Abstract
To receive an ample amount of light, plants use elongation growth in response to vegetation shade. The combined interaction of light and hormones, including jasmonic acid (JA) signaling controls this elongation. However, the detailed molecular mechanisms underlying the response are still emerging. FAR-RED INSENSITIVE 219/JASMONATE RESISTANCE 1 (FIN219/JAR1), a cytoplasmic localized JA-conjugating enzyme, integrates far-red light and JA signaling. Here, we report that FIN219/JAR1 negatively regulates shade-induced hypocotyl elongation and gene expression in Arabidopsis seedlings in response to shade. In turn, simulated shade reduces FIN219 protein accumulation. Analysis of phyA 211 fin219-2 double mutants indicated that FIN219 and phyA are synergistic in regulating shade-induced hypocotyl elongation and gene expression. Moreover, FIN219 differentially affected the expression of the shade-signaling bHLH factors PIF5 and PAR1, thereby increasing the expression of the auxin-response genes IAA29 and SAUR68 on exposure to shade. Furthermore, the protein level of CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) was affected in both fin219 mutants and overexpression lines as compared with the wild type under shade. Intriguingly, ectopic expression of FIN219 inhibited the nuclear accumulation of COP1 in response to shade. Further co-immunoprecipitation studies revealed that FIN219 interacted with COP1 and phyA under shade. Therefore, FIN219/JAR1 may play a vital role in modulating the Arabidopsis response to simulated shade via multiple layers of molecular mechanisms.
Keywords: shade avoidance response, FIN219/JAR1, jasmonates, shade-induced hypocotyl elongation, Arabidopsis, shade signaling
Introduction
Light carries most of the essential information needed for plant growth and development (Von Arnim and Deng, 1996; Fankhauser and Chory, 1997; Chory, 2010). Photoreceptors such as phytochromes, cryptochromes, and UVR8 perceive light as a developmental signal (Li et al., 2011; Liu et al., 2011; Tilbrook et al., 2013). Vegetation shade reduces plants’ access to sufficient light. Shade-intolerant plants such as Arabidopsis trigger a suite of responses, collectively called shade avoidance syndrome (SAS), including elongation of hypocotyls, stems and petioles, hyponasty (upward bending of leaves) and early flowering (Franklin, 2008; Casal, 2012). SAS is a default developmental program and suppressed under normal light by phyB along with phyD and phyE (Franklin and Quail, 2010; Casal, 2013).
Phytochromes exist in two forms: the red light (666 nm)-absorbing Pr form, and far-red light (730 nm)-absorbing Pfr form (Li et al., 2011). However, monochromatic red (R) or far-red (FR) light usually does not convert 100% of Pr to Pfr or vice versa. Thus, a dynamic equilibrium is established between the two phy forms (Pr and Pfr) depending on the quality of light (Possart et al., 2014). Under white light (high R:FR), the active phyB, Pfr, migrates to the nucleus and interacts with various PIFs and drives their degradation (Lorrain et al., 2008). The onset of low R:FR ratio shifts the steady-state equilibrium toward the inactive Pr form (Franklin and Quail, 2010). As a result, PIFs become stabilized and more abundant (Lorrain et al., 2008). Genome-wide analysis revealed that PIF4 and PIF5 preferentially bind to auxin biosynthetic and signaling gene promoters (Hornitschek et al., 2012; Leivar et al., 2012b) and activate their expression under low R:FR light. PIF7 also functions as a major regulator for shade-induced hypocotyl elongation and is dephosphorylated in response to shade (Li et al., 2012). HFR1 physically interacts with PIF4 and PIF5 and inhibits their binding to the target promoters, thus negatively regulating shade responses (Hornitschek et al., 2009).
CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) is a repressor of photomorphogenesis and encodes an RING-finger E3 ubiquitin ligase. Under darkness, COP1 accumulates in the nucleus and targets degradation of positive regulators of photomorphogenesis such as HY5, HYH, LAF1, and HFR1 (Deng et al., 1991; Ballesteros et al., 2001; Lorrain et al., 2006). As well, COP1 is required for activating shade signaling. Mutants of COP1 can barely sense vegetation shade, whereas overexpression of COP1 leads to a constitutive shade avoidance phenotype (Pacín et al., 2013). The molecular mechanisms of COP1-mediated shade responses are emerging. COP1 rapidly accumulates in the nucleus under low R:FR, and this nuclear accumulation is essential for full shade responses (Pacín et al., 2013). Further evidence indicates that COP1 enhances HFR1 degradation under shade, leading to an increase of PIF-mediated gene expression, auxin levels, and thus stem growth (Pacín et al., 2016). COP1 and SPA1 together participate in hypocotyl and leaf petiole elongation in response to low R:FR by targeting the HFR1degradation by 26S proteasome (Rolauffs et al., 2012). The cop1 bbx21 bbx22 triple mutant is responsive to the shade, which suggests that the shade insensitivity phenotype of cop1 is mediated through both BBX21 and BBX22 (Crocco et al., 2010). However, how COP1 regulates SAS at the molecular level needs further investigation.
Phytohormones play important roles in light control of plant development (Lau and Deng, 2010; Kurepin and Pharis, 2014). The interplay of jasmonates (JAs) and light signaling is accepted (Kazan and Manners, 2011; Hsieh and Okamoto, 2014). Although low R:FR light perturbs JA signaling (Moreno et al., 2009; Robson et al., 2010; Cerrudo et al., 2012; De Wit et al., 2013; Chico et al., 2014; Leone et al., 2014), modulation of shade signaling by JA or its signaling components is still under examination. Recent evidence has revealed the interplay between JAs and shade, which is critical for growth-defense balance. This process may involve resource allocation between growth and immunity (Yang et al., 2012; Ballaré, 2014; Mazza and Ballaré, 2015).
The jar1 mutants show reduced sensitivity to exogenous JA and enhanced susceptibility against soil fungus, Pythium irregulare (Staswick et al., 1992, 1998). Later, jar1-1 was mapped to the same locus as fin219-1 (Staswick et al., 2002), a suppressor of cop1 mutant under darkness (Hsieh et al., 2000). FIN219 (GH3.11) belongs to a GH3 family of proteins, and its expression is rapidly induced by auxin (Hsieh et al., 2000). FIN219/JAR1 encodes JA-amino-synthetase, which conjugates isoleucine (Ile) with jasmonic acid (JA) to form the bioactive jasmonoyl-L-isoleucine (JA-Ile) (Staswick and Tiryaki, 2004). JA-Ile is an oxylipin that profoundly affects plant developmental and stress responses (Bari and Jones, 2009; Browse, 2009).
The fin219/jar1 mutant exhibits an insensitive long-hypocotyl phenotype under continuous far-red (cFR) light, which indicates its role in phyA-mediated signaling (Hsieh et al., 2000; Chen et al., 2007). Moreover, FIN219/JAR1 physically interacts with COP1 under darkness and cFR light and negatively regulates COP1 under FR light. Artificial induction of FIN219 protein accumulation via overexpression inhibited COP1 nuclear localization and hence stabilized HY5 protein (Wang et al., 2011). In addition, FIN219 might contribute to signaling of other hormones such as auxin, ethylene, gibberellin and abscisic acid by regulating a large number of bHLH transcription factors (TFs) (Chen et al., 2015).
Under low R:FR light, both phyA and fin219 mutant seedlings show enhanced hypocotyl elongation as compared with the wild type (Johnson et al., 1994; Yanovsky et al., 1995; Robson et al., 2010). Although the phyA phenotype is attributed to a FR light-mediated high irradiance response, the molecular mechanism underlying fin219-mediated shade avoidance phenotype is still under debate.
In the present study, we examined the functional significance of FIN219 under simulated shade in Arabidopsis seedlings. With a combination of genetic and biochemical experiments, we demonstrate that FIN219 negatively regulates shade avoidance responses by modulating key shade signaling components. Moreover, FIN219-mediated shade avoidance responses were independent of phyA-mediated high irradiance responses.
Materials and Methods
Plant Materials and Growth Conditions
The fin219-2 (SALK_059774), fin219-1, jar 1-1, phyA 211, cop1-4, cop1-6, cop1-4 fin219-2, and cop1-6 fin219-2 mutants and glucocorticoid-inducible FIN219 transgenic line (pGR:FIN219; PGR219) were described previously (Hsieh et al., 2000; Chen et al., 2007; Wang et al., 2011). pGR219 seedlings were harvested in GM plates with 1 μM dexamethasone (Dex) to induce FIN219 expression. The phyA 211 fin219-2 double mutant was generated by crossing phyA 211 with the fin219-2 mutant and selecting homozygous plants in an F2 generation by T-DNA-specific primers. All mutants are in a Col-0 background. Seeds of Arabidopsis thaliana were surface-sterilized and plated on growth medium (1/2 strength Murashige and Skoog medium, Duchefa Biochemie; 0.3% sucrose, 0.5% MES, 0.5% agar). After 3 days of incubation at 4°C, seed plates were kept at 22°C, 150 μmol m-2s-1 fluorescence white light for 16 h for germination, then transferred to an LED growth chamber (HIPONT, 721FTEC, Taiwan). Germinating seedlings were grown in continuous white light (high R:FR) for 2 days, then kept in continuous white light or transferred to continuous simulated shade (low R:FR) for an additional 4 days.
