Abstract
The increase in red blood cell mass (polycythemia) due to the reduced oxygen availability (hypoxia) of residence at high altitude or other conditions is generally thought to be beneficial in terms of increasing tissue oxygen supply. However, the extreme polycythemia and accompanying increased mortality due to heart failure in chronic mountain sickness most likely reduces fitness. Tibetan highlanders have adapted to high altitude, possibly in part via the selection of genetic variants associated with reduced polycythemic response to hypoxia. In contrast, high-altitude-adapted Quechua- and Aymara-speaking inhabitants of the Andean Altiplano are not protected from high-altitude polycythemia in the same way, yet they exhibit other adaptive features for which the genetic underpinnings remain obscure. Here, we used whole-genome sequencing to scan high-altitude Andeans for signals of selection. The genes showing the strongest evidence of selection—including BRINP3, NOS2, and TBX5—are associated with cardiovascular development and function but are not in the response-to-hypoxia pathway. Using association mapping, we demonstrated that the haplotypes under selection are associated with phenotypic variations related to cardiovascular health. We hypothesize that selection in response to hypoxia in Andeans could have vascular effects and could serve to mitigate the deleterious effects of polycythemia rather than reduce polycythemia itself.
Keywords: natural selection, adaptation, hypoxia, high altitude, Aymara, Andean
Introduction
Human adaptation to high-altitude hypoxia in populations living on the Qinghai-Tibetan plateau in western China, the Semien plateau in Ethiopia, and the Andean Altiplano in South America provides one of the best examples of adaptation to an extreme environment in modern humans. Upon exposure to high altitude (>2,500 m above sea level), low-altitude humans experience a complex, plastic physiological response characterized by an immediate reduction in plasma volume, rise in ventilation taking place over days, and increase in red blood cell production over weeks to months; collectively, these serve to raise hemoglobin (Hb) concentration and help offset the reduced arterial O2 content due to the lower inspired partial pressure of O2 (pO2).1 Although in principle such changes are expected to facilitate oxygen transport, experimental evidence suggests that excessive red blood cell mass (polycythemia [MIM: 263300]) increases blood viscosity, which in turn impairs blood flow to the tissues.2 Moreover, greater viscosity exerts substantial stress on the cardiopulmonary system and contributes to a number of altitude-related pathologies, including chronic mountain sickness (CMS [MIM:616182]),3, 4 a condition characterized by polycythemia, pulmonary hypertension, right heart failure, and other symptoms.1, 5 However, there is also evidence that benefits of polycythemia lead to improved oxygen delivery to tissue when accompanied by increased blood volume, which usually coincides with most polycythemic states.6, 7, 8
Both physiological and genomic studies indicate that Andeans, Tibetans, and to some extent the Amhara of Ethiopia have genetic adaptations that alter the regulation of the production of red blood cells (erythropoiesis).9, 10, 11, 12, 13, 14, 15, 16, 17, 18 Although at altitude these populations exhibit Hb levels above sea-level values,19, 20 the increase is lower than that observed in acclimatized lowlanders, and Tibetans exhibit lower Hb levels than Andeans and perhaps also Ethiopians living at the same elevation.16, 21, 22, 23 However, the functional significance of the Tibetans’ lower Hb levels could result from their protection from CMS in comparison with Andean highlanders1, 5 or improved exercise performance24 or be the byproduct of selection for other factors determining hypoxic responses. Consistent with physiological evidence for blood-related adaptation in Tibetans, several genomic scans for natural selection have identified strong signals of natural selection focused on two genes, EPAS1 (HIF2A [MIM: 603349]) and EGLN1 (MIM: 606425), encoding two hypoxia-inducible transcription factors (HIFs) involved in regulating erythropoiesis.10, 11, 16, 17, 18, 25, 26 On the other hand, these genes and HIFs also regulate energy metabolism and other crucial physiological functions,27 and CMS primarily occurs after the completion of the reproductive period, thus limiting the fitness values of gene variants influencing its incidence. Thus, evidence from Tibetans suggests that regulatory modifications of erythropoiesis are one way that humans have adapted to the challenges of hypoxia at altitude.
Current physiological evidence suggests that selection in Andeans has targeted physiological systems other than the regulation of erythropoiesis.28 For example, Andeans (as well as Tibetans) are protected from altitude-associated fetal growth restriction29 partly because of the maintenance of high uterine artery blood flow, suggesting that vascular factors are likely to be part of the adaptive response in these populations. Another indication of the importance of vascular factors in altitude adaptation is the population variation in hypoxic pulmonary vasoconstrictor response. Tibetans exhibit a minimal pressure rise in comparison with that present in lifelong Colorado high-altitude residents, and intermediate values are seen in Andeans and perhaps also Amhara Ethiopians.29, 30 Protection from hypoxic pulmonary hypertension is also evident in well-adapted high-altitude species such as the yak (Bos grunniens), llama (Lama glama), and viscacha (Lagidium peruanum), which in the case of bovine species, is most likely due to selection for EPAS1 variants that reduce HIF2α stability.31
Several previous studies have analyzed genomic signals of high-altitude-related natural selection in Andeans as a means of investigating altitude adaptation. These studies indicate that hypoxia-signaling-pathway-related genes, including EGLN1, EDNRA (MIM: 131243), SENP1 (ANP32D [MIM: 606878]), PRKAA1 (MIM: 602739), NOS2A (MIM: 163730), and HBB (the β globin region [MIM: 141900]), are among the genomic regions under positive natural selection.11, 13, 25, 32, 33 However, these previous studies were based on SNP arrays or case-control comparisons, so a comprehensive view of natural selection across Andean highlander genomes remains unclear. Here, we used a lightweight and economic low-coverage genome sequencing approach to compare high-altitude-adapted Aymara-speaking residents of the Bolivian Andes with lowland Native American populations and Europeans to identify genomic regions targeted by natural selection in the Andeans. By using whole-genome sequencing and appropriately correcting for admixture, we were able to obtain a more complete and unbiased picture of the landscape of selection in the Andean population. As we will show, the lessons learned from genomic analysis of Andeans are quite different from those obtained in analyses of Tibetans and Amhara and Oromo Ethiopians.
Material and Methods
DNA Samples and Genome Sequencing
In total, 42 DNA samples from two cohorts were used for sequencing and analysis. For the first cohort (the Utah cohort), 20 blood samples were collected from randomly selected healthy Aymara volunteers from Tiwanaku (3,850 m) and La Paz (3,600 m) in Bolivia; their Aymara ancestry was determined by R. Amaru (a native Aymara speaker). All volunteers provided informed consent in both Aymara and Spanish, and approval was obtained from the institutional review board of San Andrés University in La Paz, Bolivia. For DNA sampling, granulocytes were separated from whole blood by centrifugation and the Histopaque (Sigma, catalog no. 10771) density-gradient method. Genomic DNA was isolated from granulocytes with the Gentra Puregene Cell Kit (QIAGEN, catalog no. 158388).
Samples were collected for the second cohort (Colorado) from 22 persons residing at 3,600–4,300 m and processed as described previously.12 Andean ancestry of these samples was confirmed by ancestry-informative genetic marker data for the original study. Approval for sample collection was obtained from the Colorado Multiple Institutional Review Board and the Colégio Medico, the Bolivian equivalent.
DNA samples of both Colorado and Utah cohorts were submitted for library construction and sequencing to the University of Utah High-Throughput Genomics and Bioinformatic Analysis Shared Resource Core. For library preparation, 1 μg of genomic DNA was sheared by a Covaris S2 Focused-ultrasonicator. Libraries (average insert size: 350 bp) were prepared with the Illumina TruSeq DNA PCR-Free Sample Preparation Kit (catalog nos. FC-121-3001 and FC-121-3002). Adaptor-ligated molecules were analyzed for quality control by quantitative RT-PCR with the Kapa Biosystems Kapa Library Quant Kit (catalog no. KK4824). Paired-end sequencing (125 cycles) was performed on an Illumina HiSeq 2500 instrument (HiSeq Control Software v.2.2.38 and Real-Time Analysis v.1.18.61) with HiSeq SBS Kit v.4 sequencing reagents (FC-401-4003).
Read Processing and Mapping
All paired-end Illumina reads were first trimmed with Trimmomatic v.0.3234 with the following settings: ILLUMINACLIP: TruSeq3-PE.fa:2:30:10, LEADING:3, TRAILING:3, SLIDINGWINDOW:4:15, and MINLEN:75. Cleaned and trimmed read pairs were then aligned to the human genome reference (GRCh37, downloaded from the 1000 Genomes ftp site) with the Burrows-Wheeler Aligner (BWA) “aln” and “sampe” algorithms;35 only alignments with a mapping quality (-q15) of at least 15 and an edit distance (-n6) of no more than six were kept. Reads in close proximity to putative indels were re-aligned with the Genome Analysist Toolkit (GATK).36 Read-group IDs were added to each BAM file, and duplicates were marked for removal with Picard Tools. Unmapped reads, reads with an unmapped mate, reads failing quality checks, and reads whose alignment was not primary were all removed from the BAM file. Lastly, base quality scores were recalibrated with GATK on the basis of a recalibration table made from a random subset of 16 BAM files from our Aymara sample and known variable sites from dbSNP (v.138).
Site Quality Filtering
We obtained a set of genomic positions deemed reliable for population genetic analysis via several approaches. First, we generated a preliminary pileup of reads from Aymara samples by using SAMtools and applied a series of filters by using the SNPcleaner Perl Script from the ngsTools package.37 Those filters are as follows:
-
1.
Read distribution among individuals: no more than 20 individuals were allowed to have fewer than two reads covering the site.
-
2.
Mapping quality: only reads with a BWA mapping quality of at least 10 were included.
-
3.
Base quality: only bases with an Illumina base quality of at least 20 were included.
-
4.
Proper pairs: only read pairs mapped in the proper paired-end orientation and within the expected distribution of insert lengths were included.
-
5.
Hardy-Weinberg proportions: expected genotype frequencies were calculated for each variable site on the basis of allele frequencies based on genotypes called with SAMtools in the preliminary pileup. Any sites with an excess of heterozygotes (minimum p = 1 × 10−6) were considered potential mapping errors and excluded.
-
6.
Heterozygous biases: sites with heterozygous genotype calls were evaluated with SAMtools for several biases with exact tests. If one of the two alternative alleles was biased with respect to the read base quality (minimum p = 1 × 10−10), read strand (minimum p = 1 × 10−4), mapping quality (minimum p = 1 × 10−4), or distance from the end of the read (minimum p = 1 × 10−4), the site was excluded.
