Abstract
This study aimed to develop a nomogram to predict the overall survival (OS) of stage IV breast cancer patients. We searched the National Cancer Database (NCDB) for stage IV breast cancer patients diagnosed between 2010 and 2013. Predictors of OS were identified and a nomogram was developed and validated using concordance index (C‐index), calibration plots, and risk group stratifications. A total of 7199 patients from the NCDB were included in the study. With a median follow‐up of 25.7 months, the 1‐year and 3‐year OS rates were 80.6% and 52.5%, respectively. Race, age, comorbidity status, T‐stage, grade, ER/PR/Her2 status, the presence of lung/liver/brain metastasis, surgery, radiotherapy, and chemotherapy were significantly associated with OS. The developed nomogram had a C‐index of 0.722 (95% CI 0.710–0.734) and 0.725 (95% CI 0.713–0.736) in the training and the validation cohorts, respectively. The predicted survival using the nomogram is well correlated with actual OS. The nomogram was able to stratify patients into different risk groups, among which the survival benefit of local therapy varied. We developed a nomogram to predict the overall survival of stage IV breast cancer patients. Prospectively designed studies with international collaborations are needed to further validate our nomogram.
Keywords: Breast cancer, NCDB, nomogram, overall survival, prediction, prognosis, stage IV
Introduction
Breast cancer is a heterogeneous disease with different subtypes, each having different prognoses. Accurately estimating the prognosis of each patient may not only benefit clinical decision‐making but also inform the individualized design of surgical follow‐up surveillance plans. For early‐stage breast cancer, several risk prediction models (RPM) have been developed and widely validated 1, including NPI 2, Adjuvant! 3, Oncotype Dx 4, and Mammaprint 5, etc. However, no RPM has been widely accepted for Stage IV breast cancer patients. The TBCRC 013 study 6 suggested that the 21‐Gene recurrence score has prognostic value in stage IV breast cancer patients. However, there were more clinicopathological features reported to be significantly associated with survival in these patients, such as progesterone receptor (PR) positivity 7, molecular subtype 8, tumor grade 8, and metastatic patterns 9. There are no prognostic models with all clinicopathological features for survival prediction of stage IV breast cancer patients. The National Cancer Database (NCDB) is hospital based, and the participating centers are required to submit data to the database, and the data covers approximately 70% of cancer patients in United States. In this study, we used the NCDB to investigate the prognostic factors of survival in stage IV breast cancer patients and developed a nomogram using these prognostic factors for survival predictions. The aim of this study was to develop a prognostic model that could be used for individualized risk assessment of stage IV breast cancer patients.
Method
We searched the NCDB database for eligible patients. Data including the site of metastasis (bone, brain, liver and lung) and ER/PR/Her2 status are only available after 2010. In this study, we only included patients with critical data available. The detailed inclusion and exclusion criteria are listed as follows:
-
Inclusion
Female breast cancer.
Diagnosed between 2010 and 2013.
American Joint Committee on Cancer (AJCC) stage IV.
Confirmed pathology.
No prior diagnosis of breast cancer.
-
Exclusion criteria
Follow‐up months equal to 0.
Phyllodes tumor.
Unknown bone, liver, lung or brain metastatic status.
Unknown ER, PR, or HER2 status
Unknown race, tumor grade, surgery, radiotherapy, and chemotherapy.
This is an epidemiological study using de‐identified data from the NCDB registries. Therefore, consent for patient participation and study publication is not required. The study approval was waived by the ethical committee of Sun Yat‐sen Memorial Hospital based on our institutional policy. This study was reported using the STARD statement guidelines 10.
The following data were collected for each patient: year of diagnosis, age, race, county, Charlson‐Deyo score, laterality, primary tumor site, tumor grade, T‐stage, histology, estrogen receptor (ER) status, progesterone receptor (PR) status, HER2 status, metastatic sites (bone/brain/lung/liver), primary surgery categorization, radiation therapy status, chemotherapy, survival month, and overall survival status. Charlson‐Deyo score is a weighted score derived from the sum of score for each of the comorbid conditions. Higher score indicates more comorbid conditions. (http://ncdbpuf.facs.org/) Patients were categorized into two age groups based on their age at diagnosis (≤60 years, >60 years), as the median age of the study population was close to 60. Radiation therapy (RT) was divided into two categories (with RT and without RT).
