Abstract
Background:
Plasma microRNAs (miRNAs) are promising non-invasive biomarkers for colorectal cancer (CRC) prognosis. However, the published studies to date have yielded conflicting and inconsistent results for specific plasma miRNAs.
Methods:
We have conducted a study using robust assays to assess a panel of nine miRNAs for CRC prognosis and early detection of recurrence. Plasma samples from 144 patients in a prospective CRC cohort study were collected at diagnosis, 6, 12, and 24 months after diagnosis. miRNAs were assayed by Taqman qRT–PCR to generate miRNA normalised copy numbers.
Results:
Preoperative high plasma miRNA levels were associated with increased recurrence risk for miR-200b (HR [95% CI]=2.04 [1.00, 4.16], P=0.05), miR-203 (HR=4.2 [1.48, 11.93], P=0.007), miR-29a (HR=2.61 [1.34,5.07], P=0.005), and miR-31 (HR=4.03 [1.76, 9.24], P=0.001). Both plasma miR-31 (AUC: 0.717) and miR-29a (AUC: 0.703) could discriminate recurrence from these patients without recurrence. In addition, high levels of miR-31 during surveillance was associated with a three-fold increased risk of recurrence across all time points. Dynamic postoperative plasma miR-141 and 16 levels correlated with recurrence in the surveillance samples.
Conclusions:
Pre-operative plasma miR-29a, 200b, 203, and 31 are potential CRC prognosis biomarkers. In addition, dynamic postoperative miR-31, 141 and 16 levels are potential biomarkers for the early detection of recurrence during CRC surveillance.
Keywords: CRC, plasma, biomarker, miR-29a, miR-31, miR-200b, miR-203
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related death in the United States, with >50 000 death annually (Siegel et al, 2014). More than half of patients with CRC will present with or develop metastasis and the median survival time of patients with metastatic CRC is approximately 24 months (Ye et al, 2013; Yuan et al, 2013). Currently, tumour-lymph nodes-metastasis stage is the most accurate predictor of CRC recurrence and prognosis. However, it is not ideal and incorrectly predicts survival in a substantial portion of patients (Kocarnik et al, 2015). Thus, there is a need for more accurate ways to determine prognosis and response to therapy in patients with CRC.
Advances in the management of patients with metastatic CRC have also created a need for more accurate prognostic assays to guide the therapy of these patients. Improved clinical outcomes have been demonstrated when recurrent metastatic CRC is treated when it is confined to one organ such as the liver or lungs and when there are a limited number of metastatic lesions in the involved organ (Rodriguez-Moranta et al, 2006; Ye et al, 2013). Thus, the early detection of recurrence can increase the chance for curative treatment and can improve survival (Young et al, 2014). However, current surveillance methods are suboptimal because they are not sensitive for early recurrent CRC (Young et al, 2014). Blood-based biomarkers have the potential to address the need for a non-invasive and highly sensitive method for the early detection of recurrent CRC.
A number of promising serum- and plasma-based molecular markers for CRC recurrence have been identified over the last several years. Circulating nucleic acids, including tumour DNA, methylated DNA, and microRNAs (miRNAs) have all emerged as potentially clinical useful biomarkers for CRC recurrence and survival (Liu and Chu, 2016). MicroRNAs are small single-strand non-coding RNAs (typically 19–22 nucleotides in size) that can suppress gene expression by base-pairing to partially complementary sites on target mRNAs, which triggers RNA degradation or impairs translation (Selbach et al, 2008). MicroRNAs regulate a variety of biological functions and are often aberrantly expressed in pathological conditions, including the majority of cancers (Visone and Croce, 2009). Interestingly, overexpressed miRNAs in tumour tissues are often detectable in circulating blood (Toiyama et al, 2013). Although the mechanism through which miRNAs enter the circulation is still under investigation, some studies show plasma miRNAs enter the blood in exosomes released into the peripheral circulation by tumours (Valadi et al, 2007). Importantly, plasma miRNAs are stable, detectable with PCR-based assays, and quantifiable (Turchinovich et al, 2011). These features have led to an intense interest in the development of serum or plasma miRNAs as diagnostic, prognostic or predictive biomarkers for CRC (Ng et al, 2009; Huang et al, 2010).
However, despite this promise and the large number of published studies on blood-based miRNAs as CRC biomarkers, no miRNA biomarkers have proven robust enough to be used in clinical care. This is because there is little consistency or reproducibility of the results of the published studies assessing miRNA biomarkers (Schubert et al, 2016). The poor reproducibility of biomarker miRNA studies is likely a consequence of small sample sizes, lack of validation cohorts, use of different assay technologies, differences in normalisation methods across studies, and differences in pre-analytical sample handling protocols (Yamada et al, 2015; Sun et al, 2016). Thus, we have conducted a strict case-case study of candidate plasma miRNA CRC biomarkers that addresses many of the factors responsible for inconsistent results.
We selected candidate circulating miRNA biomarkers based on our own published work and a review of published studies (Adams et al, 2014). Based on prior work we selected two miRNAs (miR-31 and -203) because they have low plasma levels and are present at high levels in CRC patients. We selected six additional miRNAs based on prior studies that showed strong associations with CRC prognosis or progression (miR-29a, miR-92a) (Huang et al, 2010; Adams et al, 2014), (miR-17-3p, miR-125b, miR-200b) (Ng et al, 2009; Nishida et al, 2011; Adams et al, 2014), miR-141 (Cheng et al, 2011). Finally, miR-16 was chosen because it is commonly used as a plasma control (Hu et al, 2014). We assessed this panel of nine miRNAs in both pre-operative and serial postoperative plasma samples, correlated the results with select demographic features of the CRC patients and with time to cancer recurrence.
Materials and methods
Patients
Colorectal cancer patients were recruited in an ongoing prospective colon cancer cohort study (the ColoCare Study) managed at the Fred Hutchinson Cancer Research Center (Seattle, Washington). This is an active prospective cohort study established in 2007 that collects extensive biospecimens and follow-up data at multiple time points from patients with CRC. Patients were recruited when consulting with colorectal surgeons at the Seattle Cancer Care Alliance, University of Washington Affiliated Hospitals, Swedish Medical Center or Virginia Mason Medical Center following IRB-approved protocols. Eligibility requirements include: age 18–80, English speaking, and newly diagnosed with CRC. Written informed consent was obtained from each participant. This study was approved by the local IRB committees of the respective institutions. For this study, we included all stages I–IV ColoCare participants enrolled between January 2007 and June 2011 who had a pre-operative blood sample and had current follow-up information available. Information on diagnosis, stage, and vital status was obtained from the medical records and the Colocare clinical information database. Recurrence was determined based on imaging findings or biopsy of a recurrent lesion. Clinical-pathologic information was obtained by chart abstraction performed by at least two independent clinical abstractors. Patients with stage IV patients at enrolment in the study were only included if they achieved clinical remission following radical resection of primary tumour and treatment of the concurrent metastatic disease.