Second, we applied an additional filter based on the Strict Accessibility Mask from the 1000 Genomes Project by using only sites that passed all accessibility filters. Third, we applied a mappability filter based on the 100-mer mappability track for the human genome in the UCSC Genome Browser by using only sites with mappability scores greater than or equal to 0.5. Lastly, we restricted analysis to genomic positions with data in the genotype-likelihood VCF files from the 1000 Genomes phase 3 panel. After applying these three filter sets, we obtained a set of genomic positions deemed reliable for analysis. We restricted all of our analyses to the autosomal chromosomes (excluding X and Y) given that our Aymara sample included both males and females, and we did not want to mix haploid and diploid sequences or use a smaller sample size.
Genotype Likelihoods
We calculated genotype likelihoods and genotype probabilities directly from the read data by using Analysis of Next Generation Sequencing Data (ANGSD)38 at the genomic positions defined above. Specifically, we calculated genotype likelihoods and generated Beagle-format output files by using the following ANGSD flags: -only_proper_pairs 1, -remove_bads 1, -uniqueOnly 1, -minMapQ 30, -minQ 20, -doMajorMinor 4, -GL 2, and -doMaf 2. Using BCFtools, we converted Beagle GL files to VCF files and merged them with separate VCF files containing genotype-likelihood data for 50 individuals each from six populations in the 1000 Genomes phase 3 panel (YRI [Yoruba in Ibadan, Nigeria], CEU [Utah Residents with ancestry from northern and western Europe], PEL [Peruvians from Lima, Peru], MXL [Mexican ancestry in Los Angeles, CA], PUR [Puerto Ricans from Puerto Rico], and CLM [Colombians from Medellin, Colombia]). We filtered merged VCF files to keep only di-allelic sites with a minor allele frequency of 0.05 across the entire merged panel. We converted the cleaned, merged VCF file back to a Beagle GL format file for use in downstream analysis and also used it as input to calculate genotype probabilities for downstream analysis. Specifically, we used ANGSD to calculate genotype probabilities with the -doMaf 1, -doPost 1, and -doGeno 32 flags.
Principal-Component and Admixture Analyses
We calculated the expected covariance matrix among all individuals in the merged panel by using ngsCovar in the ngsPopGen software package37 and genotype probabilities as input. We ran ngsCovar without calling genotypes and by using all sites with a minimum-allele-frequency filter of 0.05. Results were plotted with a custom R script.
We estimated global admixture proportions and admixture-corrected allele frequencies by using NGSadmix39 and genotype likelihoods as input. We ran NGSadmix by assuming first three (K = 3) and then four (K = 4) ancestral populations and using a minimum-allele-frequency filter of 0.05 and the -printInfo flag. To confirm results from NGSadmix and test whether adding an East Asian population would change the admixture results, we added 50 randomly sampled individuals from the CHB (Han Chinese in Beijing, China) population from the 1000 Genomes phase 3 panel and estimated admixture proportions by using Ohana,40 a software program that implements a model very similar to that of NGSadmix. We ran Ohana by using the same filtering thresholds and thus the same sites as for NGSadmix.
Scan for Positive Selection
We scanned the genomes of our Aymara sample by using a modification of the population branch statistic (PBS),18 which summarizes a three-way comparison of allele frequencies between a focal group (Andeans), a closely related population (lowland Native Americans), and an outgroup (Europeans). This test specifically tests for loci where allele frequencies in the focal population are especially differentiated from those in both of the other populations. We added a slight modification to the PBS in order to scale the statistic and avoid artificially high PBS values when differentiation was low or high between all groups. Specifically, we defined a new normalized version of the standard PBS as follows:
where PBS1 indicates the PBS calculated with Andeans as the focal population, PBS2 indicates the PBS calculated with lowland Native Americans as the focal population, and PBS3 indicates the PBS calculated with Europeans as the focal population. The normalizing factor in PBSn1 was especially useful when FST values were high in all comparisons (which led to exceptionally high PBS values) but should not be considered evidence of faster evolution in the target population. This scenario is more pervasive in the comparison in this study than in past PBS-based scans given that background levels of differentiation between Andeans and lowland Native Americans are higher than in previous comparisons. All PBS values and underlying FST statistics were calculated with a custom script, and admixture-corrected allele frequencies were estimated during the NGSadmix admixture analysis above as input. We calculated PBSn1 in windows of ten sites identified as variable in at least one non-African population. Windows were considered extreme outliers if they fell within the top 0.1% of windows and also harbored at least five SNPs from the top 0.1% of SNP-wise PBSn1 values (i.e., PBSn1 > 0.3892927). We considered a window a unique signal if no other window had a higher value within 1 Mb.
Searching for Archaic Haplotypes among Top Candidate Regions
We were interested in determining whether any of the top ten candidate SNPs had evidence of positive selection on haplotypes introgressed from archaic humans by using the Altai Neanderthal and Denisovan genome.41 To address this, we first computed a series of summary statistics aimed at finding more archaic ancestry in Aymara than in Africans over 100-kb windows of the genome with a 20-kb step size between windows. We used the ancestry-corrected maximum-likelihood population-frequency estimates obtained from NGSadmix39 and computed the following statistics:42, 43, 44, 45
-
1.
D(Aymara, African, Altai Neanderthal, chimpanzee)
-
2.
D(Aymara, African, Denisova, chimpanzee)
-
3.
fD(Aymara, African, Altai Neanderthal, chimpanzee)
-
4.
fD(Aymara, African, Denisova, chimpanzee)
-
5.
Q95African, Aymara, Altai Neanderthal(1%, 100%)
-
6.
Q95African, Aymara, Denisova(1%, 100%)
-
7.
Q95African, Aymara, Altai Neanderthal + Denisova(1%, 100%)
-
8.
UAfrican, Aymara, Altai Neanderthal(1%, 50%, 100%)
-
9.
UAfrican, Aymara, Denisova(1%, 50%, 100%)
-
10.
UAfrican, Aymara, Altai Neanderthal + Denisova(1%, 50%, 100%)
In Figure S1, we plotted the autosomal genome-wide distribution of these statistics as a function of the number of SNPs in each window (windows overlapping the top ten candidate SNPs are indicated with the color yellow). Among the latter set of windows, we also highlighted in red the windows that lay within the top 99% quantile of the genome-wide distribution of each statistic. We also jointly plotted the distributions of statistics G and J (Figure S2), which can be more informative of adaptive introgression than their respective individual distributions.45
We also conducted a targeted search of several candidate regions by using an hidden Markov model (HMM)46, 47 to query the 1000 Genomes Project individuals. We searched in a 2-Mb region centered on each of the three top SNPs to find whether any of these three regions had evidence of long tracts of introgression at high frequency in non-African panels from the 1000 Genomes Project.48 We assumed a 2% admixture rate49, 50 and a time of admixture of 1,900 generations and estimated the average local recombination rates in a 2-Mb region around each top SNP by using the HapMap II recombination map.51 We called an introgressed tract if the posterior probability of introgression at a site was larger than 90%.
To visualize the haplotypes, we used the program Haplostrips52 with default parameters in a 100-kb region around each of the top three selected SNPs. We selected a representative African panel (YRI), a representative European panel (CEU), a representative East Asian panel (CHB), and the four American panels (PUR, CLM, PEL, and MXL) in the 1000 Genomes dataset. We removed sites that had a private minor allele frequency lower than 5% in any panel, sites with a mapping quality lower than 30 in any of the archaic genomes, and sites with a genotype quality lower than 40 in any of the archaic genomes.
MESA Phenotypic Association Analysis
Multi-Ethnic Study of Atherosclerosis (MESA) is a study of the characteristics of subclinical cardiovascular disease (disease detected non-invasively before it has produced clinical signs and symptoms) and the risk factors that predict progression to clinically overt cardiovascular disease or progression of the subclinical disease. The cohort is a diverse, population-based sample of 6,814 asymptomatic men and women aged 45–84 years. Approximately 38% of the recruited participants are European, 28% are African American, 22% are Hispanic, and 12% are Asian (predominantly of Chinese descent). Participants were recruited during 2000–2002 from six field centers across the US (at Wake Forest University, Columbia University, Johns Hopkins University, the University of Minnesota, Northwestern University, and the University of California, Los Angeles). All underwent anthropomorphic measurement and extensive evaluation by questionnaires at baseline and then four subsequent examinations at intervals of approximately 2–4 years. Association analyses were carried out within each ethnic group. An additive genetic model was assumed in each analysis. We used logistic regression for binary traits and linear regression for continuous traits. We included age, gender, and the top two principle components within each population in the model to adjust for potential confounding effects.
Results
We sequenced the genomes of 42 high-altitude-dwelling Andeans from multiple locations in Bolivia (Material and Methods) to an average of 5× read depth (Figure S3) by using the Illumina HiSeq 2000 platform. This depth of coverage prohibits accurate genotype calling, so all analyses were based on allele frequencies estimated from genotype likelihoods instead of called genotypes (see Material and Methods). Genotype likelihoods were calculated at genomic positions passing stringent filters, and Andean genotype likelihoods were merged with genotype likelihoods from 50 Yorubans (YRI), 50 Europeans (CEU), and 200 Native Americans (50 each from CLM, MXL, PEL, and PUR) from the 1000 Genomes phase 3 panel for comparison.
Genetic Ancestry of Andeans
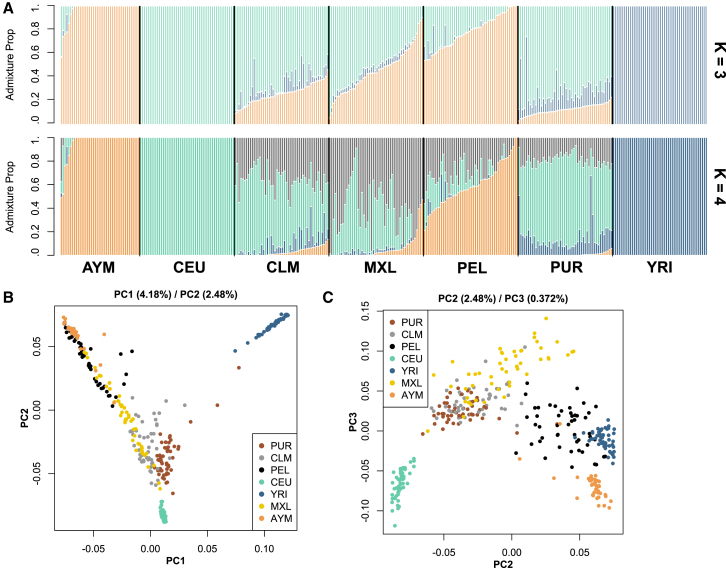
It is well known that, like many modern human populations, Central and South American populations (i.e., the reference populations used here) are admixed. In this case, they are admixed with varying degrees of African and European ancestry,53, 54, 55 which could obscure and confound scans for positive selection based on differences in population allele frequency among admixed populations. In contrast to previous efforts to identify signals of natural selection in Andeans, we obtained admixture-corrected allele frequencies and admixture proportions of ancestry from African, Europeans, and two Native American ancestral populations that we refer to as Andeans and lowland Native Americans. The Andean component was largely assigned to the Aymara and the Peruvians from Lima (PEL), whereas the lowland or non-Andean component was assigned to individuals from all American populations included here except the Aymara (Figure 1). We found that, except for seven individuals, the Andeans were not admixed and were entirely of Andean descent (Figure 1). The seven admixed individuals were sampled in the same way as the others and did not represent a distinct subset of the sample on the basis of other measures. In contrast, the CLM and MXL populations tended to carry a mixture of Andean, lowland Native American, and European ancestry with small African components. Peruvians from Lima varied in their proportion of Andean ancestry and tended to be admixed with mostly European and lowland Native American ancestry (Figure 1). We also tested whether including an East Asian component would affect the ancestry assignments, but we did not find evidence that this ancestry component changed the model in any material way (Figure S4).
