Abstract
Background
Many studies suggest that the small ubiquitin-like modifier 4 (SUMO4) M55V gene polymorphism (rs237025) may be associated with an increased risk of type 2 diabetes mellitus (T2DM). However, due to other conflicting results, a clear consensus is lacking in the matter.
Objective and methods
A meta-analysis consisting of 6,823 subjects from 10 studies was conducted to elucidate relationship between the SUMO4 M55V gene polymorphism and T2DM. Depending on the heterogeneity of the data, either a fixed or random-effects model would be used to assess the combined odds ratio (ORs) and their corresponding 95% confidence interval (CI).
Results
SUMO4 gene M55V polymorphism was significantly associated with T2DM in the whole population under allelic (OR: 1.18, 95% CI: 1.10–1.28, P = 1.63 × 10−5), recessive (OR: 1.59, 95% CI: 1.14–2.23, P = 0.006), dominant (OR: 0.815, 95% CI: 0.737–0.901, P = 6.89 × 10−5), homozygous (OR: 1.415, 95% CI: 1.170–1.710, P = 0.0003), heterozygous (OR: 1.191, 95% CI: 1.072–1.323, P = 0.001), and additive genetic models (OR: 1.184, 95% CI: 1.097–1.279, P = 1.63 × 10−5). In our subgroup analysis, a significant association was found again in the Chinese population, but not in Japanese or Iranian population.
Conclusion
SUMO4 gene M55V polymorphism may correlate with increased T2DM risk. Chinese carriers of the V allele of the SUMO4 gene M55V polymorphism may be predisposed to developing T2DM.
Keywords: small ubiquitin-like modifier 4, rs237025, polymorphism, type 2 diabetes mellitus, meta-analysis
Introduction
Type 2 diabetes mellitus (T2DM) is a long-term metabolic disorder characterized by high blood sugar, insulin resistance, and impaired insulin secretion. Recently, research found the nuclear factor-κB (NF-κB) signaling pathway to be mechanistically implicated in the pathogenesis of T2DM by increasing pancreatic β-cell apoptosis (1–3). Small ubiquitin-like modifier 4 (SUMO4), a newly discovered molecule located in insulin-dependent diabetes mellitus 5 (IDDM5), has been shown to suppress the transcription of NF-κB. SUMO4 also acts as an anti-oxidant, protecting the pancreatic β-cells from oxidative damage and promoting β-cell survival (4). Therefore, under oxidative stress by autoimmune processes, SUMO4 and the DNA damage signaling protein iKB can be mobilized to activate intracellular pathways associated with cell survival (5).
The SUMO4 gene is located at position 6q25 in the IDDM5 locus and spans 688 bp. The gene contains a single exon, encoding 95 amino acids (6). The M55V polymorphism is caused by mutation of an adenosine at the 1633d position to a guanine (rs237025, A163G). The conformational change caused by the polymorphism results in increased NF-κB transcriptional activity and, thus, increased expression of NF-κB-dependent genes.
Though researchers have found good evidence for an association between this polymorphism and T2DM susceptibility, the exact relationship is still debated, possibly due to the varying genetic backgrounds of different ethnic groups (7). In this regard, other associations with T2DM, such HMGA1 and TCFTL2 variants, have been found to be heterogeneous across different ethnic groups (8, 9).
In 2004, Bohren et al. found the SUMO4 Met55Val gene polymorphism to be associated with an increased susceptibility to type 1 diabetes mellitus (T1DM) in the Caucasian population (6). Around the same time, Guo et al. replicated the result in a Chinese population (4). In 2005, Noso et al. found Val 55 to be significantly more common in T1DM patients than in control subjects in the Japanese population (4). In 2008, Ikegami et al. has performed the genome-wide association (GWA) study and found that the M55V variant was significantly associated with T1DM in the Asian populations, but not Caucasian populations (10). Other studies have suggested the susceptibility loci for T2DM may lie in the area on Chromosome 6 that SUMO4 is located (11–14), indicating a common genetic basis for both types of diabetes. Considering the possible functional evidence for SUMO4 as a candidate gene for T2DM, Noso et al. initially explored the contribution of the SUMO4 Met55Val locus to T2DM susceptibility and found a significant association in a Japanese population (15). Ji et al. found similar results among a Chinese Han population in the Hubei region. Compared with control MM genotype, Ji et al. found that individuals with the VV or MV genotype to have higher levels of fasting insulin and Homeostasis Model Assessment-Insulin Resistance (7). A study by Pu et al. also came upon the same result in a Beijing population in China (16). However, the results on the matter are not unanimous: in 2010, Fallah et al. found that SUMO4 gene M55V variant was not associated with the susceptibility of T2DM in an Iranian population (17).
We conducted the current meta-analysis from 3,223 T2DM patients and 3,600 controls to verify the association of SUMO4 M55V gene polymorphism and T2DM.
Materials and Methods
Publication Search and Inclusion Criteria
The included studies should meet the following inclusion criteria: (a) assessment of the association of SUMO4 gene M55V polymorphism with T2DM; (b) T2DM diagnosis and classification meets guidelines proposed by the World Health Organization in 1999: fasting blood sugar no less than 7.0 mmol/l and the 2 h postprandial blood sugar no less than 11.1 mmol/l. Other metabolic conditions, such as acute and chronic complications of diabetes mellitus, T1DM, ketosis, hepatic and renal dysfunction, and other conditions were excluded. (c) Officially published case–control or cohort studies. (d) Genotype in control group follows Hardy–Weinberg equilibrium (HWE).
