Abstract
Fibronectin is a master organizer of extracellular matrices (ECMs) and promotes the assembly of collagens, fibrillin-1, and other proteins. It is also known to play roles in skeletal tissues through its secretion by osteoblasts, chondrocytes, and mesenchymal cells. Spondylometaphyseal dysplasias (SMDs) comprise a diverse group of skeletal dysplasias and often manifest as short stature, growth-plate irregularities, and vertebral anomalies, such as scoliosis. By comparing the exomes of individuals with SMD with the radiographic appearance of “corner fractures” at metaphyses, we identified three individuals with fibronectin (FN1) variants affecting highly conserved residues. Furthermore, using matching tools and the SkelDys emailing list, we identified other individuals with de novo FN1 variants and a similar phenotype. The severe scoliosis in most individuals and rare developmental coxa vara distinguish individuals with FN1 mutations from those with classical Sutcliffe-type SMD. To study functional consequences of these FN1 mutations on the protein level, we introduced three disease-associated missense variants (p.Cys87Phe [c.260G>T], p.Tyr240Asp [c.718T>G], and p.Cys260Gly [c.778T>G]) into a recombinant secreted N-terminal 70 kDa fragment (rF70K) and the full-length fibronectin (rFN). The wild-type rF70K and rFN were secreted into the culture medium, whereas all mutant proteins were either not secreted or secreted at significantly lower amounts. Immunofluorescence analysis demonstrated increased intracellular retention of the mutant proteins. In summary, FN1 mutations that cause defective fibronectin secretion are found in SMD, and we thus provide additional evidence for a critical function of fibronectin in cartilage and bone.
Keywords: fibronectin, extracellular matrix, skeletal dysplasia, spondylometaphyseal, scoliosis, cartilage, protein secretion, corner fractures, metaphyses, FN1
Main Text
Spondylometaphyseal dysplasias (SMDs), or bone dysplasias affecting the spine and growth plates, comprise a heterogeneous group of conditions from both a clinical and genetic perspective. Genetic mutations have been identified for several SMDs (in COL2A1 [MIM: 120140], TRPV4 [MIM: 605427], SBDS [MIM: 607444], GPX4 [MIM: 138322], PCYT1A [MIM: 123695], and ACP5 [MIM: 171640]), but rarer forms still escape molecular diagnosis.1 One such condition is SMD with “corner fractures” (MIM: 184255). First recognized by Sutcliffe in 1966, fewer than 25 individuals or families have been reported.2 These individuals generally show developmental coxa vara but no scoliosis (as was the case for several individuals in our cohort without FN1 [MIM: 135600] mutations; see Table S1).3, 4 At the edges of the irregular metaphyses, flake-like, triangular, or curvilinear ossification centers simulate fractures. This specifically affects the distal tibia, the distal radius (ulnar aspect), the proximal humerus, and the proximal femur. The so-called “corner fractures” are unlikely to be true fractures but instead represent irregular ossification at the growth plates and secondary ossification centers.3 These fractures tend to become larger in older children and disappear after growth has stopped. Corner fractures on radiographs can also be seen in Duetting-type SMD (or SMD type A4 [MIM: 609052]),5 Schmid (MIM: 156500) and Jansen (MIM: 156400) types of metaphyseal chondrodysplasia, Strudwick-type spondyloepimetaphyseal dysplasia (MIM: 184250),6 Blount disease (MIM: 188700), Menkes disease (MIM: 309400), nonaccidental injury, congenital contractures, rickets, and scurvy.7 Some individuals initially thought to have SMD with corner fractures were later identified to have type 2 collagenopathy.6
Fibronectin is found in the human body in both a soluble form (∼300 μg/mL in plasma) and an insoluble form as a principal component of the fibrillar extracellular matrix (ECM) of virtually all tissues.8, 9 Fibronectin contains binding sites for integrins, collagens, glycoproteins, and glycosaminoglycans, as well as self-association sites.10 It self-assembles in a cell-dependent manner upon binding to integrins and other cell-surface components9 and initiates the assembly of the ECM.11, 12, 13
We performed exome sequencing in individuals with SMD with corner fractures, who had been identified through the Texas Children’s Skeletal Dysplasia Program, International Skeletal Dysplasia Registry, Baylor-Hopkins Center for Mendelian Genetics, Skeldys emailing list of the International Skeletal Dysplasia Society (ISDS), Shriners Canada Skeletal Dysplasia Clinic, and existing collaborations. Families provided written informed consent for protocols approved by the institutional review board at Baylor College of Medicine or local institutions. The procedures followed were in accordance with the ethical standards of the relevant committees on human experimentation. Details on the exome sequencing libraries and alignment are presented in Table S2.
In two of the first few individuals sequenced, who have been previously reported, COL2A1 mutations were identified.1 In total, 13 individuals were exome sequenced, and a comparison of the rare or undescribed variants shared between these individuals revealed variants in FN1 in three of these affected individuals (two variants segregating with the disease, c.260G>T [p.Cys87Phe] [GenBank: NM_212482.2 and NP_997647.1, respectively] in family 1 and c.718T>G [p.Tyr240Asp] in family 4, and a de novo variant, c.2425_2427del [p.Thr809del] in family 6). The genetic cause of SMD is still undetermined for the other individuals. Subsequently, through the ISDS emailing list, the GeneMatcher tool, and the Shriners Canada Skeletal Dysplasia Clinic,14 four other individuals with de novo FN1 variants (c.367T>C [p.Cys123Arg] in families 2 and 7, c.675C>G [p.Cys225Trp] in family 3, and c.778T>G [p.Cys260Gly] in family 5) and SMD with corner fractures were identified. In one of them (individual 3, identified with the ISDS emailing list), only FN1 was sequenced on the basis of the initial findings. Thus, in this cohort, previously unreported FN1 variants were found in 7 of 16 families affected by SMD with corner fractures. The clinical phenotypes of the individuals with FN1 variants are detailed in Table 1, and the variants are presented in Table 2. Figure 1 shows pedigrees, Figure 2 shows photographs, Figure 3 shows selected radiographs, and Figure S1 shows additional radiographs.
