-Modulating Variants using a Multiplexed Reporter Assay. Cell. 2016;165:1519–1529. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Patwardhan R.P., Hiatt J.B., Witten D.M., Kim M.J., Smith R.P., May D., Lee C., Andrie J.M., Lee S.I., Cooper G.M. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spisák S., Lawrenson K., Fu Y., Csabai I., Cottman R.T., Seo J.H., Haiman C., Han Y., Lenci R., Li Q., GAME-ON/ELLIPSE Consortium CAUSEL: an epigenome- and genome-editing pipeline for establishing function of noncoding GWAS variants. Nat. Med. 2015;21:1357–1363. doi: 10.1038/nm.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sekar A., Bialas A.R., de Rivera H., Davis A., Hammond T.R., Kamitaki N., Tooley K., Presumey J., Baum M., Van Doren V., Schizophrenia Working Group of the Psychiatric Genomics Consortium Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 95.Fu J., Wolfs M.G., Deelen P., Westra H.J., Fehrmann R.S., Te Meerman G.J., Buurman W.A., Rensen S.S., Groen H.J., Weersma R.K. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet. 2012;8:e1002431. doi: 10.1371/journal.pgen.1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.R., Anttila V., Xu H., Zang C., Farh K., ReproGen Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Adrianto I., Wen F., Templeton A., Wiley G., King J.B., Lessard C.J., Bates J.S., Hu Y., Kelly J.A., Kaufman K.M., BIOLUPUS and GENLES Networks Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat. Genet. 2011;43:253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harismendy O., Notani D., Song X., Rahim N.G., Tanasa B., Heintzman N., Ren B., Fu X.D., Topol E.J., Rosenfeld M.G., Frazer K.A. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.