The following electronic databases were used to conduct a search: China National Knowledge Infrastructure, VIP database, Wanfang database, China Biological Medicine Database, and PubMed. Keywords used for the search were “SUMO4, rs237025, diabetes.” When searching Chinese databases, researchers employed corresponding Chinese terms. Nine publications were retrieved in our initial search of the Pubmed database and four papers met the inclusion criteria for this meta-analysis. Using keyword combination “SUMO4, Met55Val, diabetes” yielded three additional papers, all of which were eligible for our analysis. The eligible studies’ characteristics conform to the above inclusion criteria. Finally, “SUMO4, M55V, type 2 diabetes” retrieved another two additional papers. Both papers met inclusion criteria. Additional three Chinese papers were retrieved in the China National Knowledge Infrastructure database by using the keywords combination as “small ubiquitin-like modifier 4, Met55Val, diabetes.” Retrieved studies were published between 2003 and 2017.
Data Extraction
Data were extracted by three investigators using a standardized protocol (Table 1). Two investigators were responsible for identifying duplicate studies while the third acted as the mediator to resolve any disagreements between them. Studies that deviated from the major inclusion criteria were published in duplicate, or provided insufficient data were rejected. Similar data sets published in different articles by a single author group were adopted once in the meta-analysis. Items, such as the first author’s name, publication year, ethnicity, matching criteria, genotype number, and total number of cases and controls, are displayed in Table 1.
Table 1.
Characteristics of the investigated studies of the association between small ubiquitin-like modifier 4 (SUMO4) gene M55V polymorphism and T2DM.
| Reference | Year | Ethnicity | T2DM |
Control |
Matching criteria | Genotype method | Sample size (T2DM/control) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MM | MV | VV | MM | MV | VV | ||||||
| Pu et al. (16) | 2012 | Chinese | 160 | 218 | 26 | 250 | 218 | 32 | Age, sex, ethnicity | PCR-HRM | 404/500 |
| Li et al. (22) | 2011 | Chinese | 110 | 94 | 28 | 44 | 51 | 7 | Ethnicity | PCR-RFLP | 232/102 |
| Shimada et al. (23) | 2009 | Japanese | 395 | 393 | 86 | 412 | 414 | 79 | Ethnicity | TaqMan SNP genotyping assay | 874/905 |
| Lin et al. (24) | 2010 | Chinese | 275 | 254 | 45 | 186 | 115 | 22 | Ethnicity | PCR-RFLP | 574/323 |
| Noso et al. (15) | 2007 | Japanese | 146 | 166 | 43 | 200 | 158 | 40 | Ethnicity | TaqMan SNP genotyping assay | 355/398 |
| Fallah et al. (17) | 2010 | Iranian | 10 | 22 | 18 | 13 | 25 | 12 | Age, sex, ethnicity | PCR-RFLP | 50/50 |
| Ji et al. (7) | 2010 | Chinese | 195 | 185 | 46 | 143 | 123 | 15 | Age, sex, ethnicity | PCR-RFLP | 427/281 |
| Hu and Song (25) | 2009 | Chinese-Va | 59 | 31 | 6 | 68 | 33 | 3 | BMI, ALT, Cr, UA, TC, TG, HDL-C, LDL-C, ethnicity | PCR-RFLP | 96/104 |
| Hu and Song (25) | 2009 | Chinese-Lalu | 40 | 11 | 3 | 168 | 60 | 6 | Age, sex, ethnicity, BMI, WHR, SBP, DBP, HDL-C, Hypertension percentage | PCR-RFLP | 54/234 |
| Wang et al. (26) | 2015 | Chinese | 45 | 54 | 1 | 249 | 218 | 32 | Age, BMI, DBP, TG, LDL-C, ethnicity | PCR-RFLP | 100/499 |
| Zhang et al. (27) | 2017 | Chinese | 25 | 22 | 11 | 105 | 90 | 9 | Sex, BMI, Hypertension percentage, Hyperlipidemia percentage | PCR-RFLP | 58/204 |
T2DM, type 2 diabetes mellitus; TC, total cholesterol; LDL, low-density lipoprotein-cholesterol; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; PCR-HRM, polymerase chain reaction-high resolution melting curve.
PCR-RFLP and case–control study design were adopted in the above studies.
Statistical Analysis
Statistical analyses were performed by using Revman 5.0 and STATA 12.0 software (StataCorp, College Station, TX, USA). Six genetic models, allelic (V allele distribution frequency), recessive (VV vs. MV + MM), dominant (MM vs. MV + VV), homozygous (VV vs. MM), heterozygous (MV vs. MM), and additive (total V vs. total M), were used in the current meta-analysis. The association of SUMO4 gene M55V polymorphism and T2DM were compared by using the odds ratios (ORs) corresponding to its 95% confidence intervals (CIs).
The heterogeneity between the individual studies was evaluated by using the Chi-square-based Q-test with significance set at P < 0.05 level (18). If heterogeneity was detected in the studies, the random-effects model (DerSimonian and Laird method) would be used to estimate the pooled OR (19). If not, the fixed-effects model (the Mantel–Haenszel method) would be used (20). The combined OR was determined by Z-test with the significance set at P < 0.05 level.
Fisher’s exact test was used to assess HWE with the significance set at P < 0.05 level. The potential publication bias was evaluated by the funnel plot. The funnel plot symmetry was assessed by using the Egger’s linear regression test on the OR and the significance was set at P < 0.05 level (21).
Results
Studies and Populations
After the retrieval process, 10 papers met the inclusion criteria. Data were extracted from a total of 3,223 T2DM patients and 3,600 controls (Table 1) (7, 15–17, 22–27). Among the retrieved studies, one study was excluded for controls that deviated from the HWE (28). Three ethnicities were represented in the meta-analysis: Chinese, Japanese, and Iranian.