Table 1.
Clinical Features
| Family 1 | Family 2 | Family 3 | Family 4 | Family 5 | Family 6 | Family 7 | SMD Corner-Fracture Type or Sutcliffe Type (from Currarino et al.;3n = 18) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Individual in pedigree | II-2 (mother) | III-1 (first child) | III-2 (second child) | II-1 | II-1 | III-2 (child) | II-1 | II-1 | II-1 | NA |
| Gender | female | male | male | male | female | female | female | female | female | NA |
| Age at last assessment | 29 years | 13 years | 9 years | 14 years | 3 years, 9 months | 2 years, 1 month | 5 years, 9 months | 16 years, 11 months | 4 years | NA |
| Height (cm) | 147 (−3 SD) | 113 (−5.7 SD) | 107 (−4.6 SD) | 136 (−3.38 SD) | 92 (−2.5 SD) | 83.7 (−0.9 SD) | 97 (−3.3 SD) | 137 (−4.66 SD) | 89 (<3.0 SD) | NA |
| Ovoid vertebral bodies | NA | + | − | − | + | − | + | + | + | 16/18 |
| Scoliosis | + (operated) | + (operated) | + (operated) | + | − | − | + (operated) | + | + | 1/18 |
| Developmental coxa vara | − | − | − | − | − | + | − | + | + | 18/18 |
| Irregular metaphyses | NA | + | + | + | + | + | + | + | + | 16/18 |
| “Corner fractures” | − | + | + | + | + | + | + | + | + | 15/18 |
| Knee anomalies | − | − | genu varum (operated) | − | genu varum | − | genu varum (operated) | − | genu varum | one genu varum, one genum valgum |
| Chest or rib anomaly (e.g., pectus) | pectus carinatum | pectus carinatum | pectus carinatum | − | NA | − | pectus carinatum | − | − | NA |
| Other | hip surgery at 18 years, pregnancy-induced hypertension | born at term, weight 1,673 g (−4.3 SD), length 38 cm (−2.7 SD), hyponatremia at 1 month (unknown cause), hypoplasia of T12 vertebra and triangular S1, back and leg pain | born at term, weight 1,729 g (−3.9 SD), length 39 cm (−2.6 SD), leg pain | facial asymmetry and dysmorphisms (dysplastic left ear), missing teeth 34 and 44 (island of compact bone instead), intradural lipoma and megacisterna magna on MRI | born at 33 weeks, weight 1,754 g | femoral rodding | born at 36 weeks, weight 2,060 g (3rd–10th percentile), length 43.5 cm (3rd–10th percentile), OFC 29.5 cm (3rd percentile), normal renal sonography and kidney function, no signs of proteinuria or microalbuminuria, leg pain | born at 39 weeks, weight 2,280 g (<3rd percentile), length 45 cm (<3rd percentile), OFC 32 (3rd percentile), hip surgery at 2 years (both sides), multiple corrections to right hip and femur, shortening of right leg by 5 cm, bicuspid aortic valve | normal renal function, no proteinuria, no facial asymmetry or dysmorphisms | NA |
The following abbreviations are used: OFC, occipitofrontal circumference; and NA, not available.
Table 2.
Genetic Description of the FN1 Variants
| Family | Genomic Change (hg19) | Coding Change (GenBank:NM_212482.2)a | Protein Change (GenBank:NP_997647.1) | Inheritance |
|---|---|---|---|---|
| 1 | chr2: g.216299436C>A | c.260G>T | p.Cys87Phe | dominant |
| 2 | chr2: g.216298095A>G | c.367T>C | p.Cys123Arg | de novo |
| 3 | chr2: g.216295448G>C | c.675C>G | p.Cys225Trp | de novo |
| 4 | chr2: g.216293029A>C | c.718T>G | p.Tyr240Asp | dominant |
| 5 | chr2: g.216292969A>C | c.778T>G | p.Cys260Gly | de novo |
| 6 | chr2: g.216273022_216273024del | c.2425_2427del | p.Thr809del | de novo |
| 7 | chr2: g.216298095A>G | c.367T>C | p.Cys123Arg | de novo |
All variants are absent from the ExAC Browser.
