Abstract
Androgen receptor (AR) is widely expressed in different subtypes of breast cancer (BC). However, it is unclear how AR functions in HER2 positive (+) BC. Knockdown of AR with shRNAs and a new generation anti-androgen drug, Enzalutamide, were used to explore the involvement of AR on the growth of HER2 + BC cells (HCC1954 and SKBR3). AR shRNA or Enzalutamide inhibited the growth of SKBR3 cells at a similar extend compared to trastuzumab, an approved HER2 targeted drug. Combining Enzalutamide with trastuzumab further decreased the growth of HCC1954 and SKBR3 cells compared with single agent alone in vitro. Biochemical analysis revealed that inhibiting AR resulted in decreased HER2 phosphorylation and activation of Erk and Akt, without affecting the HER2 and HER3 expression. The in vivo efficacy of Enzalutamide was further tested using the HCC1954 xenograft model. Enzalutamide impaired the growth of HCC1954 tumor at a level comparable to that by trastuzumab. Enzalutamide decreased Ki67 staining and increased activated caspase3 staining compared with vehicle control in HCC1954 tumors. Our results indicate AR plays an important role in promoting the growth of HER2 + BC by cross-talking with the HER2 signaling. AR drug may be used as an alternative second line therapy for treating HER2 + BC.
Introduction
HER2 positive (+) breast cancer accounts for ~25% of breast cancer and has a poor prognosis1. Clinically, HER2 + breast cancer is identified by immunochemistry (IHC) of HER2 with a 3 + staining or fluorescent in situ hybridization (FISH) for HER2 2. HER2 mainly via heterodimerization with HER33 activates downstream signaling pathways including PI3K/Akt and Ras/Erk to drive the growth of the breast cancer cells4,5. Various HER2 targeted therapies have been developed over the years6,7. Trastuzumab (Herceptin), a humanized monoclonal antibody against HER2, when combined with chemotherapy, is very effective in treating HER2 + breast cancer, with a 50–80% response rate8. However, 20–50% of patients with HER2 + tumors do not respond to trastuzumab initially or develop acquired resistance after one year of treatment9. Thus, it is important to explore alternative second line therapies.
Androgen receptor (AR) is a ligand-activated transcription factor that belongs to the steroid hormone receptor family. AR signaling is well known to drive the growth of prostate cancer cells as reviewed10 and recent studies reveal that AR is more widely expressed than estrogen receptor (ER) or progesterone receptor (PR) in breast cancer11,12. According to the Nurses’ Health Study in 2011, AR expression can be detected in 77% of the breast cancer with the breakdown of 88% of ER+, 59% of HER2+, and 32% of triple negative breast (TNBC) expressing AR respectively12. Although as with ER, AR expression in breast has been largely associated with a better prognosis than AR negative disease,13–16, recent studies indicate that AR promotes the growth of ER + breast cancer17 and TNBC18,19 via distinct mechanisms. In ER + BC, studies from the use of a new generation of anti-androgen drug, Enzalutamide (also known as MDV3100)20, indicate that AR signaling is required for androgen and estrogen-induced tumor cell growth in vitro and in vivo 17. In the luminal AR18,21 subset of TNBC, AR activity is required for the growth of LAR-TNBC through the upregulation of HER319. In other subset of TNBC, AR activity promotes the growth of TNBC at least in part via upregulating the expression of amphiregulin22. Various clinical trials using androgen antagonists bicalutamide and Enzalutamide in treating AR + TNBC are ongoing (clinicaltrials.gov: NCT01889238, and NCT00468715).
In contrast, the role of AR in HER2 + breast cancer is less well understood. The prognostic value of AR expression have been reported to be associated with either better23 or worse14 patient survival of HER2 + breast cancer (BC). Previous studies suggested a role of AR in the molecular apocrine BC (maBC), which is a subtype of estrogen receptor negative (ER-) BC that overexpresses AR and frequently HER2 as well24. Treating MDA-MB-453 cells, a maBC line, with the first generation anti-androgen drug flutamide reduced cell proliferation and induced apoptosis of maBC in vitro 25. Furthermore, AR promoted Erk activation via upregulation of HER2 gene transcription26. Considering the role of AR and its interaction with HER2 in maBC, we hypothesize that AR expression may contribute to the growth of HER2 + BC.
In the current study, we examined the functional role of AR in HER2 + BC by blocking AR expression with shRNA and AR activity with androgen antagonist Enzalutamide in HER2 + BC cell lines, SKBR3 and HCC1954. These two cell lines are well known models of HER2 + BC that are estrogen receptor negative. Considering the reported interaction between AR and estrogen receptor in breast cancer27, our study should help dissect the role of AR in HER2 + BC independent of ER. Our data revealed that inhibiting AR impairs the growth of HER2 + breast cancer cells in vitro and vivo, in comparable to the effect of trastuzumab. AR inhibition also reduced HER2 phosphorylation and activation of Akt and Erk without affecting HER2 and HER3 protein expression. Our results suggest that AR plays a novel role in HER2 signaling and AR targeting therapy may be useful in treating HER2 + breast cancer.