Statistical analysis
We assigned the eligible patients into the training and validation study cohort, respectively (Detailed in Data S1). We conducted a descriptive analysis of the baseline clinicopathological features of the included patients and used the Chi‐square test to compare the characteristics of patients between the training and validation cohort. The median follow‐up was calculated as the median observed survival time of the entire population. Overall survival (OS) was measured as the time from diagnosis to death due to any causes. The cumulative OS rates were estimated using Kaplan–Meier analysis.
We used the unadjusted Cox regression model as a univariate analysis to screen for prognostic factors of overall survival. Factors determined to be significant by the competing‐risk analysis were incorporated into the Cox proportional hazard regression as a multivariate analysis.
We used the Cox regression model and the “rms” package in R to develop an OS prediction nomogram with 1‐year and 3‐year OS as the endpoints. To evaluate the discriminative ability of the nomogram, we used the Harrell's concordance index (C‐index) 11 with a 95% CI. To assess the accuracy of the nomogram, we used calibration plots to visualize the agreement between the predicted and actual 1‐year and 3‐year OS. All P‐values were two‐sided. P‐values less than 0.05 were considered statistically significant. The statistical analysis was performed using Stata/MP, version 13.0 (StataCorp LP, College Station, TX, USA) and R.
Results
Baseline clinicopathological features
A total of 7199 patients were included from the NCDB database. The number of patients being excluded at each step during the patient selection was summarized in Figure 1.
Figure 1.
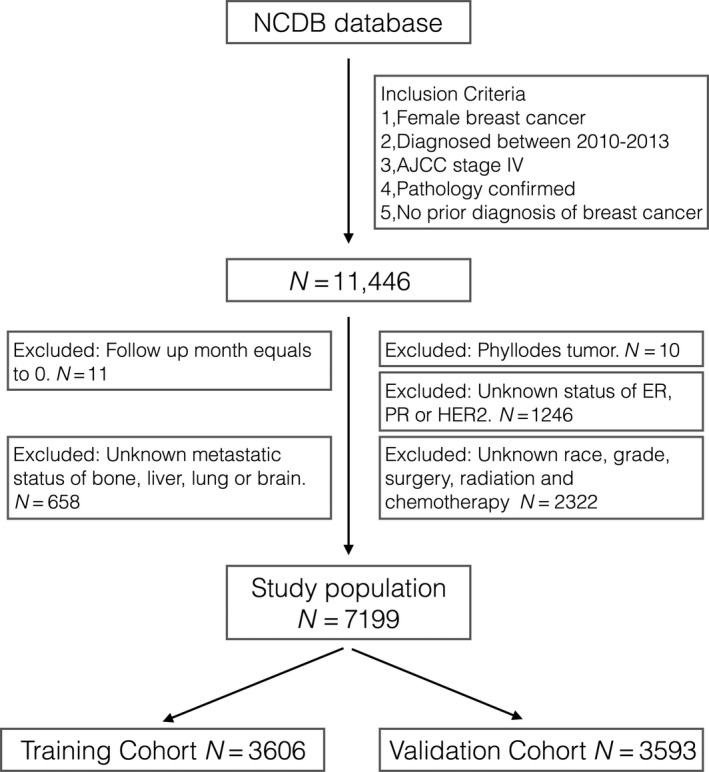
We applied the inclusion and exclusion criteria to NCDB database and enrolled 7199 patients as the study population.
The clinicopathological features of the patients are summarized in Table 1. The median age (25th–75th percentile) was 58 (49–67) years old. There were 62.7%, 24.8%, 6.0%, and 26.1% of the patients having bone, liver, brain, and lung metastasis, respectively. The median follow‐up was 25.7 months. The 1‐year and 3‐year OS rates were 80.6% and 52.5%, respectively. There were no significant differences between the training and validation cohort.
Table 1.
