Abstract
Microenvironment (or niche)-providing chemokines regulate many important biological functions of tissue-specific stem cells. However, to what extent chemokines influence human pluripotent stem cells (hPSCs) is not yet completely understood. In this study, we applied protein array to screen chemokines found within the cytokine pool in the culture supernatant of hPSCs. Our results showed that chemokines were the predominant supernatant components, and came from three sources: hPSCs, feeder cells, and culture media. Chemotaxis analysis of IL-8, SDF-1α, and IP-10 suggested that chemokines function as uniform chemoattractants to mediate in vitro migration of the hPSCs. Chemokines mediate both differentiated and undifferentiated states of hPSCs. However, balanced chemokine signaling tends to enhance their stemness in vitro. These results indicate that chemokines secreted from both stem cells and feeder cells are essential to mobilize hPSCs and maintain their stemness.
Introduction
Human pluripotent stem cells (hPSCs) are unique cell types that maintain self-renewal and pluripotency during early embryonic development1–3. Embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) are two known PSCs: the former are derived from the inner cell mass of early blastocysts, whereas the latter are dedifferentiated from mature somatic cells through genetic reprogramming2,4. Both ESCs and iPSCs have demonstrated great potential in regenerative medicine due to their unlimited capability to be differentiated into almost all types of tissues5,6. Specialized microenvironments (niches) are suggested to provide the multiple extracellular signals necessary to maintain the stemness of hPSCs7–10. Previous studies have indicated that niche-dependent extracellular matrices (ECMs) facilitate the self-renewal and pluripotency of PSCs11,12, with growth factors (such as basic fibroblast growth factor, bFGF, and epidermal growth factor, EGF) regulating their growth and survival13. Recent studies also suggest that niche-providing cytokines are possibly involved in the maintenance of PSCs14.
Chemokines are small secreted cytokines responsible for leukocyte trafficking during host defense immune response15,16. They are classified into CXC, CC, C, and CX3C subfamilies based on the conserved cysteine motif at the N-terminus of their polypeptide sequences17. Chemokine receptors are generally seven-transmembrane G protein-coupled receptors (GPCRs)18. Binding of chemokines to their receptors induces the G protein-mediated downstream signaling pathways and regulates cellular survival, proliferation, and migration19–22. Chemokines are key regulators of tissue-specific stem cells23,24. As the first identified niche-dependent chemokine, CXC chemokine SDF-1α (or CXCL12) regulates the homing and migration of hematopoietic progenitor cells (HPCs) in bone marrow25,26. CXCL12 receptor CXCR4 is expressed in HPCs27, with gene knockout of CXCR4 or blockage of CXCR4-specific antagonist AMD3100 able to inhibit the homing and migration of HPCs28,29. Our previous study found that the CXCL12-CXCR4 axis regulates the proliferation, migration, and survival of neural stem cells30. Similar roles of CXCL12-CXCR4 have been reported for many other multi-potential stem cells, such as primordial germ cells, endothelial progenitor cells, epithelial progenitor cells, neural stem cells, mesenchymal stem cells, liver oval stem cells, and cancer stem cells31–38. In addition, many other chemokines influence the biology of tissue-specific stem cells39. For example, CC chemokine MIP-1α (CCL3) inhibits the proliferation and mobilization of HPCs40,41. CXC chemokines IL-8 (CXCL8) and GRO (CXCL2) can enhance the mobilization of HPCs and human CNS stem cells42–46. In degenerative diseases, upregulated CC chemokines MCP-1 (CCL2), MIP-1α (CCL3), and RANTES (CCL5) are possibly involved in the migration of endothelial progenitor cells during tissue repair47–49, with MCP-1 and MIP-1α able to mobilize mesenchymal stem cells into ischemic brain lesions50,51. The above studies suggest that chemokines are niche-dependent signals that help maintain tissue-specific stem cells.
Recent research also suggests that chemokines maintain the stemness of hPSCs52,53. It has been reported that GROα potentially regulates the self-renewal of human ESCs in in vitro culture54. Furthermore, MCP-1 appears to cooperate with leukemia inhibitory factor to promote the stemness of mouse iPSCs through mediation of the Stat3-pathway55. SDF-1α has been reported to induce the migration of mouse ESCs and enhances their survival56. Human MCP-1 increases the expression of pluripotent marker genes through the phosphorylation of the signal transducer and activator of transcription 3 in iPSCs57. IL-8 and/or GROa also support the maintenance and proliferation of hPSCs54,58. In the current study, we performed high-throughput screening to identify three key chemokines (IL-8, IP-10, and SDF-1α) that regulate the mobilization and stemness of hPSCs.
Results
Successful establishment of new hESC cell line
We applied fertilized human eggs to generate new human embryonic stem cells (hESCs). At post-fertilization day 6, the eggs exhibited normal development of the intact inner cell mass (ICM), trophoblast cells, and pellucid zone in vitro (Fig. 1a). After pronase digestion and direct dissection, ICM cells were dissociated and cultured in Nunc 4-well plates supplied with KSR culture medium. Mitomycin-treated human foreskin fibroblasts (HFFs) were simultaneously provided as the feeder cells. We found that ICM cells could generate new clones, with cells from clones sustaining the capability to continuously form new clones for multiple generations (Fig. 1b). In addition, clone-derived cells, for example at the 10th passage (p10), demonstrated positive alkaline phosphatase activity (Fig. 1c). These observations indicate that the clone-derived cells were potential hESCs.
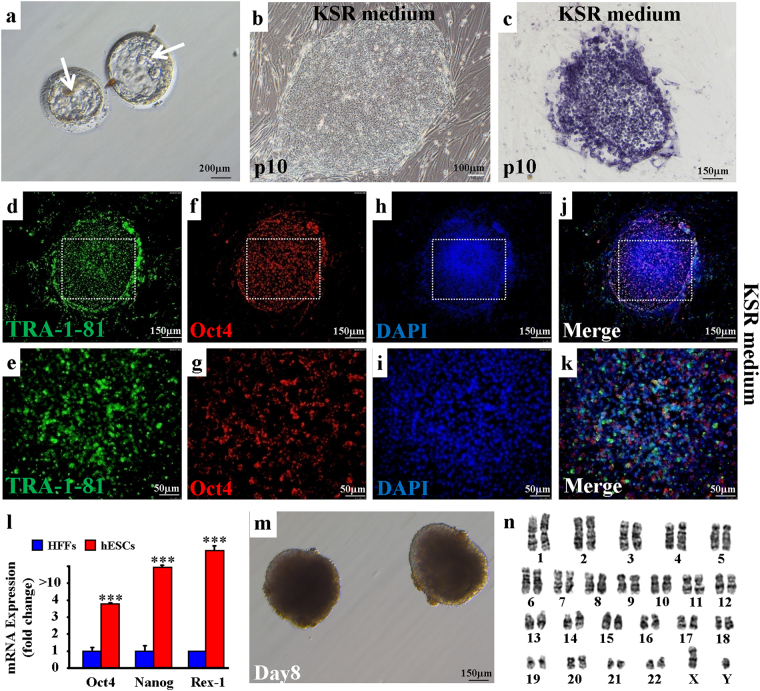
Figure 1.
Development and characterization of new hESCs. (a) Imaging of two human blastocysts at post-fertilization day 6 in vitro. Inner mass cells are indicated by arrows. Scale bar, 200 μm. (b) hESC-derived clones were obtained from inner mass cells after a 10-passage (p10) culture in the KSR medium. Scale bar, 100 μm. (c) Cells from (b) demonstrated positive staining of alkaline phosphatase. Scale bar, 150 μm. (d–k) Immunofluorescence of pluripotency markers TRA-1-81 and Oct4 expressed in p10 hESCs. TRA-1-81 (d–e); Oct4 (f,g); DAPI (h,i); and Merge (j,k). Scale bars, 150 μm (d,f,h,j) and 50 μm (e,g,i,k). (l) mRNA levels of pluripotency markers Oct4, Nanog, and Rex-1 in p10 hESCs. ***p < 0.001, n = 3 individual experiments. Error bars indicate sem. (m) hESCs could be differentiated into embryonic bodies (EBs). Scale bar, 150 μm. (n) hESCs show normal human male karyotype.