Light Measurements
The HIPONT, 721FTEC (Taiwan) growth chamber equipped with monochromatic LED light sources for white light (455 nm), red light (650 nm), and far-red light (730 nm) (Supplementary Figure S4), was used for shade avoidance study. In our study, white light and simulated shade represented high red:far-red light ratio (R5.21 μmol m-2s-1/FR2.25 μmol m-2s-1 ∼ 2.3) and low red:far-red ratio (R5.21 μmol m-2s-1/80.02 μmol m-2s-1 ∼ 0.06), respectively. Photon fluencies were estimated with use of a Li-Cor (LI-250A, LiCor Corp., Lincoln, NE, United States) and spectral distribution was measured by an Ocean Optics spectrum meter (USB2000, Florida, United States). Photosynthetically active radiation (white + red light) was kept constant at 70 μmol m-2s-1.
Hypocotyl Length Measurements and Western Blot Quantification
Digital images were analyzed by the US National Institutes of Health ImageJ software (Bethesda, MD, United States1) to measure lengths of hypocotyl and to quantify Western blot band intensities.
RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR
RNA extraction and cDNA synthesis were as described previously (Chen et al., 2015). Samples of 100 mg were ground in 0.5 ml Buffer A (1MTris-HCl, pH 7.3, 5 mMEDTA, pH 8.0, 1% SDS), then extracted twice with an equal volume of phenol and once with chloroform:isoamylalcohol (24:1). The supernatant was precipitated with LiCl (final working concentration 3 M) and incubated at -20°C overnight. After centrifugation, the pellet was dissolved completely in 0.5 ml 2% potassium acetate, then precipitated again with isopropanol. Total RNA (2 μg) was treated with DNase to prevent genomic DNA contamination, then used as a template for cDNA synthesis with the ABI cDNA transcription kit (#4368814). Real-time PCR involved CFX96 Touch Real-time PCR Detection System (Bio-Rad, United States). Gene-specific primers (Supplemental Table S1) were used for analyzing mRNA levels of ACT2 (AT3G18780), PIL1 (AT2G46970), ATHB2 (AT4G16780), FIN219 (AT2G46370), HFR1 (AT1G02340), PAR1 (AT2G42870) and PIF5 (AT3G59060), IAA29 (AT4G32280) and SAUR68 (AT1G29490) by qPCR. ACT2 was used as internal control to normalize the expression levels, then standardized to the wild-type level under white light.
Protein Extraction and Protein Gel Blot Analysis
Total protein was extracted with extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 0.1% NP-40, 1 mM PMSF and 1X protease inhibitor) as described (Hsieh et al., 2000). Total protein, 150 μg, was loaded in each lane and separated on 8% SDS-PAGE and transferred to PVDF membrane (Millipore). Protein gel blot analyses involved standard methods (Sambrook and Russell, 2001) with FIN219, PHYA and α-tubulin monoclonal, and COP1 polyclonal antibodies.
Protoplast Isolation and Subcellular Localization Study
Arabidopsis mesophyll protoplast isolation and transfection were performed as described (Yoo et al., 2007). In brief, 4-week-old well-expanded leaves were peeled (Wu et al., 2009) and incubated in enzyme solution (20 mM MES, 1.5% cellulase R10, 0.4% macerozyme R10, 0.4 M mannitol, 20 mM KCl, 10 mM CaCl2, 5 mM b-mercaptoethanol, and 0.1% bovine serum albumin, pH 5.7) for 1 h. Protoplasts were collected by centrifugation at 100 × g and washed twice with W5 solution (2 mM MES, 154 mM NaCl, 125 mM CaCl2, and 5 mM KCl, pH 5.7). Protoplasts were resuspended in W5 solution and incubated on ice for at least 30 min, then washed with MMG solution (4 mM MES, 0.4 M mannitol, and 15 mM MgCl2, pH 5.7) and resuspended at 2 × 105 cells mL-1 in MMG solution. For protoplast transfection, 200 μL protoplasts was mixed with 10 μg (∼20 μL) DNA and 220 μL PEG solution (40% polyethylene glycol 4000, 0.2 M mannitol, and 100 mM CaCl2), then transfected protoplasts were washed twice, resuspended in W5 solution with 1 μM Dex and incubated overnight in the dark. On the next day, the protoplasts were treated with white light and shade light. The GFP fusion construct p35S:GFP-COP1 was used for transfection. The nuclei of protoplasts were stained with 4′,6-diamidino-2-phenylindole. Transformed protoplasts were visualized by confocal laser scanning microscopy (Leica TCS SP5 Confocal Spectral Microscope) and image processing involved LAS AF lite software from Leica.
Co-immunoprecipitation Analysis
Co-immunoprecipitation analysis was performed as described (Chen et al., 2007). The seedlings were ground with grinding buffer (50 mM Tris-HCl, pH 5.7, 150 mM NaCl, 10 mM MgCl2, 0.1% bovine serum albumin, 0.1% Nonidet P-40, 1 mM PMSF, 2X Protease Inhibitor Cocktail [Sigma], and 50 μM MG132). A total of 2 mg of proteins was mixed with beads and incubated at 4°C for 4 h, then washed three times with same grinding buffer. Pellets were analyzed by standard SDS-PAGE and subjected to protein gel blot analysis.
Results
FIN219 Negatively Regulates Hypocotyl Elongation and Gene Expression under Simulated Shade
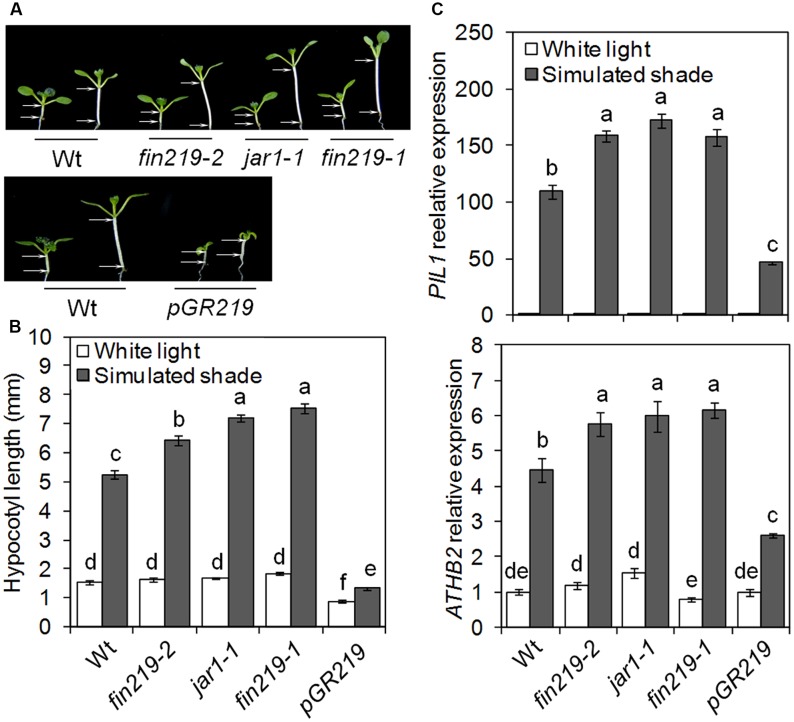
Jasmonic acid biosynthetic and signaling mutants display enhanced shade-induced hypocotyl elongation (Robson et al., 2010). We previously demonstrated that FIN219/JAR1 integrates phyA-mediated FR light and JA signaling (Wang et al., 2011; Chen et al., 2015). To further understand the role of FIN219 in the shade avoidance response, we examined hypocotyl elongation and gene expression responses under white light (high R:FR) and simulated shade (low R:FR). Seven-day-old seedlings of fin219 mutants [fin219-2 (Wang et al., 2011), jar1-1 (Staswick et al., 1998), and fin219-1 (Hsieh et al., 2000)], and overexpression of FIN219 in a fin219-2 genetic background driven by glucocorticoid-inducible promoter [pGR:FIN219, also pGR219 (Wang et al., 2011)] were compared with the corresponding wild type. Under white light, fin219 mutants showed similar hypocotyl length to that of the wild type. However, under simulated shade, the hypocotyl was longer for all fin219 mutants than the wild type [129.7% (fin219-2), 180.2% (jar1-1) and 197.6% (fin219-1), p < 0.01] (Figures 1A,B). The FIN29 overexpression line, pGR219, induced by Dex had a short-hypocotyl phenotype under white light. Although pGR219 responded to low R:FR light, hypocotyl elongation was lower than that of the wild type (Figures 1A,B). Furthermore, we quantified the expression of shade-inducible marker genes PIL1 and ATHB2. Under shade, the expression of these genes was induced in the wild type. Under white light, their expression in the fin219 mutants and pGR219 was largely similar to that of the wild type. However, under simulated shade, the expression was enhanced in the fin219 mutants but significantly reduced in pGR219 (Figures 1C,D). FIN219 may inhibit shade-induced hypocotyl elongation and gene expression.
