Abstract
Pears with red skin are attractive to consumers and provide additional health benefits. Identification of the gene(s) responsible for skin coloration can benefit cultivar selection and breeding. The use of QTL-seq, a bulked segregant analysis method, can be problematic when heterozygous parents are involved. The present study modified the QTL-seq method by introducing a |Δ(SNP-index)| parameter to improve the accuracy of mapping the red skin trait in a group of highly heterozygous Asian pears. The analyses were based on mixed DNA pools composed of 28 red-skinned and 27 green-skinned pear lines derived from a cross between the ‘Mantianhong’ and ‘Hongxiangsu’ red-skinned cultivars. The ‘Dangshansuli’ cultivar genome was used as reference for sequence alignment. An average single-nucleotide polymorphism (SNP) index was calculated using a sliding window approach (200-kb windows, 20-kb increments). Nine scaffolds within the candidate QTL interval were in the fifth linkage group from 111.9 to 177.1 cM. There was a significant linkage between the insertions/deletions and simple sequence repeat markers designed from the candidate intervals and the red/green skin (R/G) locus, which was in a 582.5-kb candidate interval that contained 81 predicted protein-coding gene models and was composed of two subintervals at the bottom of the fifth chromosome. The ZFRI 130-16, In2130-12 and In2130-16 markers located near the R/G locus could potentially be used to identify the red skin trait in Asian pear populations. This study provides new insights into the genetics controlling the red skin phenotype in this fruit.
Introduction
Pear, Pyrus L. (tribe Maleae, subfamily Amygdaloideae, family Rosaceae), is an important temperate fruit crop.1 In China (2014), the area used for pear production exceeded 1.12 million ha with a harvest of 18.10 million tons, accounting for 70.1% of global pear production (FAOSTAT, http://faostat.fao.org/). Pear fruit color is a major characteristic feature and is an important marketable trait.2 Green-, yellow- and russet brown-skinned pear cultivars are common in China, but red-skinned cultivars are rare.3–5 Previous research has indicated that consumers have a strong preference for red-skinned pears.6 In addition to the aesthetic appeal of their color, the anthocyanins in the red skin of pears have been shown to provide antioxidant benefits for human health.7,8 Compared with European pear cultivars, there are few full red-skinned Asian pear cultivars,4,9 and the red color usually appears on the side of the fruit that is exposed to the sun in these cultivars.
Although there is definite genetic governance of the trait, the extent of the red skin color of pear fruit is largely dependent on environmental conditions10 and is primarily because of the presence of anthocyanins.11–13 The biosynthesis and accumulation of anthocyanins are mainly determined by genetic factors and are influenced by light, temperature and other environmental factors.14 The peak of anthocyanin biosynthesis occurs in the middle stage of fruit development in P. communis L. and some Asian pear cultivars15–17 and at harvest in other Asian pear cultivars, a trend similar to that in apples.18,19 In pears, most of the structural and regulatory genes in the anthocyanin biosynthetic pathway have been identified and functionally characterized.20–24 Phenylalanine ammonia-lyase (PAL) and UDP-glucose: flavonoid-3-O-glucosyltransferase (UFGT) participate in the first and final steps of anthocyanin biosynthesis, respectively. The correlation between anthocyanin biosynthesis and PAL and UFGT activity is obvious in blushed pears but not in full red European pear fruits.19,25 In P. pyrifolia red-skinned ‘Mantianhong’ and ‘Aoguan’ cultivars, chalcone isomerase (CHI) activity, not PAL, was found to be closely related to anthocyanin biosynthesis and accumulation.17 A study on two other red-skinned pear cultivars, ‘Meirensu’ and ‘Yunhongli No. 1’, revealed that PAL induced anthocyanin biosynthesis and that UFGT was closely related to anthocyanin accumulation.18 Research has demonstrated that the varying levels of expression of anthocyanidin synthase (ANS), UFGT and tryptophan-aspartic acid repeat protein (WD40) genes and the coexpression of MYB10 and bHLH33 (basic helix–loop–helix 33) genes in anthocyanin biosynthesis form the basis for the differences in coloration between Asian and European pears.9,12 Additionally, the differential impact of the MYB–bHLH–WD40 complex on the expression of pear anthocyanin structural genes led to differences in the color pattern of red-skinned pears. A correlation between hypermethylation and the green-skinned pear phenotype was found by comparing the methylation levels in the PcMYB10 promoter in ‘Max Red Bartlett’ (P. communis L.) and its green-skinned mutant.25
The full red-skinned phenotype of the European pear ‘Max Red Bartlett’, a red bud mutant of ‘Bartlett’, is controlled by a dominant gene C.26 Pierantoni et al.24 mapped the position of PcMYB10 (LG9), noting that it corresponded to the same location as MdMYBa and MdMYB10 and that it controls the pigmentation of apple skin. PcMYB10, however, was not directly responsible for the red or yellow skin color in the above pear cultivars because the mutation underlying this color difference maps to a different region of the pear genome. Similarly, in the F1 hybrids of ‘BonRouge’ and ‘Packham's Triumph’ cultivars, the ratio of red- to green-skinned fruits was 1:1, which obeys Mendel's laws of inheritance.27 In addition, Dondini et al.13 analyzed seven crosses from red- and green-skinned European pears, reporting that the red-skinned phenotype in full-colored pears was controlled by a single dominant gene and that the dominant gene Red was located on linkage group four (LG4) of ‘Max Red Bartlett’. In contrast, researchers studied the F1 hybrids of two crosses from ‘Huobali’, a red-skinned Asian pear cultivar that is considered to have the genetic predisposition for russet brown, yellow and green skin coloration to be dominant over red skin coloration.28 Volz et al.29 analyzed red skin color segregation ratios in 10 Asian pear hybrid families derived from ‘Huobali’ and suggested that genetic control of blush color in this cultivar may be due to the complimentary action of at least two dominant genes. These results indicate that the genetic basis and mechanism for red skin color may differ in Asian and European pear cultivars.