To verify their stemness, we examined the protein expressions of pluripotency markers Oct4, TRA-1-81, Nanog, and TRA-1-60 in the hESCs, and found them all to be highly expressed (Figs 1d–k, 2a,b and S1). We also observed that the mRNA levels of the pluripotency genes Oct4, Nanog, and Rex-1 in the hESCs were higher than those in the HFFs (Fig. 1l). The predominant expression of pluripotency protein markers in the hESCs support that these cells maintained their stemness when cultured in KSR medium. To determine their pluripotency, we also examined whether hESC clumps could be differentiated into three germ layers in the embryoid bodies (EBs) (Fig. 1m). Firstly, the hESCs were transplanted into NOD SCID mouse leg muscles for 48-d differentiation in vivo, with the normal three germ layer-tissue differentiations then detected in the formed teratomas by histopathological analyses (Fig. S2), indicating that the established hESC line possessed pluripotency. In addition, quantitative PCR demonstrated higher mRNA expression of the three germ layer markers AFP, GATA4, GATA6, FOXA2, Sox17 (endoderm), T, MIXL1, TBX1, c-actin (mesoderm), Nestin, Sox1, NeuroD1, and PAX6 (ectoderm) in differentiated EBs than in undifferentiated hESCs (Fig. S3a). Significantly higher expressions of the Oct4 and Nanog pluripotency genes were found in the hESCs than in differentiated EBs (Fig. S3b). Consistently, we observed high protein expression of the germ layer markers AFP and GATA4 (endoderm), desmin and actin (mesoderm), and nestin and βIII-tubulin (ectoderm) in the EBs (Fig. S4). These observations suggest that pluripotent hESCs could be differentiated into endoderm, mesoderm, and ectoderm in vitro. Finally, the hESCs were found to exhibit a normal male karyotype (Fig. 1n), supporting the successful establishment of a new hESC cell line.
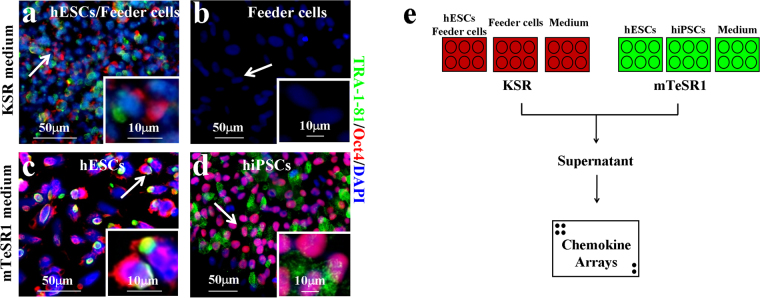
Figure 2.
Chemokine screening strategy for hPSCs. (a–d) Immunofluorescence of pluripotency markers TRA-1-81 and Oct4 on hPSCs. KSR medium (a,b); mTeSR1 medium (c,d); hESCs (a,c); hiPSCs (d); Feeder cells (b); Scale bars, 50 μm (a–d; arrows) and 10 μm (insets). Cells were counterstained with DAPI. (e) Chemokine screening strategy for hPSCs cultured in KSR (red) or mTeSR1 (green) media.
Maintenance of pluripotent stem cells (PSCs) in vitro
We next examined the stemness of the established hESCs cultured in mTeSR1 medium (Fig. S5a). Again, the hESCs demonstrated AP-positive staining, as observed in those cultured in the KSR medium (Fig. S5b). Immunofluorescence staining showed that hESCs were expressed by pluripotency markers Oct4, TRA-1-81, Nanog, and TRA-1-60 (Figs 2c, S5 c–j and S6). To compare established hESCs with other pluripotent stem cells, we cultured known human induced pluripotent stem cell line hNF C11 in mTeSR1 medium (Fig. S5k)59, and found that these cells were AP-positive (Fig. S5l) and expressed pluripotency markers Oct4, TRA-1-81, Nanog, and TRA-1-60 (Figs 2d, S5 m–t and S7). These observations further support that hPSCs, such as hESCs and hiPSCs, can be normally cultured under mTeSR1 conditions and sustain their stemness in vitro.
Identification of chemokines secreted from hPSCs and feeder cells
To screen chemokines that potentially regulate hPSC biology, we collected supernatants from the above hESC or hiPSC cultures for chemokine screening (Fig. 2e). In total, 38 chemokines had signals stronger than the background signals of the protein array membrane (Table 1 and Fig. S8). To quantify these signals, relative signal intensity (RSI) was defined by normalizing the signals with background intensities (see Methods and Materials). Under mTeSR1 conditions, 30 chemokines demonstrated obviously higher RSIs in the hESC-mTeSR1 culture compared with those in the mTeSR1 medium only (Fig. S9a). However, both the intensities and spectra of the chemokines secreted from hiPSCs exhibited decreasing expression in the hiPSC-mTeSR1 culture compared with those in the mTeSR1 medium only (Fig. S9b). These results suggest a predominant chemokine component from hPSCs themselves in the culture in vitro.
Table 1.
The relative signal intensities (RSIs) of chemokines.
| Number | Symbol | Name | Receptor | KSR medium | mTeSR1 medium | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| hESCs and Feeder cells | Feeder cells | Medium | hESCs | hiPSCs | Medium | Notes | ||||
| CC subfamily | ||||||||||
| 1 | CCL23 | CCL23 | CCR1 | — | — | 122.95 | 142.46 | — | — | G1; G2 |
| 2 | CCL28 | CCL28 | CCR3, CCR10 | — | — | — | 149.71 | 16.60 | — | F1; F2 |
| 3 | CTACK | CCL27 | CCR10 | 291.48 | 131 | 110.97 | 242.79 | 102.15 | 118.93 | H1; H2 |
| 4 | Eotaxin1 | CCL11 | CCR2, CCR3, CCR5 | 239.46 | 122.5 | 88.66 | 151.38 | 34.61 | 25.39 | K1; K2 |
| 5 | Eotaxin2 | CCL24 | CCR3 | 154.11 | 86 | 103.12 | 155.28 | 32.68 | — | L1; L2 |
| 6 | Eotaxin3 | CCL26 | CCR3 | — | 72.5 | 113.45 | — | 12.09 | 47.10 | A3; A4 |
| 7 | MCP1 | CCL2 | CCR2 | 535.33 | 289 | 130.39 | 321.38 | 237.88 | 29.56 | L3; L4 |
| 8 | MCP2 | CCL8 | CCR2B | 444.29 | 232.5 | — | — | 8.23 | 32.91 | A5; A6 |
| 9 | MCP3 | CCL7 | CCR2 | 766.17 | 309 | — | 122.40 | 6.30 | — | B5; B6 |
| 10 | MCP4 | CCL13 | CCR2, CCR3, CCR5 | 448.35 | 185 | — | 149.71 | — | — | C5; C6 |
| 11 | MDC | CCL22 | CCR4 | 199.63 | 92.5 | — | 224.40 | 15.31 | 30.40 | D5; D6 |
| 12 | MIP-1α | CCL3 | CCR1 | 276.03 | 82 | 87.84 | 241.12 | 17.88 | 38.75 | F5; F6 |
| 13 | MIP-1 β | CCL4 | CCR1, CCR5 | 289.85 | 183 | 96.51 | 241.68 | 41.04 | 73.83 | G5; G6 |
| 14 | MIP-1δ | CCL15 | CCR1, CCR3 | 121.60 | 48.5 | — | 115.15 | — | 23.72 | H5; H6 |
| 15 | MIP-3α | CCL20 | CCR6 | 403.65 | 49.5 | — | 265.09 | 10.81 | — | I5; I6 |