FIGURE 1.
FIN219 negatively regulates shade-induced hypocotyl elongation and gene expression. (A) Seedlings of fin219 mutants (fin219-2, jar1-1, fin219-1) and FIN219 overexpression line (pGR219) were grown under white light (high R:FR) or simulated shade (low R:FR) for 4 days and then photographs were taken. (B) Quantification of hypocotyl lengths of seedlings grown in A. Data are mean ± SE (n = 25). (C) Quantitative RT-PCR (qRT-PCR) analyses of PIL1 and ATHB2 expression in seedlings grown under white light or simulated shade for 4 days standardized to the wild type under white light. Data are mean ± SE from three biological replicates. Different lowercase letters represent significant differences by ANOVA at P < 0.05.
Simulated Shade Reduces FIN219 Protein Level and FIN219 Affects phyA and COP1 Protein Accumulation
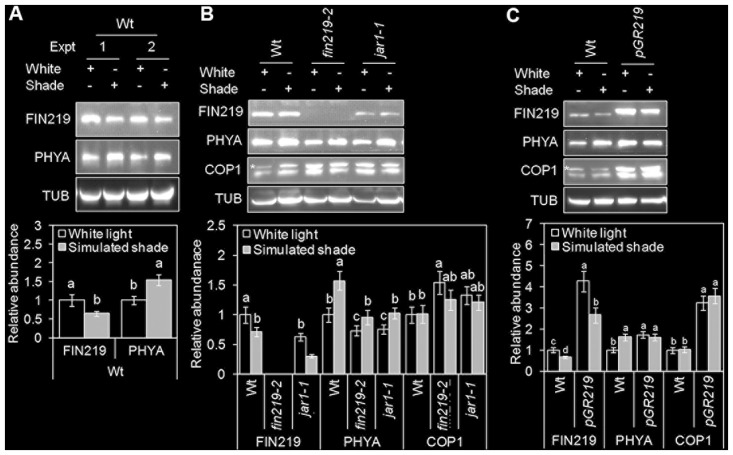
The onset of shade avoidance is coupled with transcriptional and translational reprogramming. FIN219 (AtGH11.3) is a GH3 family protein, which is rapidly upregulated in response to auxin (Hsieh et al., 2000). Recently, GH3.3 was found induced by simulated shade (Crocco et al., 2015). We used quantitative real-time PCR (qRT-PCR) to determine the effect of simulated shade on FIN219 transcript levels. Simulated shade did not alter FIN219 expression in the wild type and fin219 mutants; however, FIN219 mRNA level was induced in pGR219 (Supplementary Figure S1). Furthermore, we used protein gel blot analysis to demonstrate how shade affects FIN219 protein levels. FIN219 protein level was significantly reduced under simulated shade as compared with white light both in the wild type and pGR219 (Figures 2A–C). Under white light, FIN219 protein level was lower in the fin219 mutants (the null mutant fin219-2 and jar1-1) than in the wild type. Simulated shade further reduced FIN219 level in these mutants (Figure 2B). Thus, shade light may reduce FIN219 protein level, and overaccumulation of FIN219 protein in pGR219 (Figure 2C) may inhibit shade responses (Figures 1A–C).
FIGURE 2.
FIN219 levels reduced under simulated shade light. Gel blot analyses of FIN219, PHYA, and COP1 protein levels in wild-type Col (A), fin219 mutants (B), and pGR219 (C) in response to white and simulated shade light. Seedlings were germinated and grown under white light (high R:FR) for 2 days, then kept in white light or transferred to simulated shade (low R:FR) for 4 days. Total protein of 150 μg was loaded in each lane and probed with FIN219, PHYA and α-tubulin monoclonal and COP1 polyclonal antibodies. α-tubulin (TUB) was a loading control. Data are mean ± SE from two independent experiments. The asterisk indicates a non-specific band. Different lowercase letters represent significant differences by ANOVA at P < 0.05.
We further determined the effect of FIN219 on PHYA and COP1 protein levels under simulated shade. In agreement with previous reports (Martínez-García et al., 2014), under shade light, PHYA protein level was increased in the wild type and slightly reduced in fin219 mutants (Figures 2A,B). However, the pGR219 line showed spontaneous accumulation of high levels of PHYA under white light, with no enhanced accumulation under simulated shade (Figure 2C). Thus, pGR219 line may be insensitive to shade light.
CONSTITUTIVE PHOTOMORPHOGENIC 1 is essential for hypocotyl and petiole elongation under shade light (Rolauffs et al., 2012). COP1 level does not change with exposure to shade, and COP1 nuclear accumulation is required for full shade responses (Pacín et al., 2013). To determine how FIN219 affects COP1 level, we measured COP1 protein levels. As expected, COP1 level did not change largely under shade light; however, under white light and shade, COP1 level was greater in fin219 mutants and pGR219 than the wild type (Figures 2B,C). FIN219-altered COP1 may in turn affect the shade responses.
fin219-2-Mediated Shade Responses Are Independent of phyA-Mediated High-Irradiance Response
The phyA mutant exhibits a long hypocotyl phenotype under continuous FR light or continuous white light supplemented with FR light. This response of phyA is due to the FR high-irradiance response (HIR) (Johnson et al., 1994; Yanovsky et al., 1995; Martínez-García et al., 2014). FIN219 is a component of phyA-mediated FR light signaling (Hsieh et al., 2000). FIN219-mediated shade responses may be due to phyA-mediated HIR signaling. To clarify this possibility, we analyzed phenotypic responses of the phyA211 fin219-2 to shade (Supplementary Figures S2A,B). Shade-induced hypocotyl elongation and gene expression were measured in fin219-2 and phyA 211 single mutants and phyA211 fin219-2 double mutants under white light and simulated shade. Under simulated shade, hypocotyl elongation and the expression of shade-induced marker genes such as PIL1 and ATHB2 was greater in the double mutant than each of the single mutants (Figures 3A–C), which suggests that FIN219-mediated shade response is independent of phyA-mediated FR-HIR.
FIGURE 3.
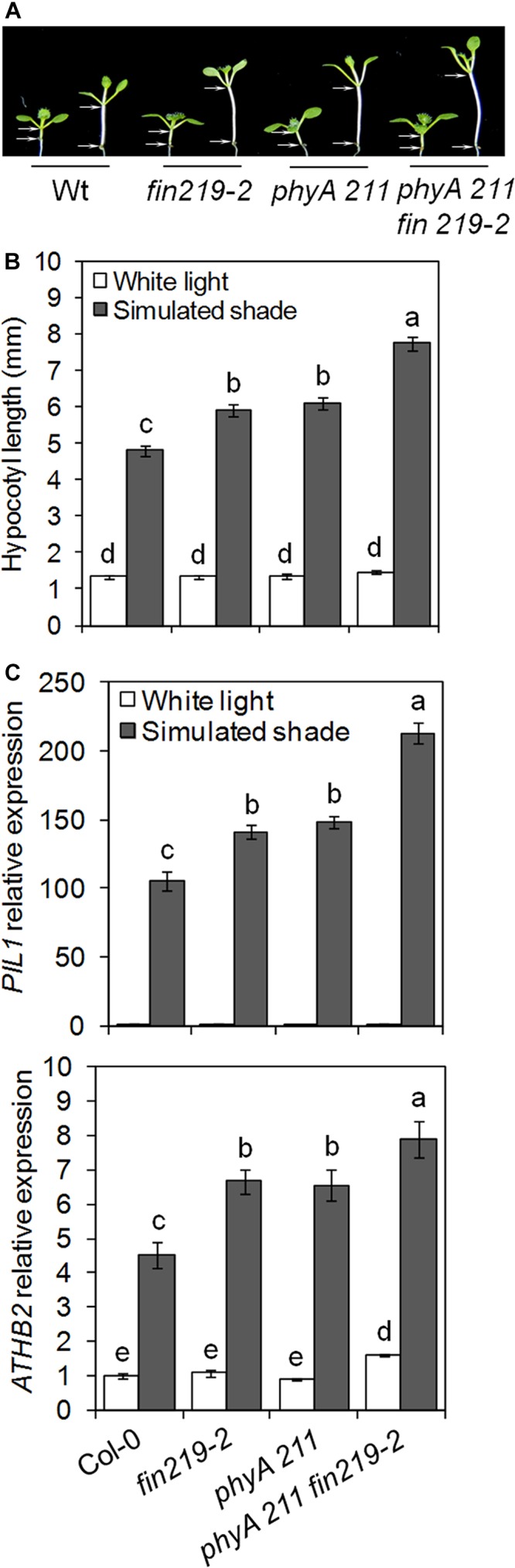
FIN219 and phyA have a synergistic effect on shade-mediated hypocotyl elongation and gene expression. (A) Seedlings of fin219-2, phyA 211 and the double mutant phyA 211 fin219-2 were grown under white light (high R:FR) or simulated shade (low R:FR) for 4 days. (B) Quantification of hypocotyl lengths of seedlings shown in A. Data are mean ± SE (n = 25). (C) qRT-PCR of PIL1 and ATHB2 expression in seedlings grown under white light or simulated shade for 4 days standardized to the wild type under white light. Data are mean ± SE from three biological replicates. Different lowercase letters represent significant differences by ANOVA at P < 0.05.
