Abstract
Objective(s):
Autophagy-related 4B (ATG4B) plays an important role in the process of autophagy induction. However, the molecular events that govern the expression of ATG4B in this process are not well known.
Materials and Methods:
Human ATG4B 3’-UTR region (1377 nt) containing miR-34a/miR-34c-5p binding site was amplified by PCR. Luciferase assay was used to assess the activity of reporter genes. Real-time PCR was used to detect the levels of miR-34a and miR-34c-5p. Western blot was used toanalyze the protein levels of ATG4B, LC3 and p62.
Results:
Both miR-34a and miR-34c-5p could directly target the 3’-UTR of ATG4B mRNA at same site. Overexpression of either miR-34a or miR-34c-5p significantly down-regulated ATG4B at both mRNA and protein levels and this effect can be reversed by ATG4B overexpression. Moreover, Rapamycin-induced autophagy is accompanied with the upregulation of ATG4B and the downregulation of miR-34a/miR-34c-5p. Ectopic expression of either miR-34a or miR-34c-5p markedly suppressed rapamycin-triggered autophagy.
Conclusion:
In the present study, we found that miR34/ATG4B signaling axis involves in rapamycin-triggered autophagy. This study may provide a new insight for understanding the mechanisms of ATG4B regulation and autophagy induction.
Keywords: Autophagy, ATG4B, miR-34a, miR-34c-5pRapamycin
Introduction
Autophagy is a tightly regulated process by which cells consume unwanted cytoplasmic macromolecular constituents and recycle them for cellular remodeling (1, 2). In this process, proteins and organelles were packaged in a double-membrane structure, named as autophagosome. Finally, autophagosome fuses with lysosome to degrade its content (2).
The molecular mechanism underlying autophagy has been extensively researched during the past years, as well as the genes participating in this process, named as ATGs, which has been found to be conserved among the different species (3). ATG4B, an autophagy-related protein, plays an important role in the formation of autophagosomes. A major event in autophagosome formation is the proteolytic cleavage of cyto-plasmic microtubule associated protein 1 light chain 3 (LC3) to generate LC3-I, which is then exposed a critical internal glycine residue that enables the conjugation to phosphatidylethanolamine, generating membrane-bound LC3-II. Proteolytic cleavage is also requiredtosubsequentlydelipidate LC3-II to LC3-I (4, 5). Thesekeystepsarecatalyzedbythecysteine protease ATG4B and its paralogs, which are necessary for membrane closure and recycling of LC3 from the autophagosomal membrane. To date, more and more researches demonstrate that elevated ATG4Bcorresponds to a rise in autophagy (6-8). However, little is known about the molecular events that govern ATG4B expression in the process of autophagy induction.
MicroRNAs (miRNAs) are endogenous 19-22 ntnon-coding RNAs, which are important post- transcriptional regulators on gene expression via specific binding to 3’-UTR of mRNA, causing inhibition of translation and/or mRNA degradation (9-12). Previous studies have shown that miR-34a could target ATG4B directly and depress autophagy (13, 14). Nevertheless, whether there are other miRNAs targeting to ATG4B is unclear.
In this work, we confirmed that not only miR-34a but also miR-34c-5p, a member of miR-34 family, could directly target ATG4B and cause thedegrada-tionof ATG4B mRNA, thereby suppresses rapamycin-induced autophagy.
Materials and Methods
Reagents
miR-34a/miR-34c-5p mimics and control miRNAs (NC oligo), miR-34a/miR-34c-5p inhibitor and control inhibitor(NC inhibitor), were synthesized by Sangon Biotech (Shanghai, China).
Cell culture and treatment
Human cancer cell lines HeLa and SKOV3 were obtained from the American Type Culture Collection (ATCC) and cultured in high-glucose DMEM (Gibco), which contained 10% fetal bovine serum (Hyclon), under a 5% CO2 atmosphere. HeLa or SKOV3 cells were seeded in 6-well plates, grown to 70% to 80% confluence. After miRNAs mimics transfection for 24hr, cells were exposed to rapamycin(0.5μM) for another 6hr or 12 hr.