| 16 | MIP-3β | CCL19 | CCR7 | 178.50 | 69.5 | 125.43 | 197.09 | 23.67 | — | J5; J6 |
| 17 | MPIF1 | CCL23 | CCR1 | 111.03 | 61 | 168.40 | 184.27 | 25.60 | 25.39 | K5; K6 |
| 18 | HCC4 | CCL16 | CCR1, CCR2, CCR5, CCR8 | 267.09 | 132 | 114.28 | 394.95 | 16.60 | 29.56 | F3; F4 |
| 19 | I-309 | CCL1 | CCR8 | 119.97 | 78 | — | 160.86 | — | — | G3; G4 |
| 20 | PARC | CCL18 | CCR8, PITPNM3, GPR30 | — | — | — | — | 9.52 | 33.74 | A7.A8 |
| 21 | RANTES | CCL5 | CCR5 | 121.60 | 45 | — | — | 17.88 | 52.11 | B7; B8 |
| 22 | TARC | CCL17 | CCR4 | — | — | — | — | — | 66.31 | E7; E8 |
| 23 | TECK | CCL25 | CCR9 | — | — | — | — | 8.23 | — | F7; F8 |
| CXC subfamily | ||||||||||
| 24 | BLC | CXCL13 | CXCR5 | — | 49.5 | — | 266.20 | 15.95 | — | E1; E2 |
| 25 | CXCL16 | CXCL16 | CXCR6 | 215.07 | 90 | — | 201.55 | 21.74 | — | I1; I2 |
| 26 | ENA78 | CXCL5 | CXCR2 | 359.76 | 209 | — | 241.12 | 201.86 | 23.72 | J1; J2 |
| 27 | GCP2 | CXCL6 | CXCR1, CXCR2 | 740.16 | 392 | 102.71 | 432.85 | 151.04 | 62.14 | C3; C4 |
| 28 | GRO | CXCL2 | CXCR1, CXCR2 | 1041.71 | 427 | — | 614.56 | 265.54 | 32.07 | D3; D4 |
| 29 | GRO-α | CXCL1 | CXCR2 | 820.63 | 377 | — | 573.31 | 51.98 | 23.72 | E3; E4 |
| 30 | IL-8 | CXCL8 | CXCR1, CXCR2 | 773.48 | 403.5 | — | 484.13 | 17.24 | — | I3; I4 |
| 31 | IP-10 | CXCL10 | CXCR3 | 585.72 | 259 | 135.35 | 466.85 | 29.46 | 56.29 | J3; J4 |
| 32 | I-TAC | CXCL11 | CXCR3, CXCR7 | 255.71 | 155.5 | 115.10 | 401.64 | 25.60 | 42.09 | H3; H4 |
| 33 | MIG | CXCL9 | CXCR3 | 164.68 | 65.5 | — | 236.66 | 14.67 | 41.26 | E5; E6 |
| 34 | SDF-1α | CXCL12 | CXCR4 | — | — | — | — | — | — | C7; C8 |
| 35 | SDF-1β | CXCL12 | CXCR4 | — | — | — | — | 11.45 | 26.22 | D7; D8 |
| 36 | NAP2 | CXCL7 | — | 256.53 | 79 | 175.01 | 239.45 | 47.47 | 32.91 | L5; L6 |
| C subfamily | ||||||||||
| 37 | XCL1 | XCL1 | XCR1 | 341.87 | 213 | 107.67 | 379.90 | 88.64 | 60.47 | K3; K4 |
| CX3 C subfamily | ||||||||||
| 38 | Fractalkine | CX3CL1 | CX3CR1 | 177.68 | 115 | 145.68 | 200.99 | 27.53 | 46.27 | B3; B4 |
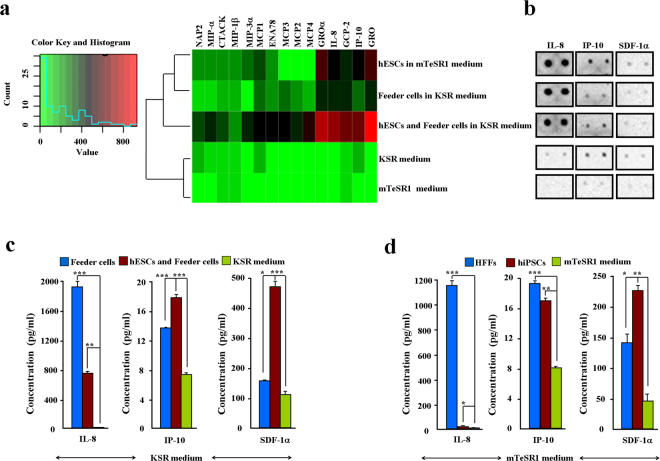
We next examined the chemokines secreted from feeder cells. Results showed that feeder cells secreted chemokines under KSR conditions (Fig. S9c). Co-culturing feeder cells with hESCs resulted in the secretion of more diverse and higher concentrations of chemokines than when feeder cells were cultured alone (Fig. S9d). These observations suggest that microenvironments represented by feeder cells significantly contributed to the chemokine components in the hESCs under KSR culture conditions. To verify this hypothesis, we compared the hierarchical clustering-secretion spectra of 15 chemokines between the two different hESC cultures (Fig. 3a). We found that hESCs and feeder cells co-cultured under KSR conditions secreted the maximal concentration of chemokines. Chemokines GRO, GROα, IL-8, GCP-2, and IP-10 were the most highly concentrated (Fig. 3a). These observations were confirmed by detecting representative chemokines IL-8, IP-10, and SDF-1α by ELISA (Fig. 3c,d). All three chemokine proteins were detected in the supernatants (Fig. 3c,d). Interestingly, results showed a relatively higher protein level of SDF-1α, though this protein was excluded in initial array screening (Fig. 3b–d). Our findings suggest that both chemokines secreted from hPSCs themselves and feeder cells are the main supernatant components in hPSC culture.
Figure 3.
Identification of chemokines secreted from hPSCs and feeder cells. (a) Heatmap of 15 identified chemokines. Color key indicates relative signal intensity (RSI) ranges from high (red) to low (green) values. (b) Duplicated signals of chemokines IL-8, IP-10, and SDF-1α on protein membranes. (c,d) Protein concentrations of chemokines IL-8, IP-10, and SDF-1α were examined by ELISA. KSR medium (c); mTeSR1 medium (d).
Migration of hPSCs functionally relies on chemokine signals
We further examined the mRNA levels of the CC and CXC subfamilies of chemokine receptors in the hPSCs. Quantitative PCR showed that chemokine receptors were expressed in the hPSCs and demonstrated variable mRNA levels (Fig. S10). It is worth noting that IL-8 receptors CXCR1/2 and SDF-1α receptor CXCR4 had higher mRNA expressions in the hESCs than in the feeder cells (Fig. S10a). Conversely, IP-10 receptor CXCR3 and SDF-1α receptor CXCR7 exhibited lower expressions in hESCs than in feeder cells. In addition, a considerable difference was found in mRNA levels between hESCs and hiPSCs, though both were cultured in mTeSR1 (Fig. S10b).
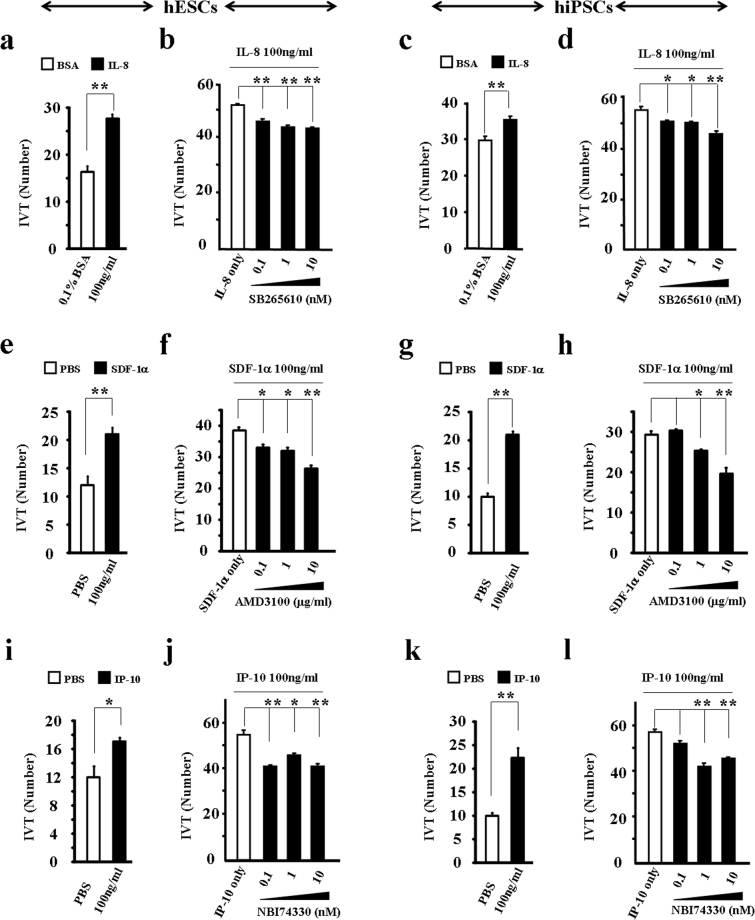
The broad expression of chemokine receptors in hPSCs indicates that chemokines might mediate their migration. To address this hypothesis, we tested the chemoattractant effects of IL-8, IP-10, and SDF-1α. We found that these three human recombinant proteins functioned as exogenous sources to induce the efficient transmigration of both hESCs and hiPSCs in vitro (Fig. 4). For instance, migrated hESCs and hiPSCs increased by 69.4% and 19.1%, respectively, after treatment with 100 ng/ml of IL-8 (Fig. 4a and c). However, the IL-8-induced transmigration of hESCs and hiPSCs significantly decreased after treatment with CXCR2-specific inhibitor SB265610 (Fig. 4b and d). Similar observations were obtained after examining the effects of SDF-1α, IP-10, and their receptor antagonists (Fig. 4e–l). Moreover, cell growth images and MTT assay demonstrated no toxicity or side effects of the antagonists on hESC survival (Fig. S11a–g). Similarly, these antagonists had no side effects on hiPSC survival (Fig. S11h–n). These results suggest that chemokine signals functionally mediate the migration of hPSCs.