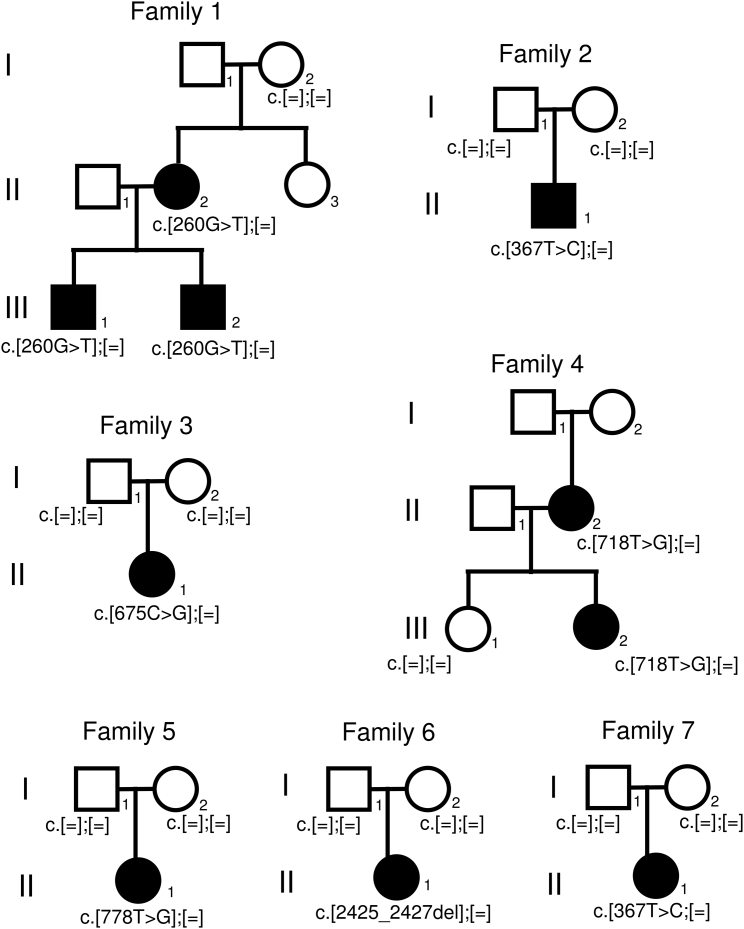
Figure 1.
Pedigrees of the Families with FN1 Mutations
Co-segregation of the variants with the trait in families 1 and 4 suggests dominant inheritance. Consistently, de novo FN1 mutations in the affected individuals of simplex families 2, 3, 5, 6, and 7 were observed.
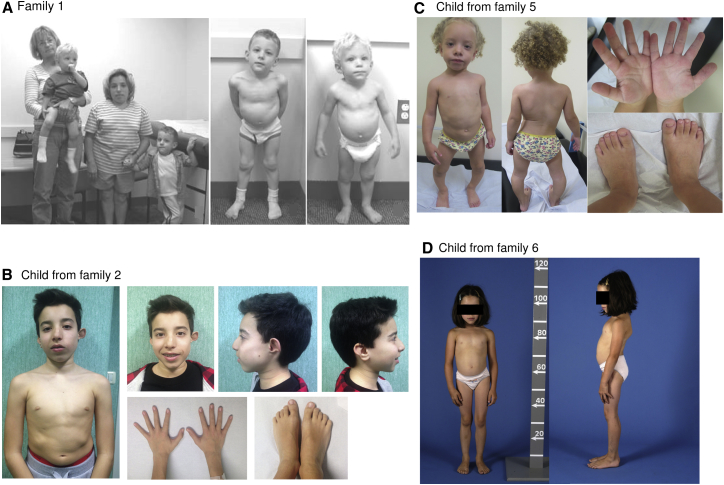
Figure 2.
Photographs of Some of the Individuals with FN1 Mutations
(A) Photographs in a figure reproduced with permission from Sutton et al.30 (copyright © 2005 Wiley-Liss, Inc.). On the left are the three affected individuals who have an FN1 mutation and the maternal grandmother. In the middle is the older child at age 5 years, and on the right is the younger child at age 3 years.
(B) Affected child from family 2 at age 13 years. Note the short trunk, facial asymmetry, dysplastic left ear, and normal hands and feet.
(C) Affected child from family 5 at age 2 years and 11 months. Note the scoliosis, genu varum, and normal hands and feet.
(D) Affected child from family 6 at age 8 years. Note the short trunk and scoliotic posture.
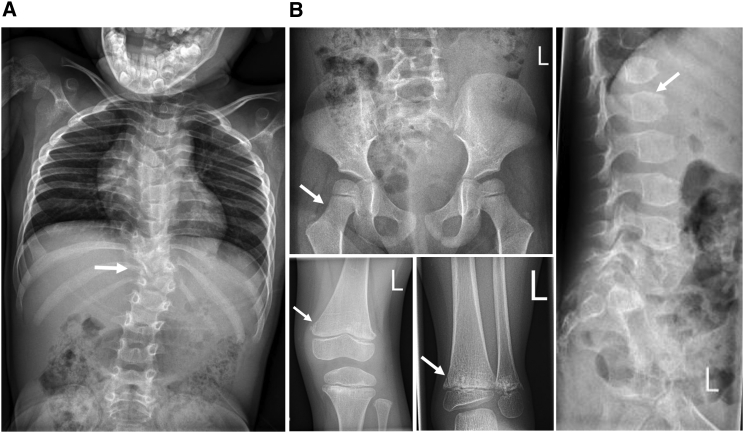
Figure 3.
Radiographs Showing the “Corner Fractures” and Other Radiological Changes
(A) Individual from family 7; note the significant scoliosis.
(B) Individual from family 3; note the absence of coxa vara, the presence of irregular metaphyses with corner fractures, and the presence of ovoid vertebral bodies.
Additional radiographs from all families are available in Figure S1.