Michelmore et al.30 developed a bulked segregant analysis (BSA) method for rapidly identifying markers linked to any specific gene or genomic region and used this method to identify three randomly amplified polymorphic DNA markers linked to downy mildew resistance in lettuce. The BSA method is applicable to qualitative trait gene and quantitative trait loci (QTL) analyses.31 With the development and widespread use of high-throughput whole-genome sequencing technology, BSA analysis has become a fast, efficient and widely used method for identifying gene mutations and their locations in the genome.32 Advances in the use of X-QTL,33,34 MutMap,35 MMAPPR32 and QTL-seq36 have been recently reported. Takagi et al.36 identified QTLs for partial resistance to rice blast disease and seedling vigor using QTL-seq on recombinant inbred lines and F2 populations of rice. They used whole-genome resequencing of two DNA bulks of progeny (20–50 individuals each) that exhibited extremely contrasting phenotypes. The DNA bulks were aligned to the reference sequence of one parental line to calculate the single-nucleotide polymorphism-index (SNP-index), and QTLs were identified as peaks or valleys in the SNP-index plot. This method can be generally applied in population genomic studies to rapidly identify genomic regions associated with specific traits. QTL-seq has been successfully applied in rice,36,37 foxtail millet,38 cucumber,39 tomato,40 chickpea41–43 and peach.44
Little information has been reported on the hereditary law of the red color trait in Asian pears. Because there is no significant correlation between the red-skinned trait and the color of young shoots and leaves, it has not been possible to make an initial selection for red-skinned fruit at the seedling stage by examining shoot or leaf color. Unfortunately, there are no molecular markers for the red- or green-skinned phenotypes. Therefore, the present study used a modified QTL-seq method to map the red or green skin trait in a segregating population of two red-skinned Asian pears. The initial focus of the study was to identify molecular markers that could be used in a marker-assisted breeding program. This study also provides a solid foundation for identifying the gene responsible for red-skinned fruit and the mechanisms involved in the development of red skin in Asian pears.
Materials and methods
Plant material
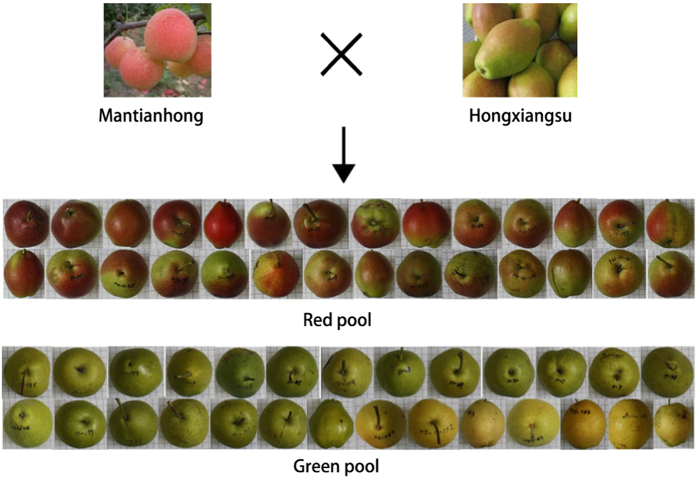
The red-skinned Asian pear cultivars ‘Mantianhong’ (P. pyrifolia), ‘Hongxiangsu’ (P. pyrifolia White Pear Group) and ‘Yuluxiang’ (P. pyrifolia White Pear Group) were grown as the parental lines in the orchard of the Zhengzhou Fruit Research Institute (ZFRI), Chinese Academy of Agricultural Sciences (CAAS) in Zhengzhou (Henan Province, China). One F1 hybrid population resulting from a cross between ‘Mantianhong’ and ‘Hongxiangsu’ (n=348) and another F1 hybrid population resulting from a cross between ‘Yuluxiang’ and ‘Mantianhong’ (n=458) were used as the study material. The populations were established in 2009 and grown at the affiliated experimental orchard of the ZFRI, CAAS in Xinxiang (Henan Province, China). Most individuals bore their first fruit in 2013. Trees bearing one or more fruits with a red blush surface (>0%) were considered the red-skinned phenotype, and trees with green to yellow coloration on all fruit surfaces were considered the green-skinned phenotype. Individuals with extreme red or green skin coloration in the population, resulting from the cross between the ‘Mantianhong’ and ‘Hongxiangsu’ cultivars, are shown in Figure 1.
Figure 1.