Figure 4.
Chemokine signaling functionally mediates the transmigration of hPSCs. Chemoattractant effect of exogenous IP-10, IL-8, and SDF-1α on hPSCs. hESCs (a,b,e,f,i,j); hiPSCs (c,d,g,h,k,l); IL-8 (a,c) and CXCR2 antagonist SB265610 (b,d); SDF-1α (e,g) and CXCR4 antagonist AMD3100 (f,h); IP-10 (i,k) and CXCR3 antagonist NBI74330 (j,l). Values on graphs represent means ± sem, n = 3 individual experiments. *P < 0.05, **P < 0.01.
Maintenance of hPSCs depends on chemokine signaling
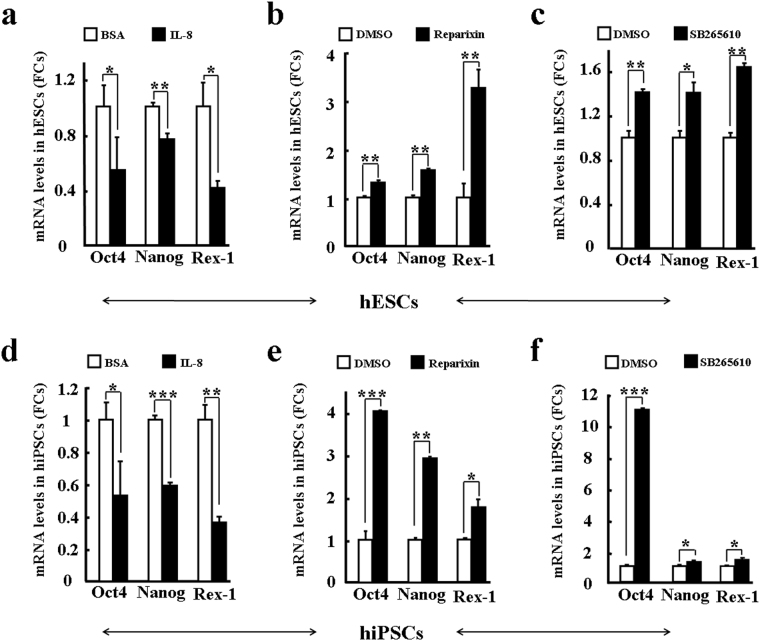
Previous studies suggest that chemokines are important signals for maintaining tissue-specific stem cells. We hypothesized that hESC-secreting or feeder cell-secreting chemokines have similar roles on hPSCs as on tissue-specific stem cells. To test this, quantitative PCR was used to examine the mRNA expressions of Oct4 and Nanog in hPSCs after treatment with chemokines or their receptor inhibitors. We found that exogenous IL-8 significantly decreased mRNA expressions of Oct4, Nanog, and Rex-1 in hESCs (Fig. 5a). However, blocking IL-8 receptors CXCR1/2 with antagonists reparixin and SB265610 increased the mRNA expressions of Oct4, Nanog, and Rex-1 in hESCs (Fig. 5b and c), with similar observations obtained for hiPSCs (Fig. 5d–f). Importantly, quantitative PCR of three differentiation genes (AFP, c-actin, and Sox1) demonstrated that exogenous IL-8 significantly increased the mRNA expressions of AFP, c-actin, and Sox1 in hPSCs (Fig. S12a,d), whereas blocking IL-8-CXCR1/2 signaling with reparixin/SB265610 decreased the mRNA expressions (Fig. S12b,c,e,f). These results strongly suggest that IL-8 and its signaling pathway might differentiate hPSCs.
Figure 5.
Chemokine IL-8 facilitates the differentiation of hPSCs. Stemness of hPSCs was evaluated by the expressions of three pluripotency genes (Oct4, Nanog, and Rex-1) after treatment with IL-8 and CXCR2 antagonists reparixin/SB265610. hESCs (a–c); hiPSCs (d–f); IL-8 (a,d); Reparixin (b,e); and SB265610 (c,f). Values on graphs represent means ± sem, n = 3 individual experiments. *P < 0.05, **P < 0.01, ***P < 0. 001.
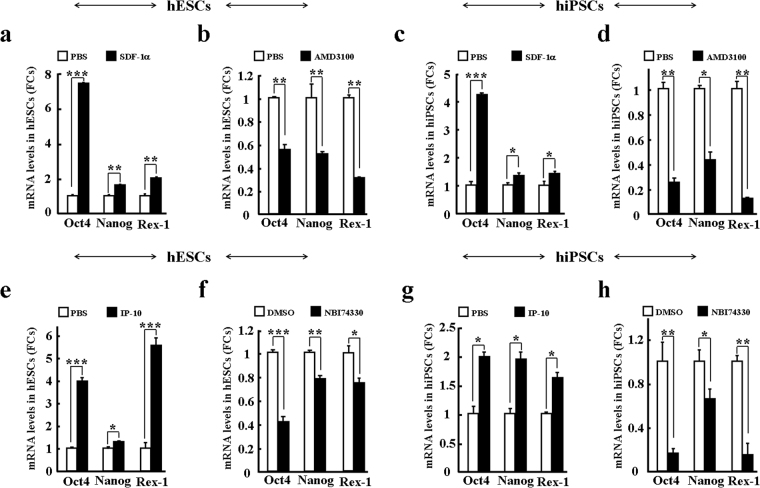
We also examined the effects of exogenous SDF-1α on the maintenance of hPSCs. Different from IL-8, we found that SDF-1α tended to enhance the mRNA expressions of Oct4, Nanog, and Rex-1 in hESCs and hiPSCs, respectively (Fig. 6a and c). These observations were confirmed by blocking the SDF-1α receptor CXCR4 with AMD3100, with mRNA levels of Oct4, Nanog, and Rex-1 significantly decreased in both hESCs and hiPSCs (Fig. 6b and d). Furthermore, exogenous SDF-1α significantly decreased the mRNA expressions of AFP, c-actin, and Sox1 in hPSCs (Fig. S13a,c), but blocking SDF-1α-CXCR4 signaling with AMD3100 increased their mRNA expressions (Fig. S13b,d). In addition, IP-10 was found to enhance the mRNA expressions of Oct4, Nanog, and Rex-1, but reduce the mRNA expressions of AFP, c-actin, and Sox1 in hPSCs (Figs 6e–h and S13e–h). The above findings strongly suggest that supernatant chemokines are essential for maintaining the stemness of hPSCs, with chemokines IL-8, IP-10, and SDF-1α possibly playing a critical role in regulating the maintenance of hPSCs.
Figure 6.