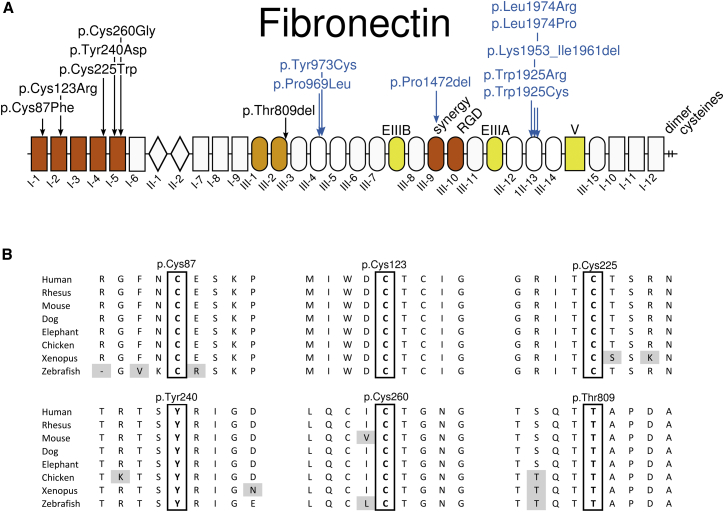
All FN1 variants discovered in this study are absent from the ExAC Browser and affect highly conserved residues (Figure 4). The majority (4/6) affect cysteine residues, all of which form disulfide bonds that are critical for the three-dimensional structure of the type I fibronectin domains.15 Five of the mutations (c.260G>T [p.Cys87Phe], c.367T>C [p.Cys123Arg], c.675C>G [p.Cys225Trp], c.718T>G [p.Tyr240Asp], and c.778T>G [p.Cys260Gly]) are located in the N-terminal assembly domain (spanning fibronectin domains I-1 through I-5), which is important to initiating assembly on the cell surface.9 The remaining mutation (c.2425_2427del [p.Thr809del]) was found in the III-2 domain, which contains a fibronectin binding site and is involved in conformational changes promoting fibronectin assembly.9 The Tyr240 residue has previously been mutated into a Ser residue in vitro and shown to be critical for fibronectin binding to fibroblasts.16
Figure 4.
Position and Conservation of Amino Acids Affected by Substitutions
(A) Location of the SMD-associated fibronectin amino acid substitutions (in black) and those underlying glomerulopathy (in blue). The fibronectin domains I, II, and III are numbered, V stands for variable domain, and EIIIA and EIIIB indicate the extra type III repeat A and B segments. These three domains in yellow are subject to alternative splicing. Domains I-1 to I-5 in red represent the N-terminal assembly domain. Domains III-9 and III-10, also in red, contain the synergy site and the RGD site. Domains III-1 and III-2, in orange, contain self-interaction sites and are involved in conformational changes promoting fibronectin assembly.
(B) Amino acid conservation of the mutated residues across vertebrates. Gray shading indicates non-conserved amino acid residues.
Heterozygous FN1 mutations have previously been reported in type 2 autosomal-dominant glomerulopathy with fibronectin deposits (MIM: 601894).17 Remarkably, all FN1 mutations previously implicated in this glomerulopathy cluster in more C-terminally-located regions important for heparin binding and integrin binding (Figure 4).17, 18, 19 In vitro, the mutations cause decreased heparin and integrin binding, reduced endothelial cell spreading, and cytoskeletal reorganization, which has been hypothesized to affect glomerular size selectivity and protein trafficking. Importantly, none of the SMD individuals in our cohort had any evidence of renal disease.
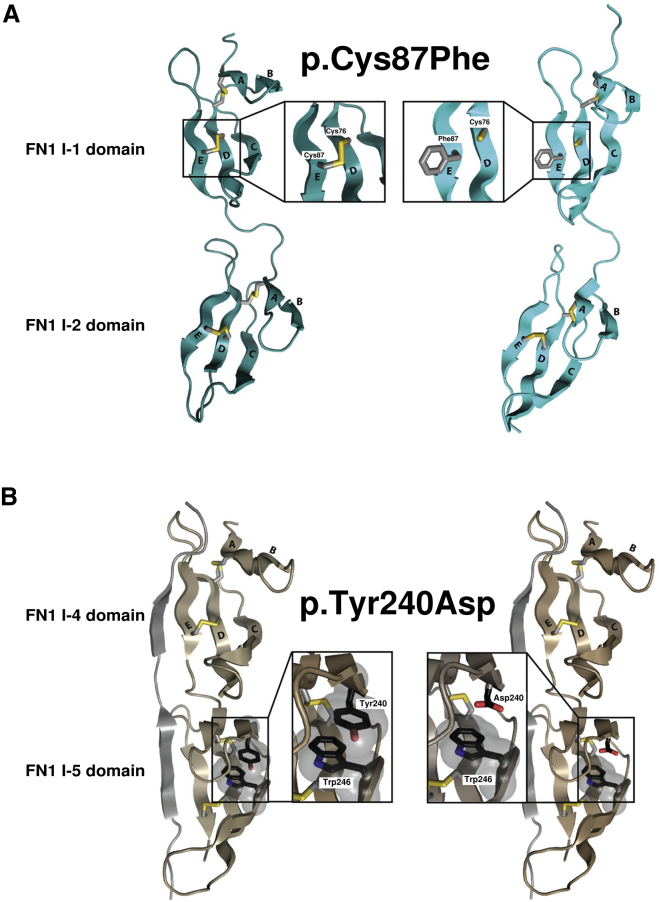
As stated above, all cysteine residues affected by mutations in our cohort are involved in disulfide bonds in fibronectin type I domains (bridges form between Cys87 and Cys76, between Cys123 and Cys135, between Cys225 and Cys213, and between Cys260 and Cys231). We selected one of these (p.Cys87Phe) to model the consequence of the mutation on the three-dimensional structure of fibronectin domains (Figure 5A). The p.Cys87Phe substitution was predicted to destabilize the structure of the I-1 domain by breaking the disulfide bond with residue Cys76 and by displaying a hydrophobic residue at the surface. A similar disruptive impact was predicted for the other missense changes involving Cys123, Cys225, and Cys260. For the two other amino acid residues affected by mutations (Tyr240 and Thr809), 3D structures adequate for modeling the mutations were available only for p.Tyr240Asp (Thr809 was found only at the C-terminal end of a solution structure of III-2,20 which limits the interpretation of its interactions). Modeling predicted that the p.Tyr240Asp variant would disrupt the pi-stacking (or π-π stacking) of the side chains of Tyr240 and Trp246 and thus destabilize the I-5 domain (Figure 5B).