Pooled Analyses
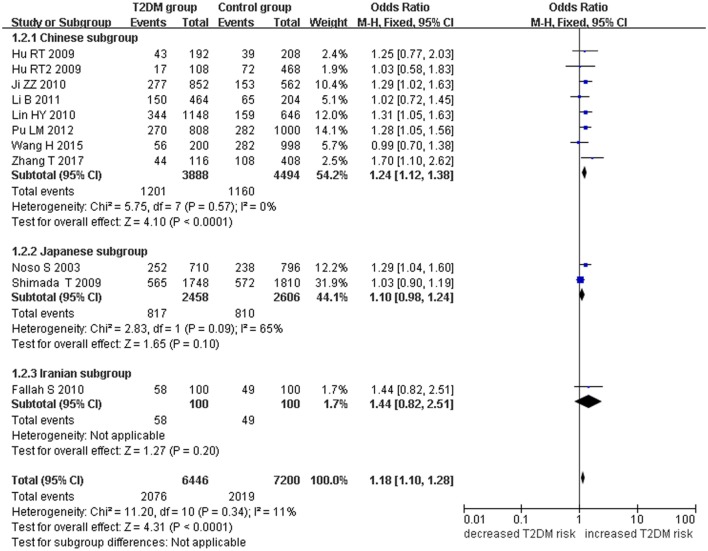
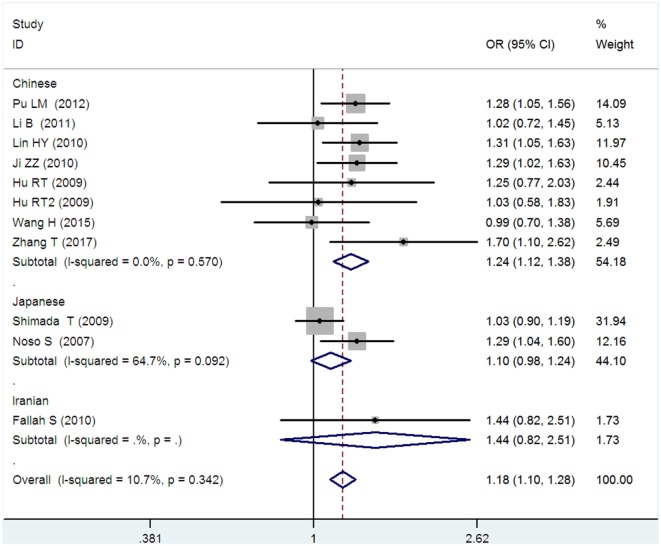
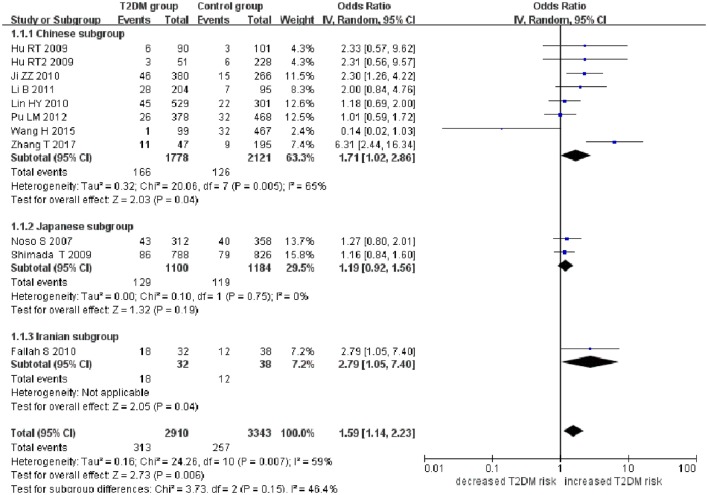
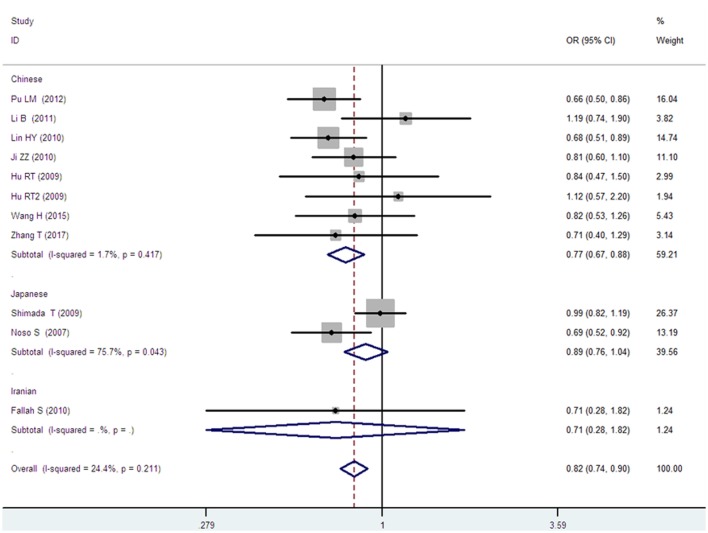
Small ubiquitin-like modifier 4 gene M55V polymorphism demonstrated a significant association with T2DM susceptibility in the whole population under allelic (OR: 1.18, 95% CI: 1.10–1.28, P = 1.63 × 10−5), recessive (OR: 1.59, 95% CI: 1.14–2.23, P = 0.006), dominant (OR: 0.815, 95% CI: 0.737–0.901, P = 6.89 × 10−5), homozygous (OR: 1.415, 95% CI: 1.170–1.710, P = 0.0003), heterozygous (OR: 1.191, 95% CI: 1.072–1.323, P = 0.001), and additive genetic models (OR: 1.184, 95% CI: 1.097–1.279, P = 1.63 × 10−5) (Figures 1–6; Table 2).
Figure 1.
Forest plot of type 2 diabetes mellitus (T2DM) associated with small ubiquitin-like modifier 4 (SUMO4) gene M55V polymorphism under an allelic genetic model (distribution of V allelic frequency of SUMO4 gene).
Figure 6.
Forest plot of type 2 diabetes mellitus associated with small ubiquitin-like modifier 4 gene M55V polymorphism under an additive genetic model (V vs. M).
Table 2.