Figure 1.
Genetic Ancestry of Native American Populations
(A) Genetic cluster analysis of whole-genome sequencing data from Aymara (AYM), Europeans (CEU), Colombians (CLM), Peruvians from Lima (PEL), Puerto Ricans (PUR), Mexicans from Los Angeles, CA (MXL), and Yoruba from Nigeria (YRI). Colors correspond to the proportion of ancestry from each ancestral population. K indicates the number of presumed ancestral populations in the model. Vertical bars represent individuals.
(B) Principal-component analysis (PCA) of the same panel of genomes. Points correspond to individuals, and colors indicate population assignments. The first and second principal components are plotted on the x and y axes, respectively.
(C) The second and third principal components from the PCA are plotted on the x and y axes, respectively.
In addition to analyzing admixture, we also compared genetic relatedness among these populations by principal-component analysis (Material and Methods). Two major clusters corresponding to African and non-African populations formed along the first principal-component axis, thus recapitulating Out-of-Africa historical demography (Figure 1). Along the second principal-component axis, the non-African cluster was further subdivided into Europeans and Andeans at the extremes and lowland Native Americans distributed between these two extremes. Inspection of the third principal-component axis revealed additional genetic variation partially separating the MXL, PUR, and CLM populations from the others, corresponding to the lowland Native American genetic component revealed in the admixture analysis above. These analyses underscore the importance of admixture in these populations, and we used admixture-corrected allele frequencies to improve sensitivity in downstream analysis of positive selection.
Genomic Targets of Natural Selection
We searched for genomic regions targeted by high-altitude-related natural selection in the Andeans by scanning Andean genomes for regions of exceptional genetic differentiation between Andean and both lowland Native Americans and Europeans. Regions of extreme differentiation unique to the Andean would be candidates for natural selection in this population. We used a modified version of the three-way PBS (PBSn1) that rescales the standard PBS to correct for saturation (see Material and Methods). These statistics allowed us to compare admixture-corrected allele frequencies in Andeans, lowland Native Americans, and Europeans and thus to identify Andean-specific signals of selection. Genome-wide genetic differentiation between Andeans and lowland Native Americans was intermediate (FST = 0.08), whereas these components were each more differentiated from Europeans (FST = 0.15 for Andeans versus Europeans; FST = 0.13 for lowland Native Americans versus Europeans). Because the ability to robustly detect outlier loci in the genome requires that background differentiation be sufficiently low,56 the intermediate genomic differentiation between the lowland Native American group and the Andeans (FST = 0.08) indicates that the FST-based scans are likely to have sufficient power to identify targets of natural selection (Figure S5).
In total, we analyzed 662,699 non-overlapping ten-SNP windows covering ∼2.27 Gb across the autosome. The mean windowed value of PBSn1 in this comparison was 0.0395 with a standard deviation of 0.0619. The top windows fell more than 5 standard deviations above the mean (PBSn1 > 0.3492), and the top window (PBSn1 = 0.5073) exceeded 7 standard deviations above the mean (PBSn1 > 0.4732), providing robust support for these loci as clear outliers in the genome. The top ten loci with the strongest evidence of positive selection are presented in Table 1. We identified candidate loci on the basis of windowed statistics, but we present the SNP with the highest PBSn1 value at each locus in Table 1 to indicate the position with the strongest evidence of selection. None of the most differentiated SNPs at each of top ten peaks fell within protein-coding regions, but the genes nearest these peaks of differentiation were, in ranked order, BRINP3 (aka FAM5C), NOS2 (MIM: 163730), SH2B1 (MIM: 608937), TBX5 (MIM: 601620), PYGM (MIM: 608455), CTAGE1 (MIM: 608856), ULBP1 (MIM: 605697), SHISA6 (MIM: 617327), TMEM38B (MIM: 611236), and PPA2 (MIM: 609988). Among these candidates, only NOS2 has been identified in previous scans for natural selection in Andeans.13 In many cases, the peak of differentiation was relatively narrow, providing good evidence for localization of the target of selection. However, in other cases, such as the third-ranked peak, the signal of differentiation was shared along a ∼0.8-Mb haplotype of chromosome 16 (Figure S6), compromising our ability to localize the underlying target of selection.
Table 1.
Top Ten Most Differentiated Genomic Regions in Andeans
| Chr | Top SNP Position | Top SNP ID | fANDa | fLNAb | fEUROc | PBSn1(Wind)d | PBSn1(SNP)e | Gene | Annotationf | Base Pairs from Exon |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 189,780,792 | rs11578671 | 0.13 | 0.82 | 0.86 | 0.5073 | 0.5596 | BRINP3 | IG | 286,069 |
| 17 | 26,133,177 | rs34913965 | 0.92 | 0.25 | 0.32 | 0.4320 | 0.4927 | NOS2 | REG | 7,100 |
| 16 | 28,871,191 | rs12448902 | 0.96 | 0.30 | 0.34 | 0.4223 | 0.5022 | multiple | DS | 3,295 |
| 12 | 114,784,040 | rs10744822 | 0.09 | 0.78 | 0.86 | 0.4002 | 0.5595 | TBX5 | INT | 396 |
| 11 | 64,546,891 | rs487105 | 0.80 | 0.21 | 0.13 | 0.3986 | 0.4860 | multiple | REG | 939 |
| 18 | 20,029,026 | rs11081933 | 0.99 | 0.49 | 0.32 | 0.3892 | 0.4358 | CTAGE1 | IG | 31,543 |
| 6 | 150,303,241 | rs4869782 | 0.91 | 0.33 | 0.06 | 0.3858 | 0.4946 | ULBP1 | DS | 8,163 |
| 17 | 11,245,308 | rs78264921 | 0.70 | 0.19 | 0.03 | 0.3794 | 0.4387 | SHISA6 | INT | 37,581 |
| 9 | 109,103,066 | rs12685887 | 0.81 | 0.25 | 0.20 | 0.3747 | 0.4494 | TMEM38B | INT | 285,888 |
| 4 | 106,437,015 | rs2214403 | 0.27 | 0.85 | 0.89 | 0.3696 | 0.4945 | PPA2 | IG | 38,969 |
Abbreviations are as follows: Chr, chromosome; DS, downstream; NC, noncoding; IG, intergenic; INT, intronic; and REG, regulatory.
Reference allele frequency in Andeans.
Reference allele in Lowland Native Americans.
Reference allele frequency in Europeans.
Ten-SNP-window-based value of PBSn1.
Highest SNP-based value of PBSn1 within the window.
Functional annotation of SNPs.
Functional Annotation of Selected Variants and Genes
To identify the putative functional consequences of genetic variation at the SNPs with the strongest evidence of natural selection, we used the Combined Annotation Dependent Depletion (CADD) pipeline57 to query a large number of functional databases (Table S1). We sorted the SNPs according to their composite score from the CADD pipeline, which provides an estimate of the functional severity of each SNP, and found that several of the candidate SNPs might have important functional consequences. Most notably, we found that three of the high-scoring selected SNPs are located within or nearby promoter or enhancer regions for the genes SLC9A31, MACROD2 (MIM: 611567), and TBX5; these regions are also highly conserved sites according to PhastCons or GERP scores. In addition, we found that a promoter or enhancer region for NOS2 is targeted by two high-scoring candidate SNPs. All 100 of the top candidate SNPs queried here are located in non-coding regions (i.e., introns, untranslated regions, and intergenic regions), suggesting that selection has targeted regulatory elements rather than protein structure in this population.
Previous studies of high-altitude-related natural selection in Tibetans and Ethiopians have implicated hypoxia-signaling-pathway-related genes as the primary targets of natural selection, but members of this pathway were notably absent from the top of our ranked list. The top of the ranked list of genes in this analysis did not include those regulating the production of HIFs, and only EGLN1 fell within the top percentile (windowed PBSn1 > 0.2284; Table 1). However, even EGLN1 was only number 96 in a ranked list of genes with proximity to candidate SNPs (see Material and Methods). Furthermore, a SNP-based Gene Ontology (GO) enrichment analysis of our top loci identified 16 categories with significant evidence of enrichment (false-discovery rate < 0.2; Table S2) but did not include support for selection in hypoxia-related categories (Table S2).
Previous selection scans on Andeans, based on SNP data, have focused on identifying the hypoxia-signaling-pathway genes with the strongest evidence of selection. For example, Bigham et al.13 identified three hypoxia-signaling-pathway genes with SNPs in the top 5% tail of at least three test statistics: EDNRA, NOS2, and PRKAA1. These genes ranked 14,681; 2; and 4,267, respectively, in our study. They also found marginal evidence of selection for EGLN1 according to one of the tests used, although the most differentiated SNP in this gene was ranked only 297 in that study.