Results
Inhibiting AR impairs the growth of HER2 + breast cancer cells
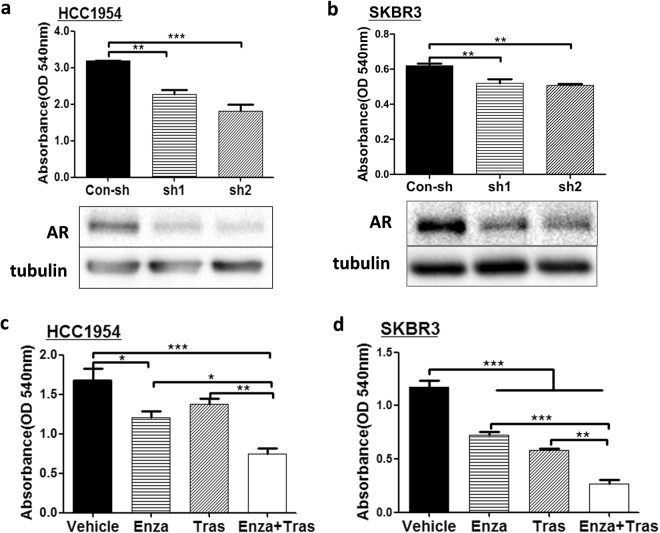
To test whether AR can drive the growth of HER2 + breast cancer, AR shRNAs were used to knockdown AR expression in HCC1954 and SKBR3. Both of the AR shRNAs significantly inhibited AR protein expression in HCC1954 and SKBR3 cells as indicated from the western blot analysis (Fig. 1a and b bottom panels). Furthermore, data from colony forming assays revealed that AR shRNA impaired the growth of HCC1954 (Fig. 1a, top panel) and SKBR3 (Fig. 1b, top panel) cells compared to control shRNA. Similar results were obtained when the growth of HCC1954 and SKBR3 was analyzed using CCK-8 reagent (Supplementary Fig. S1a,b), further supporting that knockdown of AR expression reduced the growth of HER2 + breast cancer cells.
Figure 1.
Inhibition of AR impairs the growth of HER2 breast cancer cells. (a,b) Knockdown of AR with shRNAs reduced the growth of HCC1954 and SKBR3. HCC1954 (a) and SKBR3 (b) cells were infected with lentivirues expressing control shRNA (Con-sh) and two different AR shRNAs (sh1 and sh2) and immunoblotted with anti-AR and anti-tubulin antibodies (a,b bottom panels). All cropped blots were run under the same experimental conditions. The original blots are included in Supplementary Figure S2. Colony formation assay was used to assess the growth of HCC1954 and SKBR3 cells expressing Con-sh, sh1, and sh2 (a,b top panels). AR antagonist, enzalutamide (Enza), together with trastuzumab (Tras) inhibited the growth of HCC1954 (c) and SKBR3 (d) cells. Cells were subjected to colony formation assay in the presence of Vehicle, 20 μM Enza, 20 μg/ml Tras, and 20 μM Enza + 20 μg/ml Tras for 6 days before analyzed with crystal violet staining. Data shown is the representative from three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001, one-way ANOVA.
To further test whether AR is a potential target for treatment of HER2 + breast cancer, Enzalutamide (Enza), an FDA approved AR targeting drug, was used to treat HCC1954 and SKBR3 cells in vitro using colony formation assay. In addition, the effects of trastuzumab alone or together with Enza on the growth of HCC1954 and SKBR3 cells were also assessed (Fig. 1c and d). Similarly to AR knockdown, Enza treatment alone resulted in significant inhibition of cell growth in both cell lines. In SKBR3 cells, the effect of Enza treatment was similar to the inhibitory effect of trastuzumab (Fig. 1d). Although trastuzumab alone had minimal effect on the growth of HCC1954 cell (Fig. 1c), treatment with both Enza and trastuzumab led to further reduction in cell growth compared with treatment with either single agent in both cell lines (Fig. 1c and d). Again, similar observations were seen when the effects of Enza and Trastuzumab on the growth of HCC1954 and SKBR3 were analyzed using CCK-8 reagent (Supplementary Fig. S1c,1d). These data indicate that AR plays a critical role in the growth of HER2 + breast cancer cells.