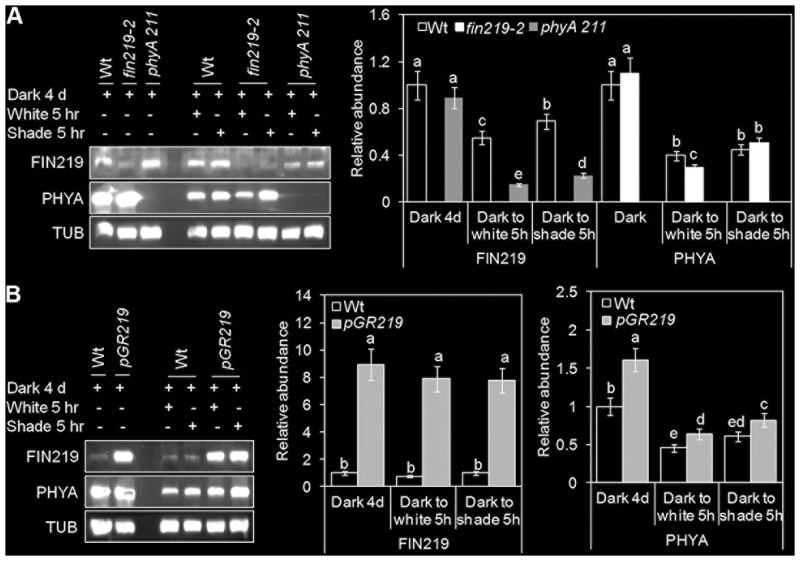
We showed reduced FIN219 level under shade light. However, in a dark-to-light transition experiment, FIN219 level in the wild type was reduced under light as compared with under dark, with a significant increase in FIN219 level under white and shade light (Figure 4A). Intriguingly, FIN219 level was lower in phyA mutants than the wild type under all light conditions (Figure 4A and Supplementary Figure 2B) except dark (Figure 4A), which implies that phyA positively regulates FIN219 under short-term shade. Similarly, in contrast to light-grown seedlings (Figures 2B,C), in dark-grown seedlings transferred to white light for 5 h, PHYA level was lower in the fin219-2 mutant than the wild type, and under shade light, it was comparable in fin219-2 and wild type (Figure 4A). However, under all light conditions examined, PHYA protein level was greater in pGR219 than the wild type (Figure 4B). FIN219 and phyA may regulate each other depending on the genotype and light condition, and output of this regulation may depend on a specific light condition.
FIGURE 4.
FIN219 and phyA regulate each other in response to shade. Gel blot analyses of FIN219 and PHYA transient levels in transition from dark to shade. Seedlings of wild-type (Wt), fin219-2 and phyA 211 (A) or Wt and pGR219 (B) were grown in the dark for 4 days, transferred to white light or simulated shade for 5 h, and then subjected to protein gel blot analyses. Data are mean ± SE protein abundance from two independent experiments. Different lowercase letters represent significant differences by ANOVA at P < 0.05.
FIN219 Differentially Alters the Expression of PIF5 and PAR1, for Altered Auxin Responses
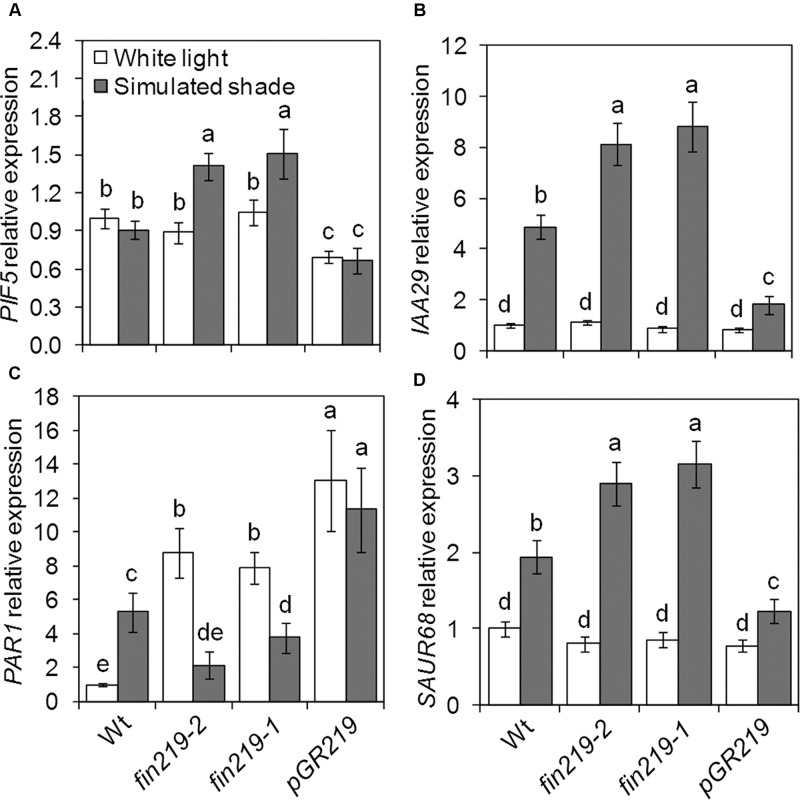
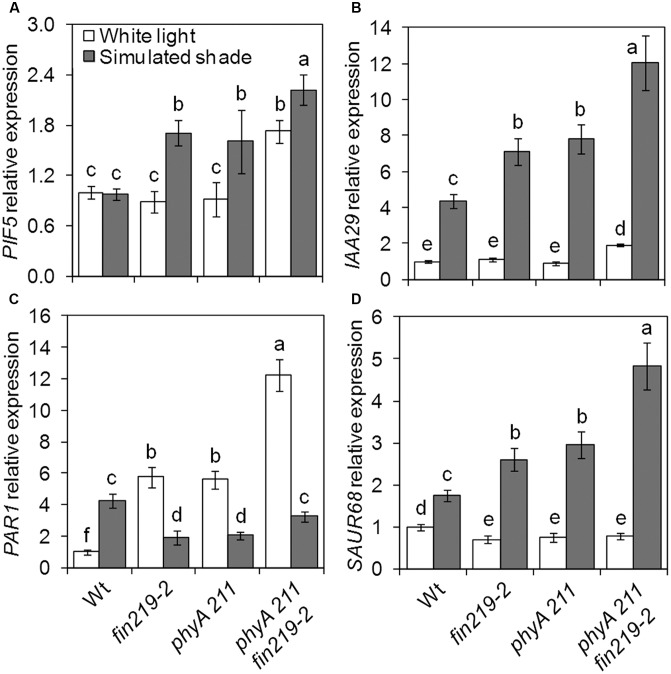
Several groups of TFs are involved in shade light signaling. In our recent study, we found that FIN219 affects a number of bHLH TFs (Chen et al., 2015). While checking the expression of shade signaling genes, we found that the bHLH TFs PIF5 and PAR1 were affected by FIN219. As expected, PIF5 expression in the wild type did not differ under simulated shade and white light (Figure 5A). Disruption of FIN219 expression by mutation (fin219-2 and fin219-1) significantly induced PIF5 expression under simulated shade, which was decreased in pGR219 regardless of light condition (Figure 5A). Similarly, the effect of FIN219 on PAR1 expression was significantly pronounced under both white and shade light. Under white light, PAR1 expression was greater in fin219 mutants and pGR219, whereas under shade light, PAR1 expression was reduced in fin219 mutants and remained unchanged in pGR219 (Figure 5C). Both PIF5 and PAR1 affect auxin biosynthesis and signaling under shade light (Roig-Villanova et al., 2007; Hornitschek et al., 2012). To check how their altered expression was translated into downstream effects, we evaluate the expression patterns of auxin-response genes IAA29 and PAR1 target SAUR68. The expression of both IAA29 and SAUR68 was enhanced in fin219 mutants and reduced in pGR219 under shade (Figures 5B,D). In addition, PIF5 showed high induction and PAR1 reduced expression in the phyA 211 fin219-2 double mutant under shade (Figures 6A,C). Moreover, the expression of both IAA29 and SAUR68 was synergistically increased in phyA 211fin219-2 under simulated shade (Figures 6B,D). Thus, FIN219 modulates auxin homeostasis under simulated shade through PIF5 and PAR1.
FIGURE 5.