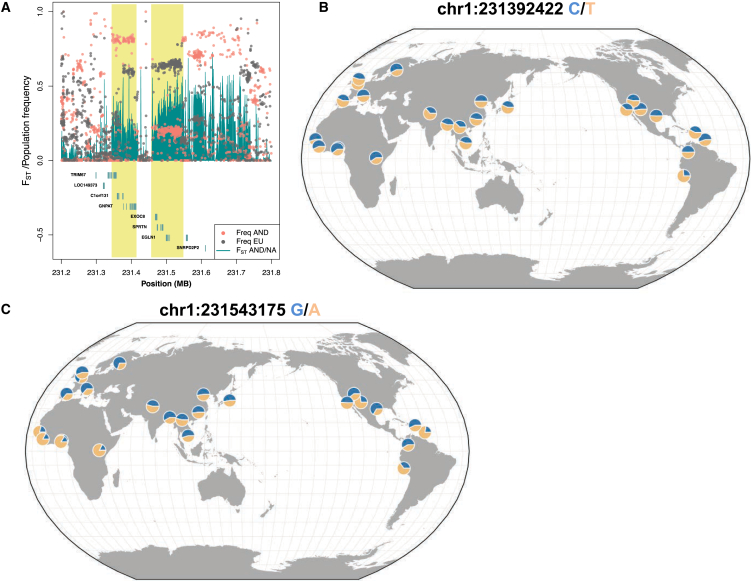
In our analyses, EGLN1 was the hypoxia-pathway gene with the strongest evidence of selection (Table 2). However, it is still not ranked among the top genes in the genome. This is in contrast to selection scans in Tibetans (based on either SNP scans or human exome sequencing), which tend to find the hypoxia-response-pathway genes EGLN1 and EPAS1 as the top ranked, or among the top ranked, genes.10, 11, 17, 18 We note that the haplotype pattern in EGLN1 is genomically quite unusual in that it includes two very long differentiated haplotypes (Figure 2) in Andeans and other populations. Both of these haplotypes exist in other populations; for example, the frequencies are approximately 0.54 and 0.61 in CEU Europeans and 0.5 and 0.52 in Han Chinese in samples from the 1000 Genomes Project. One possible explanation for this pattern is that these haplotypes segregated at an intermediate frequency in the ancestor of the Andean population, and natural selection led to a frequency shift in the Andean population when this population entered a new environment. Further analysis is needed, however, to evaluate how genomic features (such as variations in local recombination rate) that could explain the extended haplotype pattern and demographic history contribute to the unusual patterns at this locus.
Table 2.
Maximum Window-Based PBSn1 Value for HIF-Pathway Candidate Genes
| Gene | MIM Number | Maximum PBSn1(10 kb) | Maximum PBSn1(200 kb) |
|---|---|---|---|
| HIF1A | 603348 | 0.1571 | 0.1976 |
| HIF1AN | 606615 | −0.0226 | 0.0517 |
| HIF2A (EPAS1) | 603349 | 0.1373 | 0.2103 |
| HIF3A | 609976 | 0.1601 | 0.1601 |
| EGLN1 | 606425 | 0.3026 | 0.3363 |
| EGLN2 | 606424 | 0.1083 | 0.3213 |
| EGLN3 | 606426 | 0.0687 | 0.1859 |
| ARNT | 126110 | −0.0028 | −0.0020 |
| EDNRA | 131243 | 0.0783 | 0.1120 |
| EDNRB | 131244 | 0.0830 | 0.1995 |
| SENP1 | 612157 | 0.1124 | 0.1648 |
| ANP32D | 606878 | 0.2091 | 0.2091 |
| PRKAA1 | 602739 | 0.1742 | 0.2758 |
| PPARA | 170998 | 0.0624 | 0.1218 |
| PPARG | 601487 | 0.2533 | 0.2681 |
| ALDH2 | 100650 | 0.0352 | 0.0802 |
PBSn1 distribution quantiles: 25%, 0.0686; 10%, 0.1202; 5%, 0.1562; 1%, 0.2284; 0.1%, 0.3146; and 0.01%, 0.3813.
Figure 2.
Unusually Long and Differentiated Haplotypes at Hypoxia-Pathway Gene EGLN1
(A) SNP frequencies for Andeans and Europeans are plotted as points along the genomic position. Genetic differentiation (FST) between Andeans and lowland Native Americans is plotted by genomic position as a curve with the area shaded in green. RefSeq exons are plotted below. Two long haplotypes segregating at substantially different frequencies in Andeans, Europeans, and lowland Native Americans are noted by yellow bars.
(B) Frequency of the first haplotype (left) plotted for global samples from the 1000 Genomes Project with a single tag SNP (rs6674439) and the Geography of Genetic Variants Browser.
(C) Global frequency of haplotype two (right; rs6674439). The minor Andean haplotypes (blue) segregate at intermediate frequencies in nearly all other 1000 Genomes populations.
Our results suggest that the hypoxia-signaling pathway is not the primary target of hypoxia-related natural selection in Andeans, in line with previous studies where HIF-pathway genes were not over-represented among selected gene regions,13 raising the possibility that adaptation to high altitude in the Andeans involves a different set of genotypes conferring unique adaptive physiological features in the high-altitude environment. This is consistent with the evidence from comparative physiological studies suggesting that Andeans might not have high-altitude adaptations similar to those of Tibetans.1, 28, 58
To test for enrichment of the functional categories associated with the selected genes, we performed GO enrichment analysis and found evidence for 16 significant categories (Table S2). Four genes (SLC9A3R1 [MIM: 604990], ATP2A1 [MIM: 108730], PLK1 [MIM: 602098], and TBX5) were shared between at least two categories, resulting in four broad classes of GO categories. We note that the power of this analysis was limited by the fact that the significant GO categories represent only a small number of genes and are in large part single-gene and two-gene categories, but the dominance of these four genes in the enrichment results leads several themes within the significant categories to suggest that selection has targeted proteins involved in solute management, muscle control, regulation of mitosis, and cardiac development.
Top Candidate Genes
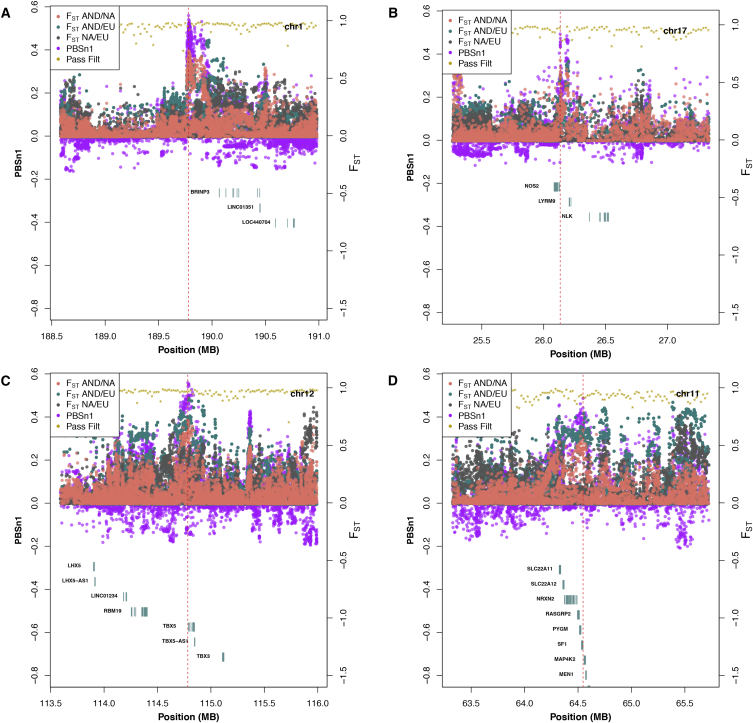
The top candidate regions are listed in Table 1; the top five are closest to BRINP3, NOS2, SH2B1, TBX5, and PYGM. In some of these, the signal of selection is quite localized and/or occurs in a region with few genes (Figure 3). These cases provide stronger confidence in the assignment of the selection signal to a specific gene than cases with a much wider selection signal in gene-rich regions (Figure S4). Importantly, the most differentiated regions in the genome do not fall in regions where the short-read mapping or genome assembly is problematic (Figure 3).
Figure 3.
Signals of Positive Natural Selection in Andean Genomes
(A) Population genetic statistics plotted by physical genomic position for the top-ranked genome-wide peak of differentiation. Two-population comparisons (FST) are plotted for three comparisons as points. Scaled three-population comparisons (PBSn1) are plotted as points for each SNP. The proportions of sites in a window passing filters are plotted (on the FST scale) as gold stars. RefSeq exons are plotted as boxes by genomic location. A large peak of Andean-specific differentiation is centered near BRINP3.
(B) The second-ranked genome-wide differentiation peak centered on the regulatory region of NOS2.
(C) The fourth-ranked peak centered near TBX5.
Of the top five ranked genes, these three are sufficiently narrow to allow convincing assignment of the signal to a gene, whereas the third- and fifth-ranked genes are substantially wider.
The most differentiated window (PBSn1 = 0.5073; Figure 3A) falls in a relatively gene-sparse region of chromosome 1 (center of window = 189,780,727) in the upstream region of BRINP3, a member of the bone morphogenetic protein/retinoic acid inducible family. Originally identified as a gene whose regulation is affected by retinoic acid levels and bone morphogenesis proteins in mice,59 variants in this gene have been associated with a number of cardiovascular phenotypes in humans, including left atrial size,60 susceptibility to atrial fibrillation,61 myocardial infarction,62 and coronary heart disease.63 Additionally, BRINP3 variants are associated with differential expression of this gene in aortic smooth muscle cells, a functional effect that is potentially related to proliferation and senescence of these cells.62 BRINP3 expression is also implicated in the modulation of reactive oxygen species production and NF-κB activity with downstream effects on leukocyte adhesion and inflammation in humans.64 Although the peak lies a considerable distance from BRINP3, it is relatively narrow and is supported by a large number of SNPs (Figure 3A), possibly including cis-regulatory elements.
The second strongest peak of differentiation in our ranked list (PBSn1 = 0.4320; Figure 3B) overlaps a known regulatory element of NOS2, which encodes NOS, one of three enzymes (encoded by different chromosomes) that are responsible for synthesizing nitric oxide (NO). NO is an inorganic gas with many biological functions, including vascular homeostasis, smooth muscle relaxation, inflammation, neurotransmission, inhibition of platelet aggregation, stimulation of angiogenesis, blood pressure reduction, and alterations in immune system functioning.65 NOS2 is expressed in cardiac myocytes, and its production increases under conditions of hypoxia in cell-culture experiments and in vivo conditions during fetal life via HIF-1-dependent mechanisms.66, 67
The third-ranked peak in our list includes a long haplotype overlapping multiple genes (Figure S6), including SH2B1, TUFM (MIM: 602389), and SULT1A2 (MIM: 601292). SH2B1 (Src-homology 2B adaptor protein 1) encodes an adaptor protein with an SH2 domain that activates various kinases in a signaling pathway. SH2B1 is also known as a negative regulator or erythropoietin-signaling pathway by physically interacting with erythropoietin receptor,68 suggesting that selection for a variant of SH2B1 might explain CMS associated with high Hb levels in some Andeans. Both male and female SH2B1-null mice are infertile; female mice have small, anovulatory ovaries with reduced numbers of follicles, and male mice exhibit small testes and sperm deficits. TUFM (Tu Translation Elongation Factor, Mitochondrial) is involved in mitochondrial protein translation, and mice with mutated TUFM are phenotypically normal. SULT1A2 (sulfotransferase family 1A member 2; aka SULT1C1 in mice) catalyzes sulfate conjugation of many hormones, neurotransmitters, drugs, and xenobiotic compounds.