Prediction of the microRNAs
Three common online bioinformatic tools (TargetScan, miRanda and PicTar) were used to predict the miRNAs that target the 3’-UTR of ATG4B mRNA. Among several predicted miRNAs, miR34a and miR34c-5p were discovered by all 3 online tools.
Plasmid construction
The full-length human ATG4B 3’-UTR region (1377 nt) containing miR-34a/miR-34c-5p binding site was amplifiedbyPCRusingHeLacell-derivedcDNA as template. Then the fragments were cloned into pMIR-REPORT vector at Mlu I and Hind III sites downstream of the luciferase gene (the structure of plasmid was shown in Suppl. Figure 1A), and the resulting plasmid was named as pMIR-ATG4B. Similarly, a cDNAfragment from ATG4B open reading frame without 3’- UTR was amplified by PCR with the cDNA of HeLa as template and cloned into the pCMV5 vector at EcoR I and Kpn I sites downstream of the CMV promotor (the structure of plasmid was shown in Suppl. Figure 1B) and the resulting plasmid was named as pCMV5-ATG4B. As for the mutant plasmid containing the 3’-UTR without the seed sequence of miR-34a/miR-34c-5p binding site was constructed by using overlap extension PCR with the cDNA of HeLa as template and cloned into the pMIR-REPORT vector at the same sites. The resulted plasmid was named as pMIR-ATG4B mu.
Figure 1.
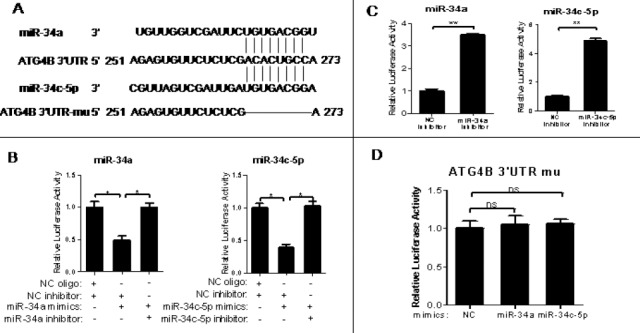
3′-Untranslational region of ATG4B mRNA contains a functional target site for miR-34a and miR-34c-5p
(A) A putative binding site of miR-34a/miR-34c-5p in the 3′-UTR of human ATG4B mRNA. The potential functional target sites have also been deleted artificially
(B-D) The wild-type ATG4B 3′-UTR reporter plasmid was transiently co-transfected with RNA oligos (NC oligo and NC inhibitor, NC inhibitor and miR-34a/miR-34c-5p mimics, miR-34a/miR-34c-5p mimics and inhibitor of itself) (B), or with miR-34a/miR-34c-5p inhibitor or inhibitor NC(C) into HeLa cells for 24hr. The mutated-type ATG4B 3’-UTR reporter plasmid was also transiently co-transfected with equal amount of miR-34a/miR-34c-5p mimics into HeLa cells (D) for 24hr. Then the luciferase assay was conducted. The data were the firefly luciferase activities normalized with the Renilla luciferase activity. The transfection experiments were performed at least 3 times in triplicate, and the data were represented as fold induction over the NC control
Transient transfection
HeLa or SKOV3 cells were seeded in 6 or 48-well plates and cultured for 16 hr. When the cells grown to 70%-80% confluence, the luciferase reporters (pMIR/ATG4B 3’UTR, pMIR/ATG4B 3’UTR mu), expression plasmids (pCMV5, pCMV5-ATG4B) and RNA oligos (NC, miR-34a/miR34c-5p mimics, miR-34a/miR-34c-5p inhibitor) were transiently co-transfected into the cells with lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions.