Fruit skin color phenotype of ‘Mantianhong’, ‘Hongxiangsu’ and the extreme phenotypes in the pooled red and green skinned individuals. Note: Both the female parent ‘Mantianhong’ and the male parent ‘Hongxiangsu’ of the population are with red skinned phenotypes. The majority of fruits of plants selected for the red pool exhibited a red blush over 50% of their fruit surface. All fruits of plants selected to represent the extreme phenotype of the green pool were obviously green to yellow and with no red blush apparent on their fruit surface.
DNA extraction and library construction
The first few young leaves that appeared at the end of the current shoot of each individual were collected at the beginning of the vegetative period of growth. The leaves were immediately frozen in liquid nitrogen and transferred to a −80 °C laboratory freezer. The genomic DNA was isolated using the cetyltrimethylammonium bromide method,45 and two libraries were constructed out of the pooled samples for QTL-seq, insertion/deletion (InDel) and simple sequence repeat analyses.
A red-skinned pool was constructed from equal amounts of DNA from 28 red-skinned pears, and a green-skinned pool was constructed from equal amounts of DNA from 27 green-skinned pears. Libraries with 350-bp inserted fragments were constructed using a TruSeq Nano DNA HT Sample Preparation Kit (Illumina Inc., San Diego, CA, USA) following the manufacturer’s protocol and sequenced using an Illumina HiSeq 4000 instrument (San Diego, CA, USA) to obtain 150-bp paired-end reads.
Sequence data analysis
Quality trimming was conducted to generate high confidence variant calling. The raw reads were processed to obtain high-quality clean reads by (1) removing reads with ⩾10% unidentified nucleotides and (2) removing reads with >50% bases having low Phred quality scores (<5). The Burrows-Wheeler Aligner46 was used to align the clean reads from each sample against the public reference genome for pears47 using ‘mem 4−k 32−M’, where k is the minimum seed length and M is an option used to mark shorter split alignment hits as secondary alignments. The resulting alignment was used to identify SNPs and InDels. Variant calling was performed on all samples using the GATK’s Unified Genotyper 3.3,48 and SNPs and InDels were filtered using GATK’s Variant Filtration with appropriate parameter settings (-Window 4, -filter ‘QD<4.0||FS>60.0||MQ<40.0 ‘, -G_filter ‘GQ<20’). Variants exhibiting segregation distortion or sequencing errors were discarded. The ANNOVAR Software Tool (Philadelphia, PA, USA)49 was used to align and annotate SNPs and InDels and to determine their physical locations in the genome.
The SNP-index was defined as the ratio between the number of reads of a mutant SNP and the number of reads corresponding to the SNP. The index is equal to 1 when all short reads differ from the reference genome and is equal to 0 when all short reads are identical to the reference genome.36 In homozygous crops, changes in the SNP-index and Δ(SNP-index) in the offspring pool are usually caused by the exchange and recombination of homologous chromosomes when the QTL-seq analysis of the genome of one of the parents is taken as the reference sequence. Using the Δ(SNP-index) for sliding window analysis, changes in the SNP-index and Δ(SNP-index) of the SNP sites within the candidate interval are consistent within the recombination section, and the candidate section and the different phenotypes correspond to the sign of the Δ(SNP-index) window (Figure 2a). When QTL-seq analysis is performed in heterozygous crops, the switch of the corresponding section of the reference sequence between two homologous chromosomes also results in changes in the SNP-index and Δ(SNP-index) of the offspring, producing an effect similar to that of homologous chromosome exchange, which we refer to as the ‘pseudoexchange effect.’ Site changes in the reference sequence of heterozygous crops are random, which may result in the opposite notation of the Δ(SNP-index) of the adjacent site. The window Δ(SNP-index) would tend to 0, and use of the Δ(SNP-index) for sliding window analysis may miss a portion of the candidate interval (Figure 2b).
Figure 2.
Demonstration of QTL-seq analysis under different scenarios. a: Scenario of chromosome recombination in a homozygous crop; b: Scenario of reference sequence change in a heterozygous crop. Case 1 in b: The ideal condition of using Δ (SNP-index) to locate the correct interval. Case 2 in b: The reference sequence is changed. Therefore, it is not possible to use Δ (SNP-index) to locate the trait. The red rectangle boxes indicate the sliding window representing QTL-seq analysis; ⟶ indicates the slide direction of the window; Window Δ (SNP-index) and Window |Δ (SNP-index)| indicates the average value of the Δ (SNP -index) and |Δ (SNP -index)| in the red rectangle boxes, respectively.
In this study, both parental lines belonging to the red-skinned pears were highly heterozygous, which was not ideal for calculating the SNP-index of the DNA pool of the F1 generations because the origin of the SNPs in the target area could not be estimated based on the positive/negative sign of the Δ(SNP-index) window. Therefore, the genome sequence of the ‘Dangshansuli’ 47 cultivar was used as the reference to calculate the SNP-index and Δ(SNP-index) between the red- and green-skinned pools to save labor. The average SNP-index of the SNPs in the window was calculated and used for the sliding window plot. Sliding window analysis was performed on the scaffold sequences in the genome file 225117_ref_Pbr_v1.0_chrUn.fa (ftp://ftp.ncbi.nlm.nih.gov/genomes/Pyrus_x_bretschneideri/CHR_Un/) and applied to the SNP-index, Δ(SNP-index) and |∆(SNP-index)| plots with 200-kb windows and 20-kb increments.