Blood samples and clinical database
Participants provided blood samples prior to surgery, tumour and normal tissues at the time of resection, and were followed at regular intervals (approximately every 6 months) for outcomes related to treatment, drug toxicities, recurrence, and survival. Blood collection occurred during clinic visits or, at times, by remote blood draw. Pre-analytic variables such as time of blood draw, time of processing or freezing, and deviations from standard operating procedures were recorded following standard protocols. All samples were given a study ID number and barcode and were tracked via barcoded specimen IDs using a customised LIMS.
Isolation of miRNA
MicroRNAs were extracted from baseline plasma (prior to surgery) using the Qiagen miRNeasy Serum/Plasma miRNA extraction kit, following modifications as previously described (Ng et al, 2009). Briefly, 250 μl of blood sample was centrifuged at 3000 g for 7.5 min at room temperature (RT). Then, 200 μl of supernatant plasma was transferred to a PhaseLock Gel 15 ml tube (5 Prime Inc.) containing 2.0 ml Qiazol (Qiagen, Hilden, Germany), inverted 10 times, and incubated for 5 min at RT. After the denaturation step and prior to extraction (described below), each sample received a well-mixed, single-use aliquot of 30 fmol of synthetic C. elegans miRNA (cel-miR-39; Integrated DNA Technologies, Coralville, IA, USA) and 1 μg of carrier rRNA (Roche Life Sciences, Penzberg, Germany, cat# 10 206 938 001) in Qiazol. The sample was inverted 10 times, chloroform (0.2 × volume) was added and the sample was inverted again 10 times, and incubated at RT for 2 min. The samples were centrifuged at 1500 g for 5 min at RT and processed following the relevant portions of miRNeasy extraction kit protocol.
Quantitation of plasma miRNA level by qRT–PCR
MicroRNAs were quantified using pre-designed TaqMan MicroRNA assays (Life Technologies, Waltham, MA, USA, Cat#4440887). Following RNA extraction, reverse transcription was completed using the Taq-Man miRNA Reverse Transcription Kit and miRNA-specific stem-loop primers in a 5 μl reaction following a previously described protocol (Ng et al, 2009). We used a fixed volume (1.67 μl), rather than a fixed mass, of eluted RNA sample and adjusted for differences in RNA recovery by normalisation to the synthetic spiked-in cel-miR-39 measured in each sample. We did not use RNA yield to determine RNA input into the reverse transcriptase reaction for two reasons (Ng et al, 2009): (1) the yield of RNA from small volumes of plasma is typically too low to accurately quantify, and (2) the 1 μg carrier rRNA added to improve recovery obscures our ability to detect miRNA mass differences between samples. In parallel with the experimental samples, we reverse transcribed a 7-point standard curve for each miRNA using synthetic miRNA targets (HPLC-purified, 5′ phosphorylated oligonucleotides, Integrated DNA Technologies) ranging from 3 × 107 to 7 × 102 copies.
After reverse transcription, quantitative real-time PCR (qRT–PCR) Taqman assays were performed in triplicate using a commercially available kit (Life Technologies, Carlsbad, CA, USA, Thermo Fisher Scientific, Waltham, MA, USA), scaled down to 5 μl for this application, as described previously (Ng et al, 2009). Assays were run on a 7900HT Sequence Detection System. Each assay plate contained the standard curve in triplicate, no template controls, external reference plasma controls, positive controls (miRNA from tissue), and inter- and intra-plate duplicates of study samples. Laboratory members were blinded to case-control status and other sample characteristics.
Normalisation of miRNA copy number
Raw qRT–PCR data were analysed with SDS Absolute Quantification Software version 2.2.3 (Applied Biosystems, Inc., Foster City, CA, USA), using the automatic cycle threshold (Ct) setting to assign baseline and threshold for Ct determination. The standard curves for each miRNA were used to calculate miRNA copy number for each miRNA for each sample. Plasma miRNA level was normalised to a C. elegans external spike-in control (cel-miR-39) and expressed as a normalised copy number, using a median normalisation procedure based on a previously published method (Ng et al, 2009). Briefly, the TaqMan qRT–PCR assay for cel-miR-39 was run in triplicate, and the mean cel-miR-39 Ct for each sample was calculated and converted into a cel-miR-39 copy number based on the cel-miR-39 standard curve. Then, the median value of all the cel-miR-39 copy numbers for all the samples in the study was calculated. For each sample, a normalisation factor was calculated by dividing the median value of the cel-miR-39 copy numbers by the cel-miR-39 copy number of the sample of interest. The copy number for the miRNA of interest was then multiplied by the normalisation factor to give the normalised copy number. Lastly, natural logarithm of copy numbers as ln (normalised copy numbers) was applied to represent miRNA absolute values.
Statistical analyses
Binary variables were summarised as proportions, continuous variables by means and median and range. Evaluation of the association between log-transformed miRNA levels over time and demographic variables was done using a linear mixed model with an exchangeable correlation structure. To evaluate the association between recurrence-free survival and miRNA levels at baseline, levels were dichotomised at the median (over all time points) and evaluated using a Cox proportional hazards model. Recurrence-free survival was defined as the duration from study enrolment to recurrence or death due to any cause, whichever came first. Patients last known to be alive and recurrence free or recurrence free at 3 years, were censored at the date of their last contact. The Cox model was used to estimate hazard ratios (and associated 95% confidence intervals, using Wald confidence interval and Schoenfeld residuals) and RFS at 1 and 2 years. Evaluation of associations between miRNA levels measured over time and RFS was done using a time-varying Cox proportional hazards model. Receiver operating characteristic curves were conducted for each miRNA for discriminating patients with recurrence or without recurrence. The area under the curve (AUC) was estimated along with 95% CI. All analyses were done using SAS/STAT software version 9.4. A significance level of two-sided 0.05 was used for all analyses.