Luciferase assay
HeLa cells were seeded in 48-well plates and grown to 70–80% confluence. Then the corresponding reporter plasmid (0.4 μg/well) and miR-34a/miR-34c-5p mimics, miR-34a/miR-34c-5p inhibitor or NC oligos (10pmol/well) were co-transfected into the cells for 24 hr. Then, the cells were lysed and the firefly and Renilla luciferase activities were measured by Dual-Luciferase Reporter System (Promega) according to the manufacturer’s instructions. Firefly luciferase activity was normalized against the Renilla luciferase activity. The transfection experiments were performed at least 3 times in triplicate. The data were represented as fold induction over the NC control.
Western blot
Protein extracts from HeLa or SKOV3 cells were separated by 10% or 12% sodium dodecyl sulfate polyacrylamidegelelectrophoresis(SDS-PAGE)and were electrophoretically transferred to apolyvinyl- idenedifluoride (PVDF) membrane (Bio-RadCorpora-tion). Then the membrane was incubated with primary antibodies against LC3 (L7543, Sigma), ATG4B (A2981, Affinity) or p62 (sc-514790, Santa Cruz), followed by horseradish peroxidase (HRP)-labeled secondary antibodies. Tublin was used as a protein loading control.
ATG4B mRNA level assay by quantitative PCR (qPCR)
Total RNA was isolated with TRIzol (Invitrogen) and reverse-transcribed into cDNA with M-MLV Reverse Transcriptase (TAKARA) following the manufacturer’s instructions. The qPCR was performed using SYBR Green Supermix (TAKARA) on the ABI 7500 Real-Time PCR Detection System, taking β-actin mRNA as internal control. The relative level of ATG4B expression was determined with the 2-ΔΔCT method.
Endogenous miR-34a/miR-34c-5p expression assay by quantitative PCR (qPCR)
miRcutemiRNA Isolation Kit (TIANGENBiotechno-logy, Beijing, China) was used to isolate snmRNA in the treated cells. Then the reverse transcription reaction and qPCR analysis were performed as described (9).
Statistical analysis
The data were expressed as mean±SD. T-test was used to analyze the variance, using GraphPad Prism software (GraphPad Software Inc). P<0.05 was defined as statistically significant.
Results
3’-UTR of ATG4B mRNA contains a functional binding site for miR-34a and miR-34c-5p
Firstly, we used three common online bioinformatic tools (TargetScan, miRanda and PicTar) to predict the microRNAs (miRNAs) that target 3’-UTR of ATG4B mRNA. The result showed that miR-34a and miR-34c-5p shared a same binding site in the 3’-UTR of human ATG4B mRNA (Figure 1A). Secondly, we constructed two luciferase reporter plasmids separately containing the wild-type and mutant 3’-UTR of ATG4B, named as pMIR-ATG4B and pMIR-ATG4B-mu, respectively (Figure 1A). Luciferase reporter assay showed that both the miR-34a and miR-34c-5p mimics could cause an over 50% reduction of luciferase activity in the cells transfected with pMIR-ATG4B (but not pMIR-ATG4B-mu) (Figure 1B and D). Meanwhile, the inhibitors of miR-34a and miR-34c-5p could increase the luciferase activity of pMIR-ATG4B through repressing endogenous miR-34a and miR-34c-5p (Figure 1C) and reversed miR-34a/ miR-34c-5p-induced luciferase activity reduction in the cells transfected with pMIR-ATG4B (Figure 1B). These results indicate that both miR-34a and miR-34c-5p can target the 3’-UTR of ATG4B mRNA.
miR-34a and miR-34c-5p downregulate the expression of ATG4B
To further validate the effect of miR-34a and miR-34c-5p on the expression of ATG4B, we assessed the mRNA level and protein level of ATG4B after miR-34a/miR-34c-5p transfection. As shown in Figure 2A and 2B, miR-34a/miR-34c-5p dramatically repressed the expression of ATG4B at both mRNA and protein levels in HeLa and SKOV3 cells, which were markedly attenuated by the co-transfection of miR-34a/miR-34c-5p inhibitors. Moreover, co-transfection of pCMV5-ATG4B (without 3’-UTR of ATG4B) could also reverse miR-34a/miR-34c-5p-induced expression downregulation of ATG4B both at mRNA level and protein level (Figure 2C and D). These results demonstrated that miR-34a/miR-34c-5p canspecifically downregulate the expression of ATG4B in cells.