Clinicopathological features of the included patients
| Study population | |||||
|---|---|---|---|---|---|
| Training | Validation | P a | |||
| N | % | N | % | ||
| Year of diagnosis | |||||
| 2010 | 851 | 23.60 | 870 | 24.21 | NS |
| 2011 | 962 | 26.68 | 991 | 27.58 | |
| 2012 | 854 | 23.68 | 884 | 24.60 | |
| 2013 | 939 | 26.04 | 848 | 23.60 | |
| County type | |||||
| Metropolitan | 2991 | 82.95 | 2950 | 82.10 | NS |
| Nonmetropolitan/unknown | 615 | 17.05 | 643 | 17.90 | |
| Race | |||||
| White | 2839 | 78.73 | 2858 | 79.54 | NS |
| African American | 614 | 17.03 | 593 | 16.50 | |
| Others | 153 | 4.24 | 142 | 3.95 | |
| Age group | |||||
| <=60 | 2046 | 56.74 | 1987 | 55.30 | NS |
| >60 | 1560 | 43.26 | 1606 | 44.70 | |
| Laterality | |||||
| Left | 1798 | 49.86 | 1803 | 50.18 | NS |
| Right | 1778 | 49.31 | 1749 | 48.68 | |
| Othersb | 30 | 0.83 | 41 | 1.14 | |
| Primary site | |||||
| Nipple/central portion | 219 | 6.07 | 243 | 6.76 | NS |
| UIQ | 274 | 7.60 | 243 | 6.76 | |
| LIQ | 147 | 4.08 | 146 | 4.06 | |
| UOQ | 979 | 27.15 | 1001 | 27.86 | |
| LOQ | 243 | 6.74 | 217 | 6.04 | |
| Overlapping/unknown | 1744 | 48.36 | 1743 | 48.51 | |
| Charlson/Deyo score | |||||
| 0 | 2950 | 81.81 | 2923 | 81.35 | NS |
| 1 | 530 | 14.70 | 516 | 14.36 | |
| 2 | 126 | 3.49 | 154 | 4.29 | |
| Histology | |||||
| IDC | 2751 | 76.29 | 2715 | 75.56 | NS |
| ILC | 346 | 9.60 | 336 | 9.35 | |
| NOS/others | 509 | 14.12 | 542 | 15.08 | |
| T‐stage | |||||
| T0–T1 | 491 | 13.62 | 479 | 13.33 | NS |
| T2 | 856 | 23.74 | 875 | 24.35 | |
| T3 | 418 | 11.59 | 387 | 10.77 | |
| T4 | 522 | 14.48 | 528 | 14.70 | |
| Tx | 1319 | 36.58 | 1324 | 36.85 | |
| N‐stage | |||||
| N0 | 372 | 10.32 | 399 | 11.10 | NS |
| N1 | 712 | 19.74 | 714 | 19.87 | |
| N2 | 522 | 14.48 | 532 | 14.81 | |
| N3 | 514 | 14.25 | 459 | 12.77 | |
| Nx | 1486 | 41.21 | 1489 | 41.44 | |
| Grade | |||||
| I | 257 | 7.13 | 244 | 6.79 | NS |
| II | 1441 | 39.96 | 1459 | 40.61 | |
| III | 1908 | 52.91 | 1890 | 52.60 | |
| ER | |||||
| Negative | 972 | 26.96 | 942 | 26.22 | NS |
| Positive | 2634 | 73.04 | 2651 | 73.78 | |
| PR | |||||
| Negative | 1449 | 40.18 | 1384 | 38.52 | NS |
| Positive | 2157 | 59.82 | 2209 | 61.48 | |
| Her‐2 | |||||
| Negative | 2668 | 73.99 | 2663 | 74.12 | NS |
| Positive | 938 | 26.01 | 930 | 25.88 | |
| Bone metastasis | |||||
| No | 1351 | 37.47 | 1332 | 37.07 | NS |
| Yes | 2255 | 62.53 | 2261 | 62.93 | |
| Lung metastasis | |||||
| No | 2672 | 74.10 | 2651 | 73.78 | NS |
| Yes | 934 | 25.90 | 942 | 26.22 | |
| Liver metastasis | |||||
| No | 2701 | 74.90 | 2736 | 76.15 | NS |
| Yes | 905 | 25.10 | 857 | 23.85 | |
| Brain metastasis | |||||
| No | 3382 | 93.79 | 3388 | 94.29 | NS |
| Yes | 224 | 6.21 | 205 | 5.71 | |
| Breast surgery | |||||
| No_surgery | 1401 | 38.85 | 1427 | 39.72 | NS |
| Bcs | 616 | 17.08 | 620 | 17.26 | |
| Mastectomy | 1589 | 44.07 | 1546 | 43.03 | |
| Radiation therapy | |||||
| No | 2123 | 58.87 | 2132 | 59.34 | NS |
| Yes | 1483 | 41.13 | 1461 | 40.66 | |
| Chemotherapy | |||||
| None | 1360 | 37.71 | 1358 | 37.80 | NS |
| Single‐agent chemotherapy | 492 | 13.64 | 504 | 14.03 | |
| Multiagent chemotherapy | 1754 | 48.64 | 1731 | 48.18 | |
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; LIQ, lower‐inner quadrant; LOQ, lower‐outer quadrant; UIQ, upper‐inner quadrant; UOQ, upper‐outer quadrant; BCS, breast‐conserving surgery; IDC, infiltrating ductal carcinoma; ILC, infiltrating lobular carcinoma; NOS, non otherwise specific.