Results
Demographic features of the study subjects
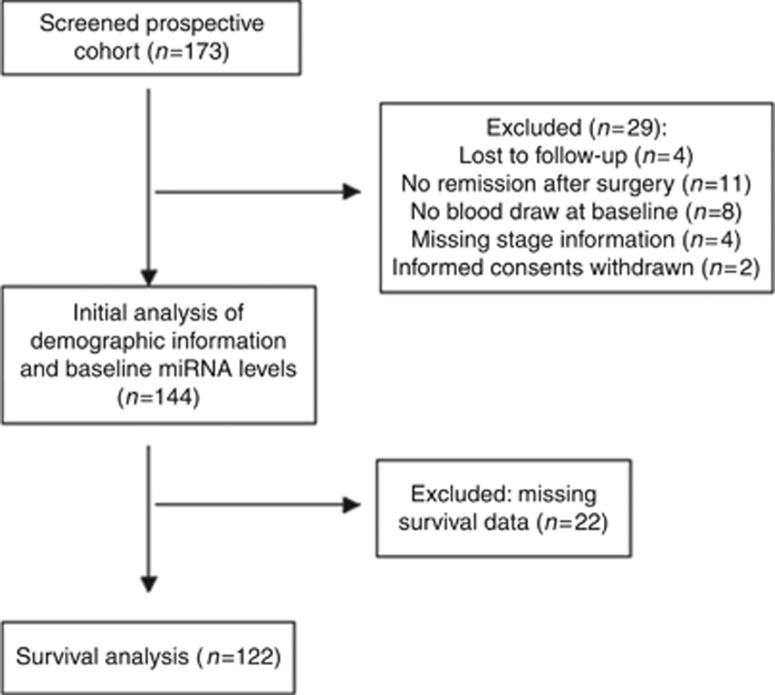
Between 2007 and 2011, 144 eligible CRC patients were enrolled into this study and had plasma samples obtained prior to surgery and at serial time points after curative surgery evaluated for miRNA levels. Of the 144 patients, 122 had recurrence-free survival information available (Figure 1). A summary of demographic information for both the total 144 patients and 122 patients with RFS data is included in Table 1. The median age of this total population was 56 years, with the subset of patients with RFS data being slightly older (median of 57 years). The majority of patients were male (56 male and 44% female), Caucasian (86%), and had advanced disease (Stage III/IV 62 vs I/II 38%). The majority of the patients had colon cancer (59%) vs rectal cancer (41%). There was no significant difference in these demographic features in the patients with RFS data available.
Figure 1.
Schematic diagram of study population.
Table 1. Demographic information.
|
All patients (N=144) |
Patients with RFS data (N=122) |
|||
|---|---|---|---|---|
| % | % | |||
| Age | N | N | ||
| Median [range] | 56.1 | [20.9, 103.5] | 57.6 | [20.9, 80.6] |
| <56 | 71 | 49% | 67 | 55% |
| ⩾56 | 73 | 51% | 55 | 45% |
| Gender | N | N | ||
| Female | 63 | 44% | 57 | 47% |
| Male | 81 | 56% | 65 | 53% |
| Race | N | N | ||
| White | 124 | 86% | 85 | 89% |
| Non-whitea | 18 | 13% | 11 | 11% |
| Height (cm) | ||||
| Median [range] | 170.6 | [147.3, 195.6] | 170 | [147.3, 195.6] |
| TNM stageb | N | N | ||
| I/II | 55 | 38% | 50 | 41% |
| III/IV | 89 | 62% | 72 | 59% |
| Tumour location | N | N | ||
| Colon | 85 | 59% | 70 | 57% |
| Rectum | 59 | 41% | 52 | 43% |
Abbreviation: RFS= recurrence-free survival; TNM=tumour-lymph nodes-metastasis.
Non-white included: Asian: n=11 (8%), Hispanic: n=1 (1%), Black: n=1 (1%), more than one: n=5 (3%); unknown: n=2 (1%) (The unknown patients were not included in the race analysis).
The TNM Classification of Malignant Tumours (tumour-lymph nodes-metastasis).
Correlation of plasma miRNA levels with clinical features of CRC patients
The miRNA (log-transformed) levels are summarised in Table 2, along with the number of patients with available data at each time point. Four miRNAs (miR-31, 203, 141, 200b) were not detected in >50% of cases and were determined to be not evaluable. It should be noted that pre-operative plasma miR-203 was detected in only six patients and was below the limits of detection in the other samples. These miRNAs had been initially selected in part because of their low levels in plasma and consequent potential to be CRC-specific circulating biomarkers. However, they were not detectable in the CRC patients in this study and thus were not assessed further.
Table 2. Summary of plasma miRNA levels at serial time points.
|
Pre-operative |
6 Months |
12 Months |
24 Months |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Mean* | Median* | (Min, Max) | N | Mean* | Median* | (Min, Max) | N | Mean* | Median* | (Min, Max) | N | Mean* | Median* | (Min, Max) | |
| miR125b | 137 | 10.32 | 10.23 | (8.16, 19.2) | 89 | 10.24 | 10.1 | (8.76, 14.55) | 77 | 10.02 | 9.92 | (8.34, 12.36) | 30 | 10.12 | 9.93 | (9, 15.33) |
| miR141 | 22 | 9.61 | 9.36 | (8.05, 12.84) | 16 | 9.91, | 9.49 | (8.77, 15.04) | 11 | 8.98 | 8.8 | (8.2, 9.89) | 2 | 8.87 | 8.87 | (8.41, 9.33) |
| miR16 | 144 | 15.24 | 15.15 | (12.67, 25.45) | 94 | 15.23 | 15.18 | (9.94, 20.33) | 79 | 15.35 | 15.36 | (11.07, 17.45) | 34 | 15.69 | 15.45 | (14.11, 20.57) |
| miR17-3p | 130 | 8.78 | 8.59 | (6.83, 19.5) | 87 | 8.66 | 8.64 | (7.09, 11.54) | 72 | 8.82 | 8.8 | (7.17, 11.23) | 26 | 9.19 | 8.89 | (7.65, 15.68) |
| miR200b | 58 | 9.35 | 9.21 | (7.15, 19.15) | 48 | 9.38 | 9.39 | (7.79, 12.14) | 42 | 9.27 | 9.21 | (7.63, 11.28) | 21 | 9.02 | 9.07 | (7.24, 12.44) |
| miR203a | 6 | 11.01 | 10.21 | (9.72, 14.27) | 9 | 10 | 9.32 | (8.45, 13.59) | 3 | 11.76 | 10.2 | (9.41, 15.66) | 2 | 10.11 | 10.11 | (8.66, 11.56) |
| miR29a | 133 | 13.93 | 13.79 | (12.21, 23.49) | 87 | 13.91 | 13.92 | (12.55, 16.6) | 76 | 13.99 | 14.01 | (12.53, 16.35) | 31 | 14.14 | 13.83 | (12.67, 19.87) |
| miR31 | 27 | 9.28 | 9.31 | (8.03, 11.58) | 18 | 9.46 | 9.41 | (8.51, 10.91) | 21 | 9.45 | 9.52 | (8.49, 11.53) | 10 | 9.19 | 9.33 | (8.3, 10.33) |
| miR92a | 144 | 15.56 | 15.58 | (8.71, 25.79) | 94 | 15.53 | 15.62 | (10.14, 18.72) | 80 | 15.81 | 15.78 | (14.06, 18.36) | 34 | 15.5 | 15.47 | (8.52, 22.45) |
*miRNA measured in ln (normalised copies).
miR-203 was detected in only six patients prior to surgery.