Summary of meta-analysis of association between small ubiquitin-like modifier 4 (SUMO4) gene M55V polymorphism and type 2 diabetes mellitus (T2DM).
| Genetic model | Pooled OR (95% CI) | Z value | P-value | Study number | T2DM size | control size | |
|---|---|---|---|---|---|---|---|
| Allelic genetic model | 1.18 (1.10–1.28) | 4.31 | 1.63 × 10−5* | 10 | 3,223 | 3,600 | 0.34 (11.0%) |
| Chinese subgroup | 1.24 (1.12–1.38) | 4.10 | 4.13 × 10−5* | 7 | 1,944 | 2,247 | 0.57 (0%) |
| Japanese subgroup | 1.10 (0.98–1.24) | 1.65 | 0.10 | 2 | 1,229 | 1,303 | 0.09 (65.0%) |
| Iranian subgroup | 1.44 (0.82–2.51) | 1.27 | 0.20 | 1 | 50 | 50 | NA |
| Recessive genetic model | 1.59 (1.14–2.23) | 2.73 | 0.006* | 10 | 3,223 | 3,600 | 0.007* (59.0%) |
| Chinese subgroup | 1.71 (1.02–2.86) | 2.03 | 0. 04* | 7 | 1,944 | 2,247 | 0.005* (65.0%) |
| Japanese subgroup | 1.19 (0.92–1.56) | 1.32 | 0.19 | 2 | 1,229 | 1,303 | 0.75 (0%) |
| Iranian subgroup | 2.79 (1.05–7.40) | 2.05 | 0.04* | 1 | 50 | 50 | NA |
| Dominant genetic model | 0.815 (0.737–0.901) | 3.98 | 6.89 × 10−5* | 10 | 3,223 | 3,600 | 0.211 (24.7%) |
| Chinese subgroup | 0.768 (0.673–0.877) | 3.89 | 0.0001* | 7 | 1,944 | 2,247 | 0.417 (1.7%) |
| Japanese subgroup | 0.888 (0.760–1.039) | 1.48 | 0.138 | 2 | 1,229 | 1,303 | 0.043* (75.7%) |
| Iranian subgroup | 0.712 (0.279–1.818) | 0.71 | 0.477 | 1 | 50 | 50 | NA |
| Homozygous genetic model | 1.415 (1.170–1.710) | 3.58 | 0.0003* | 10 | 3,223 | 3,600 | 0.103 (37.1%) |
| Chinese subgroup | 1.580 (1.205–2.071) | 3.31 | 0.001* | 7 | 1,944 | 2,247 | 0.081 (44.6%) |
| Japanese subgroup | 1.236 (0.939–1.627) | 1.51 | 0.131 | 2 | 1,229 | 1,303 | 0.384 (0%) |
| Iranian subgroup | 1.950 (0.648–5.867) | 1.19 | 0.235 | 1 | 50 | 50 | NA |
| Heterozygous genetic model | 1.191 (1.072–1.323) | 3.26 | 0.001* | 10 | 3,223 | 3,600 | 0.072 (41.5%) |
| Chinese subgroup | 1.257 (1.095–1.443) | 3.26 | 0.001* | 7 | 1,944 | 2,247 | 0.031* (66.2%) |
| Japanese subgroup | 1.104 (0.937–1.301) | 1.18 | 0.237 | 2 | 1,229 | 1,303 | 0.043* (75.6%) |
| Iranian subgroup | 1.144 (0.419–3.122) | 0.26 | 0.793 | 1 | 50 | 50 | NA |
| Additive genetic model | 1.184 (1.097–1.279) | 4.31 | 1.63 × 10−5* | 10 | 3,223 | 3,600 | 0.342 (10.7%) |
| Chinese subgroup | 1.241 (1.119–1.377) | 4.10 | 4.13 × 10−5* | 7 | 1,944 | 2,247 | 0.570 (0%) |
| Japanese subgroup | 1.104 (0.981–1.243) | 1.65 | 0.099 | 2 | 1,229 | 1,303 | 0.092 (64.7%) |
| Iranian subgroup | 1.437 (0.823–2.511) | 1.27 | 0.203 | 1 | 50 | 50 | NA |
*P ≤ 0.05.
CI, confidence interval; OR, odds ratio; T2DM size, the total number of T2DM cases; control size, the total number of control group; allelic genetic model, V allele distribution frequency; recessive genetic model, VV vs. MM + MV; dominant genetic model, MM vs. MV + VV; homozygous genetic model, VV vs. MM; heterozygous genetic model, MV vs. MM; additive genetic model, V vs. M.
Figure 2.
Forest plot of type 2 diabetes mellitus (T2DM) associated with small ubiquitin-like modifier 4 gene M55V polymorphism under a recessive genetic model (VV vs. MM + MV).
Figure 3.
Forest plot of type 2 diabetes mellitus associated with small ubiquitin-like modifier 4 gene M55V polymorphism under a dominant genetic model (MM vs. MV + VV).
Figure 4.
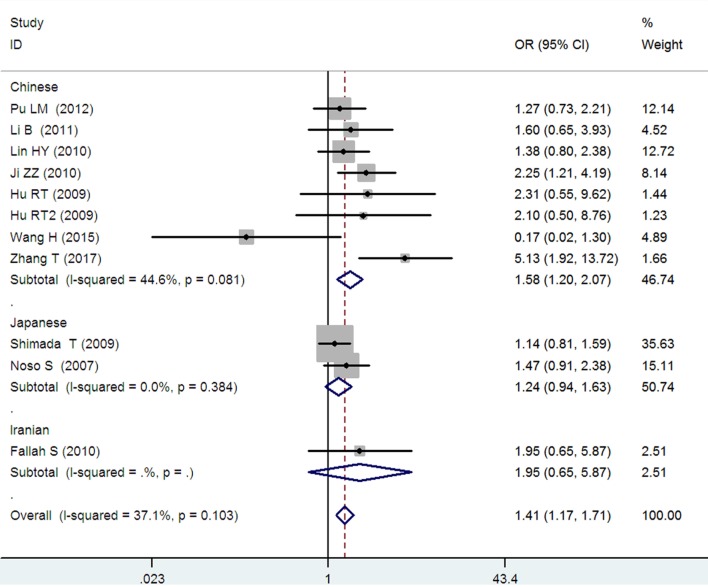
Forest plot of type 2 diabetes mellitus associated with small ubiquitin-like modifier 4 gene M55V polymorphism under a homozygous genetic model (VV vs. MM).