Figure 5.
Structural Impact of SMD-Causing Mutations
(A) Structural models of fibronectin domains I-1 and I-2 (PDB: 1O9A,31 without the S. dysgalactiae FnBP B3 peptide). On the left is the wild-type protein, and on the right is the model with the variant. The model for the p.Cys87Phe substitution was generated by the ModWeb Server.32 The disulfide bond formed between Cys76 and Cys87 stabilizes domain I-1. The p.Cys87Phe substitution is predicted to destabilize the structure by breaking the disulfide bond and by displaying a hydrophobic residue at the surface.
(B) Structural models of fibronectin domains I-4 and I-5 (PDB: 2RKY,33 without the S. aureus FnBPA peptide). To generate the model for the p.Tyr240Asp change, we replaced tyrosine with aspartic acid and manually adjusted the rotomer position to minimize the steric clash with the rest of the protein by using Coot.34 For the wild-type protein, the side chains of Tyr240 and Trp246 interact through π stacking and stabilize the fibronectin domain. This interaction is predicted to be lost in the mutant protein.
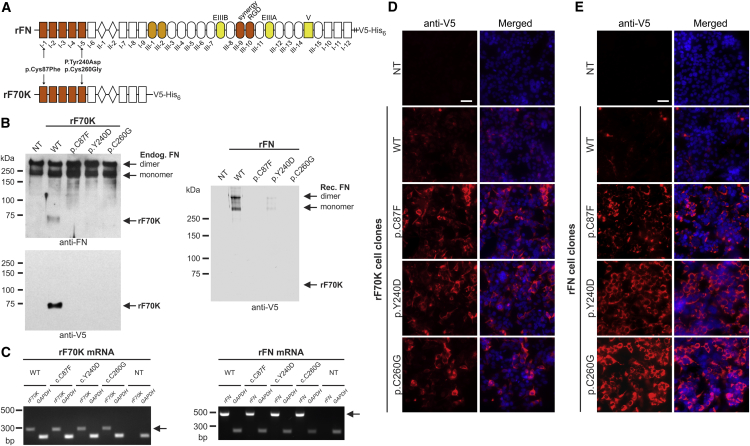
We next aimed to analyze the functional consequences of selected FN1 mutations on the fibronectin protein. Because cells from affected individuals were not available for this study, we generated a recombinant 70 kDa N-terminal fragment of fibronectin (rF70K), spanning the region where five of the six missense variants identified in this study are located, and a full-length recombinant fibronectin (rFN) (Figure 6A). Expression vectors for the human wild-type rF70K and rFN, as well as selected mutants, were generated by standard cloning procedures in the pcDNA3.1+ plasmid (Thermo Fisher Scientific, V79020) with the sequence for a C-terminal V5 epitope tag to facilitate detection and a hexa-histidine tag intended for chromatographic purification. At the 5′ ends, the vectors contained the coding sequence for either the BM40 signal peptide (rF70K) or the native fibronectin signal peptide (rFN) to ensure secretion of the recombinant protein through the secretory pathway. The rFN expression plasmid was constructed with the sequence for the alternatively spliced EDA and EDB domains. For this analysis, we selected two cysteine missense variants (p.Cys87Phe and p.Cys260Gly) because they represent the major group of the identified mutations, as well as a non-cysteine missense variant (p.Tyr240Asp). These point mutants were generated from the wild-type plasmids via the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, 200519).
Figure 6.
Analysis of Protein Secretion of Recombinant Wild-Type and SMD-Causing Mutants of rF70K and rFN
(A) Schematic overview of full-length fibronectin (rFN) and the 70 kDa N-terminal fragment of fibronectin (rF70K). To ensure secretion through the secretory pathway, the expression plasmids contain either the sequence coding for the endogenous fibronectin signal peptide (rFN) or a heterologous BM40 signal peptide (rF70K). The indicated SMD variants were engineered into the expression constructs.
(B–E) NT refers to the non-transfected HEK293 controls, WT refers to the wild-type rF70K or rFN cell clones, and the mutant cell clones are indicated in the one-letter amino acid code. (B) Western blot analysis of conditioned cell-culture medium (for 2 days) harvested from HEK293 cells transfected with rF70K (left) and rFN (right). Analysis of rF70K was performed with a rabbit anti-fibronectin antibody (top; primary antibody, Sigma, F3648; secondary goat anti-rabbit horseradish-conjugated antibody, Agilent Technologies, K4008) and a horseradish-conjugated anti-V5 monoclonal antibody (bottom, Thermo Fisher Scientific, MA5-15253-HRP), whereas the rFN analysis was performed with only an anti-V5 antibody, given that the anti-fibronectin antibody also reacts with endogenous fibronectin. All samples were analyzed under reducing conditions, the blots for rF70K were developed with the SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, 34080), and the blots for rFN were developed with 0.5 mg/mL 4-chloro-1-naphthol (Sigma, C8890) in Tris-buffered saline including 0.02% H2O2. (C) Specific RT-PCR analysis of mRNA coding for rF70K (left; 285 bp) or rFN (right; 440 bp) (excluding the endogenous FN1 mRNA). GAPDH analysis (226 bp) was included as a control. RNA was extracted with the RNeasy Plus Mini Kit (QIAGEN, 74134) according to the manufacturer’s protocol. Reverse transcription was performed with the ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs, E6560S), and PCR was performed for 40 cycles. (D and E) Immunofluorescence analysis of transfected HEK293 cells 3 days after seeding with anti-V5 antibodies (red; primary antibody, Thermo Fisher Scientific, R960-25; secondary antibody, Cy3-conjugated antibody, Thermo Fisher Scientific, A10521) for rF70K and rFN was performed according to a previously established protocol.35 Cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100; nuclei were counter-stained with DAPI (blue). The experiments were confirmed with three to four individual recombinant cell clones. The scale bar indicates 50 μm. Images were taken at 400× magnification with Zen software and an AxioImager M2 microscope (Zeiss) equipped with an ORCA-flash 4.0 camera (Hamamatsu, C11440). Note that all analyzed rF70K and rFN mutant proteins were retained intracellularly, but the wild-type proteins were not.