Chemokines SDF-1α and IP-10 enhance the stemness of hPSCs in vitro. Stemness of hPSCs was evaluated by the expressions of three pluripotency genes (Oct4, Nanog, and Rex-1) after treatment with agonists and antagonists. hESCs (a,b,e,f); hiPSCs (c,d,g,h); SDF-1α (a,c); CXCR4 antagonist AMD3100 (b,d); IP-10 (e,g); and CXCR3 antagonist NBI74330 (f,h). Values on graphs represent means ± sem, n = 3 individual experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Human pluripotent stem cells include both hESCs and hiPSCs, which are characterized by self-renewal and pluripotency reflecting their potential differentiation capability60. Under in vitro culture, these unique cells can be maintained long-term in either KSR or mTeSR1 media without loss of their stemness. In the present study, to prevent potential differentiation, inactivated HFFs were provided as feeder cells in KSR culture or were replaced by coating culture plates with 1% Matrigel in mTeSR1 culture61. So far, intracellular molecules involved in the long-term maintenance of hPSC stemness have been widely characterized62. Unfortunately, chemokines and their underlying signaling pathways have not yet been systematically studied, although these small secretion molecules represent the major cytokine portion in cell cultures. The large chemokine superfamily is generally responsible for cellular migration, proliferation, and survival18,21. Recently, increasing evidence suggests that chemokines are essential intracellular signals that help maintain the stemness of multipotent tissue-specific stem cells8,63. However, the extent of chemokines and their impact on hPSC stemness during long-term in vitro culture remain poorly understood. To explore the possible roles of chemokines related to hPSCs, we established a new hESC cell line by isolating and culturing ICM cells from fertilized human eggs. These hESCs not only expressed stem cell markers Oct4, Nanog, TRA-1-60, and TRA-1-81, but also showed potential pluripotency. For instance, hESC-derived EBs highly expressed the three germ layer markers through differentiation in vitro, with these hESCs able to form the normal germ layer tissues through transplantation into immunodeficient mice. Like hiPSCs, these hESCs could be maintained long-term in both KSR and mTeSR1 media. Furthermore, both hESCs and hiPSCs expressed pluripotency markers Oct4, TRA-1-81, Nanog, and TRA-1-60.
To determine how many chemokines were in the hPSC culture, we further applied protein arrays to screen the supernatant chemokine components of the hESCs and hiPSCs. In total, 38 chemokines were included in the array, consisting of 61% of CC, 35% of CXC, 2% of XC, and 2% of CX3C subfamilies, respectively (Table 1). Except for CXC chemokine SDF-1α, 37 chemokines showed detectable probing signals. However, the failed detection of SDF-1α might be due to its relatively low concentration since high protein concentrations of SDF-1α were detected by ELISA. Through high-throughput screening, 30 and 18 chemokines were identified from the hESCs and hiPSCs under the mTeSR1 conditions, respectively. Interestingly, 25 chemokines were directly detected from the mTeSR1 medium. However, chemokines secreted from both hiPSCs and mTeSR1 were lower than their counterparts secreted from hESCs. Under KSR culture conditions, indirect evidence indicated that 29 chemokines were secreted from hESCs. Similarly, both the feeder cells and KSR medium might provide extra chemokines. These results suggest that secretions from hPSCs and feeder cells equally contributed to the final chemokine cocktail (Fig. 7), with chemokines GROα, GRO, IL-8, GCP-2, and IP-10 demonstrating the highest supernatant concentrations in the hESC culture. In summary, our findings support the hypothesis that chemokines are important cytokines for hPSCs based on their secretion spectra and protein concentrations.
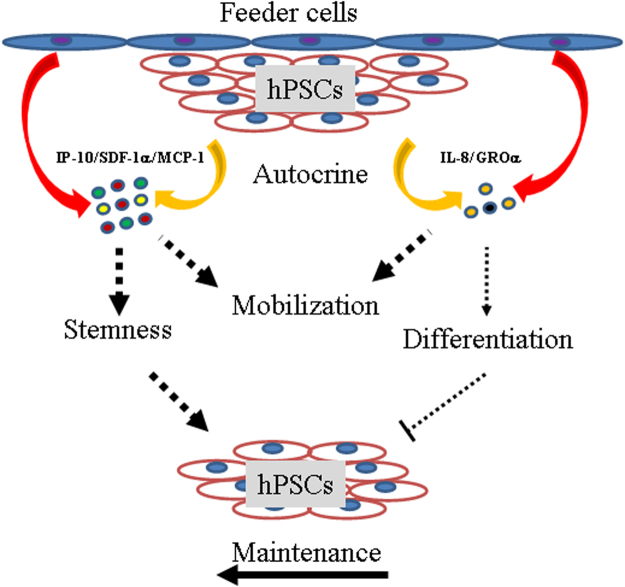
Figure 7.
Mobility and maintenance of hPSCs require multiple chemokine signals. Schematic representation of predominant chemokines from feeder cells or hPSCs themselves. 1) Released chemokines uniformly mediate the migration of hPSCs; 2) Both differentiating and maintaining chemokines were secreted in the culture supernatant. However, the final effect of chemokines tended to maintain the stemness of hPSCs in vitro.
Chemokines provide conserved chemoattractant functions for nearly all cell types. More importantly, chemokines mediate the migration of multipotent stem cells30,43,49,56,63. In the current study, we examined the chemotactic effect of three representative chemokines, that is, IL-8, SDF-1α, and IP-10. These three chemokines demonstrated very uniform induction of both hESC and hiPSC migration. Blocking their corresponding cognate receptors on hPSCs significantly reduced ligand-induced in vitro transmigration. This offers further evidence to support that chemokines and their receptors provide signaling to mediate the mobilization of hPSCs.
Microenvironment (niche)-dependent SDF-1α has been shown to enhance the maintenance of hematopoietic stem cells (HSCs) as well as other tissue-specific stem cells25. Recent studies also suggest that GROα and MCP-1 mediate the pluripotency of hPSCs53–55. For instance, GROα-CXCR2 chemotaxis has been reported to enhance the differentiation of hESCs to adopt a neuronal fate54. Conversely, it has also been suggested that IL-8 and GROα signaling through CXCR2 might support both pluripotency and proliferation of hPSCs without exogenous bFGF58. We observed that IL-8-CXCR1/2 decreased the pluripotency of hPSCs; for example, IL-8-CXCR1/2 decreased the expressions of pluripotency markers (Oct4, Nanog, and Rex-1), but increased the expressions of differentiation markers (AFP, c-actin, and Sox1). However, this signaling regulation was not necessary for the proliferation of hPSCs (Fig. S11). This evidence suggests that some chemokines, represented by IL-8 and GROα, participate in the differentiation of hPSCs. In addition, MCP-1 and CCR2 reportedly support pluripotency of both human and mouse iPSCs55,57. In the current study, we identified that SDF-1α-CXCR4 and IP-10-CXCR3 play similar roles in enhancing the pluripotency of hPSCs. Again, we did not observe an obvious effect of SDF-1α on the proliferation of hPSCs (Fig. S11). However, NBI 74330 (a CXCR3 antagonist) did inhibit the proliferation of hPSCs (Fig. S11g,n), though the potential regulatory mechanism involved in this process needs to be further studied. These findings suggest that other chemokines represented by MCP-1, SDF-1α, or IP-10 enhance the stemness of hPSCs. Therefore, we hypothesize that chemokines play two different roles to regulate the maintenance of hPSCs. The final maintenance of hPSCs depends not only on balanced chemokine secretion, but also on signaling shifts that tend to increase their pluripotency (Fig. 7).
Materials and Methods
Ethics statement
All experiments were carried out according to the relevant guidelines. All human sample studies and protocols were reviewed and approved by the Ethics Committee of the First People’s Hospital of Yunnan Province (AMEP: 2016009) and the Life Sciences Department of Yunnan University, China. Experiments were conducted following the exact regulations issued by both committees. For all human samples, legal informed consent was provided by all donors and patients.
Human specimens
Human foreskin tissue was provided by the Department of Urology from Kunming Children’s Hospital. Human blastocysts were obtained from the Department of Reproduction and Genetics of the First People’s Hospital of Yunnan Province. The final developmental stage of human blastocysts was strictly limited to post-fertilization day 6. Legal informed consent was provided by all donors and patients.