AR is important for HER2 activation and signaling, independent of HER3 protein expression, in HER2 + breast cancer cells
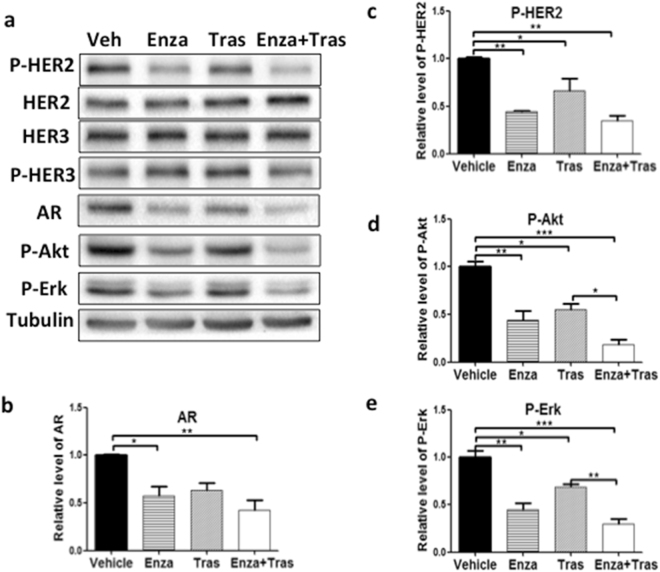
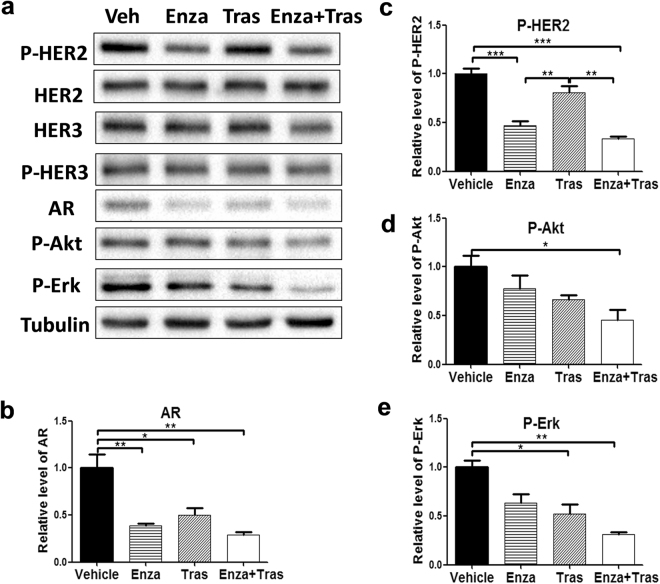
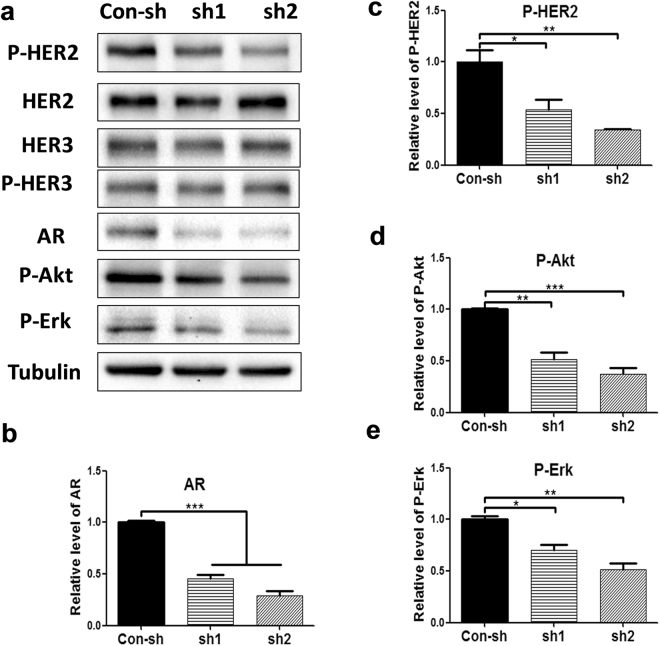
Because a previous report showed that AR activates HER2 signaling via upregulation of HER3 expression in luminal AR triple negative cells lines (MB-453 and SUM185PE)18. Western blot analysis was used to examine the phosphorylation status of HER2, HER3, and total HER2, HER3 protein expression in HER2 + breast cancer cells treated with Enza (Fig. 2). Compared with vehicle control, Enza treatment, which reduced AR protein expression, significantly decreased HER2 phosphorylation in HCC1954 and SKBR3 cells (Figs 2a,c and 3a,c). In addition, Enza treatment also reduced activation of Akt and Erk (Figs 2d,e and 3d,e), known downstream pathways of HER2. However, Enza treatment did not inhibit HER3 and HER2 total protein expression (Figs 2a and 3a). Likewise, knockdown of AR expression by shRNA impaired HER2 phosphorylation, Akt and Erk activation without affecting HER3 and HER2 protein expression in HCC1954 (Fig. 4). Interestingly, trastuzumab treatment inhibited HER2 phosphorylation as well as Akt and Erk activation in the similar extent as Enza treatment in SKBR3 cells (Fig. 2a). These data indicate that AR plays an important role in HER2 activation and signaling, without affecting HER2 and HER3 protein level.
Figure 2.
Enzalutamide inhibits HER2 phosphorylation and signaling without affecting HER3 protein expression in SKBR3 cells. Equal amounts of lysates from SKBR3 cells treated with Vehicle, 20 μM Enza, 20 μg/ml Tras, and 20 μM Enza + 20 μg/ml Tras for 4 days were immunoblotted with the indicated antibodies (a). All cropped blots were run under the same experimental conditions. The original blots are included in Supplementary Figure S3. The relative levels of AR (b), P-HER2 (c), P-Akt (d), and P-Erk (e) were quantified by normalizing with the Tubulin levels. Data shown is the average from three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001, one-way ANOVA.
Figure 3.
Enzalutamide inhibits HER2 phosphorylation and signaling without affecting HER3 protein expression in HCC1954 cells. Equal amounts of lysates from HCC1954 cells treated with Vehicle, 20 μM Enza, 20 μg/ml Tras, and 20 μM Enza + 20 μg/ml Tras for 6 days were immunoblotted with the indicated antibodies (a). All cropped blots were run under the same experimental conditions. The original blots are included in Supplementary Figure S4. The relative levels of AR (b), P-HER2 (c), P-Akt (d), and P-Erk (e) were quantified by normalizing with the Tubulin levels. Data shown is the average from three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001, one-way ANOVA.
Figure 4.
Knockdown of AR expression inhibits HER2 phosphorylation and signaling without affecting HER3 protein expression in HCC1954 cells. Equal amounts of lysates from HCC1954 cells expressing con-sh, AR sh1, and AR sh2 were immunoblotted with the indicated antibodies (a). All cropped blots were run under the same experimental conditions. The original blots are included in Supplementary Figure S5. The relative levels of AR (b), P-HER2 (c), P-Akt (d), and P-Erk (e) were quantified by normalizing with the Tubulin levels. Data shown is the average from three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001, one-way ANOVA.