The plasmids were transfected into human HEK293 cells, which synthesize and secrete endogenous fibronectin and represent an efficient system for producing a range of correctly folded and posttranslationally modified ECM proteins (subfragments as well as mutants).21, 22, 23 After the plasmids were stably transfected into human HEK293 cells as described previously,24 the secreted wild-type recombinant proteins were readily identified in the cell-culture medium (conditioned for 2 days) with fibronectin-specific or anti-V5-specific antibodies (Figure 6B). Surprisingly, all three rF70K and rFN mutants were either undetectable in the culture medium or detectable at much lower levels than the wild-type protein. Analysis of recombinant mRNA with primers excluding the endogenous fibronectin mRNA revealed that the amount of mutant mRNA was similar to that of wild-type mRNA (Figure 6C). Immunofluorescence analysis demonstrated very low amounts of the wild-type proteins within the stably transfected cells because the proteins secreted into the culture medium (Figures 6D and 6E). In contrast to this finding, all three rF70K and rFN mutants consistently showed strong accumulation within the cells. These data clearly demonstrate that p.Cys87Phe, p.Tyr240Asp, and p.Cys260Gly exert a common molecular defect in fibronectin secretion. To assess whether the intracellular retention induced the unfolded protein response in HEK293 cells, we analyzed XBP1 splicing (IRE1 pathway) and the mRNA expression levels of CHOP and ATF4 (PERK pathway). However, no indication of increased XBP1 splicing or changes in the CHOP or ATF4 mRNA expression levels was detected (Figure S2). In addition, the mutant rFN cell clones did not undergo caspase-3-mediated apoptosis (Figure S3).
Fibronectin is present throughout cartilage differentiation and persists in mature cartilaginous tissue.25 However, little is known about the functional role of fibronectin in the development and homeostasis of cartilage. Given that fibronectin has been established as a key ECM protein that guides fibrillogenesis of several other ECM proteins, including proteins present in cartilage, such as fibrillin-1,26 it is conceivable that a reduced amount of secreted fibronectin from chondrocytes could result in a deficient matrix network in cartilage. It is well known that mutations affecting another cartilage protein, cartilage oligomeric matrix protein (COMP [MIM: 600310]), lead to pseudoachondroplasia (MIM: 177170) and multiple epiphyseal dysplasia (MIM: 132400) through accumulation of mutant COMP in the endoplasmic reticulum of chondrocytes.27 It is thought that this protein accumulation impairs secretion of other matrix proteins and leads to abnormal ECM and chondrocyte death.28, 29 It is possible that a similar mechanism exists for retained mutant fibronectin and leads to the SMD phenotype observed in our cohort. Conditional double-knockout experiments of fibronectin in mouse cartilage and liver or knockin of some of the mutations are currently underway to further elucidate the molecular basis of fibronectin in the development and maintenance of cartilage.
Acknowledgments
This project was supported in part by operating grants from the Canadian Institutes for Health Research (clinician-scientist award RN315908 to P.M.C.; MOP-137091 to D.P.R.), the Fonds de Recherche du Québec - Santé clinical research scholar award 30647 to P.M.C., the Quebec Network for Oral and Bone Health Research (RSBO Emerging Collaborating Project 2014-2015 to P.M.C. and D.P.R.), the NIH (UM1 HG006542 to the Baylor Hopkins Center for Mendelian Genomics), the Heart and Stroke Foundation of Canada (G-16-00014634 to D.P.R.), the Italian Ministry of Health (Ricerca Corrente 2016 to M.T. and M.N.), and Fondazione Bambino Gesù (Vite Coraggiose grant to M.T.). This work was also supported by NIH P01 HD070394, by HD024064 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development granted to the Baylor College of Medicine (BCM) Intellectual and Developmental Disabilities Research Center (for processing samples obtained and managing clinical protocols), by the BCM Advanced Technology Cores through funding from the NIH (AI036211, CA125123, and RR024574), by the Rolanette and Berdon Lawrence Bone Disease Program of Texas, and by the BCM Center for Skeletal Medicine and Biology (to B.H.L). Analysis of individual 5 was supported by the São Paulo Research Foundation (FAPESP 2015/21783-9; Centros de Pesquisa, Inovação, e Difusão [CEPID] 2013/08028-1) and National Council for Scientific and Technological Development (CNPq 302605/2013-4 and 304130/2016-8 to D. B). E.L. is supported by grants from the German Research Foundation (CRC 1140), the German Ministry for Education and Research (MaTrOC), and the European Union (SYBIL grant agreement no. 602300; RARENET). We thank Dr. Deane Mosher and Dr. Douglas Annis for providing the FN1 pAcGP67A plasmid. We thank Dr. Reggie Hamdy for referring individual 7 to Dr. Campeau and Dr. Amélie Damphousse for the interpretation of the radiographs.
Published: November 2, 2017
Footnotes
Supplemental Data include three figures and two tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.09.019.
Contributor Information
Dieter P. Reinhardt, Email: dieter.reinhardt@mcgill.ca.