Figure 5.
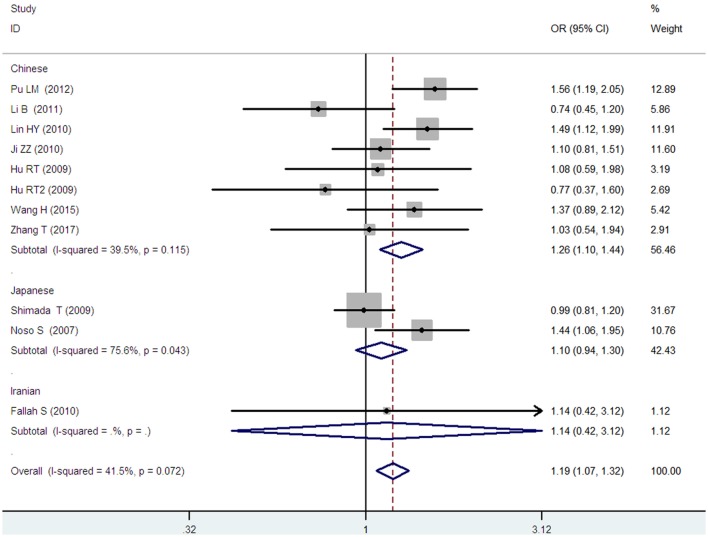
Forest plot of type 2 diabetes mellitus associated with small ubiquitin-like modifier 4 gene M55V polymorphism under a heterozygous genetic model (MV vs. MM).
In the subgroup analysis, a significant association was also found in the Chinese population under allelic (OR: 1.24, 95% CI: 1.12–1.38, P = 4.13 × 10−5), recessive (OR: 1.71, 95% CI: 1.02–2.86, P = 0.04), dominant (OR: 0.768, 95% CI: 0.673–0.877, P = 1.57 × 10−4), homozygous (OR: 1.580, 95% CI: 1.205–2.071, P = 0.001), heterozygous (OR: 1.257, 95% CI: 1.095–1.443, P = 0.001), and additive genetic models (OR: 1.241, 95% CI: 1.119–1.377, P = 4.13 × 10−5).
No significant association was detected in Japanese population under allelic (OR: 1.10, 95% CI: 0.98–1.24, P = 0.10), recessive (OR: 1.19, 95% CI: 0.92–1.56, P = 0.19), dominant (OR: 0.888, 95% CI: 0.760–1.039, P = 0.138), homozygous (OR: 1.236, 95% CI: 0.939–1.627, P = 0.131), heterozygous (OR: 1.104, 95% CI: 0.937–1.301, P = 0.237), or additive genetic models (OR: 1.104, 95% CI: 0.981–1.243, P = 0.099).
In the Iranian population, a marginally significant association was detected only under recessive genetic model (OR: 2.79, 95% CI: 1.05–7.40, P = 0.04). No significant association was detected in Iranian population under allelic (OR: 1.44, 95% CI: 0.82–2.51, P = 0.20), dominant (OR: 0.712, 95% CI: 0.279–1.818, P = 0.477), homozygous (OR: 1.950, 95% CI: 0.648–5.867, P = 0.235), heterozygous (OR: 1.144, 95% CI: 0.419–3.122, P = 0.793) or additive genetic models (OR: 1.437, 95% CI: 0.823–2.511, P = 0.203).
As no significant heterogeneity was detected under allelic, dominant, homozygous, heterozygous, or additive genetic models (Pheterogeneity > 0.05), a fixed-effect model was used to determine the pooled OR. While only under the recessive genetic model, significant heterogeneity was detected (Pheterogeneity < 0.05) and random-effect model was used (Table 2).
Bias Diagnostics
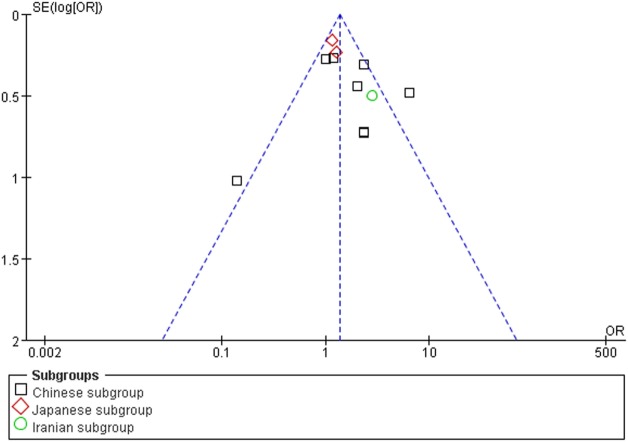
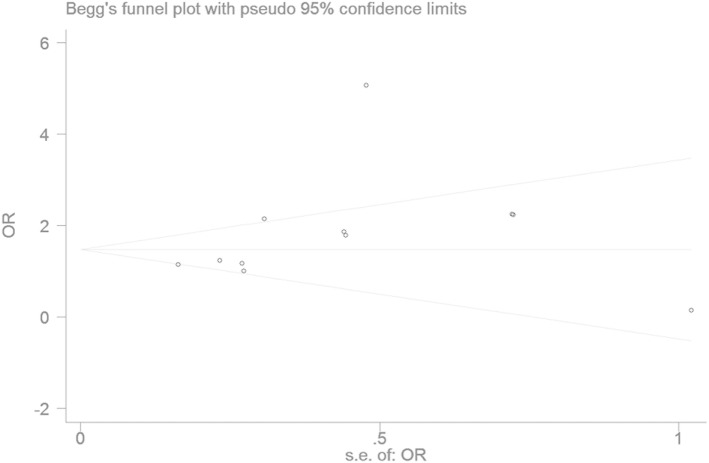
The publication bias of the individual studies was evaluated by using the funnel plot. No visual evidence for publication bias was evident in the funnel plot under the recessive genetic model (Figure 7). No significant publication bias was detected in this meta-analysis (T = 1.54, P = 0.159) under the recessive genetic model by using Egger’s test (Figure 8).