Establishment of HFFs and hESCs
Human foreskin tissue was digested by 0.25% trypsin and cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% defined fetal bovine serum (FBS) to obtain human foreskin fibroblasts (HFFs). To prepare feeder cells, HFFs were further inactivated by 10 μg/ml of mitomycin C (Sigma, USA). Human blastocysts were washed in PBS and then treated with 0.25% pronase (Sigma, USA) to remove zona pellucid. The inner cell masses (ICMs) were next dissected from the trophectoderm by immunosurgery with normal anti-human and anti-guinea pig whole serum (Jackson Immune Research, USA). Isolated ICMs were directly plated into Nunc 4-well plates with HFFs as the feeder cells and further cultured in modified KSR medium. The DMEM/F12 medium included 20% knockout serum replacement (KSR), 0.1 mM MEM non-essential amino acid, 0.1 mM 2-mercaptoethanol, and 8 ng/ml of basic fibroblast growth factor. All other reagents were purchased from Life Science Inc. (Gibco, USA). ESC-derived clones were continuously passaged for at least 10 generations to establish stable embryonic stem cell lines.
In vitro culture and treatment of hPSCs
The p27 hiPSC cell line hNF C11 was purchased from the Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences. Both newly-established hESCs and hiPSCs were seeded in 6-well plates pre-coated with 1% Matrigel (BD Biosciences) and cultured with the mTeSR1 complete kit (Stem Cell Technology, USA). Recombinant human chemokine proteins IL-8, SDF-1α, and IP-10 were purchased from R&D Systems (USA). CXCR1/CXCR2-specific antagonist reparixin and CXCR3-specific antagonist NBI74330 were bought from MCE (USA). CXCR4-specific antagonist AMD3100 and CXCR2-specific antagonist SB265610 were purchased from Sigma (USA). The hPSCs were treated by either 1 μg/ml of IL-8, 100 nM reparixin, 100 nM SB265610, 100 ng/ml of SDF-1α, 100 nM AMD3100, 50 ng/ml of IP-10, or 100 nM NBI74330 overnight.
Quantitative PCR
The mRNA expressions of chemokine cognate receptors, three germ layer markers, and pluripotency markers Oct4 and Nanog in hPSCs were examined by quantitative PCR (primers provided in Table S1) performed on an ABI 7300 sequence detection instrument (Applied Biosystems, USA) following the manufacturer’s instructions. Total RNA was extracted from human ESCs and iPSCs using total RNA isolation reagent (TaKaRa, Japan). The first-strand cDNA was synthesized using Oligo (dT) 18 primer (PrimeScript® 286 RT, TaKaRa) per the manufacturer’s protocols. Quantitative PCR was conducted following standard procedures using SYBR® 303 Premix Ex Taq (TaKaRa, Japan). Sequences of oligo primers are listed in Table S1. All quantitative PCR experiments were independently repeated three times.
Immunofluorescence staining
Both hESCs and hiPSCs were fixed in 4% phosphate-buffered paraformaldehyde (PFA). Immunofluorescence staining was carried out using standard procedures. Briefly, fixed hESCs and hiPSCs were stained by rabbit anti-human Oct4, rabbit anti-human Nanog, mouse anti-human TRA-1-81, and mouse anti-TRA-1-60 antibodies. Goat secondary antibodies anti-rabbit conjugated with Alex594, anti-mouse conjugated with Alex 488, and anti-rat conjugated with Alex 594 were chosen. TRA-1-81/TRA-1-60 primary antibodies and secondary antibodies were purchased from Millipore (USA). Primary antibodies AFP, GTATA4, desmin, actin, nestin, and β-III tubulin were purchased from Wanleibio (China).
Alkaline phosphatase staining
Alkaline phosphatase staining was conducted using an alkaline phosphatase assay kit (Beyotime Biotechnology, China) according to the standard procedures described by the manufacturer.
Embryoid body differentiation
Human ESC clumps (~500 cells) were collected and suspended in KSR medium without bFGF to form embryoid bodies (EBs). The culture medium was replaced with DFSR medium after 5 d of culture. The EBs were further differentiated in DFSR medium (KSR medium without bFGF, but supplemented with 10% FBS) on Matrigel for 10–15 d. In the EBs, mRNA expressions of germ layer markers AFP, GATA4, GATA6, Sox17, and FOXA2 (endoderm), T, MIXL1, TBX1, and c-actin (mesoderm), and Sox1, Nestin, NeuroD1, and PAX6 (ectoderm) were examined by quantitative PCR. In addition, germ layer protein markers AFP and GATA4 (endoderm), desmin and actin (mesoderm), and nestin and β-III tubulin (ectoderm) were evaluated by immunofluorescence staining.
Karyotyping
G-band karyotyping was conducted in the Genetics and Diagnosis Laboratory from the First People’s Hospital of Yunnan Province. Briefly, hESCs were treated with 0.6 mg/ml of colchicine (Gibco, USA) for 1.5 h. Next, hypotonic hESCs were prepared after treatment with 0.075 mol/L potassium chloride for 20 min in a 37 °C water bath. Hypotonic hESCs were fixed in methanol and glacial acetic acid mixture (3:1). Chromosomal spreads were obtained by dropping prepared hESCs directly onto glass slides. Giemsa staining of human chromosomes was conducted according to standard protocols.
Teratoma formation assay
The hESCs were cultured in a 6-well plate with mTeSR1 medium on 1% Matrigel. Six healthy female NOD SCID mice (6–8 weeks old) were purchased from Charles River Laboratories (USA). Human ESC clumps (~3 × 106 cells) were mixed with 0.5 g/ml Matrigel and injected intramuscularly into the NOD SCID mice. After 6–8 weeks, teratomas were observed, surgically excised, and fixed in 4% PFA overnight. Teratoma tissues were embedded in paraffin, sliced into 5-µm sections, and stained by hematoxylin and eosin according to standard protocols.
Imaging
Fluorescent, phase contrast, and bright-field pictures were taken using a Leica DMi8 automated and manual microscope (Leica, Germany). Pictures were analyzed by Leica Application Suite X software (Leica, Germany). Karyotypes were scanned by the CytoVision scanning system of an Olympus Exol microscope (Olympus, Japan).
Chemokine array
Supernatants were collected and screened using Ray-Bio Human Chemokine Antibody Array C1 kits (RayBiotech, USA) according to the standard protocols provided by the manufacturer. FluorChem E (Protein Simple, USA) was used to detect signal chemiluminescence intensities from protein array membranes. Quantitative analysis for chemiluminescent intensity was conducted using NIH image J Version 1.43. Relative signal intensity (RSI) was calculated by subtracting chemiluminescence intensities with negative control (NC) intensities. A RSI-based heatmap was constructed using R statistical software. Hierarchical clustering algorithm with correlation dissimilarity measure was applied to group sample RSIs in red-green color-coded grids to represent high and low RSIs, respectively.
Enzyme-linked immunosorbent assay (ELISA)
Supernatants were collected after 48 h of cell culture. We purchased ELISA kits for detecting human IL-8 (CXCL8) (Cusabio Biotech, China), IP-10 (CXCL10) (Shanghai Enzyme-Linked Biotech Co., Ltd, China), and SDF-1α (CXCL12) (Elisa Biotech Co., Ltd, China). ELISA experiments were conducted following the manufacturers’ instructions. Plates were read by a micro-plate reader (SpectraMax®314 340PC, USA) at a wave length of 450 nm.
In vitro transmigration
In vitro transmigration was performed using 48-well AP48 Boyden chambers (Neuro Probe, USA) according to the methods provided by the manufacturer. Briefly, 30 μl of 0.1% BSA (control) and different concentrations of recombinant human chemokine proteins were added to the lower chambers. The lower chambers were separated with 8-μm pore-sized polycarbonate membranes (Neuro Probe, USA). Then, 50 μl of cell suspension (~40,000 cells per ml) was seeded in the top wells. To block chemokine-induced transmigration, receptor-specific antagonists were added to the top wells. Boyden chambers were incubated at 37 °C with 5% CO2 for 4 h. After staining with 20 nM Hoechst 33342 (Thermo, USA), migrated cells on the filters were counted using an inverted fluorescence microscope (Olympus IX71, Japan). In addition, 0.1–10 nM SB265610, 0.1–10 µg/ml of AMD3100, and 0.1–10 nM NBI74330 were used to treat the cells. All transmigration experiments were independently repeated three times.