Philippe M. Campeau, Email: p.campeau@umontreal.ca.
Accession Numbers
The accession numbers for the variants identified in this study are ClinVar: SCV000574553–SCV000574558.
Web Resources
Baylor-Hopkins Center for Mendelian Genetics (BH-CMG), http://bhcmg.org/
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
GeneMatcher, https://www.genematcher.org
International Skeletal Dysplasia Society (ISDS), http://www.isds.ch/
International Skeletal Dysplasia Registry (ISDR), http://ortho.ucla.edu/isdr
OMIM, http://www.omim.org/
RCSB Protein Data Bank, https://www.rcsb.org/pdb/home/home.do
UniProt, http://www.uniprot.org/uniprot/
Supplemental Data
References
- 1.Machol K., Jain M., Almannai M., Orand T., Lu J.T., Tran A., Chen Y., Schlesinger A., Gibbs R., Bonafe L. Corner fracture type spondylometaphyseal dysplasia: Overlap with type II collagenopathies. Am. J. Med. Genet. A. 2017;173:733–739. doi: 10.1002/ajmg.a.38059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutcliffe J. Metaphyseal dysostosis. Ann. Radiol. (Paris) 1966;9:R215–R223. [Google Scholar]
- 3.Currarino G., Birch J.G., Herring J.A. Developmental coxa vara associated with spondylometaphyseal dysplasia (DCV/SMD): “SMD-corner fracture type” (DCV/SMD-CF) demonstrated in most reported cases. Pediatr. Radiol. 2000;30:14–24. doi: 10.1007/s002470050005. [DOI] [PubMed] [Google Scholar]
- 4.Langer L.O., Jr., Brill P.W., Ozonoff M.B., Pauli R.M., Wilson W.G., Alford B.A., Pavlov H., Drake D.G. Spondylometaphyseal dysplasia, corner fracture type: a heritable condition associated with coxa vara. Radiology. 1990;175:761–766. doi: 10.1148/radiology.175.3.2343127. [DOI] [PubMed] [Google Scholar]
- 5.Duetting T., Schulze A., Troeger J., Spranger J. A rare form of spondylometaphyseal dysplasia-type A4. Am. J. Med. Genet. 1998;78:61–66. doi: 10.1002/(sici)1096-8628(19980616)78:1<61::aid-ajmg13>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 6.Walter K., Tansek M., Tobias E.S., Ikegawa S., Coucke P., Hyland J., Mortier G., Iwaya T., Nishimura G., Superti-Furga A., Unger S. COL2A1-related skeletal dysplasias with predominant metaphyseal involvement. Am. J. Med. Genet. A. 2007;143A:161–167. doi: 10.1002/ajmg.a.31516. [DOI] [PubMed] [Google Scholar]
- 7.Kozlowski K., Beighton P. Springer-Verlag; 1984. Gamut index of skeletal dysplasias: an aid to radiodiagnosis. [Google Scholar]
- 8.Mosher D.F. Plasma fibronectin concentration: a risk factor for arterial thrombosis? Arterioscler. Thromb. Vasc. Biol. 2006;26:1193–1195. doi: 10.1161/01.ATV.0000223342.15969.7a. [DOI] [PubMed] [Google Scholar]
- 9.Singh P., Carraher C., Schwarzbauer J.E. Assembly of fibronectin extracellular matrix. Annu. Rev. Cell Dev. Biol. 2010;26:397–419. doi: 10.1146/annurev-cellbio-100109-104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pankov R., Yamada K.M. Fibronectin at a glance. J. Cell Sci. 2002;115:3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 11.Sabatier L., Chen D., Fagotto-Kaufmann C., Hubmacher D., McKee M.D., Annis D.S., Mosher D.F., Reinhardt D.P. Fibrillin assembly requires fibronectin. Mol. Biol. Cell. 2009;20:846–858. doi: 10.1091/mbc.E08-08-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sottile J., Hocking D.C. Fibronectin polymerization regulates the composition and stability of extracellular matrix fibrils and cell-matrix adhesions. Mol. Biol. Cell. 2002;13:3546–3559. doi: 10.1091/mbc.E02-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallas S.L., Sivakumar P., Jones C.J., Chen Q., Peters D.M., Mosher D.F., Humphries M.J., Kielty C.M. Fibronectin regulates latent transforming growth factor-beta (TGF beta) by controlling matrix assembly of latent TGF beta-binding protein-1. J. Biol. Chem. 2005;280:18871–18880. doi: 10.1074/jbc.M410762200. [DOI] [PubMed] [Google Scholar]
- 14.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baron M., Norman D., Willis A., Campbell I.D. Structure of the fibronectin type 1 module. Nature. 1990;345:642–646. doi: 10.1038/345642a0. [DOI] [PubMed] [Google Scholar]
- 16.Sottile J., Schwarzbauer J., Selegue J., Mosher D.F. Five type I modules of fibronectin form a functional unit that binds to fibroblasts and Staphylococcus aureus. J. Biol. Chem. 1991;266:12840–12843. [PubMed] [Google Scholar]
- 17.Ohtsubo H., Okada T., Nozu K., Takaoka Y., Shono A., Asanuma K., Zhang L., Nakanishi K., Taniguchi-Ikeda M., Kaito H. Identification of mutations in FN1 leading to glomerulopathy with fibronectin deposits. Pediatr. Nephrol. 2016;31:1459–1467. doi: 10.1007/s00467-016-3368-7. [DOI] [PubMed] [Google Scholar]