Because the SNP-index was based on a reference genome47 rather than the genome of the parental line, the origin of each SNP or the linkage around its neighboring region could not be estimated. Positive or negative values of the |Δ(SNP-index)| did not correspond to the phenotype of the parental lines. Reorganization and ‘pseudoexchange’ can change the positive and negative direction of the Δ (SNP-index) but cannot change its absolute value (i.e., |Δ(SNP-index)|). Although use of |Δ(SNP-index)| in a sliding window analysis ignores the positive and negative direction of Δ(SNP-index), it had good positioning ability, especially in locating trait loci in heterozygous crops (Figure 2b). Therefore, the |Δ(SNP-index)| was used as the main parameter to identify the target phenotype. As the length of the pear genetic map is ~1500–2000 cM and the resolution of an initial QTL mapping is typically ~5–10 cM, translating into ~0.5% of the genetic map, the top 0.5% SNP intervals with the highest |Δ(SNP-index)| were selected as candidate gene intervals.
Mapping to linkage groups
The location of the scaffolds containing candidate gene intervals was determined using the high-density genetic map of ‘Bayuehong’בDangshansuli’ published by Wu et al.50 Use of the congruent relationships between the scaffold names in the file ‘scaffold_names’ (ftp://ftp.ncbi.nlm.nih.gov/genomes/Pyrus_x_bretschneideri/) provided the ability to determine the linkage groups of the scaffolds that contain candidate intervals based on their SNP markers in ‘Supplementary Table S4’.50 This information was compared with the corresponding positions for the red-skinned trait loci in Asian pears reported by Wu et al.50 to confirm the results.
Linkage verification
To verify the accuracy of the candidate interval from QTL-seq using linkage analysis, several primers for InDel and simple sequence repeat markers within the candidate intervals were designed using the Primer3 Software (Tartu, Estonia).51 The primers for these markers were then screened on nine red-skinned and seven green-skinned genotypes from the red-skinned and green-skinned pools, respectively. Twelve markers that were polymorphic between the nine red-skinned and seven green-skinned genotypes were selected and successfully amplified in the F1 population resulting from the ‘Mantianhong’בHongxiangsu’ cultivar cross. The polymerase chain reactions were carried out in a final reaction volume of 20 μL, containing 20 ng of DNA, 1× polymerase chain reaction buffer (Mg2+ plus), 0.2 mmol/L of each dNTPs, 6 pmol of each primer and 0.5 U Taq DNA polymerase (TaKaRa Biotechnology, Dalian, China), using the following amplification program: initial denaturation for 3 min at 95 °C, 30 cycles for 30 s at 95 °C, 30 s at 55 °C and 45 s at 72 °C, and a final 10-min extension at 72 °C. The amplicons were evaluated using 8% non-denaturing polyacrylamide gel electrophoresis. Joinmap 4.052 was used to perform the linkage analysis. A logarithm of odds score of 4.0 was set for grouping, and the genetic distances were calculated using the Kosambi function.
Results
Inheritance of the red/green-skinned phenotype in Asian pears
The red- and green-skinned phenotypes segregated in the F1 populations of the two Asian parental lines. An analysis of the 2014 and 2015 data indicated that there were 117 and 182 red- and green-skinned pear individuals, respectively, among the 299 ‘Mantianhong’בHongxiangsu’ F1 hybrids. Among the 310 ‘Yuluxiang’בMantianhong’ F1 hybrids, there were 148 and 162 red- and green-skinned individuals. The segregation data are presented in Table 1.
Table 1. Segregation of the red/green-skinned trait in individuals from the two crosses in this study.
| Group | Year | Fruited individuals | Red individuals | Green individuals | Red:green |
|---|---|---|---|---|---|
| ‘Mantianhong’בHongxiangsu’ | 2014 | 177 | 80 | 97 | 4:5 |
| 2015 | 279 | 111 | 168 | 2:3 | |
| Total | 299 | 117 | 182 | 2:3 | |
| ‘Yuluxiang’בMantianhong’ | 2014 | 189 | 101 | 88 | 8:7 |
| 2015 | 262 | 121 | 141 | 6:7 | |
| Total | 310 | 148 | 162 | 8:9 |
Genes responsible for the red/green-skinned phenotype narrowed down to a 1.86-Mb interval using QTL-seq
Illumina high-throughput sequencing resulted in 78 220 374 and 93 410 888 reads from the red-skinned (19.65×depth coverage or 94.74% coverage) and green-skinned (22.98×depth coverage or 95.15% coverage) DNA pools, respectively. The resulting reads were aligned to the pear reference genome,47 and SNPs were identified. An SNP-index was calculated for each identified SNP. Low-quality SNPs with an SNP-index value <0.3 and a read depth <7 or those without an SNP-index in one of the F1 hybrid pools were excluded to decrease the influence of sequencing and alignment error. A total of 2 280 509 SNPs were identified within the two DNA pools.