MTT assay
A MTT (3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide) assay was conducted according to the standard procedures described by the manufacturer’s instructions. Briefly, human ESC or iPSC clumps (~500 cells) were collected and seeded into 96-well plates (~1 × 104 cells/well) in mTeSR1 medium on 1% Matrigel. The toxicities of 0.1–100 nM antagonists, including reparixin, SB265610, AMD3100, and NBI 74330, were tested according to the standard procedures described by the manufacturer. The plate was read using a micro-plate reader (SpectraMax®314 340PC, USA) at a wave length of 490 nm. The MTT solution (Cell Titer 96® Aqueous One Solution Reagent) was purchased from Program (USA).
Statistics
All experiments were conducted in duplicate and data were expressed as means ± sem (standard error of mean). Statistical analyses were performed by one-way analysis of variance (ANOVA) with Dunnett’s test. Values of P < 0.05 were considered statistically significant.
Electronic supplementary material
Acknowledgements
We thank the Department of Urology from Kunming Children’s Hospital and Department of Reproduction and Genetics from the First People’s Hospital of Yunnan Province for kindly providing human specimens. We thank the Genetics and Diagnosis lab from the First People’s Hospital of Yunnan Province for karyotyping and G-band analysis. This work was financially supported by the National Natural Science Foundation of China (No. 81271330, No. 31270131, No. 81660247, No. 31260276, and No. 31260226), Talent Program of Yunnan Province (No. W8110305), Key Project of Yunnan Education Department (No. ZD2015002), Applied Basic Research Self-Raised Fund Key Projects of Yunnan Province (No.2013FZ262), and Yunnan University’s Research Innovation Fund for Graduate students (ynuy2016013). These funders had no roles in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
Z.J., M.L., Y.L., Y.M.: conception, design, interpretation, and manuscript writing; Z.J., Y.T., X.J., H.Y., L.D., M.Y., Q.C., M.Z.: collection, assembly, analysis, and interpretation of data; M.L., Y.L., Y.M., Z.J.: financial and administrative support. Final approval of manuscript (all authors).
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Zongmin Jiang and Yonggang Li contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15081-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yanping Ma, Email: mayanpinghome@163.com.
Meizhang Li, Email: mzhli@ynu.edu.cn.
References
- 1.Hasegawa K, Pomeroy JE, Pera MF. Current technology for the derivation of pluripotent stem cell lines from human embryos. Cell Stem Cell. 2010;6:521–531. doi: 10.1016/j.stem.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Choi J, et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol. 2015;33:1173–1181. doi: 10.1038/nbt.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Liang G, Zhang Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell. 2013;13:149–159. doi: 10.1016/j.stem.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludwig TE, et al. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 6.Inoue H, et al. iPS cells: a game changer for future medicine. EMBO J. 2014;33:409–417. doi: 10.1002/embj.201387098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peerani R, et al. Niche-mediated control of human embryonic stem cell self-renewal and differentiation. EMBO J. 2007;26:4744–4755. doi: 10.1038/sj.emboj.7601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagers AJ. The Stem Cell Niche in Regenerative Medicine. Cell Stem Cell. 2012;4:362–369. doi: 10.1016/j.stem.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 9.Ledran MH, et al. Efficient hematopoietic differentiation of human embryonic stem cells on stromal cells derived from hematopoietic niches. Cell Stem Cell. 2008;3:85–98. doi: 10.1016/j.stem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Bendall SC, et al. IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature. 2007;448:1015–1021. doi: 10.1038/nature06027. [DOI] [PubMed] [Google Scholar]
- 11.Gattazzo F, Urciuolo A, Bonaldo P. Extracellular matrix: a dynamic microenvironment for stem cell niche. Biochim Biophys Acta. 2014;1840:2506–2519. doi: 10.1016/j.bbagen.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Luo X, Leighton J. Extracellular matrix and integrins in embryonic stem cell differentiation. Biochem Insights. 2015;8:15–21. doi: 10.4137/BCI.S30377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawksworth OA, Coulthard LG, Taylor SM, Wolvetang EJ, Woodruff TM. Brief report: complement C5a promotes human embryonic stem cell pluripotency in the absence of FGF2. Stem Cells. 2014;32:3278–3284. doi: 10.1002/stem.1801. [DOI] [PubMed] [Google Scholar]
- 14.Peterson H, et al. Qualitative modeling identifies IL-11 as a novel regulator in maintaining self-renewal in human pluripotent stem cells. Front Physiol. 2013;4:303. doi: 10.3389/fphys.2013.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarris M, Olekhnovitch R, Bousso P. Manipulating leukocyte interactions in vivo through optogenetic chemokine release. Blood. 2016;127:35–41. doi: 10.1182/blood-2015-11-684852. [DOI] [PubMed] [Google Scholar]
- 16.Bardina SV, et al. Differential Roles of Chemokines CCL2 and CCL7 in Monocytosis and Leukocyte Migration during West Nile Virus Infection. J Immunol. 2015;195:4306–4318. doi: 10.4049/jimmunol.1500352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36:705–716. doi: 10.1016/j.immuni.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/S1074-7613(00)80165-X. [DOI] [PubMed] [Google Scholar]
- 19.Rabenstein M, et al. Osteopontin mediates survival, proliferation and migration of neural stem cells through the chemokine receptor CXCR4. Stem Cell Res Ther. 2015;6:99. doi: 10.1186/s13287-015-0098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Boyle G, et al. Chemokine receptor CXCR3 agonist prevents human T-cell migration in a humanized model of arthritic inflammation. Proc Natl Acad Sci USA. 2012;109:4598–4603. doi: 10.1073/pnas.1118104109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lokensgard JR, Mutnal MB, Prasad S, Sheng W, Hu S. Glial cell activation, recruitment, and survival of B-lineage cells following MCMV brain infection. J Neuroinflammation. 2016;13:114. doi: 10.1186/s12974-016-0582-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, et al. Optogenetic control of chemokine receptor signal and T-cell migration. Proc Natl Acad Sci USA. 2014;111:6371–6376. doi: 10.1073/pnas.1319296111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Liang R, Lu Y, Wang M, Li Z. RTN3 Regulates the Expression Level of Chemokine Receptor CXCR4 and is Required for Migration of Primordial Germ Cells. Int J Mol Sci. 2016;17:382. doi: 10.3390/ijms17040382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 25.Carbajal KS, Schaumburg C, Strieter R, Kane J, Lane TE. Migration of engrafted neural stem cells is mediated by CXCL12 signaling through CXCR4 in a viral model of multiple sclerosis. Proc Natl Acad Sci USA. 2010;107:11068–11073. doi: 10.1073/pnas.1006375107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan RN, Psaila B, Lyden D. Niche-to-niche migration of bone-marrow-derived cells. Trends Mol Med. 2007;13:72–81. doi: 10.1016/j.molmed.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 27.De Clercq E. Recent advances on the use of the CXCR4 antagonist plerixafor (AMD3100, Mozobil™) and potential of other CXCR4 antagonists as stem cell mobilizers. Pharmacol Ther. 2010;128:509–518. doi: 10.1016/j.pharmthera.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Li M, et al. Chemokine receptor CXCR4 signaling modulates the growth factor-induced cell cycle of self-renewing and multipotent neural progenitor cells. Glia. 2011;59:108–118. doi: 10.1002/glia.21080. [DOI] [PubMed] [Google Scholar]
- 29.Juarez JG, et al. Sphingosine-1-phosphate facilitates trafficking of hematopoietic stem cells and their mobilization by CXCR4 antagonists in mice. Blood. 2012;119:707–716. doi: 10.1182/blood-2011-04-348904. [DOI] [PubMed] [Google Scholar]