Figure 2.
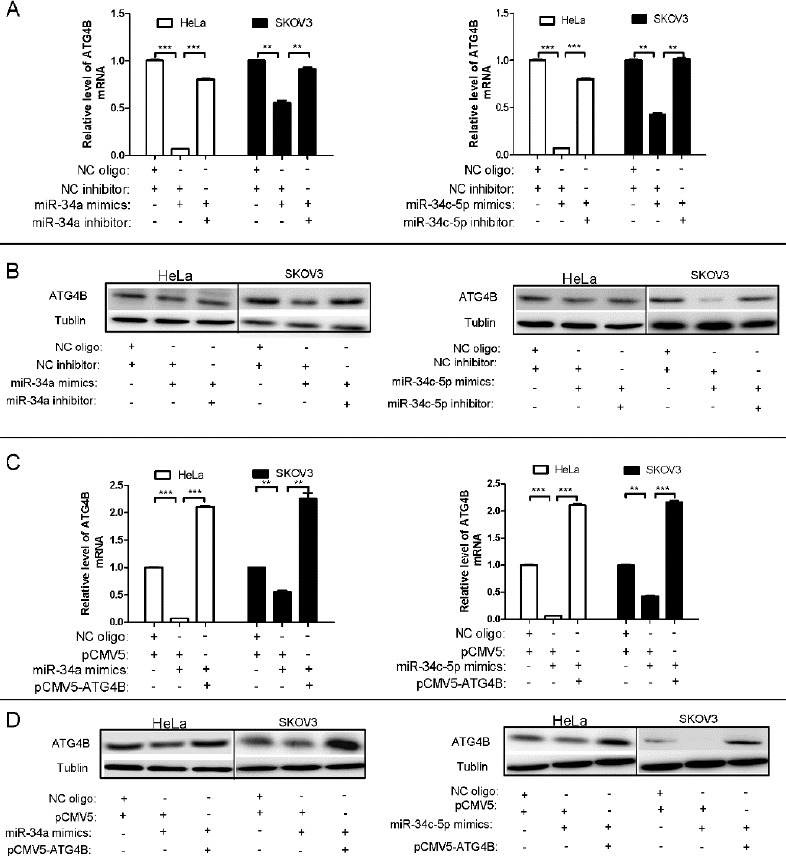
miR-34a and miR-34c-5p downregulate the expression of ATG4B
(A, C) qPCR analysis for ATG4B mRNA levels in HeLa and SKOV3 cells after transfection with equal amount of indicated plasmids and/or RNA oligos for 24 hr. ATG4B mRNA level was normalized with β-actin mRNA, and the data were represented as fold induction over the NC control
(B, D)Western blot analysis for ATG4B protein level in HeLa and SKOV3 cells after transfection with equal amount of indicated plasmids and/or RNA oligos for 24hr
Rapamycin-induced autophagy is accompanied with the upregulation of ATG4B and the downregulation of miR-34a/miR-34c-5p
To investigate the role of the miR-34a/miR-34c-5p/ATG4B signaling pathway in autophagy induction, we measured the levels of ATG4B and mature miR-34a/miR-34c-5p in rapamycin-treated cells. Rapamy-cin treatment obviously enhanced the accumulation of LC3-II and the degradation of p62, which were well-used autophagic marker (Figure 3A). Meanwhile, rapamycin also significantly up-regulated theexpre-ssion of ATG4B at both mRNA and protein levels (Figure 3A and B) while down-regulated the levels of mature miR-34a and miR-34c-5p in HeLa and SKOV3 cells (Figure 3C). The results suggested that the upregulation of ATG4B and the downregulation of miR-34a/miR-34c-5p may play an important role in rapamycin-induced autophagy.