FIN219 differentially regulates transcription factors PIF5 and PAR1 under shade. qRT-PCR analysis of PIF5 (A), IAA29 (B), PAR1 (C), and SAUR68 (D) expression in seedlings grown under white light or simulated shade for 4 days standardized to the wild type under white light. Data are mean ± SE from three biological replicates. Different lowercase letters represent significant differences by ANOVA at P < 0.05.
FIGURE 6.
FIN219 and phyA regulates key shade regulators. qRT-PCR analysis of PIF5 (A), IAA29 (B), PAR1 (C), and SAUR68 (D) expression in seedlings of wild type (Wt), fin219-2, phyA 211, and fin219-2phyA 211 grown under white light or simulated shade for 4 days standardized to the wild type under white light. Data are mean ± SE from three biological replicates. Different lowercase letters represent significant differences by ANOVA at P < 0.05.
FIN219 Reduces Nuclear Accumulation of COP1 through Direct Physical Interaction under Shade
The cop1 mutants show very short hypocotyls under high and low R:FR conditions. The cop1-4 mutant still shows a residual shade response (Pacín et al., 2013), which is not observed in cop1-6 (Supplementary Figures S3A,B). To demonstrate how fin219 mutation affects cop1 mutants under simulated shade, we assessed the double mutants cop1-4 fin219-2 and cop1-6 fin219-2 in response to simulated shade. Shade-induced hypocotyl elongation was significantly enhanced although slightly in cop1-4 fin219-2 as compared with cop1-4, with no difference between cop1-6 fin219-2 and cop1-6 (Supplementary Figures S3A,B).
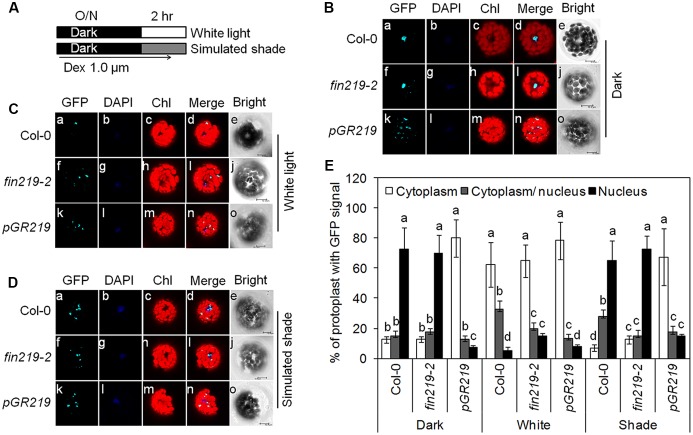
Enhanced COP1 nuclear accumulation is essential for shade responses (Pacín et al., 2013). Previous studies showed that FIN219 interacted with COP1 under dark and FR light. Moreover, FIN219 overexpression can exclude COP1 from the nucleus to the cytoplasm even in the dark (Wang et al., 2011). To further investigate the effect of FIN219 on COP1 nuclear localization under white and shade light, we performed protoplast transient assays using Arabidopsis mesophyll protoplasts from the wild type, fin219-2 and pGR219 and the construct p35S: GFP-COP1 in a binary vector. Under the dark, GFP-COP1 localized in the nucleus in wild-type protoplasts and fin219-2 (Figures 7Ba–i,E) but mostly in the cytoplasm in pGR219 protoplasts (Figures 7Bk–n,E). Under white light, GFP-COP1 signal is mostly abundant in the cytoplasm in wild type, fin219-2 and pGR219 protoplasts (Figures 7Ca–d,f–i,k–n,E). Upon exposure to simulated shade for 1 h, GFP-COP1 signals mainly concentrated in the nucleus in wild type and fin219-2 (Figures 7Da–d,f–i,E). However, overexpression of FIN219 in pGR219 inhibited COP1 nuclear accumulation (Figures 7Dk–n,E). Therefore, FIN219 overexpression can induce COP1 accumulation in the cytoplasm in response to simulated shade.
FIGURE 7.
FIN219 overexpression is able to exclude GFP-COP1 from the nucleus to the cytoplasm in response to shade. (A) A scheme to illustrate experimental conditions of the protoplasts. Transformed Arabidopsis mesophyll protoplasts by 35S:GFP-COP1 were incubated at 22°C overnight in the dark (B) and the next day transferred to white light (C) or simulated shade (D) for 1 h. Bars = 10 μm. (E) Quantification of the percentage of protoplasts with GFP signals in different cellular compartments. Data are mean ± SE from two biological replicates. Different lowercase letters represent significant differences by ANOVA at P < 0.05.
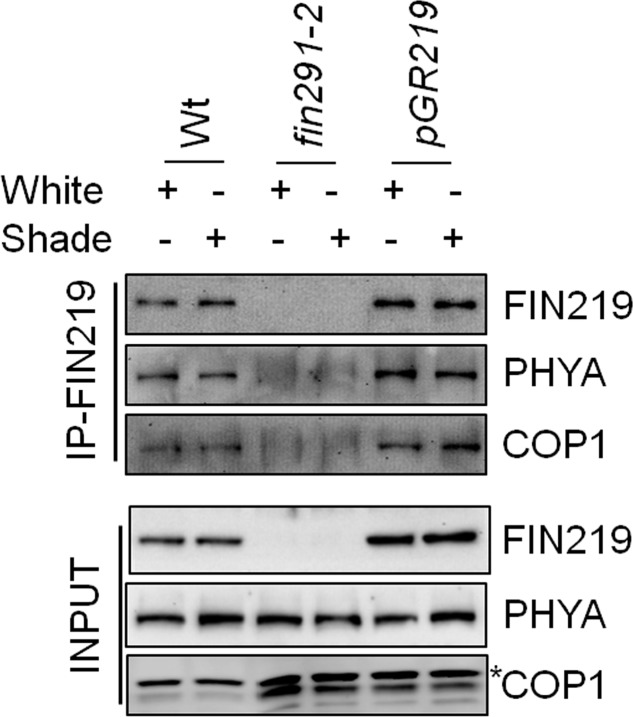
Furthermore, to determine the possibility of physical interaction between FIN219 and COP1 under shade, we used co-immunoprecipitation with wild type, fin219-2 and pGR219 seedlings grown under simulated shade. FIN219 directly interacted with COP1 and phyA (Figure 8). Interaction of both molecules was stronger in the FIN219 overexpression line pGR219, which suggests that FIN219 abundance may affect the degree of interactions (Figure 8). Hence, FIN219 can regulate the subcellular localization of COP1 under white and simulated shade, and lack of FIN219 regulation of COP1 in fin219 mutants compared to the wild type makes it more sensitive to shade light.
FIGURE 8.
FIN219 physically interacts with phyA and COP1 under shade light. Co-immunoprecipitation analysis of FIN219 interacting with phyA and COP1 in indicated genotypes under shade light. In total, 2 mg protein extracts from Col-0 (Wt), fin219-2 and pGR219 seedlings grown under white and shade light for 2 days were mixed with FIN219 monoclonal antibodies and then immunoprecipitated. The pellets were used for SDS-PAGE and protein gel blot analysis. The probes are FIN219 and PHYA monoclonal antibodies and COP1 polyclonal antibody. The asterisk indicates a non-specific band.
Discussion
Hypocotyl elongation is a key adaptation of Arabidopsis seedlings to avoid shade light. Here, we report that FIN219/JAR1, a JA-conjugating enzyme, plays a negative role in regulating shade responses such as hypocotyl elongation and expression of shade components. The double mutant phyA 211fin219-2 had a synergistic effect in response to simulated shade, which suggests that phyA and FIN219/JAR1 work in parallel pathways to regulate shade signaling, which is consistent with the additive levels of TFs PIL1 and ATHB2 and auxin-response genes IAA29 and SAUR68 under the same condition. Moreover, FIN219/JAR1 and PHYA levels were down- and upregulated by shade, respectively. Intriguingly, PHYA and COP1 levels were affected in the fin219 mutants under simulated shade. In particular, FIN219/JAR1 overexpression under shade could change COP1 subcellular localization from the nucleus to the cytoplasm. Further Co-IP studies under shade revealed that FIN219/JAR1, phyA and COP1 interacted with each other. These data indicate that FIN219/JAR1 plays a vital role in regulating shade responses likely by modulating the expression and subcellular location of shade components.