Figure 7.
Funnel plot for studies of the association of type 2 diabetes mellitus and small ubiquitin-like modifier 4 gene M55V polymorphism under a recessive genetic model (VV vs. MM + MV). The horizontal and vertical axis correspond to the OR and confidence limits. OR, odds ratio.
Figure 8.
Begg’s funnel plot for studies of the association of type 2 diabetes mellitus and small ubiquitin-like modifier 4 gene M55V polymorphism under a recessive genetic model (VV vs. MM + MV). The horizontal and vertical axis correspond to the OR and confidence limits. OR, odds ratio.
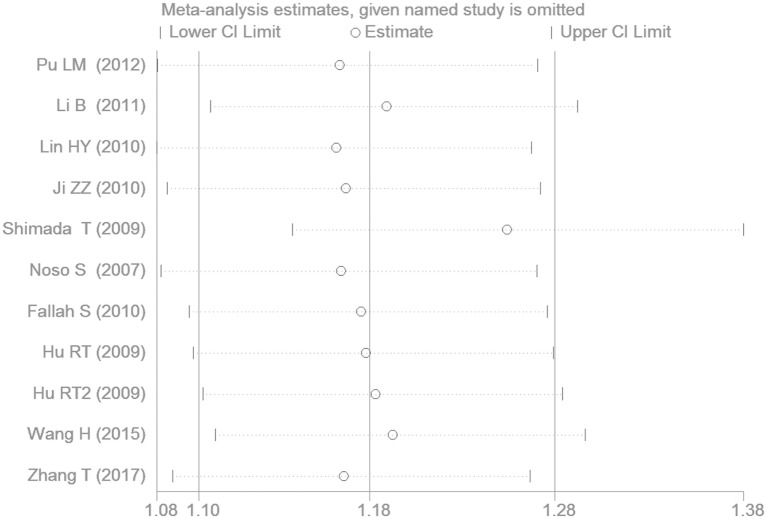
Sensitivity Analysis
The removal of any one study from the meta-analysis did not change the significant association between SUMO4 gene M55V polymorphism and T2DM under the additive genetic model, suggesting that the results are stable and robust (Figure 9).
Figure 9.
The sensitivity analysis results under the additive genetic model (V vs. M).
Discussion
In the current meta-analysis, a significant association was detected between SUMO4 gene M55V polymorphism and T2DM under the allelic (OR: 1.18), recessive (OR: 1.59), dominant (OR: 0.815), homozygous (OR: 1.415), heterozygous (OR: 1.191), and additive (OR: 1.184) genetic models. In the subsequent subgroup analysis, a significant association was only detected in the Chinese population, but not in the Japanese or Iranian population. Significant heterogeneity was only found under the recessive genetic model. Heterogeneity was reduced in the Chinese and Japanese populations, suggesting that ethnicity was the main source for the heterogeneity. As only one study of the Iranian population was present in our analysis, the heterogeneity detection was not applicable.
The SUMO4 gene M55V polymorphism is located in the evolutionarily crucial CUE structural domain and involves the substitution of the highly conserved Met (ATG) for Val (GTG). This substitution could modify site at which PKC phosphorylates the protein. The change in molecular conformation and functional activity reduces its ability to regulate NF-κB, resulting in increased NF-κB activation enhancement and overexpression of NF-κB-dependent gene products. In 2004, Guo et al. found that the M55V substitution resulted in 5.5 times greater NF-κB transcriptional activity and approximately two times greater expression of IL12B (4).
Nuclear factor-κB activation is an important molecule in the inflammatory response (29), upregulation of endothelin (30), and apoptosis (31). NF-κB activation may also promote apoptosis in both pericytes and endothelial cells in the pathogenesis of diabetes retinopathy (32).
In recent years, T2DM has been increasingly associated with mild, but systemic chronic inflammation that is characteristic of obesity and metabolic syndrome. The disruption of the internal equilibrium by factors, such as stress and poor nutrition, activates the inflammatory response as a protective response. Initially, the body is in a peri-inflammatory state, but long-term activation of this state can result in chronic inflammation (33). NF-κB is an important inflammatory factor involved in IR, but also acts as transcription factor for other pro-inflammatory proteins (34). The pivotal studies have also found that the SUMO4 cytosis substrate included the anti-oxidative stress proteins, the regulation proteins for DNA repair and synthesis, protein degradation associated proteins, glycometabolism-associated proteins under stress conditions induced by starvation (35). These findings point toward the functional complexity of SUMO4, which warrants further investigation and discussion.
In 2012, Tang et al. performed a meta-analysis on the relationship between SUMO4 gene M55V polymorphism and T2DM where they found a significant association between them in the Asian population (P < 0.05) (36). However, only six individual studies were included and only four genetic models were adopted in their meta-analysis. In addition, with no subgroup analysis stratified by ethnicity, their conclusion may be more limited to the analysis we offer in this current study. In 2017, Zhang et al. performed a meta-analysis on this subject and came to a similar conclusion (37). However, only five Chinese individual studies published before 2012 were included in Zhang’s meta-analysis and five genetic models were merely used. While in the current meta-analysis, six genetic models were used and the publications in 2015 and 2017 were also included. Hence, our study increased the number of studies to seven individual studies in the Chinese population and offers an updated and more comprehensive result.
This study is not without limitations. Many factors can influence plasma concentrations of SUMO4, such as SUMO4 rs237024 and rs600739 polymorphisms, diet, smoking, and hypertension (16). In addition, the current meta-analysis lacks large-scale studies on the subject. This variant has been studied only in Chinese, Japanese, and Iranian subjects. No study in other ethnicities has been found. Furthermore, no data from GWA studies on the region that includes this variant have been found. Much remains to be clarified on how the SUMO4 gene M55V polymorphism affects patient T2DM susceptibility.