Chi‐square test.
Bilateral/side unspecified/unknown included.
Screen for prognostic factors for OS
Prognostic factors including year of diagnosis, tumor location (Quadrants), and tumor laterality are theoretically not associated with survival, and were excluded from this analysis. We used unadjusted Cox regression and observed that race (White vs. African American), age (≤60 years vs. >60 years), Charlson score (1 or 2 vs. 0), T‐stage, tumor grade(III vs. I), ER/PR/HER2 status, lung metastasis (Yes vs. No), liver metastasis (Yes vs. No), brain metastasis (Yes vs. No), breast surgery(BCS/Mastectomy vs. No), radiation therapy (Yes vs. No), and chemotherapy (Multiagent vs. None) were significantly associated with OS. The presence of bone metastasis, N‐stage (N2 vs. N0), and histology (IDC, ILC, Others) were not associated with OS (Table S1). We noticed that some subgroup of patients had similar OS, and therefore we combined them as one category in the multivariate analysis, including T0‐1 and T2, T4 and Tx, Grade I and II, no chemotherapy and single‐agent chemotherapy, BCS and mastectomy. In the multivariate analysis, all of these factors were significantly correlated with OS (Table 2).
Table 2.
Multivariate Cox regression
| Features | HR (95% CI) | P |
|---|---|---|
| Race | ||
| White | 1 | |
| African American | 1.14 (1.01–1.29) | 0.032 |
| Others/unknown | 0.59 (0.44–0.78) | <0.001 |
| Age group | ||
| <=60 | 1 | |
| >60 | 1.22 (1.10–1.34) | <0.001 |
| Charlson/Deyo score | ||
| 0 | 1 | |
| 1 | 1.37 (1.21–1.55) | <0.001 |
| 2 | 2.04 (1.63–2.56) | <0.001 |
| T‐stage | ||
| T0–T2 | 1 | |
| T3 | 1.28 (1.08–1.51) | 0.004 |
| T4/Tx | 1.51 (1.32–1.72) | <0.001 |
| Grade | ||
| I–II | 1 | |
| III | 1.26 (1.13–1.40) | <0.001 |
| ER | ||
| Negative | 1 | |
| Positive | 0.64 (0.56–0.74) | <0.001 |
| PR | ||
| Negative | 1 | |
| Positive | 0.61 (0.54–0.70) | <0.001 |
| HER‐2 | ||
| Negative | 1 | |
| Positive | 0.52 (0.46–0.59) | <0.001 |
| Lung metastasis | ||
| No | 1 | |
| Yes | 1.34 (1.20–1.49) | <0.001 |
| Liver metastasis | ||
| No | 1 | |
| Yes | 1.54 (1.38–1.72) | <0.001 |
| Brain metastasis | ||
| No | 1 | |
| Yes | 1.62 (1.36–1.94) | <0.001 |
| Breast surgery | ||
| No_surgery | 1 | |
| Surgery | 0.74 (0.65–0.84) | <0.001 |
| Radiation therapy | ||
| No | 1 | |
| Yes | 0.87 (0.78–0.96) | 0.007 |
| Chemotherapy | ||
| None/single‐agent chemotherapy | 1 | |
| Multiagent chemotherapy | 0.72 (0.65–0.80) | <0.001 |
ER, estrogen receptor; PR, progesterone receptor; HER‐2, human epidermal growth factor 2.
Nomogram development and validation
A nomogram was developed using the training cohort (Fig. 2). Each prognostic factor used to create the nomogram was given a score. By adding up these scores, we can calculate a total score. Then we can draw a straight line down from the total point scale to estimate the 1‐year and 3‐year OS. The C‐index of the nomogram were 0.722 (95% CI 0.710–0.734) and 0.725 (95% CI 0.713–0.736) in the training and validation cohort, respectively. The C‐index of the nomogram is higher than ER status, PR status, HER2 status, and the presence of liver/lung/brain metastases (Table S2). The calibration plots suggested that the accuracy of the predicted 1‐year and 3‐year OS using the nomogram is excellent in both the training and validation cohorts (Fig. 3).