FIN219/JAR1 is a JA-conjugating enzyme, responsible for the formation of JA-Ile and involved in the regulation of plant development and defense responses likely via crosstalk with different hormones and TF-mediated signaling pathways (Chen et al., 2015). Mutation in the FIN219/JAR1 locus resulted in enhanced shade-induced hypocotyl elongation with different degrees (Figures 1A,B) (Robson et al., 2010), whereas its overexpression led reduced elongation responses (Figures 1A,B), which suggests a negative role of FIN219 in shade signaling. Since FIN219 is an auxin and JA inducible gene and its function in JA signaling is well known, how it modulates other hormone signaling pathways is largely unknown. Our previous studies also indicated that FIN219 could crosstalk with other different hormones (Chen et al., 2015). The fin219-2 is a T-DNA insertion knockout mutant (Wang et al., 2011), whereas jar1-1 is an EMS mutant (Staswick et al., 2002) and fin219-1 is a mis-regulated mutant with changes of methylation status in the promoter of its gene (Hsieh et al., 2000). Thus, jar1-1 and fin219-1 mutants contain reduced levels of proteins compared to wild type. It could be possible that in these mutant lines (jar1-1 and fin219-1), FIN219 perturbs other hormone signaling pathways and makes plants more sensitive to shade. This is our hypothesis that needs further investigation to clarify. In addition, COP1 is necessary for hypocotyl and petiole elongation under shade light (Rolauffs et al., 2012). Furthermore, B-box-containing 21 (BBX21) acts downstream of COP1 to negatively regulate the shade response (Crocco et al., 2010). FIN219/JAR1 appears to affect COP1 levels under shade (Figures 2B,C). Intriguingly, BBX21 negatively regulated FIN219 transcript levels under canopy shade (Crocco et al., 2010), so FIN219-COP1-BBX21 may form a regulatory feedback loop in response to shade environments. Recent studies revealed that phyB and PIFs form a mutually negative feedback loop under continuous red but not shade light (Leivar et al., 2012a). Thus, a local loop regulation initiated by FIN219-COP1-BBX21, leading to reduced levels of active JA-Ile, may play a vital role in fine-tuning shade light signaling.
FIN219/JAR1 acts as a major enzyme for the formation of physiologically active JA-Ile to regulate plant growth and defense responses (Staswick et al., 2002; Wang et al., 2011). Its protein levels should be strictly modulated in response to developmental and environmental cues. So far, multiple photoreceptors, including phyA, and different hormones such as auxin and JAs can regulate FIN219 levels. Current work indicated that pGR219 with FIN219 overexpression has always severely stunted phenotype (Figure 1). The expression levels of IAA29 and SAUR68 in pGR219 are always similar with Col-0 under white light (Figures 5B,D). Moreover, PIF5 and PAR1 have opposing effects on shade responses, which suggests that FIN219 may trigger major effects on shade-responsive genes mainly under shade conditions. Besides, PIF5 positively regulates and PAR1 negatively affects shade responses. In pGR219, the PIF5 expression is less and PAR1 expression is greater than wild type (Figures 5A,C). However, these gene expressions do not alter with response to shade in pGR219. In addition, our previous studies indicated that FIN219 overexpression could exclude COP1 from the nucleus to the cytoplasm even in the dark without greatly altering COP1 level (Wang et al., 2011). Shade also resulted in COP1 accumulation in the nucleus (Figures 7D,E). In contrast, pGR219 under shade showed more COP1 accumulation in the cytoplasm and substantial reduction of COP1 in the nucleus compared to wild-type Col-0 under the same condition, which suggests that the accuracy of COP1 subcellular location is critical for shade responses in addition to expression of shade components. Thus, the PGR219 with pronounced short-hypocotyl phenotype under white light and shade conditions may have substantial effects on plant growth and development in addition to light signaling.
Shade light illumination affects a number of genes, including TFs (Sessa et al., 2005; Roig-Villanova et al., 2006). PIF-related TFs are essential for shade-induced responses (Lorrain et al., 2008; Casal, 2013). As expected, wild-type PIF5 transcript levels did not change greatly under shade light. However, FIN219 negatively regulated PIL1 and PIF5 gene expression under low R:FR light (Figures 1C, 5A). As well, the expression of several TFs such as HFR1, ATHB2, PIL1, and PAR1/2 were rapidly induced under shade light (Carabelli et al., 1996; Salter et al., 2003; Sessa et al., 2005; Roig-Villanova et al., 2006). HFR1 and PAR1/2 negatively regulate shade responses (Roig-Villanova et al., 2007; Hornitschek et al., 2009), whereas ATHB2 and PIL1 are positive (Steindler et al., 1999; Salter et al., 2003) and negative regulators, respectively (Li et al., 2014). PAR1 expression was positively regulated by FIN219 under shade (Figure 5C). Therefore, FIN219 may regulate the expression of early shade-induced genes positively and negatively. In addition, PIF5 and PAR1 modulate auxin biosynthesis and signaling (Roig-Villanova et al., 2007; Hornitschek et al., 2012). In agreement, the expression of auxin-response genes IAA29 and PAR1 target SAUR68 is associated with PIF5 and PAR1 expression, respectively. As well, the PIF5 and PAR1 expression was higher in the double mutant phyA211 fin219-2 than in each single mutant under shade (Figures 6A,C), so FIN219 may act independently of phyA-mediated signaling to modulate auxin signaling in response to shade.
In addition, PAR1 transcripts are repressed by PHYA and PHYB overexpression under simulated shade (Roig-Villanova et al., 2006). Its transcripts are also suppressed by phyA and phyB under FR and R light, respectively. The PHYA protein level in fin219 mutants was similar to that in wild type (Figure 2B) and it was greater in PGR219 under white light (Figure 2C), which suggests that PAR1 transcripts would be less than wild type under white light. However, in Figure 5C, PAR1 expression was even greater in both fin219 mutants and PGR219 than in wild type under white light, which implies that PAR1 expression patterns in fin219 mutants and PGR219 may involve hormone effects such as JAs and auxin. The detailed mechanisms remain elusive.
Phytochromes were shown to play vital roles in shade signaling (Devlin et al., 1999; Franklin and Whitelam, 2005; Casal, 2012). phyB is stable in light and functions as a major photoreceptor in suppression of shade-mediated hypocotyl elongation. Under a high R:FR ratio, the phenotype of the phyB mutant was similar to the wild type under shade. Furthermore, phyD and phyE were redundant to phyB in repressing shade responses, whereas phyC did not play a role in shade responses (Franklin and Quail, 2010). Functional roles of phyA involved in the repression of shade responses remain largely unknown. Here, we found that PHYA levels were increased by simulated shade (low R:FR ∼ 0.05) (Figures 2, 4), which agrees with the report by Martínez-García et al. (2014). An increase in PHYA level on exposure to shade even for 5 h (Figure 4) is likely related to the phyA function in fine-tuning the chlorophyll biosynthesis in response to partial shading (Brouwer et al., 2014). Moreover, phyA appears to positively regulate FIN219 levels in short-term exposure to shade (Figure 4A), rather than long-term exposure to shade (Supplementary Figure S2B). In turn, increased FIN219 levels leading to enhanced JA-Ile levels resulted in reduced the chlorophyll content under shade. This speculation is consistent with JAs being able to decrease chlorophyll content and reduce photosynthesis in light (He et al., 2002; Zhai et al., 2007). In contrast, with longer exposure to shade, FIN219 level was reduced in the wild type (Figure 2 and Supplementary Figure S2B) and positively regulated PHYA level (Figure 2B). Thus, reduced FIN219 level under shade may involve other mechanisms, and both FIN219 and phyA may regulate each other in response to low R:FR light.
CONSTITUTIVE PHOTOMORPHOGENIC 1 is a negative regulator of photomorphogenesis and encodes an E3 ubiquitin ligase. Under the dark, COP1 accumulates in the nucleus and results in the degradation of positive regulators such as HY5 of photomorphogenesis. Upon light exposure, COP1 is inactivated by migrating to the cytoplasm and other unknown mechanisms. Shade light can trigger COP1 accumulation rapidly in the nucleus, and it positively modulates the shade responses (Pacín et al., 2013). Moreover, COP1 physically interacts with FIN219 under the dark and continuous FR light and restricts its nuclear accumulation on overexpression of FIN219 (Wang et al., 2011). We found COP1 mainly accumulated in the cytoplasm and unable to migrate to the nucleus in pGR219 under the conditions examined, including shade for 1 h (Figures 7B–E), which significantly affects hypocotyl elongation (Figures 1A,B). Thus, abundant FIN219 protein affects the total content of active COP1 protein in the nucleus. Ethylene promotes hypocotyl elongation in the light by triggering COP1 nuclear accumulation, which enhances HY5 degradation (Yu et al., 2013). FIN219 levels responsible for the formation of JA-Ile, an active form of JA, are likely tightly regulated in response to low R:FR to modulate the subcellular localization of COP1.
FIN219/JAR1 is a cytoplasmic protein in FR light and remains in the same location even under shade (Hsieh et al., 2000; Figure 7). Co-IP studies indicated that FIN219/JAR1 dose-dependently interacted with phyA and COP1 proteins under shade (Figure 8). FIN219 overexpression excluded COP1 from the nucleus to the cytoplasm under FR light (Wang et al., 2011). As well, ectopic expression of FIN219 under low R:FR light changed the COP1 subcellular location from the nucleus to the cytoplasm because COP1 was localized in the nucleus under shade light. However, FIN219 level in the wild type was reduced by shade light (Figure 2) and was increased by 5-h shade as compared with white light (Figure 4). FIN219 and COP1 interaction likely occurred in the cytoplasm at the early stage of shade light exposure. This speculation is consistent with transient expression at 1-h shade that resulted in more accumulation of GFP-COP1 in the cytoplasm by ectopic expression of FIN219 (pGR219) (Figure 7).