In conclusion, we found a significant association between SUMO4 gene M55V polymorphism and T2DM risk in the current meta-analysis. Individuals with the Val allele of SUMO4 gene M55V polymorphism may be more susceptible to T2DM in the Chinese population. This association in Japanese or Iranian population needs to be further verified in the larger samples studies. This conclusion may help researchers formulate an individual treatment strategy to prevent T2DM in the future.
Author Contributions
Conceived and designed the meta-analysis: Y-yL and HW. Performed the meta-analysis: Y-yL, X-xY, H-yG, and GG. Analyzed the data: Y-yL. Contributed material/analysis tools: Y-yL. Wrote the manuscript: Y-yL and HK. Reference collection and data management: Y-yL and J-jW. Statistical analyses and paper writing: Y-yL, Y-hZ, and HK. Study design: Y-yL and J-jW.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer MH and handling editor declared their shared affiliation.
Acknowledgments
This work was funded by the National Natural Science Foundation of China (NSFC 81100073 to Y-yL), Excellent Young and Middle-Aged Teachers Assistance Program of Nanjing Medical University for Y-yL (2013–2015, JX2161015034) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). This work was also funded by the Natural Science Foundation of Jiangsu Province (BK 2012648 to HW), “six talent peaks” project in Jiangsu Province (2015-WSN-033). Thank all our colleagues working in the Department of geriatrics, the First Affiliated Hospital of Nanjing Medical University.
References
- 1.Maedler K, Sergeev P, Ris F, Oberholzer J, Joller-Jemelka HI, Spinas GA, et al. Glucose-induced beta cell production of IL-1 beta contributes to glucotoxicity in human pancreatic islets. J Clin Invest (2002) 110:851–60. 10.1172/JCI200215318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai D, Yuan M, Frantz DF, Melendez PA, Hansen L, Lee J, et al. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat Med (2005) 11:183–90. 10.1038/nm1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, et al. Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes (2006) 55:2993–3003. 10.2337/db06-0477 [DOI] [PubMed] [Google Scholar]
- 4.Guo D, Li M, Zhang Y, Yang P, Eckenrode S, Hopkins D, et al. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat Genet (2004) 36:837–41. 10.1038/ng1391 [DOI] [PubMed] [Google Scholar]
- 5.Manza LL, Codreanu SG, Stamer SL, Smith DL, Wells KS, Roberts RL, et al. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem Res Toxicol (2004) 17:1706–15. 10.1021/tx049767l [DOI] [PubMed] [Google Scholar]
- 6.Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J Biol Chem (2004) 279:27233–8. 10.1074/jbc.M402273200 [DOI] [PubMed] [Google Scholar]
- 7.Ji ZZ, Dai J, Xu YC. Association between small ubiquitin-like modifier 4 M55V polymorphism with type 2 diabetes and related factors. Chin J Diabetes Mellitus (2010) 2:344–8. 10.3760/cnm.j.issn.1674-5809.2010.05.006 [DOI] [Google Scholar]
- 8.Pullinger CR, Goldfine ID, Tanyolaç S, Movsesyan I, Faynboym M, Durlach V, et al. Evidence that an HMGA1 gene variant associates with type 2 diabetes, body mass index, and high-density lipoprotein cholesterol in a Hispanic-American population. Metab Syndr Relat Disord (2014) 12:25–30. 10.1089/met.2013.0086 [DOI] [PubMed] [Google Scholar]
- 9.Saxena R, Saleheen D, Been LF, Garavito ML, Braun T, Bjonnes A, et al. Genome-wide association study identifies a novel locus contributing to type 2 diabetes susceptibility in Sikhs of Punjabi origin from India. Diabetes (2013) 62:1746–55. 10.2337/db12-1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikegami H, Noso S, Babaya N, Hiromine Y, Kawabata Y. Genetic basis of type 1 diabetes: similarities and differences between east and west. Rev Diabet Stud (2008) 5:64–72. 10.1900/RDS.2008.5.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silander K, Scott LJ, Valle TT, Mohlke KL, Stringham HM, Wiles KR, et al. A large set of Finnish affected sibling pair families with type 2 diabetes suggests susceptibility loci on chromosomes 6, 11, and 14. Diabetes (2004) 53:821–9. 10.2337/diabetes.53.3.821 [DOI] [PubMed] [Google Scholar]
- 12.Xiang K, Wang Y, Zheng T, Jia W, Li J, Chen L, et al. Genome-wide search for type 2 diabetes/impaired glucose homeostasis susceptibility genes in the Chinese: significant linkage to chromosome 6q21-q23 and chromosome 1q21-q24. Diabetes (2004) 53:228–34. 10.2337/diabetes.53.1.228 [DOI] [PubMed] [Google Scholar]
- 13.Sale MM, Freedman BI, Langefeld CD, Williams AH, Hicks PJ, Colicigno CJ, et al. A genome-wide scan for type 2 diabetes in African-American families reveals evidence for a locus on chromosome 6q. Diabetes (2004) 53:830–7. 10.2337/diabetes.53.3.830 [DOI] [PubMed] [Google Scholar]
- 14.Duggirala R, Blangero J, Almasy L, Arya R, Dyer TD, Williams KL, et al. A major locus for fasting insulin concentrations and insulin resistance on chromosome 6q with strong pleiotropic effects on obesity-related phenotypes in nondiabetic Mexican Americans. Am J Hum Genet (2001) 68:1149–64. 10.1086/320100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noso S, Fujisawa T, Kawabata Y, Asano K, Hiromine Y, Fukai A, et al. Association of small ubiquitin-like modifier 4 (SUMO4) variant, located in IDDM5 locus, with type 2 diabetes in the Japanese population. J Clin Endocrinol Metab (2007) 92:2358–62. 10.1210/jc.2007-0031 [DOI] [PubMed] [Google Scholar]