The associations between miRNA levels over time and patient characteristics are presented in Table 3. For the majority of the miRNAs, we did not observe any associations with plasma miRNA levels and clinicopathological features of the CRCs, except for increased miR-125b levels in older patients. Higher miR-125b levels were associated with older age (mean difference of −0.45 for age⩾56 years vs<56 years; P=0.03). Elevated miR-29a levels were observed in stage IV, compared to stage II (P=0.042) or stage III (P=0.023) CRC patients (Supplementary Figure S1A). The other eight plasma miRNA levels did not show any associations with stages (Supplementary Figure S2).
Table 3. The associations between plasma miRNA levels over time with patient characteristics.
|
miR125b† |
miR141† |
miR16† |
miR17-3p† |
miR200b† |
miR29a† |
miR31† |
miR92a† |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate§ (95% CI) | P-val | Estimate (95% CI) | P-val | Estimate (95% CI) | P-val | Estimate (95% CI) | P-val | Estimate (95% CI) | P-val | Estimate (95% CI) | P-val | Estimate (95% CI) | P-val | Estimate (95% CI) | P-val | |
| Intercept | 10.54 (9.68, 11.4) | 9.5 (7.03, 11.96) | 15.38 (14.62, 16.13) | 8.96 (7.95, 9.96) | 9.65 (8.26, 11.03) | 13.59 (12.75, 14.43) | 8.56 (7.57, 9.56) | 15.81 (14.95, 16.66) | ||||||||
| Age | 0.03 | 0.19 | 0.13 | 0.09 | 0.08 | 0.26 | 0.40 | 0.28 | ||||||||
| ⩾56 | −0.45 (−0.87, −0.04) | 0.93 (−0.66, 2.52) | −0.29 (−0.66, 0.09) | −0.44 (−0.93, 0.06) | −0.62 (−1.32, 0.07) | −0.24 (−0.65, 0.18) | −0.21 (−0.74, 0.32) | −0.23 (−0.65, 0.19) | ||||||||
| <56a | ||||||||||||||||
| Gender | 0.37 | 0.93 | 0.55 | 0.46 | 0.17 | 0.42 | 0.97 | 0.91 | ||||||||
| Male | −0.19 (−0.62, 0.23) | 0.06 (−1.65, 1.77) | 0.12 (−0.26, 0.49) | −0.19 (−0.7, 0.31) | −0.5 (−1.21, 0.21) | −0.17 (−0.59, 0.25) | −0.01 (−0.49, 0.47) | 0.02 (−0.4, 0.45) | ||||||||
| Femalea | ||||||||||||||||
| Race | 0.78 | 0.61 | 0.91 | 0.73 | 0.74 | 0.23 | 0.31 | 0.87 | ||||||||
| White | 0.09 (−0.55, 0.73) | −0.45 (−2.58, 1.68) | −0.03 (−0.59, 0.52) | 0.13 (−0.62, 0.88) | −0.17 (−1.21, 0.87) | 0.38 (−0.24, 1.01) | 0.41 (−0.43, 1.26) | 0.05 (−0.57, 0.68) | ||||||||
| Non-whitea | ||||||||||||||||
| Stage | 0.37 | 0.70 | 0.83 | 0.27 | 0.47 | 0.09 | 0.20 | 0.69 | ||||||||
| III/IV | 0.2 (−0.23, 0.62) | 0.32 (−1.68, 2.31) | 0.04 (−0.35, 0.43) | 0.29 (−0.22, 0.81) | 0.28 (−0.49, 1.04) | 0.37 (−0.06, 0.81) | 0.36 (−0.21, 0.93) | −0.09 (−0.53, 0.35) | ||||||||
| I/IIa | ||||||||||||||||
| Tumour locat | 0.77 | 0.84 | 0.99 | 0.58 | 0.28 | 0.87 | 0.59 | 0.45 | ||||||||
| Colon | −0.06 (−0.48, 0.36) | 0.14 (−1.54, 1.81) | 0 (−0.38, 0.37) | −0.14 (−0.65, 0.36) | 0.4 (−0.32, 1.12) | 0.04 (−0.39, 0.46) | 0.15 (−0.43, 0.72) | −0.16 (−0.59, 0.26) | ||||||||
| Rectuma | ||||||||||||||||
| Timepoint | 0.10 | 0.61 | 0.16 | 0.27 | 0.74 | 0.00 | 0.39 | 0.49 | ||||||||
|
Baselinea | ||||||||||||||||
| 6 Mo | −0.14 (−0.39, 0.1) | −0.07 (−1.09, 0.96) | 0.01 (−0.29, 0.31) | −0.13 (−0.41, 0.14) | 0.08 (−0.33, 0.49) | 0.03 (−0.22, 0.27) | 0.28 (−0.16, 0.73) | −0.02 (−0.36, 0.32) | ||||||||
| 12 Mo | −0.28 (−0.54, −0.02) | −0.49 (−1.5, 0.51) | 0.15 (−0.17, 0.47) | 0.04 (−0.25, 0.33) | 0.24 (−0.19, 0.66) | 0.12 (−0.13, 0.37) | 0.28 (−0.14, 0.7) | 0.23 (−0.13, 0.59) | ||||||||
| 24 Mo | 0.12 (−0.28, 0.51) | −0.31 (−1.8, 1.18) | 0.51 (0.05, 0.97) | 0.33 (−0.12, 0.79) | 0.08 (−0.49, 0.65) | 0.37 (−0.01, 0.74) | −0.02 (−0.65, 0.61) | 0.23 (−0.29, 0.75) | ||||||||
Abbreviations: CI=confidence interval; P-val=P-value; locat=location; Mo=month. † miR203 analysis not done due to small sample size; miRNA measured in ln (normalised copies).
Reference.
Of note, we analysed another data set of circulating miRNA in CRC patients (NCBI, GEO database, GSE67075), which contains samples from 8 healthy subjects, 8 subjects with polyps, 16 subjects with adenomas, 8 subjects with stages I/II early CRC and 8 subjects with stages III/IV CRC. We found that plasma levels of miR-29, miR-200b, and miR-203 in patients with CRC were greatly increased when compared to subjects with polyps or healthy control subjects (Supplementary Figure S3).