Figure 2.
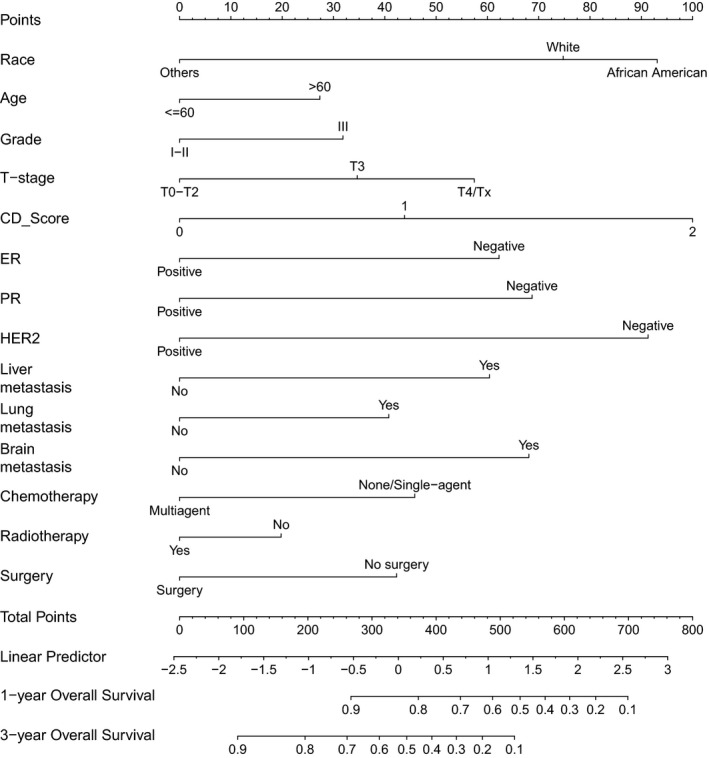
Nomogram to predict the 1‐year and 3‐year overall survival. For each patient, we calculated the points of the corresponding clinicopathological features, and summed up the points to obtain the total points. The predicted 1‐year and 3‐year OS can be estimated based on the total points of each patient.
Figure 3.
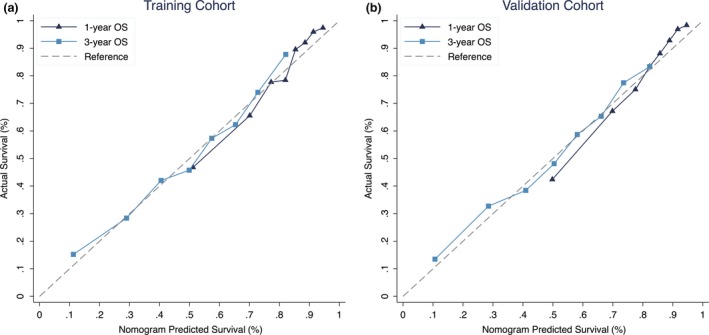
Calibration plots suggested that the predicted 1‐year and 3‐year OS were in agreement with the actual OS in the (A) training and (B) validation cohort.
Risk stratifications using the new nomogram
The distribution of the predicted 1‐year and 3‐year OS was shown in Figure S1. We assigned the patients into different subgroups based on the quartile of the predicted 3‐year OS. Using our model to stratify patients with only bone metastases and ER+/HER2‐ diseases into four different groups allowed for significant distinctions between the KM curves (Figure S2). The first, second, third, and fourth quartile subgroups had 98.2%, 93.5%, 80.1%, and 68.0% 1‐year OS, and 82.2%, 63.3%, 49.6%, and 25.4% 3‐year OS, respectively. Similarly, in patients with visceral metastases (lung, liver, or brain) and ER‐/PR‐ disease (N = 5 patients in the first quartile group and were excluded), three subgroups of patients also had significant distinctions among the KM curves (Figure S2). The second, third, and fourth quartile subgroups had 98.1%, 84.0%, and 52.1% 1‐year OS, and 71.1%, 44.6%, and 19.0% 3‐year OS, respectively.
Exploratory analysis: estimation of the survival benefit of local surgery in stage IV breast cancer patients
With the nomogram, we can estimate the survival benefit of surgery for each patient, by calculating the difference of the predicted OS when the patient was considered as received and did not receive surgery. The median (25–75th percentile) benefit of surgery of 1‐yr and 3‐yr OS was 0.8% (0.3–1.6%) and 1.6% (0.7–3.0%), respectively.