- 18.Castelletti F., Donadelli R., Banterla F., Hildebrandt F., Zipfel P.F., Bresin E., Otto E., Skerka C., Renieri A., Todeschini M. Mutations in FN1 cause glomerulopathy with fibronectin deposits. Proc. Natl. Acad. Sci. USA. 2008;105:2538–2543. doi: 10.1073/pnas.0707730105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ertoy Baydar D., Kutlugun A.A., Bresin E., Piras R. A case of familial glomerulopathy with fibronectin deposits caused by the Y973C mutation in fibronectin. Am. J. Kidney Dis. 2013;61:514–518. doi: 10.1053/j.ajkd.2012.08.050. [DOI] [PubMed] [Google Scholar]
- 20.Vakonakis I., Staunton D., Rooney L.M., Campbell I.D. Interdomain association in fibronectin: insight into cryptic sites and fibrillogenesis. EMBO J. 2007;26:2575–2583. doi: 10.1038/sj.emboj.7601694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirschner R., Hubmacher D., Iyengar G., Kaur J., Fagotto-Kaufmann C., Brömme D., Bartels R., Reinhardt D.P. Classical and neonatal Marfan syndrome mutations in fibrillin-1 cause differential protease susceptibilities and protein function. J. Biol. Chem. 2011;286:32810–32823. doi: 10.1074/jbc.M111.221804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKee K.K., Harrison D., Capizzi S., Yurchenco P.D. Role of laminin terminal globular domains in basement membrane assembly. J. Biol. Chem. 2007;282:21437–21447. doi: 10.1074/jbc.M702963200. [DOI] [PubMed] [Google Scholar]
- 23.Fox J.W., Mayer U., Nischt R., Aumailley M., Reinhardt D., Wiedemann H., Mann K., Timpl R., Krieg T., Engel J. Recombinant nidogen consists of three globular domains and mediates binding of laminin to collagen type IV. EMBO J. 1991;10:3137–3146. doi: 10.1002/j.1460-2075.1991.tb04875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin G., Tiedemann K., Vollbrandt T., Peters H., Batge B., Brinckmann J., Reinhardt D.P. Homo- and heterotypic fibrillin-1 and -2 interactions constitute the basis for the assembly of microfibrils. J. Biol. Chem. 2002;277:50795–50804. doi: 10.1074/jbc.M210611200. [DOI] [PubMed] [Google Scholar]
- 25.Singh P., Schwarzbauer J.E. Fibronectin and stem cell differentiation - lessons from chondrogenesis. J. Cell Sci. 2012;125:3703–3712. doi: 10.1242/jcs.095786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keene D.R., Jordan C.D., Reinhardt D.P., Ridgway C.C., Ono R.N., Corson G.M., Fairhurst M., Sussman M.D., Memoli V.A., Sakai L.Y. Fibrillin-1 in human cartilage: developmental expression and formation of special banded fibers. J. Histochem. Cytochem. 1997;45:1069–1082. doi: 10.1177/002215549704500805. [DOI] [PubMed] [Google Scholar]
- 27.Maddox B.K., Keene D.R., Sakai L.Y., Charbonneau N.L., Morris N.P., Ridgway C.C., Boswell B.A., Sussman M.D., Horton W.A., Bächinger H.P., Hecht J.T. The fate of cartilage oligomeric matrix protein is determined by the cell type in the case of a novel mutation in pseudoachondroplasia. J. Biol. Chem. 1997;272:30993–30997. doi: 10.1074/jbc.272.49.30993. [DOI] [PubMed] [Google Scholar]
- 28.Hecht J.T., Montufar-Solis D., Decker G., Lawler J., Daniels K., Duke P.J. Retention of cartilage oligomeric matrix protein (COMP) and cell death in redifferentiated pseudoachondroplasia chondrocytes. Matrix Biol. 1998;17:625–633. doi: 10.1016/s0945-053x(98)90113-5. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto Y., Tomiyama T., Yamano Y., Mori H. Mutation (D472Y) in the type 3 repeat domain of cartilage oligomeric matrix protein affects its early vesicle trafficking in endoplasmic reticulum and induces apoptosis. Am. J. Pathol. 2003;163:101–110. doi: 10.1016/S0002-9440(10)63634-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutton V.R., Hyland J.C., Phillips W.A., Schlesinger A.E., Brill P.W. A dominantly inherited spondylometaphyseal dysplasia with “corner fractures” and congenital scoliosis. Am. J. Med. Genet. A. 2005;133A:209–212. doi: 10.1002/ajmg.a.30567. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz-Linek U., Werner J.M., Pickford A.R., Gurusiddappa S., Kim J.H., Pilka E.S., Briggs J.A., Gough T.S., Höök M., Campbell I.D., Potts J.R. Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature. 2003;423:177–181. doi: 10.1038/nature01589. [DOI] [PubMed] [Google Scholar]
- 32.Pieper U., Webb B.M., Barkan D.T., Schneidman-Duhovny D., Schlessinger A., Braberg H., Yang Z., Meng E.C., Pettersen E.F., Huang C.C. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2011;39:D465–D474. doi: 10.1093/nar/gkq1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bingham R.J., Rudiño-Piñera E., Meenan N.A., Schwarz-Linek U., Turkenburg J.P., Höök M., Garman E.F., Potts J.R. Crystal structures of fibronectin-binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains. Proc. Natl. Acad. Sci. USA. 2008;105:12254–12258. doi: 10.1073/pnas.0803556105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Hubmacher D., Sabatier L., Annis D.S., Mosher D.F., Reinhardt D.P. Homocysteine modifies structural and functional properties of fibronectin and interferes with the fibronectin-fibrillin-1 interaction. Biochemistry. 2011;50:5322–5332. doi: 10.1021/bi200183z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.