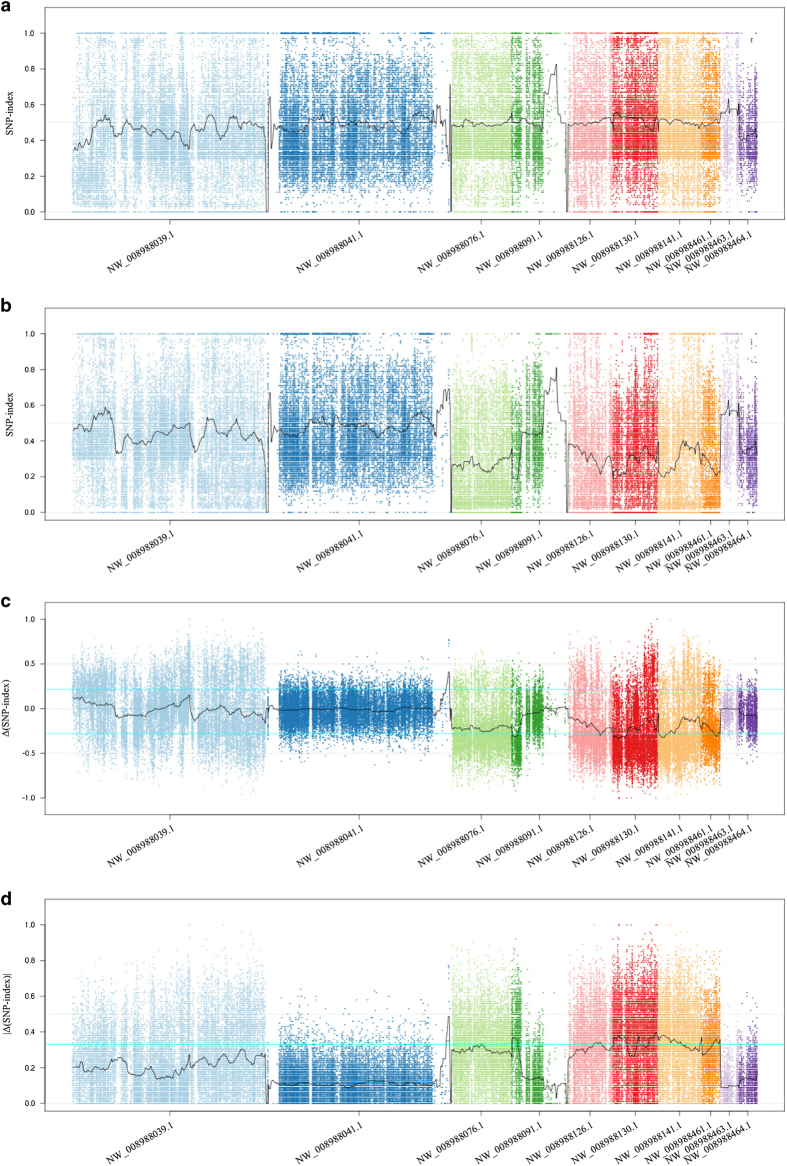
The top 0.25% and lowermost 0.25% threshold lines for the ∆(SNP-index) plot (Figure 3) were set at 0.215 and −0.280, respectively. The threshold line for the top 0.5% |∆(SNP-index)| plot was set at 0.33, which was greater than the 0.215 and 0.280 values previously indicated. This was primarily due to the inability to obtain allele linkage information of the adjacent SNP loci from the highly heterozygous pear genome. Therefore, the plus or minus sign of the ∆(SNP-index) may have been opposite to the actual linkage. This problem eventually resulted in an offset within the window through averaging of the ∆(SNP-index) values, and the ∆(SNP-index) value of the window tended to be zero (Figure 2).
Figure 3.
SNP-index graphs of the red pool (a), green pool (b), Δ(SNP-index) (c) and |Δ(SNP-index)| (d) derived from the QTL-seq analysis. The X-axis indicates the position of the ten scaffolds and the Y-axis indicates the SNP-index.
The candidate intervals derived from thresholds of −0.280 and 0.215 based on the ∆(SNP-index) resulted in the partial overlap of some intervals from the |∆(SNP-index)| using a threshold of 0.33. Based on the |∆(SNP-index)| method, the candidate interval was 2.18 Mb. However, based on the ∆(SNP-index) method, the candidate interval was 1.72 Mb, indicating a 1.1 Mb overlap. Although both methods had a similar distribution on some scaffolds, the candidate interval estimated by the ∆(SNP-index) method was relatively smaller, and most intervals were in the subset of the |∆(SNP-index)| method. The influence of the ‘pseudoexchange effect’ caused many of the intervals predicted by the |∆(SNP-index)| method and verified in the linkage analysis as candidate intervals to be missing in the ∆(SNP-index) method results. This was the case for the intervals located between 500 001 and 580 000 bp and between 760 001 and 960 000 bp on scaffold NW_008988130.1 and between 880 001 and 940 000 bp on scaffold NW_008988141.1 (Figure 3), which indicated that the |∆(SNP-index)| method was much better than the ∆(SNP-index) method for locating trait loci in heterozygous crops (Figure 2b). The size of the interval estimated by the |∆(SNP-index)| method was reduced from 2.18 to 1.86 Mb after filtering the intervals, with only a few SNPs that were widely separated (Table 2).