Discussion
There is an increasing interest in stage IV breast cancer patients, as the proportion of these patients seems to be higher in recent years, when compared with the past. Systemic use of advanced imaging methods, such as PET‐CT scanning, should be the underlying reasons, and this phenomenon was called as stage migration, that is some patients who previously would have been classified as early‐stage, would be reclassified to late‐stage, due to the advanced imaging examinations. The clinical decision‐making for these patients was highly dependent on the predicted survival. However, different metastatic patterns at initial diagnosis (bone, brain, liver lung, etc.) combined with different molecular disease subtypes (HR+/HER2‐, HR+/HER2+, ER‐/HER2+, ER‐/HER2‐) may lead to varied overall survival in these patients 7, 8, 9. Accurately predicting the survival of these patients is of paramount importance. Many RPMs are widely used in early‐stage breast cancer 1; however, there are only a few RPMs that have been reported in advanced/metastatic breast cancer. Lee et al. 12 developed a nomogram to predict the survival time in women with advanced breast cancer using data from clinical trials conducted by the Australia & New Zealand Breast Cancer Trials Group. Prognostic factors used to create that nomogram included ECOG status, ER status, neutrophil levels, age, number of metastatic sites, hemoglobin levels, and alkaline phosphatase levels. Lee et al. 13 used multicenter data from hospitals in South Korea and developed a PMOS system that utilized stage, HR status, Ki67 index, distant metastasis‐free interval, symptoms, and number of metastatic sites to predict the survival of patients with metastatic breast cancer. Both of these studies focused on predicting survival of patients with metastatic breast cancer who present with distant metastatic events a period of time after the treatment for the primary breast tumor. These patients are different from de novo stage IV breast cancer patients, as they are reported to have more favorable outcomes 14. In this study, we used the NCDB database to retrieve data from all stage IV breast cancer patients, developed a nomogram predicting the 1‐year and 3‐year OS rates for these patients.
The value of this nomogram can be seen in the stratification analysis. As per the recommendations from current guidelines 15, hormone receptor‐positive advanced breast cancer patients without visceral crisis should receive endocrine therapy as a first‐line therapy. However, our study showed that from these patients the new nomogram was able to identify a high‐risk subgroup who might need more intensive therapy (e.g., chemotherapy) first. In patients with only bone metastasis and ER+/HER2‐ diseases, the 3‐year OS rates were 82.2% and 25.4% in the first and fourth quartile subgroups, respectively. Therefore, this new model can identify high‐risk patients who were considered to have a favorable prognosis based on the current standards. Similarly, in patients with visceral metastases and ER‐/PR‐ diseases, the new nomogram can also identify low‐risk patients, with 1‐year and 3‐year OS of 98.1% and 71.1%, respectively. Prognosis stratifications using our new model would be informative and helpful for clinical decision‐making. It could inform the risks and benefits of certain treatment plans, aid in designing an appropriate surveillance plan, and provide psychological/sentimental support.
In this study, we noticed that the predicted benefits of local surgery on OS in stage IV breast cancer patients were very low (<2%). This is consistent with the Tata trial 16, in which stage IV breast cancer patients were randomized to locoregional treatment versus no locoregional treatment group with 2‐year OS of 41.9% versus 43.0%, respectively. Similarly, the MF07‐01 trial 17 also revealed that at 54 months, the survival rate was 35% and 31% in the surgery and no surgery group, respectively. In addition, they reported that the benefit of surgery was more significant in patients with bone metastasis only. In contrast, we did not observe any association between the benefit of surgery with any known clinicopathological features. Including treatment variables as predictors may lead to bias, such as confounding by indications. However, we suggested that in real world, whether a treatment was implemented or not may also have prognostic role as well. Some patients, even if suitable for surgery or chemotherapy, may refuse the treatments due to some reasons such as insurance coverage or religion belief. These patients may possibly have inferior survival. Thus, we suggested that treatment variables should be included as predictors.