Association of plasma miRNA levels with recurrence-free survival
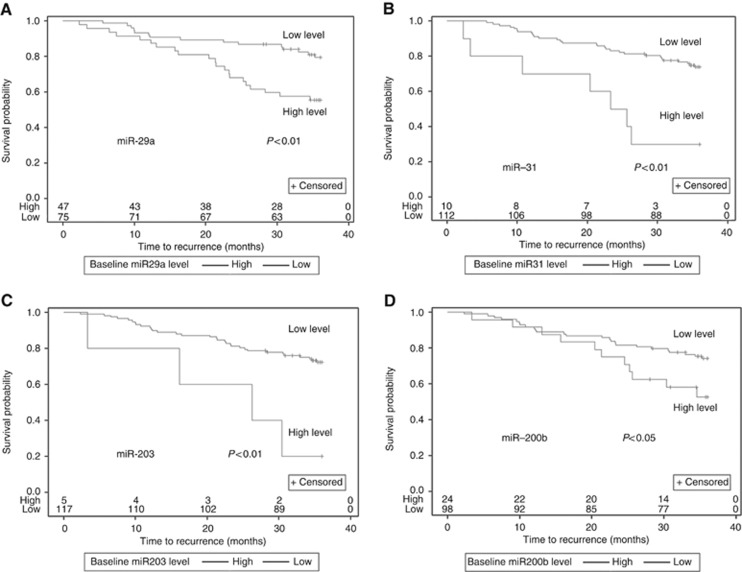
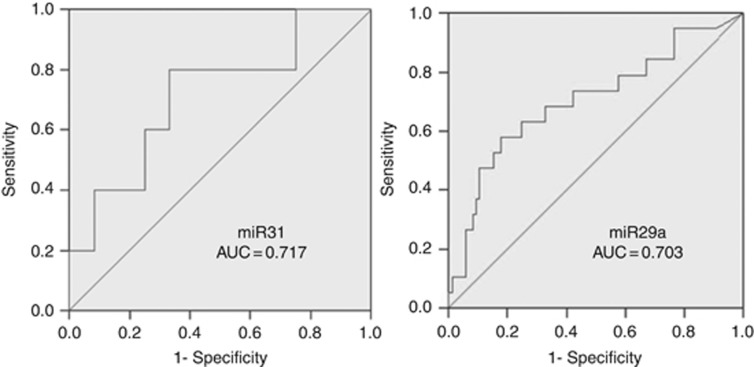
Pre-operative (Baseline) plasma miRNA levels above the median were associated with an increased risk of recurrence for miR-200b (HR=2.04 [95% CI: 1.00, 4.16], P=0.05), miR-203 (HR=4.2 [95% CI: 1.48, 11.93], P=0.007), miR-29a (HR=2.61 [95% CI: 1.34, 5.07], P=0.005), and miR-31 (HR=4.03 [95% CI: 1.76, 9.24], P=0.001). The associations between pre-operative miRNA levels and RFS are shown in Figure 2 and Supplementary Figure S4. We also performed receiver operating characteristic analysis of these plasma miRNAs for detecting tumour recurrence. Both plasma miR-31 (AUC: 0.717 [95% CI: 0.439, 0.995]) and miR-29a (AUC: 0.703 [95% CI: 0.562, 0.845]) showed promising prognostic significance and could discriminate patients with recurrence from those without recurrence (Figure 3A and B). Other miRNAs did not show prognostic significance, including miR200b (AUC: 0.578 [95% CI: 0.390, 0.766]), miR125b (AUC: 0.531, [95% CI: 0.381, 0.682]), miR16 (AUC: 0.557 [95% CI: 0.432, 0.682]), miR17-3p (AUC: 0.502 [95% CI: 0.346, 0.659]), and miR92a (AUC: 0.475 [95% CI: 0.318, 0.632]) (Supplementary Figure S5). In addition, we found elevated pre-operative levels of miR-29a were observed in recurrent patients of all stages (P=0.010) (Supplementary Figure S1B) and stage III patients in subgroup analysis (P=0.042) (Supplementary Figure 1C). Pre-operative levels of the other eight miRNAs did not show a significant association with recurrence (Supplementary Figure S6).
Figure 2.
The 3-year recurrence-free survival (RFS) analyses for pre-operative plasma miRNAs. Kaplan–Meier analysis was made in CRC patients (N=122). (A–D) Higher pre-operative plasma miR-29a (P<0.01), miR-203 (P<0.01), miR-31 (P<0.01) and miR-200b (P=0.05) predict a shorter RFS. For each miRNA, the median value was used to divide the samples into those with high or low levels. The number of patients at risk is shown below each curve.
Figure 3.
Preoperative plasma miR31 and 29a can discriminate patients developing recurrence. Analyses of receive operating characteristic (ROC) curve showed prognostic power of plasma miR31 (AUC: 0.717 (95% CI: 0.439, 0.995)) and miR29a (AUC: 0.703 (95% CI: 0.562, 0.845)), which could discriminate patients with recurrence from these without recurrence.
Dynamic plasma miRNAs changes can predict tumour recurrence
The associations between RFS and serial plasma miRNA levels at baseline (pre-operative), 6, 12, and 24 months are shown in Table 4. Higher levels of miR-31 were associated with a three-fold increased risk of recurrence across all time points (HR=3.11 [95% CI: 1.11, 8.76], P=0.03). Elevated miR-29a (HR=1.32 [95% CI: 0.97, 1.8], P=0.08) and miR-200b (HR=1.37 [95% CI: 0.95, 1.96], P=0.09) also showed associations with higher risks of CRC recurrence, although the results did not reach statistical significance.
Table 4. Hazard ratios for recurrence-free survival (RFS) using plasma miRNA levels obtained at serial time points (time-varying Cox model).
| miRNAa | N | CRC recurrences or deaths (N)b | HR (95% CI)c | P-Value |
|---|---|---|---|---|
| miR125b | 122 | 17 | 1.3 (0.93, 1.82) | 0.13 |
| mirR141 | 122 | 3 | 1.71 (0.75, 3.9) | 0.20 |
| miR16 | 122 | 18 | 1.22 (0.86, 1.72) | 0.27 |
| miR17-3p | 122 | 15 | 1.21 (0.83, 1.77) | 0.33 |
| miR200b | 122 | 11 | 1.37 (0.95, 1.96) | 0.09 |
| miR203 | 122 | 2 | 0.45 (0.04, 4.8) | 0.51 |
| miR29a | 122 | 17 | 1.32 (0.97, 1.8) | 0.08 |
| miR31 | 122 | 6 | 3.11 (1.11, 8.76) | 0.03 |
| miR92a | 122 | 18 | 1.2 (0.85, 1.71) | 0.31 |
Abbreviations: CI=confidence interval; CRC= colorectal cancer; HR=hazard ratio.
Variables are included as continuous (log-transformed).
Recurrence or Death: event based on whichever came first.
The hazard ratio represents the increased/decreased risk of RFS for a 1−log increase in the miRNA level.