The fourth-ranked peak (PBSn1 = 0.4092; Figure 3C) is relatively narrow and centered very closely to TBX5. T-box 5, the protein encoded by this gene, is a transcription factor that is involved in cardiac development and specification of limb identity.69 Mutations in this gene have been associated with risk of atrial fibrillation70 (similarly to BRINP3 mutations), blood pressure levels,71 and electrocardiography (EKG) phenotypes including PR and QRS intervals.72
The fifth-ranked peak is close to multiple genes, making assignment of candidate genes somewhat uncertain. But we note that the peak falls close to PYGM, which encodes the muscle isoform of glycogen phosphorylase, the major rate-determining enzyme for glycogen mobilization in many normal cells under physical exercise73 and in oxygen-deprived cancer cells.74 Pescador et al.75 found a modest reduction in glucogen phosphorylase in cells exposed to hypoxia as part of a hypoxia-mediated increased level of glucogen accumulation. Genetic variants in PYGM could be relevant for glycogen mobilization and energy storage and utilization in cold environments and for glycogen mobilization and accumulation in response to hypoxia and should be interrogated in future studies of Aymaran evolutionary adaptations.
The fact that three of the top candidate genes (BRINP3, NOS2, and TBX5) are related to cardiac function and the fact that the significant GO categories include, for example, “muscle control” and “cardiac development” raises the hypothesis that natural selection might target the cardiovascular system in the Andeans to compensate for the substantial stress on the vascular and pulmonary system associated with high-altitude living.
Evolutionary Origins of Positively Selected Haplotypes
Several recent studies14, 46, 76 have shown that haplotypes with adaptive alleles at high frequency in some modern human populations were introduced through introgression with archaic hominid species. We examined the evidence of archaic introgression in our top candidates by several different methods. First, we calculated several different summary statistics to measure archaic ancestry (Material and Methods). From these summary statistics, we identified three candidate regions (BRINP3, TBX5, and SHISA6, corresponding to SNPs rs11578671, rs10744822, and rs78264921, respectively) with high putative archaic ancestry (Figures S1 and S2) and then investigated these further.
We used a HMM aimed at detecting archaic introgressed tracts46, 47 to search for evidence of introgressed tracts at high frequency in non-African individuals in the 1000 Genomes Project panel.48 Figures S7–S12 show the output of the HMM in four super-populations (AMR, SAS, EAS, and EUR) from the 1000 Genomes Project in a 1-Mb region around the three top candidate SNPs under the assumption that the source population was the population to which either the Altai Neanderthal (Figures S7, S9, and S11) or the Denisovan (Figures S8, S10, and S12) individual belonged. The SHISA6 region (Figures S7 and S8) contained only short tracts in a few individuals, and they did not overlap the top Aymara SNP when either the Altai Neanderthal genome or the Denisovan genome was used as the source. The BRINP3 region (Figures S9 and S10) contained a long archaic tract overlapping the top SNP when the Neanderthal was used as the source, and this tract was found only in a few individuals in the American, European, and South Asian super-panels. The TBX5 region (Figures S11 and S12) contained several short tracts, some of which overlapped the top SNP, at high frequency in all super-panels when the Neanderthal was used as the source. Many of these short tracts occurred on the same chromosome. This suggests that they could belong to the same haplotype but that the HMM might have failed to call a single long tract, perhaps because the input source population was not a good proxy for the true source population.
To understand the haplotype structure in the region, we used the program Haplostrips52 to sort the haplotypes by similarity to the Altai Neanderthal genome. In the cases of SHISA6 and TBX5, we observed no evidently introgressed haplotype at medium or high frequency in present-day humans (Figures S13–S15). In the case of BRINP3, we observed a much stronger haplotype structure with three highly differentiated haplotype groups, one of which shared strong similarity to the Altai Neanderthal genome (Figure S15). This haplotype, however, was present at substantial frequencies in YRI, suggesting that this was most likely not part of the Neanderthal-into-non-African introgression event but could have segregated in the present-day human population before the out-of-Africa expansion.
In conclusion, eight of the top ten regions (including the SHISA6 region) showed no evidence of adaptive introgression. The regions containing genes BRINP3 and TBX5 had very weak evidence of selection for archaic alleles, but the pattern observed could also be consistent with an ancestrally segregating haplotype in the modern human population during a time before the out-of-Africa expansion. Furthermore, none of these regions appeared in a recent scan for adaptive introgression performed with a variety of summary statistics.77 Although it is very difficult to positively exclude the possibility that introgression could have affected these regions, we found no statistical evidence supporting introgression similarly to that observed in studies of TBX15 in Inuit46 or EPAS1 in Tibetans.18
Effect of Selected Alleles on Gene Expression
We queried the Genotype-Tissue Expression (GTEx) Portal for associations between genotypes and gene expression across a large panel of human tissues (see Web Resources) for evidence of cis-expression quantitative trait loci (eQTLs) (Table S3) at each of the candidate SNPs with highest differentiation in NOS2 (rs34913965), TBX5 (rs10744822), and BRINP3 (rs11578671). We also interrogated an additional TBX5 SNP (rs2555030), which tags a second, equally highly differentiated haplotype at this locus. After we accounted for multiple testing across tissues and SNPs, none of the SNPs passed the p value cutoff (0.05/126 or 0.000397). Nevertheless, we report here the lowest p values (p < 0.01), which for the expression of NOS2 (rs34913965) were reduced for the Andean allele in the tibial nerve (p = 0.00072, negative effect), thyroid (p = 0.0046, negative effect), and skeletal muscle (p = 0.0062, negative effect) but increased in the prostate (p = 0.0077, positive effect). For BRINP3 (rs11578671), the lowest p value corresponded to the sigmoid colon (p = 0.0074, positive effect for the selected Andean allele). For TBX5 (rs2555030), the lowest p value corresponded to the aorta (p = 0.0044, positive effect), whereas for TBX5 (rs10744822), it corresponds to the left ventricle of the heart (p = 0.0056, positive effect for the selected Andean allele). Notably, the left ventricle and the aorta were among the tissues with the highest overall expression of TBX5 (Figure S16).
We also tested for trans-eQTL effects of these SNPs (Table S4) with the following genes that are downstream of our top candidates in regulatory pathways: FGA (MIM: 134820), FGB (MIM: 134830), FGG (MIM: 134850), NPPB (MIM: 600295), PRKAA1, and PRKAA2 (MIM: 600497). After we accounted for multiple testing across tissues and SNPs, none of the SNPs were significant (p value cutoff = 0.05/834 = 5.995 × 10−5). The lowest p values (p < 0.01) corresponded to PRKAA2 (rs2555030) in brain hypothalamus (p = 0.0011, negative effect for the alternative allele), PRKAA2 (rs10744822) in pancreas (p = 0.0013, positive effect), PRKAA2 (rs34913965) in thyroid (p = 0.0063, positive effect), NPPB (rs34913965) in brain substantia nigra (p = 0.0065, negative effect), PRKAA1 (rs10744822) in pancreas (p = 0.0097, positive effect), and FGG (rs2555030) in unexposed suprapubic skin (p = 0.0097, positive effect).
Association Analyses
To further investigate the physiological effects of genetic variants of the three top genes (BRINP3, NOS2, and TBX5), we identified a panel of phenotypes relevant to cardiovascular function and blood physiology (Table S5) and tested for possible associations between the candidate variants and these phenotypes. Direct measurements in cohorts of the Andean high-altitude residents would be most appropriate, but such data are currently not available. However, the candidate SNPs are of relatively high frequency in other populations as well, and associations in these populations could provide important clues regarding the phenotypic effects of the selected SNPs. Previous studies70, 72 have shown that the selected SNPs at TBX5 are in high linkage disequilibrium with markers associated with protection against atrial fibrillation (r2 = 0.83, odds ratio = 0.88 for protective allele; r2 = 0.62, relative risk = 1.12 for the non-Andean allele) and affect EKG phenotypes including PR, QRS, and QT intervals in genome-wide association studies (GWASs) of Europeans72 (r2 = 0.83, effect percentage = 7.35%, 7.36%, and 5.88%, respectively).
To further investigate the possible phenotypic effects of the candidate SNPs, we took advantage of the extensive phenotypic data available in the MESA study.78 The MESA study has recruited participants from four different ethnic groups, including those of European descent (EUR), African Americans (AA), Hispanic Americans (HA), and Chinese Americans (CHN). Although we expected substantial haplotype sharing between the Andeans and the other non-African cohorts, we did not expect the same level of haplotype sharing with Africans (or African Americans). As such, the AA cohort was intentionally excluded from this analysis, thus reducing the multiple-testing burden. We therefore tested associations between four candidate SNPs (one BRINP3 haplotype, one NOS2 haplotype, and two TBX5 haplotypes) and a panel of 42 phenotypes from the EUR, HA, or CHN cohort from the MESA study (Table S6).
Although genetic variants at BRINP3 have been associated with several cardiac phenotypes, including left atrial size and risk of atrial fibrillation in the Framingham Heart Study60, 61 and risk of coronary heart disease in a cohort of women of unknown ethnicity in the US,63 we did not find significant associations between the Andean haplotype and cardiac phenotypes in the MESA cohorts (all had an uncorrected p value > 0.0167; Table S6). Power analysis showed that we had sufficient power (80% or greater) to detect association at the 0.0167 (0.05/3 genes) significance level if a variant explained 0.4% of phenotypic variance with 2,652 EUR samples. A sample size of 1,490 (MESA HA sample) would identify only associated genetic variants that explained 0.7% of the phenotypic variance, and the 775 CHN samples would be able to identify a SNP explaining 1.4% of phenotypic variance. Under this framework, we discovered a significant association between the Andean BRINP3 haplotype and blood fibrinogen levels in the CHN cohort (uncorrected p = 9.64 × 10−6, Bonferroni-corrected p = 0.0051). The non-Andean allele was associated with a decrease in fibrinogen levels (β = −0.1369). Fibrinogen is an essential part of hemostasis because it is the principal component of blood clots (fibrin). Fibrin also interacts with components of the inflammatory pathway and augments chronic inflammation.