Figure 3.
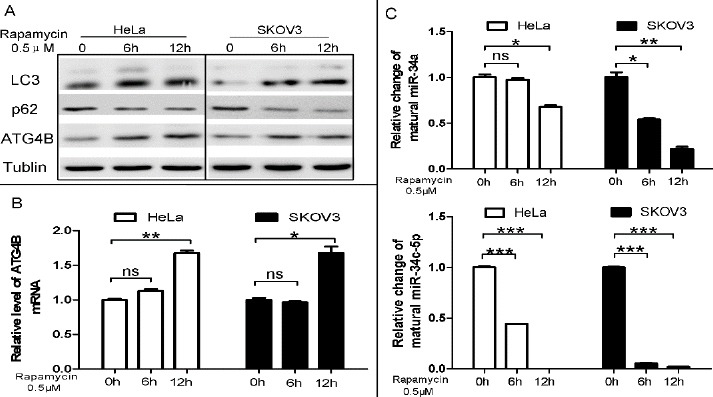
Rapamycin-induced autophagy is accompanied with the upregulation of ATG4B and the downregulation of miR-34a/miR-34c-5p
(A) HeLa and SKOV3 cells were treated with 0.5μMrapamycin for 6h or 12hr. Total protein was extracted and Western blot was used to detect the protein level of LC3 62 and ATG4B, taking tubulin as a loading control
(B, C) qPCR analysis for ATG4B mRNA and miR-34a/miR-34c-5p expression level. The relative level was determined with the 2-ΔΔCT method, and the data were represented as fold induction over the NC control
Overexpression of either miR-34a or miR-34c-5p suppresses rapamycin-triggered autophagy
To further determine the role of the miR-34a/miR-34c-5p/ATG4B signaling pathway in rapamycin-induced autophagy, we overexpressed miR-34a/miR-34c-5p in presence or absence of the inhibitors in rapamycin-treated cells. As shown in Figure 4A, rapamycin-induced LC3-II accumulation and p62 degradatonwere reduced by miR-34a/miR-34c-5p overexpression, which can be reversed by miR-34a/miR-34c-5p inhibitor co-transfection. Moreover, ectopic expression of ATG4B could also rescue miR-34a/miR-34c-5p-induced autophagy reduction in rapamycin-treated cells (Figure 4B). These results demonstrate that miR-34a/miR-34c-5p-induced downregulation of ATG4B can suppress rapamycin-triggered autophagy.
Figure 4.
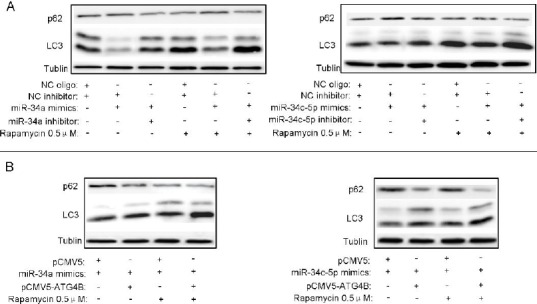
Overexpression of either miR-34a or miR-34c-5p represses rapamycin-triggered autophagy
(A, B) HeLa and SKOV3 cells were seeded in 6-well plates and transfected with indicated plasmids and/or RNA oligos. After 12hr, the cells were treated with 0.5μMrapamycin for additional 12hr. Then the protein levels of p62 and LC3 were detected by Western blot, taking Tubulin as a loading control
Discussion
Autophagy is a conserved homeostatic process that degrades organelles and proteins (15, 16). Moderate autophagy will help cells survive extreme environment, denoted protective autophagy (17, 18). However, excessive autophagy will lead autophagic cell death (19). Under stress, the modulation of autophagy is very important for cells (20). ATG4B, a member of ATG4 family, is necessary for the formation of autophagosome in mammalian cells (21). Therefore, the regulation of ATG4B is a key point to modulate autophagy in cells. Previous studies showed that C/EBPβ promotes autophagy through transactivation of ATG4B (22), while RNF5 restrains autophagy by ubiquitinationof ATG4B (23). In the present study, we find out two miRNAs, miR-34a and miR-34c-5p, targeting ATG4B and therefore regulating rapamycin-induced autophagy.