Enzalutamide is effective in inhibiting HER2 + breast cancer in xenograft model
Because of its efficacy in reducing the growth of HER2 + breast cancer cells in vitro, the effect of Enza on HER2 + tumor growth was examined in HCC1954 xenograft model. HCC1954 cells mixed with matrigel were injected into both flanks of the Balbc/nude mice. Mice were monitored for tumor growth by palpation and caliper measurement, and randomized into four groups for treatment with Vehicle, Enza, Trastuzumab, or Enza + Trastuzumab two weeks after cell injection.
While HCC1954 tumors treated with vehicle control grew gradually, Enza treatment significantly decreased the tumor growth. Likewise Trastuzumab treatment dramatically reduced HCC1954 tumor growth (Fig. 5a). Addition of Enza to Trastuzumab did not significantly further reduce the growth of HCC1954 tumors compared with Trastuzumab treatment alone. This indicates that Enza efficacy is similar to that of Trastuzumab (Tras) in preclinical model of HER2 breast cancer (Fig. 5a, and b).
Figure 5.
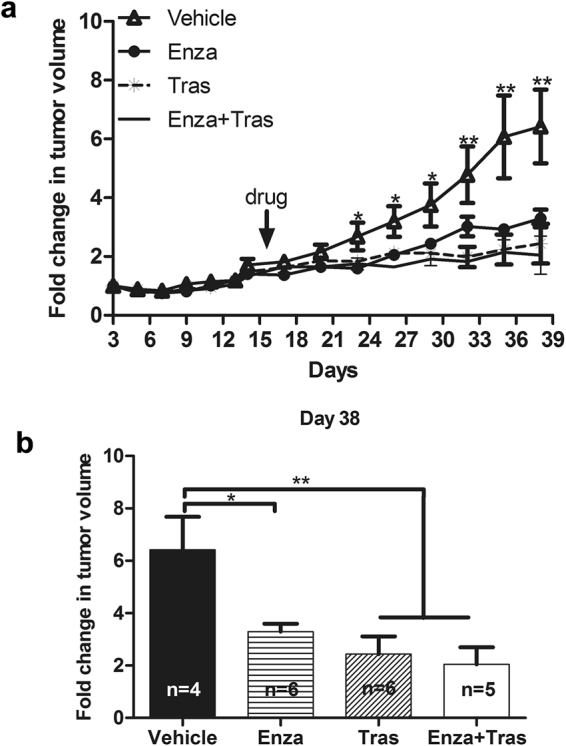
Enzalutamide inhibits HER2 + tumor growth in HCC1954 xenograft model. HCC 1954 cells were injected into the flanks of Balbc/nude mice. Sixteen days later, mice were randomized and treated with Vehicle, Enza, Tras, and Enza + Tras for 21 days. Tumor growth was monitored by caliper measurement every 3 days (a). (b) Relative tumor growth on day 36 after initial HCC1954 cell injection into the mice. Relative tumor growth was shown as fold change in tumor volume compared with the tumor size on day 3 after initial cell injection. n = the numbers of mice used in each treatment group. *p < 0.05, and **p < 0.01, one-way Anova. Data shown is representative from two independent experiments.
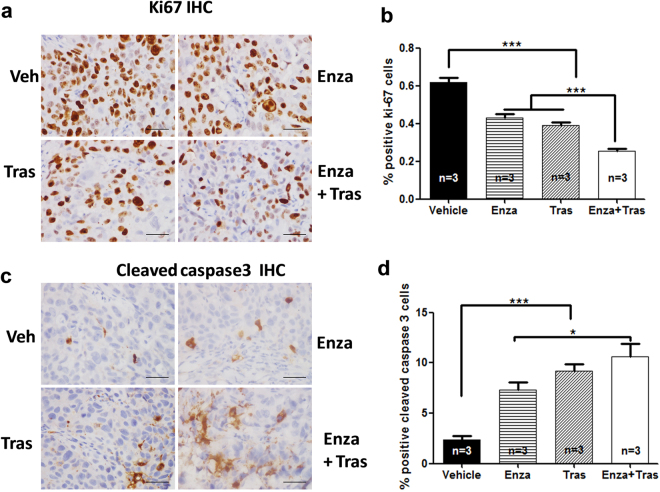
Immunohistochemistry (IHC) using antibodies against Ki67 and activated caspase 3 (Casp3) was used to examine the effects of Enza on cell proliferation and apoptosis respectively in xenograft tumors (Fig. 6). Compared with vehicle control, Enza and Tras treatments decreased Ki67 staining by ~33% (p < 0.001) and ~35% (p < 0.001) respectively whereas Enza plus Tras treatment reduced Ki67 staining by ~50% (Fig. 6a and b). While vehicle control tumors displayed ~2.5% Casp3 stained (+) cells, tumors treated with Enza and Tras showed ~7% (p < 0.001) and 9% (p < 0.001) Casp3 + cells. Addition of Enza to Tras further increased Casp3 + cells compared with Enza alone (p < 0.05) (Fig. 6c and d).These data revealed that Enza inhibits HCC1954 tumor growth via reduced cell proliferation and increased cell death, similar to the effects of trastuzumab on HCC1954.