We did not identify clear associations, after correcting for multiple tests, between the Andean allele at NOS2 and the limited range of phenotypes tested here, so the functional consequences associated with the high-frequency Andean variants remained unclear at this point. However, we discovered significant associations between the Andean haplotype at the TBX5 locus and two blood-related phenotypes, Hb A1c levels (uncorrected p = 1.88 × 10−7, Bonferroni-corrected p = 9.93 × 10−5) and fasting insulin levels (Bonferroni-corrected p = 0.0227), in the EUR cohort. In both cases, the alternative, non-Andean allele was associated with an increase in blood levels of fasting insulin (β = 68.8857) and Hb (HgB) A1c (β = 1.6024). HgB A1C reflects the mean level of glycemia (serum level of glucose in blood) during the lifespan of erythrocytes, and elevated HgB A1C indicates prediabetes or diabetes mellitus. This finding is consistent with previous studies that found a reduced prevalence of diabetes at high altitude in the Andes, despite a high prevalence of obesity and dyslipidemia in some cases,79, 80, 81 whereas the opposite is true in Tibet, where researchers have found a significant excess of glucose intolerance.82
Discussion
In Tibetans, natural selection related to high-altitude adaptation seems to have acted on genes in the hypoxia-response pathway to modulate erythropoiesis, possibly to avoid or reduce polycythemia.10, 11, 17, 18, 25 This might also be the case in Ethiopians, although relatively few studies have been performed, and differing results have been obtained regarding Hb levels.9, 16, 26 In Andeans, there is marginal evidence of selection for EGLN1 in both this and previous studies11 and evidence that the genomic regions targeted by selection in Andeans help preserve a normal rise in uteroplacental blood flow during pregnancy and fetal growth at high altitude.12 Nonetheless, our whole-genome sequencing revealed that the SNPs with the strongest differentiation in the Andeans are not involved in the regulation of erythropoiesis. Rather, our strongest candidate genes were BRINP3, NOS2, and TBX5, which are associated with a number of important processes in the cardiovascular system, but not erythropoiesis. This is in accordance with observations made by Beall,28 who found that Tibetans and Ethiopians in high altitude have Hb concentrations that are not much different from those observed at sea level, whereas Andeans have much higher Hb concentrations at high altitude. Beall28 argued that selection associated with hypoxia in high altitude had resulted in two very different physiological outcomes in the Andes and Tibet.
Our association study revealed a statistically significant association between BRINP3, the gene showing the strongest evidence of selection, and fibrinogen levels. Fibrinogen levels are used as a general marker of inflammation, and high levels are associated with cardiovascular disease.83 Increased fibrinogen levels are also associated with peripheral arterial narrowing,84 and furthermore, there is a well-established positive correlation between fibrinogen levels and blood viscosity. 85 The reduced fibrinogen levels associated with the adaptive BRINP3 allele, therefore, suggest a mitigating effect of the allele on the negative fitness effects of polycythemia. Whether the effect of the BRINP3 allele on fibrinogen works indirectly by affecting conditions leading to vascular inflammation or directly by regulating fibrinogen expression remains speculative. However, recent evidence indicates an association between a fibrinogen splice variant, gamma prime (γ′) fibrinogen, and cardiovascular morbidity and mortality.86, 87 We intend to determine in future studies whether the proportion of γ′ fibrinogen is altered in Aymaras carrying the selected allele.
Our limited association-mapping study did not find any associations between NOS2 and the investigated cardiac phenotypes. However, there is a well-established connection between NO production and cardiac health. Perhaps because of the more transient nature of NOS protein activation, studies of NO production under conditions of high altitude have focused largely on neuronal and endothelial NOS (NOS1 and NOS3).88 Activation of these isoforms results in the production of small amounts of NO, whereas NOS2 is activated over hours, can remain active over days, and produces up to 1,000-fold greater amounts of NO than NOS1 or NOS3.89 Importantly, chronic hypoxia selectively upregulates myocardial NOS2 activation and NO generation during fetal life via HIF-α-dependent mechanisms.67 NOS2-derived NO contributes to ischemic preconditioning in adult rat hearts,90 suggesting a cardio-protective effect, but on the other hand, NOS2 is a major pathophysiologic mediator of inflammatory or ischemia-reperfusion-induced cardiac injury.91 Future studies are needed to determine whether the allele under selection in Andeans affect NO production and, if so, its direct functional consequences.
We have found an association between the selected Andean allele in TBX5 and decreased Hb A1C and insulin. Chronic exposure to high altitude leads to lower fasting glycemia.79 We hypothesize that selection has favored genetic variants that restore blood glucose homeostasis, i.e., that increase glycemia to normal levels. Such variants would be associated with increased Hb A1C levels, as found in this study. The effect would be similar to that observed in other genetic adaptations affecting physiology in response to altered environmental conditions, e.g., the downregulation of erythropoiesis in Tibetans in high altitude and the decrease in endogenous synthesis of certain long-chained poly-unsaturated fatty acids (PUFAs) in Inuit populations with a diet rich in these PUFAs from fish and marine mammals. In these cases, selection acts on the phenotype in the opposite direction of that induced by the altered environment, consistent with the effect we observed for the selected variants in TBX5 on Hb A1C levels in this study.
We note that the presently described phenotypic associations of TBX5 and BRINP3 SNPs were interrogated at ambient oxygen concentrations. It will be essential to evaluate the association of these selected genotype-phenotype relations in people of Andean ancestry at low altitude, such as Santa Cruz, Bolivia (altitude 350 m; population 3.4 million in 2015; >60% indigenous population), and high altitude, such as La Paz (altitude 3,650 m; population 1.7 million; 25% Aymara) and El Alto (altitude 4,150; population > 90,000; 76% Aymara) in Bolivia.
We have shown here that detailed population genetic analysis of low-coverage whole-genome sequencing data provides an economic, efficient, and powerful approach to discovering novel candidate-gene phenotypes that might be adaptive via GWASs of lowland samples. Our evolutionary genomic analysis in Andeans suggests that, in contrast to Tibetans, Andeans have adapted to high-altitude living by mitigating the effects of polycythemia by increasing the cardiovascular tolerance to this condition, consistent with Beall’s21 hypothesis of two different biological outcomes in Tibet and the Andes of the selection imposed by high-altitude living.
Acknowledgments
For the Colorado cohort, we thank the Instituto Boliviano de Biologia de Altura investigators and the local physicians and other health-care personnel who helped with the study, the many subjects who generously participated, and grant support from the NIH (HLBI 079647 and TW 001188). The Multi-Ethnic Study of Atherosclerosis (MESA) is supported by NIH contracts HHSN2682015000031, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, and N01-HC-95169 and by grants UL1-TR-000040, UL1-TR-001079, and UL1-RR-025005 from the National Center for Research Resources. Funding for MESA SHARe genotyping was provided by NHLBI contract N02-HL-6-4278. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, the Clinical and Translational Science Institute (grant UL1TR000124), and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (grant DK063491 to the Southern California Diabetes Endocrinology Research Center). We also thank two anonymous reviewers for their comments, which helped to improve the manuscript.
Published: November 2, 2017
Footnotes
Supplemental Data include 16 figures and 6 tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.09.023.
Contributor Information
Josef T. Prchal, Email: josef.prchal@hsc.utah.edu.
Rasmus Nielsen, Email: rasmus_nielsen@berkeley.edu.
Accession Numbers
All short-read data were submitted to the NCBI Sequence Read Archive under BioProject accession number PRJNA393593.
Web Resources
1000 Genomes Genome Accessibility Masks, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/supporting/accessible_genome_masks/StrictMask/
1000 Genomes Phase 3 Genotype Likelihoods, ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20130502/supporting/genotype_likelihoods/shapeit2/
BCFtools, https://samtools.github.io/bcftools
Combined Annotation Dependent Depletion (CADD), http://cadd.gs.washington.edu/
ENCODE Mappability Downloadable Files, http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeMapability
Geography of Genetic Variants Browser, http://popgen.uchicago.edu/ggv/
GTEx Portal, https://www.gtexportal.org/home/
Multi-Ethnic Study of Atherosclerosis (MESA), https://www.mesa-nhlbi.org
Mutagenetix, http://mutagenetix.utsouthwestern.edu/home.cfm
NCBI BioProject, https://www.ncbi.nlm.nih.gov/bioproject/
ngsPopGen, https://github.com/mfumagalli/ngsPopGen
OMIM, http://www.omim.org
Picard Tools, http://broadinstitute.github.io/picard/
Supplemental Data
The SNPs with the highest per-SNP PBSn1 value within the top 100 peaks in the genome were submitted for annotation via the CADD online portal at http://cadd.gs.washington.edu/. The 25 SNPs with the strongest evidence of functional consequences are bolded. Additional information on the annotation pipeline and output can be found on the website.
We tested a representative SNP for our top genes. Each SNP was interrogated for effects on expression in the GTeX panel of tissues via the online portal at http://gtexportal.org/home/.
Same as Table S3 but interrogating known downstream genes.