- 16.Pu LM, Nan N, Yang Z, Jin ZN. Association between SUMO4 polymorphisms and type 2 diabetes mellitus. Yi Chuan (2012) 34:315–25. 10.3724/SP.J.1005.2012.00315 [DOI] [PubMed] [Google Scholar]
- 17.Fallah S, Jafarzadeh M, Hedayati M. No association of the SUMO4 polymorphism M55V variant in type 2 diabetes in Iranian subjects. Diabetes Res Clin Pract (2010) 90:191–5. 10.1016/j.diabres.2010.05.033 [DOI] [PubMed] [Google Scholar]
- 18.Cochran WG. The effectiveness of adjustment by subclassification in removing bias in observational studies. Biometrics (1968) 24:295–313. 10.2307/2528036 [DOI] [PubMed] [Google Scholar]
- 19.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials (1986) 7:177–88. 10.1016/0197-2456(86)90046-2 [DOI] [PubMed] [Google Scholar]
- 20.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst (1959) 22:719–48. [PubMed] [Google Scholar]
- 21.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (1997) 315:629–34. 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, Li HF, Wang YM, Zhou XX, Zhang X, Song DP. The association of SUMO4 gene 163 A/G polymorphism with type 2 diabetic nephropathy. Chin J Diabetes (2011) 19:726–8. 10.3969/j.issn.1006-6187.2011.10.002 [DOI] [Google Scholar]
- 23.Shimada T, Furukawa Y, Furuta H, Yasuda K, Matsuno S, Kusuyama A, et al. SUMO4 Met55Val polymorphism is associated with coronary heart disease in Japanese type 2 diabetes individuals. Diabetes Res Clin Pract (2009) 85:85–9. 10.1016/j.diabres.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 24.Lin HY, Li SL, Yu ML, Hsiao PJ, Hsieh MC, Lin KD, et al. Small ubiquitin-like modifier-4 Met55Val polymorphism is associated with glycemic control of type 2 diabetes mellitus in Taiwan. J Endocrinol Invest (2010) 33:401–5. 10.3275/6624 [DOI] [PubMed] [Google Scholar]
- 25.Hu RT, Song DP. The Association of SUMO4 Gene 163 A/G Polymorphism and Dysglycemia in Va and Lalu Population. Kunming: Kunming Medical University; (2009). [Google Scholar]
- 26.Wang H, Dong H, Wang YX, Wang W, Song MS, Zhang J, et al. Association between polymorphisms in SUMO4 gene and susceptibility to type 2 diabetes: a case-control study in north Chinese population and a meta-analysis. The 4th Postgraduate Academic Symposium Essays of School of Public Health. Beijing: Capital Medical University; (2015). p. 23–36. [Google Scholar]
- 27.Zhang T, Liu Y, Hu Y, Zhang X, Zhong L, Fan J, et al. Association of donor and recipient SUMO4 rs237025 genetic variant with new-onset diabetes mellitus after liver transplantation in a Chinese population. Gene (2017) 627:428–33. 10.1016/j.gene.2017.06.060 [DOI] [PubMed] [Google Scholar]
- 28.Sozen S, Horozoglu C, Bireller ES, Karaali Z, Cakmakoglu B. Association of SUMO4 M55V and -94ins/del gene variants with type-2 diabetes. In Vivo (2014) 28:919–23. [PubMed] [Google Scholar]
- 29.Morigi M, Angioletti S, Imberti B, Donadelli R, Micheletti G, Figliuzzi M, et al. Leukocyte-endothelial interaction is augmented by high glucose concentrations and hyperglycemia in a NF-kB-dependent fashion. J Clin Invest (1998) 101:1905–15. 10.1172/JCI656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quehenberger P, Bierhaus A, Fasching P, Muellner C, Klevesath M, Hong M, et al. Endothelin 1 transcription is controlled by nuclear factor-kappaB in AGE-stimulated cultured endothelial cells. Diabetes (2000) 49:1561–70. 10.2337/diabetes.49.9.1561 [DOI] [PubMed] [Google Scholar]
- 31.Du X, Stocklauser-Färber K, Rösen P. Generation of reactive oxygen intermediates, activation of NF-kappaB, and induction of apoptosis in human endothelial cells by glucose: role of nitric oxide synthase? Free Radic Biol Med (1999) 27:752–63. 10.1016/S0891-5849(99)00079-9 [DOI] [PubMed] [Google Scholar]
- 32.Mizutani M, Kern TS, Lorenzi M. Accelerated death of retinal microvascular cells in human and experimental diabetic retinopathy. J Clin Invest (1996) 97:2883–90. 10.1172/JCI118746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medzhitov R. Origin and physiological roles of inflammation. Nature (2008) 454:428–35. 10.1038/nature07201 [DOI] [PubMed] [Google Scholar]
- 34.Yuan M, Konstantopoulos N, Lee J, Hansen L, Li ZW, Karin M, et al. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of IKKbeta. Science (2001) 293:1673–7. 10.1126/science.1061620 [DOI] [PubMed] [Google Scholar]
- 35.Guo D, Han J, Adam BL, Colburn NH, Wang MH, Dong Z, et al. Proteomic analysis of SUMO4 substrates in HEK293 cells under serum starvation-induced stress. Biochem Biophys Res Commun (2005) 337:1308–18. 10.1016/j.bbrc.2005.09.191 [DOI] [PubMed] [Google Scholar]
- 36.Tang ST, Peng WJ, Wang CJ, Tang HQ, Zhang Q. Polymorphism M55V in gene encoding small ubiquitin-like modifier 4 (SUMO4) protein associates with susceptibility to type 1 (and type 2) diabetes. Diabetes Metab Res Rev (2012) 28:679–87. 10.1002/dmrr.2335 [DOI] [PubMed] [Google Scholar]
- 37.Zhang Q, Liu D, Zhao ZY, Sun Q, Ding LX, Wang YX. Association between the SUMO4 M55V polymorphism and susceptibility to type 2 diabetes mellitus: a meta-analysis. Biomed Environ Sci (2017) 30:288–95. 10.3967/bes2017.038 [DOI] [PubMed] [Google Scholar]