Figure 6.
Enzalutamide inhibits cell proliferation and increases cell death in HER2 + xenograft tumors. Tumors from mice treated with Vehicle, Enza, Tras, Enza + Tras for 21 days were subjected to IHC staining with Ki67 antibody (a,b) and cleaved Casp3 antibody (c,d). n = 3 mice for each group. Scale bar = 50 μm. *p < 0.05, and ***p < 0.001, one-way ANOVA.
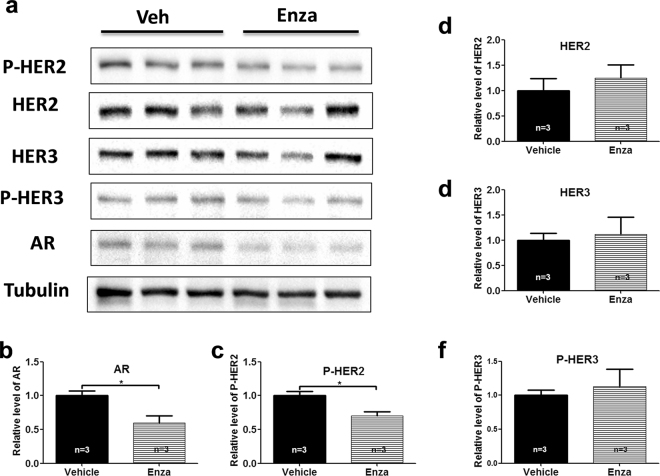
Western blot analysis was used to examine the effect of Enza on HER2 signaling in xenograft tumors. Similarly to what was found in vitro (Fig. 3), Enza treatment reduced AR expression, and inhibited HER2 phosphorylation without affecting HER2 and HER3 protein expression compared with vehicle treatment in HCC1954 xenografts (Fig. 7).
Figure 7.
Enzalutamide inhibits HER2 phosphorylation and signaling without affecting HER3 protein expression in HER2 + xenograft tumors. Tumor lysates from mice (three mice each group) treated with Vehicle and Enza for 21 days were immunoblotted with the indicated antibodies (a). All cropped blots were run under the same experimental conditions. The original blots are included in Supplementary Figure S6. The relative levels of AR (b), P-HER2 (c), HER2 (d), HER3 (e), and P-HER3 (f) were quantified by normalizing with the Tubulin levels. *p < 0.05, two-tailed student’s t-test.
Discussion
AR is widely expressed in HER2 + breast cancer; however, its function in HER2 + breast cancer is unclear. Results from this study suggest AR can in part activate HER2 signaling to drive the growth of HER2 + breast cancer cells in vitro and in vivo. Targeting AR can be used as an alternative therapy to treat HER2 + breast cancer.
Our data suggest that AR acts in a distinctive mechanism to promote the growth of HER2 + breast cancer cells. Our data reveal that AR contributes to HER2 phosphorylation and the activation of its downstream signaling pathway including Akt and Erk. In addition, AR inhibition did not affect HER2 and HER3 protein expression in HER2 + breast cancer cells (Figs 2, 3, 4, and 7). This is different from the action of AR in LAR-TNBC (i.e. MDA-MB-453 and SUM185PE) where AR enhances HER2 phosphorylation and signaling by increasing the expression of HER3 protein19. Specially, AR signaling promoted the transcription of WNT7B, which activates Wnt signaling. Then, AR complexed with the Wnt-activated beta-catenin upregulated transcription of HER3 19 in LAR-TNBC cells. In addition, AR signaling had been shown to enhance the transcription of HER2 gene in maBC26. In contrast, our results support a model that AR activation may be important for maximal HER2 activation without affecting the expression of HER2 and HER3 in a subset of HER2 + breast cancer.
There are several mechanisms by which AR may activate HER2 in HER2 + breast cancer cells. One is that AR signaling may increase the expression of a ligand for HER3, which is a heterodimeric partner for HER2. Consistent with such a hypothesis, AR can contribute to the expression of amphiregulin, a ligand for EGFR, which is highly expressed in some TNBC22. It is also possible that AR signaling may inhibit the expression and activities of specific protein tyrosine phosphatases that can dephosphorylate and inactivate HER228,29. Future studies are certainly required to uncover the detail mechanism of AR action in HER2 + breast cancer.
Two well-known HER2 + breast cancer cell lines, SKBR3 and HCC1954, were used in our study. Our data showed that SKBR3 cells were sensitive (Fig. 1d) whereas HCC1954 cells were resistant (Fig. 1c) to trastuzumab treatment in vitro, which are in agreement with published studies by other group30,31. However, trastuzumab was quite effective in inhibiting the growth of HCC1954 tumor in our xenograft model (Fig. 5a), which is not consistent with other published studies. The unique xenograft model, HCC1954 injected into the flank of BALB/c-nu/nu mice, used in our study might be account for this discrepancy. In studies with reported trastuzumab resistance by the HCC1954 models, different strains of immune deficient host mice were used, which include nu/nu: Alpk 32 and CB17 SCID-beige 33 mice, or HCC1954 cells were injected into different location (i.e mammary fat pad) in the host mice31,33. In any event, because the efficacy of Enza in inhibiting HCC1954 cell growth in vitro and in vivo, our data suggest that Enza may be used to overcome trastuzumab resistance.