In addition, the levels of PHYA were increased and stabilized by continuous and transient shade light (Figures 2, 4), which leads to suppression of hypocotyl elongation, with an antagonistic effect on phyB deactivation by low R:FR light. PhyA and phyB likely function dynamically to modulate hypocotyl elongation in response to changes in R:FR ratios in natural environments. In addition, FIN219 and phyA positively regulated each other under shade (Figures 2, 4, 8). phyA is activated by FR light and migrates to the nucleus. Whether FIN219 affects phyA subcellular localization, leading to their interaction in the cytoplasm upon exposure to shade light, remains to be elucidated.
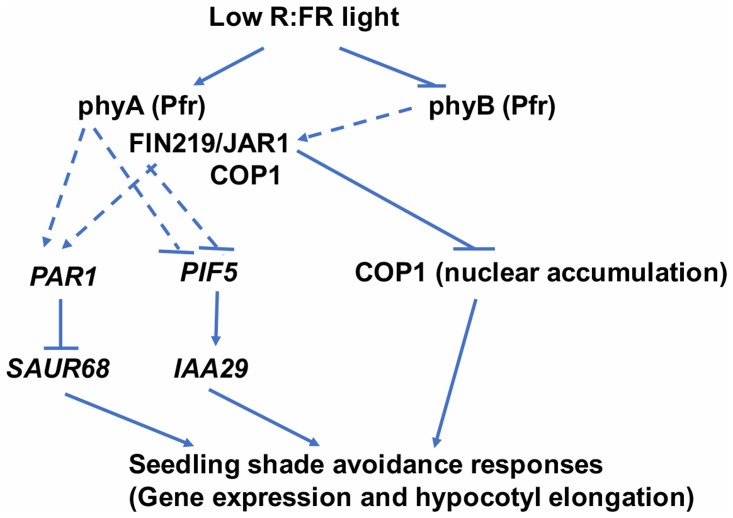
FIN219/JAR1 is a JA-conjugating enzyme and interacts with multiple partners, including COP1 (Chen et al., 2007; Wang et al., 2011; Figure 8). These data suggest that FIN219 may have a dual function with an enzymatic and protein–protein interaction activities. Collectively, our data show that FIN219/JAR1, a JA-conjugating enzyme, functions as a negative regulator in shade signaling and may work with phyA and COP1 in response to shade. Alternatively, low R:FR ratio may inactivate phyB by reducing its abundance in the nucleus and decrease FIN219/JAR1 levels, thereby leading to increased PIF5 level and reduced PAR1 level and increased COP1 accumulation in the nucleus. These events activate downstream genes such as IAA29 and SAUR68 and shade responses, including hypocotyl elongation (Figure 9).
FIGURE 9.
A model to illustrate FIN219 functions in regulating shade responses. Low R:FR light perceived by the photoreceptor phyB regulates specific gene expression likely via FIN219 associated complex (phyA-FIN219/JAR1-COP1)-mediated positive or negative modulation of transcription factors, including PAR1 and PIF5. As well, FIN219 regulated by phyA through direct interaction interacts with COP1 and inhibits its nuclear accumulation. All these events lead to shade-induced gene expression and hypocotyl elongation. Regular arrow, positive effect; inverted T, negative effect. Dashed lines represent an indirect regulation.
Author Contributions
SS and H-LH designed the experiments; SS and H-WJ performed experiments; SS and H-LH did data analyses; SS and H-LH wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the Arabidopsis Biological Research Center (Ohio State University, Columbus) for the fin219-2 and jar1-1 seeds. We thank the Technology Commons staff in the College of Life Science, National Taiwan University, for technical assistance with confocal microscopy and real-time PCR analyses. SS is a postdoctoral fellow supported by the Excellence Research Program of National Taiwan University (104R4000).
Funding. This work was supported by the National Science Council, Taiwan (NSC 101-2311-B-002-002-MY3) and Ministry of Science and Technology (MOST 104-2311-B-002-035-MY3).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01901/full#supplementary-material
References
- Ballaré C. L. (2014). Light regulation of plant defense. Annu. Rev. Plant Biol. 65 335–363. 10.1146/annurev-arplant-050213-040145 [DOI] [PubMed] [Google Scholar]
- Ballesteros M. L., Bolle C., Lois L. M., Moore J. M., Vielle-Calzada J. P., Grossniklaus U., et al. (2001). LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev. 15 2613–2625. 10.1101/gad.915001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Jones J. D. G. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69 473–488. 10.1007/s11103-008-9435-0 [DOI] [PubMed] [Google Scholar]
- Brouwer B., Gardeström P., Keech O. (2014). In response to partial plant shading, the lack of phytochrome A does not directly induce leaf senescence but alters the fine-tuning of chlorophyll biosynthesis. J. Exp. Bot. 65 4037–4049. 10.1093/jxb/eru060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60 183–205. 10.1146/annurev.arplant.043008.092007 [DOI] [PubMed] [Google Scholar]
- Carabelli M., Morellit G., Whitelam G., Ruberti I. D. (1996). Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc. Natl. Acad. Sci. U.S.A. 93 3530–3535. 10.1073/pnas.93.8.3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J. J. (2012). Shade avoidance. Arabidopsis Book 10:e0157. 10.1199/tab.0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J. J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64 403–427. 10.1146/annurev-arplant-050312-120221 [DOI] [PubMed] [Google Scholar]
- Cerrudo I., Keller M. M., Cargnel M. D., Demkura P. V., de Wit M., Patitucci M. S., et al. (2012). Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol. 158 2042–2052. 10.1104/pp.112.193359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-J., Chen C.-L., Hsieh H.-L. (2015). Far-red light-mediated seedling development in Arabidopsis involves FAR-RED INSENSITIVE 219/JASMONATE RESISTANT 1-dependent and -independent pathways. PLOS ONE 10:e0132723. 10.1371/journal.pone.0132723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.-C., Huang I.-C., Liu M.-J., Wang Z.-G., Chung S.-S., Hsieh H.-L. (2007). Glutathione S-transferase interacting with far-red insensitive 219 is involved in phytochrome A-mediated signaling in Arabidopsis. Plant Physiol. 143 1189–1202. 10.1104/pp.106.094185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico J.-M., Fernández-Barbero G., Chini A., Fernández-Calvo P., Díez-Díaz M., Solano R. (2014). Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26 1967–1980. 10.1105/tpc.114.125047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J. (2010). Light signal transduction: an infinite spectrum of possibilities. Plant J. 61 982–991. 10.1111/j.1365-313X.2009.04105.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocco C. D., Holm M., Yanovsky M. J., Botto J. F. (2010). AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J. 64 551–562. 10.1111/j.1365-313X.2010.04360.x [DOI] [PubMed] [Google Scholar]
- Crocco C. D., Locascio A., Escudero C. M., Alabadı D., Blázquez M. A., Botto J. F. (2015). The transcriptional regulator BBX24 impairs DELLA activity to promote shade avoidance in Arabidopsis thaliana. Nat. Commun. 6:6202. 10.1038/ncomms7202 [DOI] [PubMed] [Google Scholar]
- De Wit M., Spoel S. H., Sanchez-Perez G. F., Gommers C. M., Pieterse C. M., Voesenek L. A., et al. (2013). Perception of low red: far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis. Plant J. 75 90–103. 10.1111/tpj.12203 [DOI] [PubMed] [Google Scholar]
- Deng X. W., Caspar T., Quail P. H. (1991). COP1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis Genes Dev. 5 1172–1182. 10.1101/gad.5.7.1172 [DOI] [PubMed] [Google Scholar]
- Devlin P. F., Robson R. H., Patel S. R., Goosey P. L., Sharrock R. A., Whitelam G. C. (1999). Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol. 119 909–915. 10.1104/pp.119.3.909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Chory J. (1997). Light control of plant development. Annu. Rev. Cell Dev. Biol. 13 203–229. 10.1146/annurev.cellbio.13.1.203 [DOI] [PubMed] [Google Scholar]
- Franklin K. A. (2008). Shade avoidance. New Phytol. 179 930–944. 10.1111/j.1469-8137.2008.02507.x [DOI] [PubMed] [Google Scholar]
- Franklin K. A., Quail P. H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61 11–24. 10.1093/jxb/erp304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K. A., Whitelam G. C. (2005). Phytochromes and shade-avoidance responses in plants. Ann. Bot. 96 169–175. 10.1093/aob/mci165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Fukushige H., Hildebrand D., Gan S. (2002). Methyl jasmonate enhances anthocyanin accumulation and modifies production of phenolics and pigments in “Fuji” apples. J. Am. Soc. Hortic. Sci. 128 876–884. [Google Scholar]
- Hornitschek P., Kohnen M. V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., et al. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71 699–711. 10.1111/j.1365-313X.2012.05033.x [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28 3893–3902. 10.1038/emboj.2009.306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh H., Okamoto H., Wang M., Ang L., Matsui M., Goodman H., et al. (2000). FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 14 1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Hsieh H. L., Okamoto H. (2014). Molecular interaction of jasmonate and phytochrome A signalling. J. Exp. Bot. 65 2847–2857. 10.1093/jxb/eru230 [DOI] [PubMed] [Google Scholar]