- 30.Li M, Ransohoff RM. Multiple roles of chemokine CXCL12 in the central nervous system: a migration from immunology to neurobiology. Prog Neurobiol. 2008;84:116–131. doi: 10.1016/j.pneurobio.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen SR, et al. Loss of Gata4 in Sertoli cells impairs the spermatogonial stem cell niche and causes germ cell exhaustion by attenuating chemokine signaling. Oncotarget. 2015;6:37012–37027. doi: 10.18632/oncotarget.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doitsidou M, et al. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111:647–659. doi: 10.1016/S0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- 33.Kawakami Y, et al. SDF-1/CXCR4 axis in Tie2-lineage cells including endothelial progenitor cells contributes to bone fracture healing. J Bone Miner Res. 2015;30:95–105. doi: 10.1002/jbmr.2318. [DOI] [PubMed] [Google Scholar]
- 34.Xie HT, Chen SY, Li GG, Tseng SC. Limbal epithelial stem/progenitor cells attract stromal niche cells by SDF-1/CXCR4 signaling to prevent differentiation. Stem Cells. 2011;29:1874–1885. doi: 10.1002/stem.743. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz CM, et al. Stromal factors SDF1α, sFRP1, and VEGFD induce dopaminergic neuron differentiation of human pluripotent stem cells. J Neurosci Res. 2012;90:1367–1381. doi: 10.1002/jnr.23064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin Y, et al. Induction of Regulatory B-Cells by Mesenchymal Stem Cells is Affected by SDF-1α-CXCR7. Cell Physiol Biochem. 2015;37:117–130. doi: 10.1159/000430338. [DOI] [PubMed] [Google Scholar]
- 37.Mavier P, et al. Expression of stromal cell-derived factor-1 and of its receptor CXCR4 in liver regeneration from oval cells in rat. Am J Pathol. 2004;165:1969–1977. doi: 10.1016/S0002-9440(10)63248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izumi D, et al. CXCL12/CXCR4 activation by cancer-associated fibroblasts promotes integrin β1 clustering and invasiveness in gastric cancer. Int J Cancer. 2016;138:1207–1219. doi: 10.1002/ijc.29864. [DOI] [PubMed] [Google Scholar]
- 39.Almeida CR, Caires HR, Vasconcelos DP, Barbosa MA. NAP-2 Secreted by Human NK Cells Can Stimulate Mesenchymal Stem/Stromal Cell Recruitment. Stem Cell Reports. 2016;6:466–473. doi: 10.1016/j.stemcr.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baba T, et al. MIP-1α/CCL3-expressing basophil-lineage cells drive the leukemic hematopoiesis of chronic myeloid leukemia in mice. Blood. 2016;127:2607–2617. doi: 10.1182/blood-2015-10-673087. [DOI] [PubMed] [Google Scholar]
- 41.Uyangaa E, et al. Distinct Upstream Role of Type I IFN Signaling in Hematopoietic Stem Cell-Derived and Epithelial Resident Cells for Concerted Recruitment of Ly-6Chi Monocytes and NK Cells via CCL2-CCL3 Cascade. PLoS Pathog. 2015;11:e1005256. doi: 10.1371/journal.ppat.1005256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuett A, et al. IL-8 as mediator in the microenvironment-leukaemia network in acute myeloid leukeamia. Sci Rep. 2015;5:18411. doi: 10.1038/srep18411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelland EE, Gilmore W, Weiner LP, Lund BT. The dual role of CXCL8 in human CNS stem cell function: multipotent neural stem cell death and oligodendrocyte progenitor cell chemotaxis. Glia. 2011;59:1864–1878. doi: 10.1002/glia.21230. [DOI] [PubMed] [Google Scholar]
- 44.Jing L, et al. Adenosine signaling promotes hematopoietic stem and progenitor cell emergence. J Exp Med. 2015;212:649–663. doi: 10.1084/jem.20141528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang CH, et al. Junctional Adhesion Molecule-A Is Highly Expressed on Human Hematopoietic Repopulating Cells and Associates with the Key Hematopoietic Chemokine Receptor CXCR4. Stem Cells. 2016;34:1664–1678. doi: 10.1002/stem.2340. [DOI] [PubMed] [Google Scholar]
- 46.Mendt M, Cardier JE. Role of SDF-1 (CXCL12) in regulating hematopoietic stem and progenitor cells traffic into the liver during extramedullary hematopoiesis induced by G-CSF, AMD3100 and PHZ. Cytokine. 2015;76:214–221. doi: 10.1016/j.cyto.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Tei N, et al. Expression of MCP-1 and fractalkine on endothelial cells and astrocytes may contribute to the invasion and migration of brain macrophages in ischemic rat brain lesions. J Neurosci Res. 2013;91:681–693. doi: 10.1002/jnr.23202. [DOI] [PubMed] [Google Scholar]
- 48.d’Audigier C, et al. Thrombin receptor PAR-1 activation on endothelial progenitor cells enhances chemotaxis-associated genes expression and leukocyte recruitment by a COX-2-dependent mechanism. Angiogenesis. 2015;18:347–359. doi: 10.1007/s10456-015-9471-8. [DOI] [PubMed] [Google Scholar]
- 49.Ishida Y, et al. Pivotal role of the CCL5/CCR5 interaction for recruitment of endothelial progenitor cells in mouse wound healing. J Clin Invest. 2012;122:711–721. doi: 10.1172/JCI43027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoo SW, et al. Immune following suppression mesenchymal stem cell transplantation in the ischemic brain is mediated by TGF-β. Neurobiol Dis. 2013;58:249–257. doi: 10.1016/j.nbd.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Shen B, et al. CCR2 Positive Exosome Released by Mesenchymal Stem Cells Suppresses Macrophage Functions and Alleviates Ischemia/Reperfusion-Induced Renal Injury. Stem Cells Int. 2016;2016:1240301. doi: 10.1155/2016/1240301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Southgate TD, et al. CXCR4 mediated chemotaxis is regulated by 5T4 oncofetal glycoprotein in mouse embryonic cells. PLoS One. 2010;5:e9982. doi: 10.1371/journal.pone.0009982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang H, et al. CCR5 Disruption in Induced Pluripotent Stem Cells Using CRISPR/Cas9 Provides Selective Resistance of Immune Cells to CCR5-tropic HIV-1 Virus. Mol Ther Nucleic Acids. 2015;4:e268. doi: 10.1038/mtna.2015.42. [DOI] [PubMed] [Google Scholar]
- 54.Krtolica A, et al. GROα regulates human embryonic stem cell self-renewal or adoption of a neuronal fate. Differentiation. 2011;81:222–232. doi: 10.1016/j.diff.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hasegawa Y, et al. CC chemokine ligand 2 and leukemia inhibitory factor cooperatively promote pluripotency in mouse induced pluripotent cells. Stem Cells. 2011;29:1196–1205. doi: 10.1002/stem.673. [DOI] [PubMed] [Google Scholar]
- 56.Guo Y, Hangoc G, Bian H, Pelus LM, Broxmeyer HE. SDF-1/CXCL12 enhances survival and chemotaxis of murine embryonic stem cells and production of primitive and definitive hematopoietic progenitor cells. Stem Cells. 2005;23:1324–1332. doi: 10.1634/stemcells.2005-0085. [DOI] [PubMed] [Google Scholar]
- 57.Hasegawa Y, et al. CCL2 enhances pluripotency of human induced pluripotent stem cells by activating hypoxia related genes. Sci Rep. 2014;4:5228. doi: 10.1038/srep05228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jung JH, et al. CXCR2 and its related ligands play a novel role in supporting the pluripotency and proliferation of human pluripotent stem cells. Stem Cells Dev. 2015;24:948–961. doi: 10.1089/scd.2014.0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W, et al. Modeling abnormal early development with induced pluripotent stem cells from aneuploid syndromes. Hum Mol Genet. 2012;21:32–45. doi: 10.1093/hmg/ddr435. [DOI] [PubMed] [Google Scholar]
- 60.Esteban MA, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 61.Ludwig TE, et al. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 62.Heasley LE, Petersen BE. Signalling in stem cells: meeting on signal transduction determining the fate of stem cells. EMBO Rep. 2004;5:241–244. doi: 10.1038/sj.embor.7400098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beider K, Abraham M, Peled A. Chemokines and chemokine receptors in stem cell circulation. Front Biosci. 2008;13:6820–6833. doi: 10.2741/3190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.