Our result also reveals that AR antagonist Enzalutamide has efficacy in reducing HER2 + tumor growth in preclinical models. Interestingly, in these preclinical models Enzalutamide as a single agent is just as effective as trastuzumab at inhibiting the growth of HER2 + breast cancer in vitro (Fig. 1d) and vivo (Fig. 5). Enzalutamide was approved by the FDA to treat castration-resistant prostate cancer in 2012. Considering the promising results of the multiple ongoing clinical trials of Enzalutamide in TNBC and ongoing trial in ER + BC, targeting AR using Enzalutamide could be a useful second line therapy in a subset of HER2 + breast cancer that fail to respond to trastuzumab or develop trastuzumab resistance.
Materials and Methods
Cell Culture and Reagents
293T17 cells and breast cancer cell line SKBR3 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone) supplemented with 5% fetal bovine serum (FBS) (Sigma), 1 mM sodium pyruvate, 100 units/ml penicillin and 100 μg/ml streptomycin (Hyclone). Breast cancer cell line HCC1954 cells were grown in RPMI-1640 medium (Hyclone) supplemented with 5% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin. All cell lines were cultured in 37 °C humidified tissue incubator with 5% CO2 and 95% air. Enzalutamide was purchased from Selleck and dissolved in DMSO. Trastuzumab (Herceptin) was purchased from Roche.
Plasmids, lentivirus production, and infection of HCC1954 cells
Lentiviral vector PLKO.1 expressing control-shRNA and two different AR-shRNAs (1 and 2), and packaging plasmids pHR8.9deltaR and CMV-VSVG were purchased from Sigma. 293T17 cells plated in 6-cm dish at 2.5 × 106 cells 24 hours before were cotransfected with 1.88 μg pHR8.9deltaR, 1.25 μg CMV-VSVG, 2.5 μg pLKO1 shRNA using 16.8 μg PEI. Virus supernatants were harvested between 48–72 hours post-transfection, filtered through 0.45 μm low protein-binding sterile filter, and used to infect HCC1954 and SKBR3 cells in the presence of 8 μg of polybrene (Sigma). Pools of HCC1954 and SKBR3 cells expressing shRNAs were selected in the presence of 1.5 μg/ml puromycin (TaKaRa) for 3 days before using for further experiments. Sequences for AR shRNAs are:
ShRNA1: 5′-GAGCGTGGACTTTCCGGAAAT-3′;
ShRNA2: 5′-CACCAATGTCAACTCCAGGAT-3′.
Colony formation assay
Cells were seeded in 24 well plates at 5000/well in quadruplicate and treated with vehicle control, 20 μM enzalutamide, 20 μg/ml trastuzumab, and combined enzalutamide with trastuzumab. Fresh medium containing the drugs were replaced every two days. Six days later, cells were fixed and stained with 0.5% crystal violet for 20 minutes at room temperature. Crystal violet dye was extracted with 10% acetic acid and quantitated at OD 540 nm using Varioskan flash plate reader (Thermo scientific).
Western blot analysis
Breast cancer cell lines were lysed in 1X SDS sample (62.5 mM Tris-HCl pH 6.8,2% SDS, 0.002% Bromophenol Blue, 50 mM DTT, 10% glycerol). Tumors from xenografts were homogenized in modified RIPA lysis buffer [1% Triton X-100, 50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 150 mM NaCl, 0.25%Na-deoxycholate, 0.05%SDS, 10 mM NaF, 1 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail (sigma)] using Brinkmann Polytron PT2100. Tumor lysates were clarified by centrifugation (>15 K rpm) at 4 °C for 20 minutes. Protein concentrations in clarified lysates were quantified using BCA Protein Assay Kit (Beyotime Institute of Biotechnology) according to manufacturer’s protocol. Equal amounts of cell lysateswere resolved by 8% SDS-PAGE and transferred to Immobilon-P membrane (Millipore Inc.), immunoblotted using the appropriate primary and the HRP-conjugated secondary antibodies (Beyotime Institute of Biotechnology), and developed using enhanced chemiluminescence (ECL) reagent (Thermo Scientific). ECL signals were captured by the ChemiDoc MP imaging system (Bio-Rad) and were analyzed using the Image Lab 4.0 software (Bio-Rad). Alpha -Tubulin (Abcam) was used as the loading control. Monoclonal antibody (mAb) against ErbB2 was purchased from Santa Cruz Biotechnology. Antibodies recognizing phospho-HER2 (Tyr1221/1222) (P-HER2), HER3, phospho-HER3(Tyr1197) (P-HER3), cleaved Caspase-3(9G10), phospho-Akt (P-Akt) (Ser473) and phospho-P44/42 MAPK (Erk1/2) (Thr202/Tyr204) (P-Erk) were from Cell Signaling Technology. Antibodies recognizing AR (06–680) was from Merck Millipore.