Previous studies have shown that miR-34a could bind with the 3’-UTR of ATG4B mRNA directly (13, 14). In our study, we confirmed that not only miR-34a, but also miR-34c-5p, could target the 3’-UTR of ATG4B mRNA in the same binding site. Our data showed that the levels of miR-34a/miR-34c-5p were decreased in rapamycin-treated cells, accompanied with the upregulation of ATG4B and autophagy induction. Overexpression of miR-34a/miR-34c-5p could suppress rapamycin-induced ATG4B upregulation and autophagy. These results indicate that the miR-34a/ miR-34c-5p/ATG4B axis plays a key role in rapamycin-induced autophagy.
The underlying mechanisms by which rapamycin down-regulates the expression of miR-34a/miR-34c-5p is not clear. Investigation has demonstrated that methylation-induced silencing of miR-34a throughAMPK/ mTOR pathway could upregulate ATG4B directly in prostate cancer (24). Though miR-34a and miR-34c-5p both belong to the family of miR-34, the gene locations are totally different. In the human genome, miR-34a is encoded on chromosome1, while miR-34c-5p is encoded on chromosome 11 (25). So in rapamycin-treated cells, whether miR-34c-5p is also affected by methylation is still unknown. In addition, rapamycin may downregulatemiR-34a/miR-34c-5p through regulating its promoter activity, but it will take a long time to confirm this hypothesis because it is complicated to identify the promoter of miR-34a/miR-34c-5p (26). On the other way, rapamycinmay also downregulate miR-34a/miR-34c-5pthroughregulating the level of some long non-coding RNAs (lncRNAs). LncRNAs can sponge miRNAs andsubse-quentregulate their function or monitor their expression (27). Bioinformatics tools such as starBase v2.0 predict several lncRNAs that may target miR-34a/miR-34c-5p, such as NUTM2A-AS1 (data not shown). But whether these lncRNAs play an important role in the rapamycin-induced downregulation of miR-34a/miR-34c-5p requires further investigation.
Conclusion
In summary, our study confirmed that, not only miR-34a, but also miR-34c-5p can target the 3’-UTR of ATG4B mRNA and monitor the expression of ATG4B and autophagy induction. These findings provide a new insight for understanding the mechanisms of autophagy induction.
Acknowledgment
Grant support: This work received the support of the National Natural Science Foundation of China (81572375 and 81472436) and the Chongqing Natural Science Foundation (cstc2014jcyjA10110).
Conflict of interest
The authors declared that they have no conflicts of interest.