Limitations
There were several limitations in this study. The first major limitation stemmed from the lack of information concerning the use of endocrine therapy and anti‐HER2 therapy. Different treatment regimens and patient responses to these therapies 7 are possibly strong predictors for OS. Furthermore, effect modifications may exist between these therapies and the ER/PR/Her2 status or metastatic patterns. Therefore, future studies incorporating these predictors may improve our nomogram. Furthermore, lack of information about the metastatic tumor (ER/PR/HER2 status) was also one of the limitations of the nomogram. Second, cancer registry data may be miscoded, which could bring significant bias to our analysis 18. However, the large sample sizes and well represented patient groups offsets many of the disadvantages of these databases. In our study, we cannot distinguish the de novo stage IV breast cancer patients from those who progressed to stage IV after adjuvant therapies. This is a major limitation. Several studies have showed that these two subsets of patients had different survival, and for the latter ones, more variables (disease free interval, adjuvant therapies, etc.) could be incorporated in our nomogram to enhance the performance. More studies are needed. Third, we randomly separate the population into two cohorts (training and validation cohort). We still need another population from different country to externally validate this nomogram. In addition, we need to be aware that the use of our nomogram in populations from randomized clinical trials will be the gold standard of its validation, and observational data are likely to provide misleading estimates of treatment effects 19.
Summary
In this study, we developed a novel nomogram predicting the 1‐year and 3‐year OS of stage IV breast cancer patients using national cancer database. The new nomogram can stratify patients into different risk subgroups. A prospective, internationally collaborative study is needed to further validate the new nomogram.
Conflict of Interest
The authors have no conflict of interest to disclose.
Supporting information
Figure S1. We used the kernel density plot to illustrate the distribution of the predicted (A) 1‐year OS and (B) 3‐year OS of our study population.
Figure S2. Patients were categorized into four subgroups, based on quartile of their predicted OS. (A) In patients with ER+/HER2‐ and bone metastasis only, the four subgroups of patients had significantly diverged KM curves. (B) In patients with visceral metastasis, ER‐ and PR‐negative diseases, only five were assigned in the first quartile subgroup (Highest predicted OS) and they were excluded for analysis. The remaining subgroups also had significantly diverged KM curves.
Data S1. Determine the training and validation cohort.
Table S1. Univariate Cox regression analysis.
Table S2. C‐index of the nomogram and clinicopathological features.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant# 81402201, Grant# 81372817), National Natural Science Foundation of Guangdong Province (Grant# 2014A030310070), and Grant [2013] 163 from the Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology.
The NCDB is a joint project of the Commission on Cancer of the American College of Surgeons and the American Cancer Society. The data used in the study are derived from a de‐identified NCDB file. The American College of Surgeons and the Commission on Cancer have not verified and are not responsible for the analytic or statistical methodology employed, or the conclusions drawn from these data by the investigator.
Cancer Medicine 2017; 6(11):2586–2594
Contributor Information
Liling Zhu, Email: liling.zhu@yale.edu, Email: zhuliling@mail.sysu.edu.cn.
Kai Chen, Email: chenkai23@mail.sysu.edu.cn.
References
- 1. Engelhardt, E. G. , Garvelink M. M., de Haes J. H., van der Hoeven J. J., Smets E. M., Pieterse A. H., et al. 2014. Predicting and communicating the risk of recurrence and death in women with early‐stage breast cancer: a systematic review of risk prediction models. J Clin Oncol. 32:238–250. [DOI] [PubMed] [Google Scholar]
- 2. Haybittle, J. L. , Blamey R. W., Elston C. W., Johnson J., Doyle P. J., Campbell F. C., et al. 1982. A prognostic index in primary breast cancer. Br J Cancer. 45:361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ravdin, P. M. , Siminoff L. A., Davis G. J., Mercer M. B., Hewlett J., Gerson N., et al. 2001. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J Clin Oncol. 19:980–991. [DOI] [PubMed] [Google Scholar]
- 4. Paik, S. , Shak S., Tang G., Kim C., Baker J., Cronin M., et al. 2004. A multigene assay to predict recurrence of tamoxifen‐treated, node‐negative breast cancer. N Engl J Med. 351:2817–2826. [DOI] [PubMed] [Google Scholar]