Table 2. Potential chromosomal regions of red/green-skinned trait loci in Asian pears.
| Scaffold | Start | End | Region length |
|---|---|---|---|
| NW_008988041.1 | 3 800 001 | 3 880 000 | 80 kb |
| NW_008988076.1 | 640 001 | 660 000 | 20 kb |
| NW_008988091.1 | 1 | 140 000 | 140 kb |
| NW_008988126.1 | 560 001 | 720 000 | 160 kb |
| NW_008988130.1 | 1 | 300 000 | 300 kb |
| NW_008988130.1 | 500 001 | 580 000 | 80 kb |
| NW_008988130.1 | 760 001 | 960 000 | 200 kb |
| NW_008988141.1 | 1 | 500 000 | 500 kb |
| NW_008988141.1 | 880 001 | 940 000 | 60 kb |
| NW_008988461.1 | 240 001 | 400 000 | 160 kb |
| NW_008988478.1 | 300 001 | 400 000 | 100 kb |
| NW_008988581.1 | 300 001 | 320 000 | 20 kb |
| NW_008989660.1 | 1 | 20 000 | 20 kb |
| NW_008989715.1 | 1 | 20 000 | 20 kb |
| Total | 1.86 Mb |
Linkage group location of the major number of scaffolds
Most scaffolds containing candidate intervals were located on the bottom portion of the fifth linkage group (LG5). The exceptions that did not map to a linkage group included NW_008988041.1, NW_008988581.1 and NW_008989660.1. The remaining nine scaffolds, NW_008988039.1 (scaffold1.0.1), NW_008988076.1 (scaffold40.0), NW_008988091.1 (scaffold56.0.1), NW_008988126.1 (scaffold92.0), NW_008988130.1 (scaffold97.0.1), NW_008988141.1 (scaffold110.0), NW_008988461.1 (scaffold430.0.1), NW_008988478.1 (scaffold448.0.1) and NW_008989715.1 (scaffold1650.0), were adjacent to each other on LG5 and concentrated in the area between 111.9 and 177.1 cM (Table 3). These results indicate the accuracy of the modified QTL-seq analysis.
Table 3. Linkage groups of scaffolds containing the candidate intervals.
| RefSeq accession file | Genome center name | Linkage group (Wu et al. 50) |
|---|---|---|
| NW_008988039.1 | Scaffold1.0.1 | LG5 |
| NW_008988041.1 | Scaffold3.0 | LG15 |
| NW_008988076.1 | Scaffold40.0 | LG5 |
| NW_008988091.1 | Scaffold56.0.1 | LG5 |
| NW_008988126.1 | Scaffold92.0 | LG5 |
| NW_008988130.1 | Scaffold97.0.1 | LG5 |
| NW_008988141.1 | Scaffold110.0 | LG5 |
| NW_008988461.1 | Scaffold430.0.1 | LG5 |
| NW_008988478.1 | Scaffold448.0.1 | LG5 |
| NW_008988581.1 | Scaffold543.0.1 | Unknown |
| NW_008989660.1 | Scaffold1606.0 | Unknown |
| NW_008989715.1 | Scaffold1650.0 | LG5 |
Linkage analysis to narrow the candidate interval
InDel (231) and simple sequence repeat (55) primer pairs (Supplementary Table 1) were used to screen for polymorphisms in the candidate intervals. Twelve primer pairs produced clear bands and performed the best in red/green skin identification. Therefore, these 12 pairs of primers were used to perform a linkage analysis using the F1 hybrid individuals resulting from the ‘Mantianhong’בHongxiangsu’ cross (Figure 4). Nine individuals were determined to be non-parental-line cross-hybrids and were removed from the data set, and 339 F1 hybrids were maintained in the subsequent analyses. A linkage group with a total length of 21.0 cM was obtained, containing 11 molecular markers and red/green skin trait loci (Figure 5 and Table 4). Three markers, In2130-12, ZFRI130-16, In2130-12 and In2130-16, were located near the red/green (R/G) locus at a genetic distance of 2.5 cM. The markers also showed an accuracy rate of >90% in 310 fruited individuals of ‘Yuluxiang’ and ‘Mangtianhong’ and accuracy rates of 80.85%, 85.11% and 97.87%, respectively, in 47 plants (34 accessions derived from the male parent ‘Hongxiangsu’ and 13 cultivars) in an additional validation (Xue, 2016, unpublished data). These could be used as the molecular markers for breeding selection of red-skinned fruit in Asian pears. Marker In5141-1, which could accurately distinguish the fruit skin color of 89.30% of the offspring, was not located in this linkage group because its segregation did not conform to the ‘CP’ population model (Supplementary Figure 1).
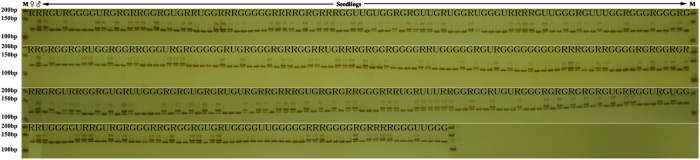
Figure 4.
PCR amplification of the InDel marker, ZFRI130-16 in the F1 individuals of the ‘Mantianhong’בHongxiangsu’ cross. Results conform to the CP pattern nn × np. M: DL2000 Marker, ♀: ‘Mantianhong’, ♂: ‘Hongxiangsu’, R: Red skin phenotype, G: Green skin phenotype, U: Unknown phenotype.