HCC1954 Xenograft model
Protocols for the animal studies were approved by the Institutional Animal Care and Use Committee of Wenzhou Medical University. All animal experiments were performed in accordance with the relevant guidelines and regulations. BALB/c female nude mice (6 to 8 week old) purchased from Shanghai SLAC laboratory animal center were used for the in vivo experiments. HCC1954 cells (2 × 106/100 μl) were mixed with 100 ul growth factor reduced Matrigel (BD Bioscience) and injected subcutaneously at the flanks of the mice. Tumor growth was measured by caliper measurement every 3 days. The modified ellipsoid formula was used to calculate the tumor volume: (width)2 × (length)/2. Once all the tumors reached the tumor volume of 300–500 mm3 (~15 days after injection), four group of mice were randomized and treated with the following four regiments: 1) vehicle control: 2.7% DMSO + 30% PEG300 plus PBS; 2) 25 mg Enzalutamide/kg plus PBS; 3) 2.7% DMSO + 30% PEG300 plus 10 mg trastuzumab/kg; 4) 25 mg Enzalutamide/kg plus 10 mg trastuzumab/kg. Enzalutamide was dissolved in 2.7% DMSO, formulated in 30% PEG300 and administrated daily to mice by oral gavage. Trastuzumab was dissolved in PBS and injected intraperitoneally twice a week. Twenty one days after treatments, the animals were sacrificed after the last measurement of tumor size. Tumor growth was presented as fold change in tumor volume, which was calculated by dividing the tumor volume on a given day with the tumor volume on day 3 after injection of cells into mouse flank. The tumor tissues were dissected, fixed in 10% formalin for 24 hours and processed for immunohistochemistry or flash-frozen in liquid nitrogen and stored at −80 °C for western blot analysis later.
Hematoxylin and Eosin (H&E) Staining and Immunohistochemistry
Sections (5 μm thickness) of the formalin-fixed, paraffin-embedded (FFPE) tumor tissues were deparaffinized in xylene, rehydrated with a series of graded ethanols, then stained in Hematoxylin solution for 15 min and counterstained in eosin solution using the Hematoxylin and Eosin Staining Kit (Solarbio).
For immunohistochemistry, sections were deparaffinized, rehydrated as above, and subjected to heat induced epitope retrieval in 10 mM citrate buffer, pH 6.0 using Supor Pressure Cooker (at 125 °C for 5 min). Slides were immersed in 3% H2O2 for 8 min at room temperature (RT), then blocked with 10% normal goat serum (Solarbio) diluted in PBS for 20 min at RT, and incubated with Ki-67 mouse mAb (1:1200) (Cell signaling technology) and cleaved caspase 3 rabbit mAb (1:400) (Cell signaling technology) overnight at 4 °C. Normal mouse IgG (Beyotime#A7028) (1:1200) and rabbit IgG (Beyotime#A7016) (1:200) were used as negative controls. For detection of Ki-67, polyperoxidase conjugated anti-mouse/rabbit IgG Fab (ZSGB-BIO) was used followed by incubation with 3, 3′-diaminobenzidine chromogen DAB detection Kit (ZSGB-BIO). For detection of cleaved caspase 3, biotin labeled anti-rabbit secondary antibodies was used followed by incubation with streptavidin-horseradish peroxidase (SABC kit SA1052) (Boster) and 3, 3′-diaminobenzidine chromogen DAB detection Kit. Sections were counterstained with diluted hematoxylin for 2 min before mounted. For quantification of Ki-67 and cleaved caspase 3 IHC, images from ten random representative 400 × fields per mouse tumor were captured by Nikon Eclipse 80i Microscope. Ki-67 and cleaved caspase 3 staining positive cells in each field were counted and the average of positive staining cells per field was presented. The IHC data shown was representative from three independent experiments.
Statistical Analysis
Statistical analyses were performed using SPSS, version 19.0. Quantitative data from at least three independent experiments were expressed as mean ± S.E.M. Student’s t-test was used to compare data between two groups. One-way ANOVA with Bonferroni’s multiple comparison test correction was used to analyze data among multiple groups. All statistical tests were two-sided and P < 0.05 was considered statistically significant.
Electronic supplementary material
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (81772819, 81372826, 81572980, and 81400125), Natural Science Foundation of Zhejiang Province, China (LY17H160058), startup fund from Wenzhou Medical University, BC120183 W81XWH-13-1-0090 DOD BCRP Clinical Translational Award (to JKR), and National Institutes of Health/National Cancer Institute R01 CA187733-01A1 (to JKR).
Author Contributions
Conceived project: J.K.R., H.Gu. Performed Experiments: L.H., Z.D., H.M., X.X., H.Gao., J.C., Z.Z., T.L. Analyzed data: L.H., Z.D., H.L., K.Y., G.C., J.K.R., H.Gu. Wrote manuscript: L.H., Z.D., J.K.R., H.Gu.
Competing Interests
Enzalutamide used in some of the in vitro studies was obtained through an MTA from Astellas, Inc and Medivation Inc (Medivation, Inc. was acquired by Pfizer, Inc. in September 2016).
Footnotes
Licai He and Zhuanyun Du contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14607-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jennifer K. Richer, Email: Jennifer.Richer@ucdenver.edu
Haihua Gu, Email: haihuagu@wmu.edu.cn.