References
- 1.Behrends C, Sowa ME, Gygi SP. Harper JW: Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Till A, Subramani S. A balancing act for autophagin. J Clin Invest. 2010;120:2273–2276. doi: 10.1172/JCI43238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–3678. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 4.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 5.Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem. 2003;278:51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- 6.Akin D, Wang SK, Habibzadegah-Tari P, Law B, Ostrov D, Li M, et al. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy. 2014;10:2021–2035. doi: 10.4161/auto.32229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabrera S, Fernandez AF, Marino G, Aguirre A, Suarez MF, Espanol Y, et al. ATG4B/autophagin-1 regulates intestinal homeostasis and protects mice from experimental colitis. Autophagy. 2013;9:1188–1200. doi: 10.4161/auto.24797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Luo Q, Yuan L, Miao C, Mu X, Xiao W, et al. JNK-dependent Atg4 upregulation mediates asperphenamate derivative BBP-induced autophagy in MCF-7 cells. Toxicol Appl Pharmacol. 2012;263:21–31. doi: 10.1016/j.taap.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 9.Zhou P, Huang G, Zhao Y, Zhong D, Xu Z, Zeng Y, et al. MicroRNA-363-mediated downregulation of S1PR1 suppresses the proliferation of hepatocellular carcinoma cells. Cell Signal. 2014;26:1347–1354. doi: 10.1016/j.cellsig.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 10.Shukla GC, Singh J, Barik S. MicroRNAs processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong D, Huang G, Zhang Y, Zeng Y, Xu Z, Zhao Y, et al. MicroRNA-1 and microRNA-206 suppress LXRalpha-induced lipogenesis in hepatocytes. Cell Signal. 2013;25:1429–1437. doi: 10.1016/j.cellsig.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Liu XJ, Hong Q, Wang Z, Yu YY, Zou X, Xu LH. MicroRNA-34a suppresses autophagy in tubular epithelial cells in acute kidney injury. Am J Nephrol. 2015;42:168–175. doi: 10.1159/000439185. [DOI] [PubMed] [Google Scholar]
- 14.Rothe K, Lin H, Lin KB, Leung A, Wang HM, Malekesmaeili M, et al. The core autophagy protein ATG4B is a potential biomarker and therapeutic target in CML stem/progenitor cells. Blood. 2014;123:3622–3634. doi: 10.1182/blood-2013-07-516807. [DOI] [PubMed] [Google Scholar]
- 15.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun WL, Chen J, Wang YP, Zheng H. Autophagy protects breast cancer cells from epirubicin-induced apoptosis and facilitates epirubicin-resistance development. Autophagy. 2011;7:1035–1044. doi: 10.4161/auto.7.9.16521. [DOI] [PubMed] [Google Scholar]
- 18.Li K, Chen X, Liu C, Gu P, Li Z, Wu S, et al. Pirarubicin induces an autophagic cytoprotective response through suppression of the mammalian target of rapamycin signaling pathway in human bladder cancer cells. Biochem Biophys Res Commun. 2015;460:380–385. doi: 10.1016/j.bbrc.2015.03.042. [DOI] [PubMed] [Google Scholar]
- 19.Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell. 2011;42:23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Grander D, Panaretakis T. Autophagy: cancer therapy's friend or foe?Future Med Chem. 2010;2:285–297. doi: 10.4155/fmc.09.155. [DOI] [PubMed] [Google Scholar]
- 21.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 22.Guo L, Huang JX, Liu Y, Li X, Zhou SR, Qian SW, et al. Transactivation of Atg4b by C/EBPbeta promotes autophagy to facilitate adipogenesis. Mol Cell Biol. 2013;33:3180–3190. doi: 10.1128/MCB.00193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuang E, Okumura CY, Sheffy-Levin S, Varsano T, Shu VC, Qi J, et al. Regulation of ATG4B stability by RNF5 limits basal levels of autophagy and influences susceptibility to bacterial infection. PLoS Genet. 2012;8:e1003007. doi: 10.1371/journal.pgen.1003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao H, Xiao Y, Hu Y, Xiao Y, Yin Z, Liu L, et al. Methylation-induced silencing of miR-34a enhances chemoresistance by directly upregulating ATG4B-induced autophagy through AMPK/mTOR pathway in prostate cancer. Oncol Rep. 2016;35:64–72. doi: 10.3892/or.2015.4331. [DOI] [PubMed] [Google Scholar]
- 25.Li XJ, Ren ZJ, Tang JH. MicroRNA-34a: a potential therapeutic target in human cancer. Cell Death Dis. 2014;5:e1327. doi: 10.1038/cddis.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song G, Wang L. Nuclear receptor SHP activates miR-206 expression via a cascade dual inhibitory mechanism. PLoS One. 2014;4:e6880. doi: 10.1371/journal.pone.0006880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]