Figure 5.
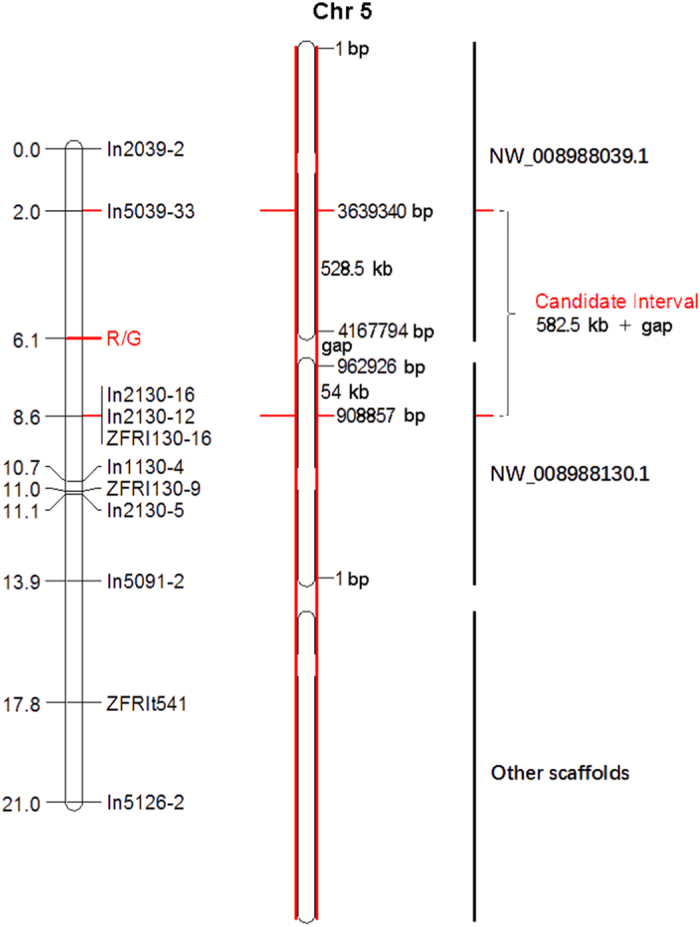
Molecular markers and the red/green skin locus on LG5. The left one is the result of R/G locus linkage position, the middle one is the diagram of the chromosome, the black vertical bar on the right indicates the corresponding position of the scaffolds on the chromosome map, the red line on the right indicates R/G locus and the candidate interval.
Table 4. Twelve R/G locus-linked markers in Asian pears.
| Locus | Primer sequences (5′→3′) | Scaffold | Scaffold position | Mapping position |
|---|---|---|---|---|
| In2039-2 | F: CCTTACCATCTCAAACTCCA R: GTCAAAGGGTTACTGGTTCA | NW_008988039.1 | 3 280 367 | 0.0 |
| In5039-33 | F: ATTTGGTGAGATCATCATGG R: TAACTTCACACGGACTTCCT | NW_008988039.1 | 3 639 340 | 2.0 |
| R/G | — | — | — | 6.1 |
| In2130-12 | F: TTGGAGATGTTACTGTGACG R: GATTACATGCATTGACGTTG | NW_008988130.1 | 908 857 | 8.6 |
| In2130-16 | F: AATAGAAATTTAACACCTACGC R: ACATATAGCACCCTTAGGAAA | NW_008988130.1 | 920 222 | 8.6 |
| ZFRI130-16 | F: CACTTTCGAAGCCAACAAAT R: GGGCAAACTGGGAAACTG | NW_008988130.1 | 922 094 | 8.6 |
| In1130-4 | F: GGCATTACTCTAATCTTCAATC R: AGAAACCCGAGACTAATCAT | NW_008988130.1 | 154 093 | 10.7 |
| ZFRI130-9 | F: ACCAGCGTAGTTTGATAAGA R: AAGGTCGTCAAGTGAGAAG | NW_008988130.1 | 435 305 | 11.0 |
| In2130-5 | F: CCGATCCGTAAGTTCCTAT R: TCTAAAGATTGCCCAGGATA | NW_008988130.1 | 113 643 | 11.1 |
| In5091-2 | F: TTACGAGGGGAAATCAATTA R: GAAATTGAAATTGCTCGTCT | NW_008988091.1 | 15 549 | 13.9 |
| ZFRIt541 | F: AAAAACCGAAAACAAAAACCTC R: CGGAGCCCTGTGCTACTAAT | NW_008988130.1 | 916 907 | 17.8 |
| In5126-2 | F: GCTATTTTGAAATGCCGTAT R: TCCTTCCCAAGTTATTTGAA | NW_008988126.1 | 50 318 | 21.0 |
| In5141-1 | F: CAAAACAGGAAGACACATGA R: AACCATGCGTTGACTAAAAT | NW_008988141.1 | 7013 | — |
The R/G locus in Asian pears was located between marker In5039-33 and markers ZFRI130-16, In2130-12 and In2130-16, which were identified in scaffolds NW_008988039.1 (scaffold1.0.1) and NW_008988130.1 (scaffold97.0.1), respectively. These two scaffolds were adjacent to each other on the bottom of LG5. Markers In2039-2 and In5039-33 from scaffold NW_008988039.1 and markers ZFRI130-16, In2130-12 and In2130-16 from scaffold NW_008988130.1 determined the direction of the two scaffolds. Therefore, the R/G locus should be in the 582.5 -kb candidate interval comprising two subintervals at the bottom of LG5: a 528.5-kb interval from 3 639 340 to 4 167 794 bp in scaffold NW_008988039.1 and a 54-kb interval from 908 857 to 962 926 bp in scaffold NW_008988130.1 (Figure 5 and Table 4). This candidate interval also includes a gap of unknown size between scaffolds NW_008988039.1 and NW_008988130.1. The markers in the same scaffold were staggered, and there was no linear relationship between the physical position and the map position. Therefore, the range of position of the red/green skin trait on LG5 of Asian pear could not be narrowed further.