- Johnson E., Bradley M., Harberd N. P., Whitelam G. C. (1994). Photoresponses of light-grown phyA mutants of Arabidopsis (phytochrome A is required for the perception of daylength extensions). Plant Physiol. 105 141–149. 10.1104/pp.105.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J. M. (2011). The interplay between light and jasmonate signalling during defence and development. J. Exp. Bot. 62 4087–4100. 10.1093/jxb/err142 [DOI] [PubMed] [Google Scholar]
- Kurepin L. V., Pharis R. P. (2014). Light signaling and the phytohormonal regulation of shoot growth. Plant Sci. 229 280–289. 10.1016/j.plantsci.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Lau O. S., Deng X. W. (2010). Plant hormone signaling lightens up: integrators of light and hormones. Curr. Opin. Plant Biol. 13 571–577. 10.1016/j.pbi.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Leivar P., Monte E., Cohn M. M., Quail P. H. (2012a). Phytochrome signaling in green Arabidopsis seedlings: impact assessment of a mutually negative phyB-PIF feedback loop. Mol. Plant 5 734–749. 10.1093/mp/sss031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J. M., Cohn M. M., Monte E., Al-Sady B., Erickson E., et al. (2012b). Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24 1398–1419. 10.1105/tpc.112.095711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone M., Keller M. M., Cerrudo I., Ballar C. L. (2014). To grow or defend? Low red: far-red ratios reduce jasmonate sensitivity in Arabidopsis seedlings by promoting DELLA degradation and increasing JAZ10 stability. New Phytol. 204 355–367. 10.1111/nph.12971 [DOI] [PubMed] [Google Scholar]
- Li J., Li G., Wang H., Deng X. W. (2011). Phytochrome signaling mechanisms. Arabidopsis Book 9:e0148. 10.1199/tab.0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Ljung K., Breton G., Schmitz R. J., Pruneda-Paz J., Cowing-Zitron C., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26 785–790. 10.1101/gad.187849.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhang Q., Pedmale U. V., Nito K., Fu W., Lin L., et al. (2014). PIL1 participates in a negative feedback loop that regulates its own gene expression in response to shade. Mol. Plant 7 1582–1585. 10.1093/mp/ssu068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Liu B., Zhao C., Pepper M., Lin C. (2011). The action mechanisms of plant cryptochromes. Trends Plant Sci. 16 684–691. 10.1016/j.tplants.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P. D., Whitelam G. C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53 312–323. 10.1111/j.1365-313X.2007.03341.x [DOI] [PubMed] [Google Scholar]
- Lorrain S., Genoud T., Fankhauser C. (2006). Let there be light in the nucleus! Curr. Opin. Plant Biol. 9 509–514. 10.1016/j.pbi.2006.07.013 [DOI] [PubMed] [Google Scholar]
- Martínez-García J. F., Gallemí M., Molina-Contreras M. J., Llorente B., Bevilaqua M. R. R., Quail P. H. (2014). The shade avoidance syndrome in Arabidopsis: the antagonistic role of phytochrome A and B differentiates vegetation proximity and canopy shade. PLOS ONE 9:e109275. 10.1371/journal.pone.0109275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza C. A., Ballaré C. L. (2015). Photoreceptors UVR8 and phytochrome B cooperate to optimize plant growth and defense in patchy canopies. New Phytol. 207 4–9. 10.1111/nph.13332 [DOI] [PubMed] [Google Scholar]
- Moreno J. E., Tao Y., Chory J., Ballaré C. L. (2009). Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc. Natl. Acad. Sci. U.S.A. 106 4935–4940. 10.1073/pnas.0900701106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacín M., Legris M., Casal J. J. (2013). COP1 re-accumulates in the nucleus under shade. Plant J. 75 631–641. 10.1111/tpj.12226 [DOI] [PubMed] [Google Scholar]
- Pacín M., Semmoloni M., Legris M., Finlayson S. A., Casal J. J. (2016). Convergence of CONSTITUTIVE PHOTOMORPHOGENESIS 1 and PHYTOCHROME INTERACTING FACTOR signalling during shade avoidance. New Phytol. 211 967–979. 10.1111/nph.13965 [DOI] [PubMed] [Google Scholar]
- Possart A., Fleck C., Hiltbrunner A. (2014). Shedding (far-red) light on phytochrome mechanisms and responses in land plants. Plant Sci. 21 36–46. 10.1016/j.plantsci.2013.11.013 [DOI] [PubMed] [Google Scholar]
- Robson F., Okamoto H., Patrick E., Harris S.-R., Wasternack C., Brearley C., et al. (2010). Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22 1143–1160. 10.1105/tpc.109.067728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou J., Sorin C., Devlin P. F., Martínez-García J. F. (2006). Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 141 85–96. 10.1104/pp.105.076331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou-Torrent J., Galstyan A., Carretero-Paulet L., Portolés S., Rodríguez-Concepción M., et al. (2007). Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 26 4756–4767. 10.1038/sj.emboj.7601890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolauffs S., Fackendahl P., Sahm J., Fiene G., Hoecker U. (2012). Arabidopsis COP1 and SPA genes are essential for plant elongation but not for acceleration of flowering time in response to a low R:FR ratio. Plant Physiol. 160 2015–2027. 10.1104/pp.112.207233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter M. G., Franklin K. A., Whitelam G. C. (2003). Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426 680–683. 10.1038/nature02174 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. (2001). Molecular Cloning: A Laboratory Manual: SDS-Polyacrylamide gel Electrophoresis of Proteins, 3rd Edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 40–48. [Google Scholar]
- Sessa G., Carabelli M., Sassi M., Ciolfi A., Mittempergher F., Becker J., et al. (2005). A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev. 19 2811–2815. 10.1101/gad.364005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P. E., Su W., Howell S. H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. U.S.A. 89 6837–6840. 10.1073/pnas.89.15.6837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P. E., Tiryaki I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16 2117–2127. 10.1105/tpc.104.023549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P. E., Tiryaki I., Rowe M. L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14 1405–1415. 10.1105/tpc.000885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P. E., Yuen G. Y., Lehman C. C. (1998). Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 15 747–754. 10.1046/j.1365-313X.1998.00265.x [DOI] [PubMed] [Google Scholar]
- Steindler C., Matteucci A., Sessa G., Weimar T., Ohgishi M., Aoyama T., et al. (1999). Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126 4235–4245. [DOI] [PubMed] [Google Scholar]
- Tilbrook K., Arongaus A. B., Binkert M., Heijde M., Yin R., Ulm R. (2013). The UVR8 UV-B photoreceptor: perception, signaling and response. Arabidopsis Book 11:e0164. 10.1199/tab.0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Arnim A., Deng X. W. (1996). Light control of seedling development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 215–243. 10.1146/annurev.arplant.47.1.215 [DOI] [PubMed] [Google Scholar]
- Wang J.-G., Chen C.-H., Chien C.-T., Hsieh H.-L. (2011). FAR-RED INSENSITIVE219 modulates CONSTITUTIVE PHOTOMORPHOGENIC1 activity via physical interaction to regulate hypocotyl elongation in Arabidopsis. Plant Physiol. 156 631–646. 10.1104/pp.111.177667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.-H., Shen S.-C., Lee L.-Y., Lee S.-H., Chan M.-T., Lin C.-S. (2009). Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5:16. 10.1186/1746-4811-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.-L., Yao J., Mei C.-S., Tong X.-H., Zeng L.-J., Li Q., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 109 E1192–E1200. 10.1073/pnas.1201616109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky M., Casal J., Whitelam G. (1995). Phytochrome A, phytochrome B and HY4 are involved in hypocotyl growth responses to natural radiation in Arabidopsis: weak de-etiolation of the phyA mutant under dense canopies. Plant Cell Environ. 18 788–794. 10.1111/j.1365-3040.1995.tb00582.x [DOI] [Google Scholar]
- Yoo S. D., Cho Y. H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Yu Y., Wang J., Zhang Z., Quan R., Zhang H., Deng X. W., et al. (2013). Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLOS Genet. 9:e1004025. 10.1371/journal.pgen.1004025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q., Li C.-B., Zheng W., Wu X., Zhao J., Zhou G., et al. (2007). Phytochrome chromophore deficiency leads to overproduction of jasmonic acid and elevated expression of jasmonate-responsive genes in Arabidopsis. Plant Cell Physiol. 48 1061–1071. 10.1093/pcp/pcm076 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.