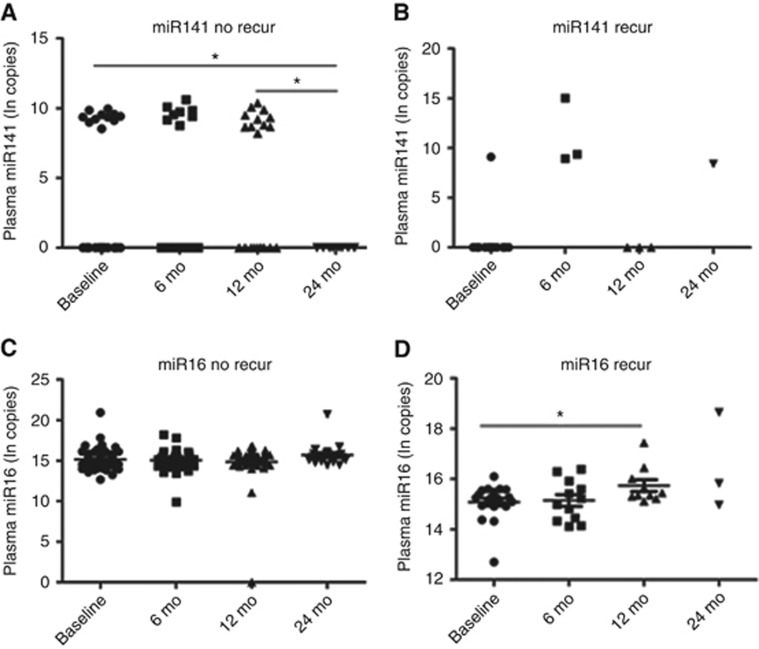
We also evaluated for changes in plasma miRNAs levels after surgery in patients with recurrence or without recurrence. The majority of miRNA levels did not show significant changes over time, predominantly because they were not detectable in plasma (e.g., miR-203) or were detectable in a small number of subjects (Supplementary Figures S7 and S8). Plasma miR141 was decreased 24 months after surgery, comparing to baseline (P=0.035, paired t-test, n=9 pairs) and 12 months after surgery (P=0.014, paired t-test, n=9 pairs) in patients without recurrence compared to baseline (Figure 4A and B), which means that decreased post-operative plasma miR-141 suggesting post-operative levels may be useful for predicting recurrence. Plasma miR-16 was increased 12 months after surgery compared to baseline (P=0.036, paired t-test, n=10 pairs) in the recurrence group (Figure 4C and D). The median RFS of these 10 patients were 17.5 months (8–33 months), and most of the recurrences happened after the increased plasma miR16 was detected. There were not enough evaluable patients with a 24-month time-point measurement to assess this time point. These results suggest post-operative levels of miR-16 may be useful for predicting CRC recurrence.
Figure 4.
Dynamic changes of plasma miR141,16 at different time points in patients with or without recurrence. Plasma miR141 was decreased 24 months after surgery, comparing to baseline (P=0.035, paired t-test, n=9 pairs) and 12 months after surgery (P=0.014, paired t-test, n=9 pairs) in patients without recurrence (A and B). Plasma miR-16 increased 12 months after surgery comparing to baseline (P=0.036, paired t-test, n=10 pairs) in recurrence group (C and D). recur=recurrence. *P<0.05.
The source of circulating miRNAs
To address the question of the possible source of the circulating miRNAs, we analysed tissue miRNA levels in one available online data set (GSE28364), which consists of 40 CRC patients and matched normal control tissues, and we found tissue miR-29a (P<0.001), miR-31 (P<0.001), and miR-203 (P=0.001) were also greatly increased in CRC tissues compared to normal control tissues (Supplementary Figure S9). Thus, it is plausible that the increased plasma miRNAs may come from the primary tumours. Interestingly, in the GSE28364 samples there was no association between plasma miR-29a and gene mutations in APC, TP53, and KRAS, with Chr18q loss of heterozygosity or with traditional serum biomarkers of CRC including CEA and CA199 (Supplementary Table S1).
We also analysed another online data set (GSE39833), which used miRNA microarrays to detect circulating exosomal miRNAs in 88 CRC patients and 11 healthy control subjects. In this study, plasma exosomal miR-29a, 200b and 31 could discriminate CRC patients from normal controls. Thus, it is also possible that circulating miRNAs may be derived from shed exosomes from the CRCs (Supplementary Figure S10).
Discussion
Current staging and surveillance protocols for CRC patients using the tumour-lymph nodes-metastasis staging system and serum CEA, CT imaging studies and endoscopy are suboptimal for determining prognosis and for maximising survival in the setting of CRC recurrence. The traditional serum biomarker CEA has a modest sensitivity for CRC detection, which varies from 43 to 69% (Hundt et al, 2007; Brunet Vega et al, 2013). This has led to an intense effort to develop biomarker assays that can more accurately determine the prognosis and enhance our ability to detect CRC recurrence early. Plasma miRNAs are an emerging class of non-invasive biomarkers in CRC. However, to date the studies of plasma miRNA biomarkers have shown inconsistent and poorly reproducible results (Yamada et al, 2015; Sun et al, 2016). This inconsistency led us to conduct a study using a carefully collected set of plasma samples from CRC patients that assessed candidate miRNA using a robust assay method.
The primary emphasis of this study was to identify potential candidate miRNAs in the plasma of CRC patients using a set of candidate miRNAs that had been selected on the basis of low levels in the plasma of healthy control subjects and high expression in primary CRCs. We found that increased circulating miRNAs of certain miRNAs levels prior to surgery could predict shorter RFS (miR-29a, 31, 203, 200b). Both miR-31 (AUC: 0.717) and miR-29a (AUC: 0.703) could discriminate patients with recurrence from these without recurrence. These results suggest that these plasma miRNAs could be useful prognostic biomarkers. In addition, higher plasma levels of miR-31 in the post-operative setting at all time-points assessed was associated with a three-fold increased risk of recurrence. We also found that changes in postoperative plasma miR-141 and 16 levels associated with tumour recurrence. We did not observe a correlation between plasma levels of the other candidate miRNAs and recurrence. It is unclear if this is a consequence of an insufficient sample size or other technical issues, or a true lack of prognostic significance of these circulating miRNAs. More sensitive and precise technologies, such as droplet digital PCR (ddPCR), could address these possibilities (Hindson et al, 2013). Our results do support the potential for a subset of miRNAs to be used as circulating biomarkers for CRC. Indeed, prior studies from our team using a non-invasive blood test based on circulating miRNAs in a case–case study design demonstrated that the miRNAs may be useful in the post-diagnosis for CRC surveillance (Adams et al, 2014).
Our results as well as those of others that have found elevated plasma miRNA levels in CRC patients raise the question of the source of plasma miRNAs. We did not directly assess the levels of these miRNAs in primary tumour tissues in our study; however, previously published studies have shown elevated miR-31 levels in CRC tissue (Reid et al, 2012). We assessed publically available data sets to attempt to address this question and found that tissue miR-29a, 31 and 203 were greatly increased in CRC tissues compared normal control tissues. By analysing these public data sets, we found plasma miR-29a, 31 and 203 were increased in CRC, compared to healthy controls. These above results revealed that increased plasma miRNAs may come from primary tumours, but additional studies are needed to assess the association between circulating plasma miRNA levels with the levels in matched CRC tissues.