References
- 1.Heath D., Williams D.R. Butterworth Scientific; 1989. High-Altitude Medicine and Pathology; pp. 102–114. [Google Scholar]
- 2.Guyton A.C., Richardson T.Q. Effect of hematocrit on venous return. Circ. Res. 1961;9:157–164. doi: 10.1161/01.res.9.1.157. [DOI] [PubMed] [Google Scholar]
- 3.Sime F., Banchero N., Penaloza D., Gamboa R., Cruz J., Marticorena E. Pulmonary hypertension in children born and living at high altitudes. Am. J. Cardiol. 1963;11:143–149. doi: 10.1016/0002-9149(63)90054-7. [DOI] [PubMed] [Google Scholar]
- 4.Gonzales G.F., Steenland K., Tapia V. Maternal hemoglobin level and fetal outcome at low and high altitudes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009;297:R1477–R1485. doi: 10.1152/ajpregu.00275.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.León-Velarde F., Maggiorini M., Reeves J.T., Aldashev A., Asmus I., Bernardi L., Ge R.-L., Hackett P., Kobayashi T., Moore L.G. Consensus statement on chronic and subacute high altitude diseases. High Alt. Med. Biol. 2005;6:147–157. doi: 10.1089/ham.2005.6.147. [DOI] [PubMed] [Google Scholar]
- 6.Prchal J.T. Chapter 34: Clinical manifestations and classification of erythrocyte disorders. In: Kaushansky K., Lichtman M.A., Prchal J.T., Levi M.M., Press O.W., Burns L.J., Caligiuri M., editors. Williams Hematology. McGraw Hill; 2015. pp. 503–512. [Google Scholar]
- 7.Erslev A.J., Caro J., Schuster S.J. Is there an optimal hemoglobin level? Transfus. Med. Rev. 1989;3:237–242. doi: 10.1016/s0887-7963(89)70084-5. [DOI] [PubMed] [Google Scholar]
- 8.Murray J.F., Gold P., Johnson B.L., Jr. The circulatory effects of hematocrit variations in normovolemic and hypervolemic dogs. J. Clin. Invest. 1963;42:1150–1159. doi: 10.1172/JCI104800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alkorta-Aranburu G., Beall C.M., Witonsky D.B., Gebremedhin A., Pritchard J.K., Di Rienzo A. The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet. 2012;8:e1003110. doi: 10.1371/journal.pgen.1003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beall C.M., Cavalleri G.L., Deng L., Elston R.C., Gao Y., Knight J., Li C., Li J.C., Liang Y., McCormack M. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. USA. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bigham A., Bauchet M., Pinto D., Mao X., Akey J.M., Mei R., Scherer S.W., Julian C.G., Wilson M.J., López Herráez D. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bigham A.W., Julian C.G., Wilson M.J., Vargas E., Browne V.A., Shriver M.D., Moore L.G. Maternal PRKAA1 and EDNRA genotypes are associated with birth weight, and PRKAA1 with uterine artery diameter and metabolic homeostasis at high altitude. Physiol. Genomics. 2014;46:687–697. doi: 10.1152/physiolgenomics.00063.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bigham A.W., Mao X., Mei R., Brutsaert T., Wilson M.J., Julian C.G., Parra E.J., Akey J.M., Moore L.G., Shriver M.D. Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum. Genomics. 2009;4:79–90. doi: 10.1186/1479-7364-4-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huerta-Sánchez E., Jin X., Asan, Bianba Z., Peter B.M., Vinckenbosch N., Liang Y., Yi X., He M., Somel M. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–197. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenzo F.R., Huff C., Myllymäki M., Olenchock B., Swierczek S., Tashi T., Gordeuk V., Wuren T., Ri-Li G., McClain D.A. A genetic mechanism for Tibetan high-altitude adaptation. Nat. Genet. 2014;46:951–956. doi: 10.1038/ng.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheinfeldt L.B., Soi S., Thompson S., Ranciaro A., Woldemeskel D., Beggs W., Lambert C., Jarvis J.P., Abate D., Belay G., Tishkoff S.A. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 2012;13:R1. doi: 10.1186/gb-2012-13-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simonson T.S., Yang Y., Huff C.D., Yun H., Qin G., Witherspoon D.J., Bai Z., Lorenzo F.R., Xing J., Jorde L.B. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 18.Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z.X.P., Pool J.E., Xu X., Jiang H., Vinckenbosch N., Korneliussen T.S. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hultgren H.N. Hultgren Publications; 1997. High Altitude Medicine; pp. 87–92. [Google Scholar]
- 20.Wu T., Wang X., Wei C., Cheng H., Wang X., Li Y., Ge-Dong, Zhao H., Young P., Li G., Wang Z. Hemoglobin levels in Qinghai-Tibet: different effects of gender for Tibetans vs. Han. J. Appl. Physiol. 2005;98:598–604. doi: 10.1152/japplphysiol.01034.2002. [DOI] [PubMed] [Google Scholar]
- 21.Beall C.M., Brittenham G.M., Macuaga F., Barragan M. Variation in hemoglobin concentration among samples of high-altitude natives in the Andes and the Himalayas. Am. J. Hum. Biol. 1990;2:639–651. doi: 10.1002/ajhb.1310020607. [DOI] [PubMed] [Google Scholar]
- 22.Winslow R.M., Chapman K.W., Gibson C.C., Samaja M., Monge C.C., Goldwasser E., Sherpa M., Blume F.D., Santolaya R. Different hematologic responses to hypoxia in Sherpas and Quechua Indians. J. Appl. Physiol. 1989;66:1561–1569. doi: 10.1152/jappl.1989.66.4.1561. [DOI] [PubMed] [Google Scholar]
- 23.Beall C.M., Decker M.J., Brittenham G.M., Kushner I., Gebremedhin A., Strohl K.P. An Ethiopian pattern of human adaptation to high-altitude hypoxia. Proc. Natl. Acad. Sci. USA. 2002;99:17215–17218. doi: 10.1073/pnas.252649199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simonson T.S., Wei G., Wagner H.E., Wuren T., Qin G., Yan M., Wagner P.D., Ge R.L. Low haemoglobin concentration in Tibetan males is associated with greater high-altitude exercise capacity. J. Physiol. 2015;593:3207–3218. doi: 10.1113/JP270518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bigham A.W., Wilson M.J., Julian C.G., Kiyamu M., Vargas E., Leon-Velarde F., Rivera-Chira M., Rodriquez C., Browne V.A., Parra E. Andean and Tibetan patterns of adaptation to high altitude. Am. J. Hum. Biol. 2013;25:190–197. doi: 10.1002/ajhb.22358. [DOI] [PubMed] [Google Scholar]
- 26.Huerta-Sánchez E., Degiorgio M., Pagani L., Tarekegn A., Ekong R., Antao T., Cardona A., Montgomery H.E., Cavalleri G.L., Robbins P.A. Genetic signatures reveal high-altitude adaptation in a set of Ethiopian populations. Mol. Biol. Evol. 2013;30:1877–1888. doi: 10.1093/molbev/mst089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semenza G.L. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu. Rev. Pathol. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 28.Beall C.M. Andean, Tibetan, and Ethiopian patterns of adaptation to high-altitude hypoxia. Integr. Comp. Biol. 2006;46:18–24. doi: 10.1093/icb/icj004. [DOI] [PubMed] [Google Scholar]
- 29.Niermeyer S., Andrade-M M.P., Vargas E., Moore L.G. Neonatal oxygenation, pulmonary hypertension, and evolutionary adaptation to high altitude (2013 Grover Conference series) Pulm. Circ. 2015;5:48–62. doi: 10.1086/679719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groves B.M., Droma T., Sutton J.R., McCullough R.G., McCullough R.E., Zhuang J., Rapmund G., Sun S., Janes C., Moore L.G. Minimal hypoxic pulmonary hypertension in normal Tibetans at 3,658 m. J. Appl. Physiol. 1993;74:312–318. doi: 10.1152/jappl.1993.74.1.312. [DOI] [PubMed] [Google Scholar]
- 31.Newman J.H., Holt T.N., Cogan J.D., Womack B., Phillips J.A., 3rd, Li C., Kendall Z., Stenmark K.R., Thomas M.G., Brown R.D. Increased prevalence of EPAS1 variant in cattle with high-altitude pulmonary hypertension. Nat. Commun. 2015;6:6863. doi: 10.1038/ncomms7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou D., Udpa N., Ronen R., Stobdan T., Liang J., Appenzeller O., Zhao H.W., Yin Y., Du Y., Guo L. Whole-genome sequencing uncovers the genetic basis of chronic mountain sickness in Andean highlanders. Am. J. Hum. Genet. 2013;93:452–462. doi: 10.1016/j.ajhg.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole A.M., Petousi N., Cavalleri G.L., Robbins P.A. Genetic variation in SENP1 and ANP32D as predictors of chronic mountain sickness. High Alt. Med. Biol. 2014;15:497–499. doi: 10.1089/ham.2014.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fumagalli M., Vieira F.G., Linderoth T., Nielsen R. ngsTools: methods for population genetics analyses from next-generation sequencing data. Bioinformatics. 2014;30:1486–1487. doi: 10.1093/bioinformatics/btu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korneliussen T.S., Albrechtsen A., Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skotte L., Korneliussen T.S., Albrechtsen A. Estimating individual admixture proportions from next generation sequencing data. Genetics. 2013;195:693–702. doi: 10.1534/genetics.113.154138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng J.Y., Mailund T., Nielsen R. Fast admixture analysis and population tree estimation for SNP and NGS data. Bioinformatics. 2017;33:2148–2155. doi: 10.1093/bioinformatics/btx098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer M., Kircher M., Gansauge M.-T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prüfer K., de Filippo C. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durand E.Y., Patterson N., Reich D., Slatkin M. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 2011;28:2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin S.H., Davey J.W., Jiggins C.D. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol. Biol. Evol. 2015;32:244–257. doi: 10.1093/molbev/msu269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Racimo F., Marnetto D., Huerta-Sánchez E. Signatures of Archaic Adaptive Introgression in Present-Day Human Populations. Mol. Biol. Evol. 2017;34:296–317. doi: 10.1093/molbev/msw216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Racimo F., Gokhman D., Fumagalli M., Ko A., Hansen T., Moltke I., Albrechtsen A., Carmel L., Huerta-Sánchez E., Nielsen R. Archaic Adaptive Introgression in TBX15/WARS2. Mol. Biol. Evol. 2017;34:509–524. doi: 10.1093/molbev/msw283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seguin-Orlando A., Korneliussen T.S., Sikora M., Malaspinas A.-S., Manica A., Moltke I., Albrechtsen A., Ko A., Margaryan A., Moiseyev V. Paleogenomics. Genomic structure in Europeans dating back at least 36,200 years. Science. 2014;346:1113–1118. doi: 10.1126/science.aaa0114. [DOI] [PubMed] [Google Scholar]
- 48.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M., Patterson N., Li H., Zhai W., Fritz M.H.-Y. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marnetto D., Huerta-Sánchez E. Haplostrips: revealing population structure through haplotype visualization. Methods Ecol. Evol. 2017;8:1389–1392. [Google Scholar]