References
- 1.Carey LA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. Jama. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 2.Couturier J, Vincent-Salomon A, Mathieu MC, Valent A, Bernheim A. [Diagnosis of HER2 gene amplification in breast carcinoma] Pathologie-biologie. 2008;56:375–379. doi: 10.1016/j.patbio.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Holbro T, et al. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nature reviews. Molecular cell biology. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 5.Olayioye MA. Update on HER-2 as a target for cancer therapy: intracellular signaling pathways of ErbB2/HER-2 and family members. Breast cancer research: BCR. 2001;3:385–389. doi: 10.1186/bcr327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nature reviews. Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 7.Gajria D, Chandarlapaty S. HER2-amplified breast cancer: mechanisms of trastuzumab resistance and novel targeted therapies. Expert review of anticancer therapy. 2011;11:263–275. doi: 10.1586/era.10.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baselga J, Albanell J. Mechanism of action of anti-HER2 monoclonal antibodies. Annals of oncology: official journal of the European Society for Medical Oncology. 2001;12(Suppl 1):S35–41. doi: 10.1093/annonc/12.suppl_1.S35. [DOI] [PubMed] [Google Scholar]
- 9.Rexer BN, Arteaga CL. Intrinsic and acquired resistance to HER2-targeted therapies in HER2 gene-amplified breast cancer: mechanisms and clinical implications. Critical reviews in oncogenesis. 2012;17:1–16. doi: 10.1615/CritRevOncog.v17.i1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y, Bolton EC, Jones JO. Androgens and androgen receptor signaling in prostate tumorigenesis. Journal of molecular endocrinology. 2015;54:R15–29. doi: 10.1530/JME-14-0203. [DOI] [PubMed] [Google Scholar]
- 11.Moinfar F, et al. Androgen receptors frequently are expressed in breast carcinomas: potential relevance to new therapeutic strategies. Cancer. 2003;98:703–711. doi: 10.1002/cncr.11532. [DOI] [PubMed] [Google Scholar]
- 12.Collins LC, et al. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the Nurses’ Health Study. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24:924–931. doi: 10.1038/modpathol.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Angulo AM, et al. Androgen receptor levels and association with PIK3CA mutations and prognosis in breast cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009;15:2472–2478. doi: 10.1158/1078-0432.CCR-08-1763. [DOI] [PubMed] [Google Scholar]
- 14.Park S, et al. Androgen receptor expression is significantly associated with better outcomes in estrogen receptor-positive breast cancers. Annals of oncology: official journal of the European Society for Medical Oncology. 2011;22:1755–1762. doi: 10.1093/annonc/mdq678. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y, Jae E, Yoon M. Influence of Androgen Receptor Expression on the Survival Outcomes in Breast Cancer: A Meta-Analysis. Journal of breast cancer. 2015;18:134–142. doi: 10.4048/jbc.2015.18.2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu Q, Mao Y, Fei XC, Shen KW. The impact of androgen receptor expression on breast cancer survival: a retrospective study and meta-analysis. PloS one. 2013;8:e82650. doi: 10.1371/journal.pone.0082650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cochrane DR, et al. Role of the androgen receptor in breast cancer and preclinical analysis of enzalutamide. Breast Cancer Research. 2014;16:R7. doi: 10.1186/bcr3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of clinical investigation. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni M, et al. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer cell. 2011;20:119–131. doi: 10.1016/j.ccr.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran C, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters KM, et al. Androgen receptor expression predicts breast cancer survival: the role of genetic and epigenetic events. BMC cancer. 2012;12:132. doi: 10.1186/1471-2407-12-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barton VN, et al. Multiple molecular subtypes of triple-negative breast cancer critically rely on androgen receptor and respond to enzalutamide in vivo. Molecular cancer therapeutics. 2015;14:769–778. doi: 10.1158/1535-7163.MCT-14-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang HS, et al. Androgen receptor expression predicts different clinical outcomes for breast cancer patients stratified by hormone receptor status. Oncotarget. 2016;7:41285–41293. doi: 10.18632/oncotarget.9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farmer P, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 25.Naderi A, Hughes-Davies L. A functionally significant cross-talk between androgen receptor and ErbB2 pathways in estrogen receptor negative breast cancer. Neoplasia. 2008;10:542–548. doi: 10.1593/neo.08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chia KM, Liu J, Francis GD, Naderi A. A feedback loop between androgen receptor and ERK signaling in estrogen receptor-negative breast cancer. Neoplasia. 2011;13:154–166. doi: 10.1593/neo.101324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Amato NC, et al. Cooperative Dynamics of AR and ER Activity in Breast Cancer. Mol Cancer Res. 2016;14:1054–1067. doi: 10.1158/1541-7786.MCR-16-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu JH, et al. Protein tyrosine phosphatase PTPN13 negatively regulates Her2/ErbB2 malignant signaling. Oncogene. 2008;27:2525–2531. doi: 10.1038/sj.onc.1210922. [DOI] [PubMed] [Google Scholar]
- 29.Yuan T, Wang Y, Zhao ZJ, Gu H. Protein-tyrosine phosphatase PTPN9 negatively regulates ErbB2 and epidermal growth factor receptor signaling in breast cancer cells. The Journal of biological chemistry. 2010;285:14861–14870. doi: 10.1074/jbc.M109.099879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tormo E, et al. The role of miR-26a and miR-30b in HER2 + breast cancer trastuzumab resistance and regulation of the CCNE2 gene. Sci Rep. 2017;7:41309. doi: 10.1038/srep41309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong L, et al. CD147 knockdown improves the antitumor efficacy of trastuzumab in HER2-positive breast cancer cells. Oncotarget. 2016;7:57737–57751. doi: 10.18632/oncotarget.10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crafter C, et al. Combining AZD8931, a novel EGFR/HER2/HER3 signalling inhibitor, with AZD5363 limits AKT inhibitor induced feedback and enhances antitumour efficacy in HER2-amplified breast cancer models. Int J Oncol. 2015;47:446–454. doi: 10.3892/ijo.2015.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beyer I, et al. Controlled extracellular matrix degradation in breast cancer tumors improves therapy by trastuzumab. Mol Ther. 2011;19:479–489. doi: 10.1038/mt.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.