Furthermore, of relevance to our findings, recent studies have shown tumours can actively release exosomes containing miRNAs into the peripheral circulation to exchange information with distant cells, and thus mediate tumour metastasis (Valadi et al, 2007; Masuda and Izpisua Belmonte, 2014; Tkach and Thery, 2016). In this study, by analysing publically available datasets, we found that plasma exosomal miR-29a, 200b and 31 could discriminate CRC patients from normal control subjects, which also reveals a plausible mechanism through which plasma miRNA could promote CRC recurrence or metastasis by the exosomal delivery of the miRNAs to distant sites (Ogata-Kawata et al, 2014).
Our results add to a rapidly accumulating knowledge base about miRNAs in CRC prognosis and prediction. Increased plasma miR-203 (Hur et al, 2015; Sun et al, 2016) and miR-200b (Adams et al, 2014; Sun et al, 2016) have been shown to discriminate patients with CRC from healthy control subjects and to predict prognosis (Adams et al, 2014). Tissue levels of miR-31 have been reported to be upregulated in CRC tissue (Kanaan et al, 2012) and to correlate with mutations in BRAF, KRAS and APC (Reid et al, 2012; Nosho et al, 2014). We now add to our understanding of these miRNAs in CRC patients by showing elevated plasma miR-31, miR-29a, miR-203, and miR-200b levels can predict shortened RFS in CRC patients.
With regards to miR-29a, plasma miR-29a has been shown to be elevated in patients with CRC compared to healthy control subjects (Huang et al, 2010; Adams et al, 2014; Yamada et al, 2015). Furthermore, Yamada et al recently reported the combination of plasma miR-29a, miR-21 and miR-125b had substantial diagnostic power to discriminate patients with colorectal neoplasms from healthy control subjects (AUC=0.83) (Yamada et al, 2015). In this study, we provide additional evidence that plasma miR-29a may be a promising biomarker for prognosis and early detection of recurrence. Interestingly though, in our analysis of publically available cohorts, we found no associations between miR-29a and gene mutations in APC, TP53, or KRAS, or with CEA levels. There are many reasons for this lack of association including the fact that APC and KRAS mutations usually occur early in the normal–adenoma–carcinoma sequence (Davies et al, 2005), while miR-29a is increased mainly in the late stages of CRC (Brunet Vega et al, 2013).
Notably, our study has several strengths related to the study design and analysis methods. We have used the most robust methods available for the collection of our study population. We obtained our samples from a prospective cohort study that includes all stages of CRC patients, which limits confounding effects related to uncontrolled pre-analytical factors. Methodologically, we used a validated and sensitive TaqMan qPCR assay, which is more reliable than most microarray methods or SYBR Green assays (Pritchard et al, 2012a). In the present study, we used the synthetic spiked-in cel-miR-39 for data normalisation, which has been shown to lead to more reproducible results (Sun et al, 2016). Finally, because plasma miRNAs are often at low concentration and not easily detectable, we used absolute quantification of plasma miRNA copy numbers instead of fold change of relative expression to reduce variation and increase the reproducibility of our results (Hindson et al, 2013).
Our present study also has some limitations. First, miR-17-3p, miR-29a, and miR-92 are all expressed at high levels in blood cells. Pritchard et al reported that plasma miRNAs may be confounded by blood contaminants secondary to differential haemolysis or other uncontrolled pre-analytic variables (Pritchard et al, 2012b). They further found that 58% of proposed plasma miRNAs biomarkers are highly expressed in blood cells. Most plasma miRNA studies to date have not adequately addressed this issue, and thus the results of these studies and our study need to be interpreted with caution. Second, we only assayed nine miRNAs in the plasma and did not conduct an unbiased screen for candidate plasma miRNAs. Consequently, we may not have identified the best performing prognostic plasma miRNAs. We also wish to note that our candidate miRNAs were selected in 2014, prior to more recent publications on other potential miRNA biomarkers, such as plasma miR-200c (Toiyama et al, 2014), miR-21 (Rokkas et al, 2015), miR-96 (Sun et al, 2016), and panels of other miRNAs (Maierthaler et al, 2017). Thus, these miRNAs are not included in our study. Third, the sample size of our study may have led to an inability to detect plasma miRNAs that are modest prognostic markers. Fourth, our study included both colon and rectal cancer patients despite the fact that tumours in the colon and rectum have many distinct features. The inclusion of both colon and rectal cancers may have lessened our ability to detect true associations between some of the miRNA plasma levels and CRC recurrence. Finally, we did not analyse the associations between all pathological parameters and miRNAs levels, which could have resulted in missing some potentially significant findings. Finally, our study followed patients for 3 years after surgical resection and may have misclassified some recurrence-free patients who will eventually develop CRC recurrence.
In conclusion, our results suggest plasma miR-29a, 200b, 203, and 31 are potential CRC prognostic biomarkers. In addition, dynamic postoperative plasma miR-31, 141, and 16 appear to be potential biomarkers for the early detection of recurrence and in postoperative surveillance. Also, it is possible that with more technically sensitive assays that we may be able to detect those miRNAs that were not detectable in our study, which would allow us to assess these candidate miRNA biomarkers as recurrence markers in CRC surveillance. Large-scale prospective trials are now needed to validate these findings and to determine the potential value of these plasma miRNAs in clinical practice.
Acknowledgments
The work was supported by following grants: NIH grants (P30CA15704, UO1CA152756, R01CA194663, U54CA143862, P01CA077852), RACE Charities, and a Burroughs Wellcome Fund Translational Research Award for Clinician Scientist; R03 CA165153, R01 CA112516, R01 CA114467, R01 CA120523, U24 CA074794, Listwin Family Foundation, Fred Hutchinson Cancer Research Center and the International Program Fund for PhD. Candidates of Chinese Sun Yat-Sen University, National Natural Science Foundation of China (81372566 to L Wang), National Key Technology R&D Program for the 12th Five-Year Plan of China (2014BAI09B06 to L Wang). We acknowledge the technical contributions of Christine Rimorin. We thank the ColoCare Seattle staff, including Shannon Rush, Rachel Wilbur, Sara Bates, Brian Cooper and Kathy Vickers for their contributions.