- 53.Wang S., Ray N., Rojas W., Parra M.V., Bedoya G., Gallo C., Poletti G., Mazzotti G., Hill K., Hurtado A.M. Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet. 2008;4:e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Homburger J.R., Moreno-Estrada A., Gignoux C.R., Nelson D., Sanchez E., Ortiz-Tello P., Pons-Estel B.A., Acevedo-Vasquez E., Miranda P., Langefeld C.D. Genomic Insights into the Ancestry and Demographic History of South America. PLoS Genet. 2015;11:e1005602. doi: 10.1371/journal.pgen.1005602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bryc K., Velez C., Karafet T., Moreno-Estrada A., Reynolds A., Auton A., Hammer M., Bustamante C.D., Ostrer H. Colloquium paper: genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc. Natl. Acad. Sci. USA. 2010;107(Suppl 2):8954–8961. doi: 10.1073/pnas.0914618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crawford J.E., Nielsen R. Detecting adaptive trait loci in nonmodel systems: divergence or admixture mapping? Mol. Ecol. 2013;22:6131–6148. doi: 10.1111/mec.12562. [DOI] [PubMed] [Google Scholar]
- 57.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moore L.G., Niermeyer S., Zamudio S. Human adaptation to high altitude: regional and life-cycle perspectives. Am. J. Phys. Anthropol. 1998;107(Suppl 27):25–64. doi: 10.1002/(sici)1096-8644(1998)107:27+<25::aid-ajpa3>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 59.Kawano H., Nakatani T., Mori T., Ueno S., Fukaya M., Abe A., Kobayashi M., Toda F., Watanabe M., Matsuoka I. Identification and characterization of novel developmentally regulated neural-specific proteins, BRINP family. Brain Res. Mol. Brain Res. 2004;125:60–75. doi: 10.1016/j.molbrainres.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 60.Vasan R.S., Larson M.G., Aragam J., Wang T.J., Mitchell G.F., Kathiresan S., Newton-Cheh C., Vita J.A., Keyes M.J., O’Donnell C.J. Genome-wide association of echocardiographic dimensions, brachial artery endothelial function and treadmill exercise responses in the Framingham Heart Study. BMC Med. Genet. 2007;8(Suppl 1):S2. doi: 10.1186/1471-2350-8-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larson M.G., Atwood L.D., Benjamin E.J., Cupples L.A., D’Agostino R.B., Sr., Fox C.S., Govindaraju D.R., Guo C.-Y., Heard-Costa N.L., Hwang S.-J. Framingham Heart Study 100K project: genome-wide associations for cardiovascular disease outcomes. BMC Med. Genet. 2007;8(Suppl 1):S5. doi: 10.1186/1471-2350-8-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Connelly J.J., Shah S.H., Doss J.F., Gadson S., Nelson S., Crosslin D.R., Hale A.B., Lou X., Wang T., Haynes C. Genetic and functional association of FAM5C with myocardial infarction. BMC Med. Genet. 2008;9:33. doi: 10.1186/1471-2350-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cline J.L., Beckie T.M. The relationships between FAM5C SNP (rs10920501) variability and metabolic syndrome and inflammation in women with coronary heart disease. Biol. Res. Nurs. 2013;15:160–166. doi: 10.1177/1099800411424487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sato J., Kinugasa M., Satomi-Kobayashi S., Hatakeyama K., Knox A.J., Asada Y., Wierman M.E., Hirata K., Rikitake Y. Family with sequence similarity 5, member C (FAM5C) increases leukocyte adhesion molecules in vascular endothelial cells: implication in vascular inflammation. PLoS ONE. 2014;9:e107236. doi: 10.1371/journal.pone.0107236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moncada S., Higgs E.A. Endogenous nitric oxide: physiology, pathology and clinical relevance. Eur. J. Clin. Invest. 1991;21:361–374. doi: 10.1111/j.1365-2362.1991.tb01383.x. [DOI] [PubMed] [Google Scholar]
- 66.Jung F., Palmer L.A., Zhou N., Johns R.A. Hypoxic regulation of inducible nitric oxide synthase via hypoxia inducible factor-1 in cardiac myocytes. Circ. Res. 2000;86:319–325. doi: 10.1161/01.res.86.3.319. [DOI] [PubMed] [Google Scholar]
- 67.Thompson L., Dong Y., Evans L. Chronic hypoxia increases inducible NOS-derived nitric oxide in fetal guinea pig hearts. Pediatr. Res. 2009;65:188–192. doi: 10.1203/PDR.0b013e31818d6ad0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Javadi M., Hofstätter E., Stickle N., Beattie B.K., Jaster R., Carter-Su C., Barber D.L. The SH2B1 adaptor protein associates with a proximal region of the erythropoietin receptor. J. Biol. Chem. 2012;287:26223–26234. doi: 10.1074/jbc.M112.382721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boogerd C.J., Evans S.M. TBX5 and NuRD Divide the Heart. Dev. Cell. 2016;36:242–244. doi: 10.1016/j.devcel.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sinner M.F., Tucker N.R., Lunetta K.L., Ozaki K., Smith J.G., Trompet S., Bis J.C., Lin H., Chung M.K., Nielsen J.B., METASTROKE Consortium. AFGen Consortium Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation. 2014;130:1225–1235. doi: 10.1161/CIRCULATIONAHA.114.009892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu X., Wang L., Lin X., Huang J., Charles Gu C., He M., Shen H., He J., Zhu J., Li H. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Hum. Mol. Genet. 2015;24:865–874. doi: 10.1093/hmg/ddu478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holm H., Gudbjartsson D.F., Arnar D.O., Thorleifsson G., Thorgeirsson G., Stefansdottir H., Gudjonsson S.A., Jonasdottir A., Mathiesen E.B., Njølstad I. Several common variants modulate heart rate, PR interval and QRS duration. Nat. Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 73.Jensen T.E., Richter E.A. Regulation of glucose and glycogen metabolism during and after exercise. J. Physiol. 2012;590:1069–1076. doi: 10.1113/jphysiol.2011.224972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Favaro E., Bensaad K., Chong M.G., Tennant D.A., Ferguson D.J.P., Snell C., Steers G., Turley H., Li J.-L., Günther U.L. Glucose utilization via glycogen phosphorylase sustains proliferation and prevents premature senescence in cancer cells. Cell Metab. 2012;16:751–764. doi: 10.1016/j.cmet.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 75.Pescador N., Villar D., Cifuentes D., Garcia-Rocha M., Ortiz-Barahona A., Vazquez S., Ordoñez A., Cuevas Y., Saez-Morales D., Garcia-Bermejo M.L. Hypoxia promotes glycogen accumulation through hypoxia inducible factor (HIF)-mediated induction of glycogen synthase 1. PLoS ONE. 2010;5:e9644. doi: 10.1371/journal.pone.0009644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mendez F.L., Watkins J.C., Hammer M.F. A haplotype at STAT2 Introgressed from neanderthals and serves as a candidate of positive selection in Papua New Guinea. Am. J. Hum. Genet. 2012;91:265–274. doi: 10.1016/j.ajhg.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Racimo F., Sankararaman S., Nielsen R., Huerta-Sánchez E. Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 2015;16:359–371. doi: 10.1038/nrg3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bild D.E., Bluemke D.A., Burke G.L., Detrano R., Diez Roux A.V., Folsom A.R., Greenland P., Jacob D.R., Jr., Kronmal R., Liu K. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am. J. Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 79.Woolcott O.O., Ader M., Bergman R.N. Glucose homeostasis during short-term and prolonged exposure to high altitudes. Endocr. Rev. 2015;36:149–173. doi: 10.1210/er.2014-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Woolcott O.O., Castillo O.A., Gutierrez C., Elashoff R.M., Stefanovski D., Bergman R.N. Inverse association between diabetes and altitude: a cross-sectional study in the adult population of the United States. Obesity (Silver Spring) 2014;22:2080–2090. doi: 10.1002/oby.20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santos J.L., Pérez-Bravo F., Carrasco E., Calvillán M., Albala C. Low prevalence of type 2 diabetes despite a high average body mass index in the Aymara natives from Chile. Nutrition. 2001;17:305–309. doi: 10.1016/s0899-9007(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 82.Okumiya K., Sakamoto R., Ishimoto Y., Kimura Y., Fukutomi E., Ishikawa M., Suwa K., Imai H., Chen W., Kato E. Glucose intolerance associated with hypoxia in people living at high altitudes in the Tibetan highland. BMJ Open. 2016;6:e009728. doi: 10.1136/bmjopen-2015-009728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stec J.J., Silbershatz H., Tofler G.H., Matheney T.H., Sutherland P., Lipinska I., Massaro J.M., Wilson P.F.W., Muller J.E., D’Agostino R.B., Sr. Association of fibrinogen with cardiovascular risk factors and cardiovascular disease in the Framingham Offspring Population. Circulation. 2000;102:1634–1638. doi: 10.1161/01.cir.102.14.1634. [DOI] [PubMed] [Google Scholar]
- 84.Lowe G.D., Fowkes F.G., Dawes J., Donnan P.T., Lennie S.E., Housley E. Blood viscosity, fibrinogen, and activation of coagulation and leukocytes in peripheral arterial disease and the normal population in the Edinburgh Artery Study. Circulation. 1993;87:1915–1920. doi: 10.1161/01.cir.87.6.1915. [DOI] [PubMed] [Google Scholar]
- 85.Matsuda T., Murakami M. Relationship between fibrinogen and blood viscosity. Thromb. Res. 1976;8(2 suppl):25–33. doi: 10.1016/0049-3848(76)90044-x. [DOI] [PubMed] [Google Scholar]
- 86.Nienaber-Rousseau C., de Lange Z., Pieters M. Homocysteine influences blood clot properties alone and in combination with total fibrinogen but not with fibrinogen γ′ in Africans. Blood Coagul. Fibrinolysis. 2015;26:389–395. doi: 10.1097/MBC.0000000000000256. [DOI] [PubMed] [Google Scholar]
- 87.Appiah D., Heckbert S.R., Cushman M., Psaty B.M., Folsom A.R. Lack of association of plasma gamma prime (γ′) fibrinogen with incident cardiovascular disease. Thromb. Res. 2016;143:50–52. doi: 10.1016/j.thromres.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Erzurum S.C., Ghosh S., Janocha A.J., Xu W., Bauer S., Bryan N.S., Tejero J., Hemann C., Hille R., Stuehr D.J. Higher blood flow and circulating NO products offset high-altitude hypoxia among Tibetans. Proc. Natl. Acad. Sci. USA. 2007;104:17593–17598. doi: 10.1073/pnas.0707462104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maul H., Longo M., Saade G.R., Garfield R.E. Nitric oxide and its role during pregnancy: from ovulation to delivery. Curr. Pharm. Des. 2003;9:359–380. doi: 10.2174/1381612033391784. [DOI] [PubMed] [Google Scholar]
- 90.Wang Y., Chang C.F., Morales M., Chiang Y.H., Hoffer J. Protective effects of glial cell line-derived neurotrophic factor in ischemic brain injury. Ann. N Y Acad. Sci. 2002;962:423–437. doi: 10.1111/j.1749-6632.2002.tb04086.x. [DOI] [PubMed] [Google Scholar]
- 91.Moncada S., Higgs A. The L-arginine-nitric oxide pathway. N. Engl. J. Med. 1993;329:2002–2012. doi: 10.1056/NEJM199312303292706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The SNPs with the highest per-SNP PBSn1 value within the top 100 peaks in the genome were submitted for annotation via the CADD online portal at http://cadd.gs.washington.edu/. The 25 SNPs with the strongest evidence of functional consequences are bolded. Additional information on the annotation pipeline and output can be found on the website.
We tested a representative SNP for our top genes. Each SNP was interrogated for effects on expression in the GTeX panel of tissues via the online portal at http://gtexportal.org/home/.
Same as Table S3 but interrogating known downstream genes.