- 5. van ‘t Veer, L. J. , Dai H., van de Vijver M. J., He Y. D., Hart A. A., Mao M., et al. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 415:530–536. [DOI] [PubMed] [Google Scholar]
- 6. King, T. A. , Lyman J. P., Gonen M., Voci A., De Brot M., Boafo C., et al. 2016. Prognostic Impact of 21‐Gene Recurrence Score in Patients With Stage IV Breast Cancer: TBCRC 013. J Clin Oncol. 34:2359–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kawano, A. , Shimizu C., Hashimoto K., Kinoshita T., Tsuda H., Fujii H., et al. 2013. Prognostic factors for stage IV hormone receptor‐positive primary metastatic breast cancer. Breast Cancer. 20:145–151. [DOI] [PubMed] [Google Scholar]
- 8. Bertaut, A. , Mounier M., Desmoulins I., Guiu S., Beltjens F., Darut‐Jouve A., et al. 2015. Stage IV breast cancer: a population‐based study about prognostic factors according to HER2 and HR status. Eur J Cancer Care (Engl). 24:920–928. [DOI] [PubMed] [Google Scholar]
- 9. Leone, B. A. , Vallejo C. T., Romero A. O., Machiavelli M. R., Perez J. E., Leone J., et al. 2017. Prognostic impact of metastatic pattern in stage IV breast cancer at initial diagnosis. Breast Cancer Res Treat. 161:537–548. [DOI] [PubMed] [Google Scholar]
- 10. Bossuyt, P. M. , Reitsma J. B., Bruns D. E., Gatsonis C. A., Glasziou P. P., Irwig L., et al. 2015. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Clin Chem. 61:1446–1452. [DOI] [PubMed] [Google Scholar]
- 11. Harrell F. E. Jr. 2001. Regression modeling strategies: with applications to linear models, logistic regression, and survival analysis (Springer series in statistics). Springer, New York, NY. [Google Scholar]
- 12. Lee, C. K. , Hudson M., Stockler M., Coates A. S., Ackland S., Gebski V., et al. 2011. A nomogram to predict survival time in women starting first‐line chemotherapy for advanced breast cancer. Breast Cancer Res Treat. 129:467–476. [DOI] [PubMed] [Google Scholar]
- 13. Lee, E. S. , Jung S. Y., Kim J. Y., Kim J. J., Yoo T. K., Kim Y. G., et al. 2016. Identifying the potential long‐term survivors among breast cancer patients with distant metastasis. Ann Oncol. 27:828–833. [DOI] [PubMed] [Google Scholar]
- 14. Lobbezoo, D. J. , van Kampen R. J., Voogd A. C., Dercksen M. W., van den Berkmortel F., Smilde T. J., et al. 2015. Prognosis of metastatic breast cancer: are there differences between patients with de novo and recurrent metastatic breast cancer? Br J Cancer. 112:1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cardoso, F. , Costa A., Norton L., Senkus E., Aapro M., Andre F., et al. 2014. ESO‐ESMO 2nd international consensus guidelines for advanced breast cancer (ABC2)dagger. Ann Oncol. 25:1871–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Badwe, R. , Hawaldar R., Nair N., Kaushik R., Parmar V., Siddique S., et al. 2015. Locoregional treatment versus no treatment of the primary tumour in metastatic breast cancer: an open‐label randomised controlled trial. Lancet Oncol. 16:1380–1388. [DOI] [PubMed] [Google Scholar]
- 17. Atilla, S. , Vahit O., Serdar O., Hasan K., Mahmut M., Abdullah I.., et al. 2016. A randomized controlled trial evaluating resection of the primary breast tumor in women presenting with de novo stage IV breast cancer: Turkish Study (Protocol MF07‐01). J Clin Oncol. 34:(suppl; abstr 1005). [Google Scholar]
- 18. Jagsi, R. , Abrahamse P., Morrow M., Hawley S. T., Griggs J. J., Graff J. J., et al. 2010. Patterns and correlates of adjuvant radiotherapy receipt after lumpectomy and after mastectomy for breast cancer. J Clin Oncol. 28:2396–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Henson, K. E. , Jagsi R., Cutter D., McGale P., Taylor C., and Darby S. C.. 2016. Inferring the effects of cancer treatment: divergent results from early breast cancer trialists’ collaborative group meta‐analyses of randomized trials and observational data from SEER registries. J Clin Oncol. 34:803–809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. We used the kernel density plot to illustrate the distribution of the predicted (A) 1‐year OS and (B) 3‐year OS of our study population.
Figure S2. Patients were categorized into four subgroups, based on quartile of their predicted OS. (A) In patients with ER+/HER2‐ and bone metastasis only, the four subgroups of patients had significantly diverged KM curves. (B) In patients with visceral metastasis, ER‐ and PR‐negative diseases, only five were assigned in the first quartile subgroup (Highest predicted OS) and they were excluded for analysis. The remaining subgroups also had significantly diverged KM curves.
Data S1. Determine the training and validation cohort.
Table S1. Univariate Cox regression analysis.
Table S2. C‐index of the nomogram and clinicopathological features.