Author contributions
Study concept and design: Zixu Yuan, Scott V Adams, Karen Makar, William M Grady; provision of clinical specimen and information: Kathy Vickers, Brandon Dickinson, William M Grady, Karen Makar, Michele Wurshur; acquisition and analysis of data: Zixu Yuan, Scott V Adams, William M Grady; Kelsey Baker, Karen Makar, Mary Redman, Neli Ulrich, Jürgen Böhm, Ming Yu; statistical analysis: Zixu Yuan, Kelsey Baker, Mary Redman; manuscript writing and revision: Zixu Yuan, William M Grady, Scott V Adams, Lei Wang.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Adams SV, Newcomb PA, Burnett-Hartman AN, Wurscher MA, Mandelson M, Upton MP, Zhu LC, Potter JD, Makar KW (2014) Rare circulating microRNAs as biomarkers of colorectal neoplasia. PLoS One 9: e108668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet Vega A, Pericay C, Moya I, Ferrer A, Dotor E, Pisa A, Casalots A, Serra-Aracil X, Oliva JC, Ruiz A, Saigi E (2013) microRNA expression profile in stage III colorectal cancer: circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep 30: 320–326. [DOI] [PubMed] [Google Scholar]
- Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W (2011) Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One 6: e17745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies RJ, Miller R, Coleman N (2005) Colorectal cancer screening: prospects for molecular stool analysis. Nat Rev Cancer 5: 199–209. [DOI] [PubMed] [Google Scholar]
- Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M (2013) Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 10: 1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wang Z, Liao BY, Yu L, Gao X, Lu S, Wang S, Dai Z, Zhang X, Chen Q, Qiu SJ, Wu Y, Zhu H, Fan J, Zhou J, Wang J (2014) Human miR-1228 as a stable endogenous control for the quantification of circulating microRNAs in cancer patients. Int J Cancer 135: 1187–1194. [DOI] [PubMed] [Google Scholar]
- Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X (2010) Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 127: 118–126. [DOI] [PubMed] [Google Scholar]
- Hundt S, Haug U, Brenner H (2007) Blood markers for early detection of colorectal cancer: a systematic review. Cancer Epidemiol Biomarkers Prev 16: 1935–1953. [DOI] [PubMed] [Google Scholar]
- Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, Goel A (2015) Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 66: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, Galandiuk S (2012) Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg 256: 544–551. [DOI] [PubMed] [Google Scholar]
- Kocarnik JM, Shiovitz S, Phipps AI (2015) Molecular phenotypes of colorectal cancer and potential clinical applications. Gastroenterol Rep 3: 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chu KM (2016) Circulating cell-free DNAs and miRNAs as promising non-invasive biomarkers for early detection of gastric cancer. Neoplasma 63: 1–9. [DOI] [PubMed] [Google Scholar]
- Maierthaler M, Benner A, Hoffmeister M, Surowy H, Jansen L, Knebel P, Chang-Claude J, Brenner H, Burwinkel B (2017) Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int J Cancer 140: 176–187. [DOI] [PubMed] [Google Scholar]
- Masuda S, Izpisua Belmonte JC (2014) Re: Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst 106: djt457. [DOI] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ (2009) Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58: 1375–1381. [DOI] [PubMed] [Google Scholar]
- Nishida N, Yokobori T, Mimori K, Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y, Kuwano H, Mori M (2011) MicroRNA miR-125b is a prognostic marker in human colorectal cancer. Int J Oncol 38: 1437–1443. [DOI] [PubMed] [Google Scholar]
- Nosho K, Igarashi H, Nojima M, Ito M, Maruyama R, Yoshii S, Naito T, Sukawa Y, Mikami M, Sumioka W, Yamamoto E, Kurokawa S, Adachi Y, Takahashi H, Okuda H, Kusumi T, Hosokawa M, Fujita M, Hasegawa T, Okita K, Hirata K, Suzuki H, Yamamoto H, Shinomura Y (2014) Association of microRNA-31 with BRAF mutation, colorectal cancer survival and serrated pathway. Carcinogenesis 35: 776–783. [DOI] [PubMed] [Google Scholar]
- Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N (2014) Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 9: e92921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Cheng HH, Tewari M (2012. a) MicroRNA profiling: approaches and considerations. Nat Rev Genet 13: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, Tewari M (2012. b) Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 5: 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JF, Sokolova V, Zoni E, Lampis A, Pizzamiglio S, Bertan C, Zanutto S, Perrone F, Camerini T, Gallino G, Verderio P, Leo E, Pilotti S, Gariboldi M, Pierotti MA (2012) miRNA profiling in colorectal cancer highlights miR-1 involvement in MET-dependent proliferation. Mol Cancer Res 10: 504–515. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Moranta F, Salo J, Arcusa A, Boadas J, Pinol V, Bessa X, Batiste-Alentorn E, Lacy AM, Delgado S, Maurel J, Pique JM, Castells A (2006) Postoperative surveillance in patients with colorectal cancer who have undergone curative resection: a prospective, multicenter, randomized, controlled trial. J Clin Oncol 24: 386–393. [DOI] [PubMed] [Google Scholar]
- Rokkas T, Kothonas F, Rokka A, Koukoulis G, Symvoulakis E (2015) The role of circulating microRNAs as novel biomarkers in diagnosing colorectal cancer: a meta-analysis. Eur J Gastroenterol Hepatol 27: 819–825. [DOI] [PubMed] [Google Scholar]
- Schubert M, Junker K, Heinzelmann J (2016) Prognostic and predictive miRNA biomarkers in bladder, kidney and prostate cancer: where do we stand in biomarker development? J Cancer Res Clin Oncol 142: 1673–1695. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63. [DOI] [PubMed] [Google Scholar]
- Siegel R, Ma J, Zou Z, Jemal A (2014) Cancer statistics, 2014. CA Cancer J Clin 64: 9–29. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liu Y, Cogdell D, Calin GA, Sun B, Kopetz S, Hamilton SR, Zhang W (2016) Examining plasma microRNA markers for colorectal cancer at different stages. Oncotarget 7: 11434–11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach M, Thery C (2016) Communication by extracellular vesicles: where we are and where we need to go. Cell 164: 1226–1232. [DOI] [PubMed] [Google Scholar]
- Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A (2014) Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg 259: 735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A (2013) Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst 105: 849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A, Weiz L, Langheinz A, Burwinkel B (2011) Characterization of extracellular circulating microRNA. Nucleic Acids Res 39: 7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654–659. [DOI] [PubMed] [Google Scholar]
- Visone R, Croce CM (2009) MiRNAs and cancer. Am J Pathol 174: 1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H, Kou T, Kusaka T, Sasaki Y, Yagi M, Higurashi T, Yukawa N, Amanuma Y, Kikuchi O, Muto M, Ueno Y, Nakajima A, Chiba T, Boland CR, Goel A (2015) Serum miR-21, miR-29a, and miR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin Cancer Res 21: 4234–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye LC, Liu TS, Ren L, Wei Y, Zhu DX, Zai SY, Ye QH, Yu Y, Xu B, Qin XY, Xu J (2013) Randomized controlled trial of cetuximab plus chemotherapy for patients with KRAS wild-type unresectable colorectal liver-limited metastases. J Clin Oncol 31: 1931–1938. [DOI] [PubMed] [Google Scholar]
- Young PE, Womeldorph CM, Johnson EK, Maykel JA, Brucher B, Stojadinovic A, Avital I, Nissan A, Steele SR (2014) Early detection of colorectal cancer recurrence in patients undergoing surgery with curative intent: current status and challenges. J Cancer 5: 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZX, Wang XY, Qin QY, Chen DF, Zhong QH, Wang L, Wang JP (2013) The prognostic role of BRAF mutation in metastatic colorectal cancer receiving anti-EGFR monoclonal antibodies: a meta-analysis. PLoS One 8: e65995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.