Based on the reference pear genome, 81 gene models were predicted in the candidate 582.5 kb genomic region (Supplementary Table 2). Among these, 10 were not annotated with a known function, and 71 genes had a predicted gene function. No structural genes associated with the anthocyanin biosynthetic pathway, such as PAL, CHS, CHI, DFR (dihydroflavonol 4-reductase), ANS and UFGT, were found within the candidate region. The MYB transcription factor that forms the MBW complex, the basic helix–loop–helix (bHLH) and the WD40 classes were not detected within the candidate region. Several genes, including the NAC domain class transcription factor, the WRKY domain class transcription factor and ABC transporter B family member 9-like, all of which are related to anthocyanin synthesis and accumulation, were found in the candidate region but require further identification and confirmation.
Discussion
Whole-genome sequencing is a fast and cost-effective approach for identifying genetic variants.53–56 The pool sequencing (Pool-seq) strategy, which combines BSA and whole-genome sequencing, can rapidly identify linked genomic regions to target trait loci in mutation discovery57,58 and has been used in model organisms including Arabidopsis thaliana,59,60 nematodes,61,62 zebrafish63–65 and mice.66 Most studies using Pool-seq identified regions of mutations based on allele frequency and SNP density, but the SNP-index approaches used in rice were simpler and more efficient.35–37 The parental lines used in Pool-seq studies have typically been homozygous, and thus the linkage relationships among SNPs have been unambiguous. Variant regions have been identified by determining which SNP genotypes originated from each parent. A few studies used Pool-seq without a parental genome sequence because the genome in question was highly heterozygous or for other reasons. For example, Hill et al.32 used the Euclidean distance to reduce the noise inherent in RNA sequencing data sets when identifying recessive mutations in zebrafish without parental strain information. In the present study, we used the absolute value of ∆(SNP-index) to shield against background noise caused by the highly heterozygous parental lines and multiple levels of sampling. Both the introduction of the Euclidean distance and |∆(SNP-index)| reduced the dependence of Pool-seq on parental genome sequences and may thus be used in a wider scope of applications.
QTL-seq has been successfully used to conduct a BSA analysis of a segregated population of homozygous parental lines with opposite phenotypes. Calculation of the ∆(SNP-index) for the DNA pool from filial generations based on the genome of one of the parental lines revealed the parental origins of the SNPs in a hybrid pool, which are easy to relate to the target phenotype.36,39,41,42,67 In our study, we were not able to identify the linkage among filial and parental SNPs because the parental genomes were highly heterozygous. Estimating the parental origins of the SNPs according to the ∆(SNP-index) could generate errors. The positive or negative sign of the ∆(SNP-index) may have been opposite to the actual condition, resulting in a ∆(SNP-index) with an opposite direction offset within the window, with the ∆(SNP-index) value of the window approaching 0, reducing the QTL-seq detection sensitivity (detectability) of the candidate interval with target phenotypes (Figure 2). For example, the three nearest markers to the R/G locus were located within the 760 001 to 960 000 bp interval on scaffold NW_008988130.1, but this interval was not predicted by the ∆(SNP-index).
If candidate intervals are dispersed on the scaffold, the target trait intervals are interlaced with non-target trait intervals, which decreases the average value of the |∆(SNP-index)| in a window. In the current study, predicting the interval linked with target traits proved impossible. The downstream 2 500 000 bp section on scaffold NW_008988039.1 fluctuated widely, and the peaks were near the threshold. Therefore, this section could not be the candidate interval (Figure 3d). To avoid missing the candidate interval, we designed primers for the target interval and conducted polymerase chain reaction analyses. A strong linkage relationship was found between relevant sites in this interval and the R/G locus in Asian pears (Figure 5). Accordingly, we used |∆(SNP-index)| instead of the ∆(SNP-index) to conduct the QTL-seq analysis and successfully obtained the interval linked to the red/green skin phenotype. Therefore, we believe that the analytic strategy used in the present study could be expanded to identify the loci for other traits.
Dondini et al.13 analyzed seven crosses in European pears with full red bud sport and green-skinned phenotypes. Their study showed that the former phenotype in European pears was controlled by a single dominant gene, Red. This gene was located between two amplified fragment length polymorphism markers on the LG4 of the ‘Max Red Bartlett’ map, which was 13.5 cM from marker E31M56-7 and 18.2 cM from marker E33M48-5.13 In ‘Bayuehong,’ a red-blushed cultivar derived from a cross between ‘Clapp’s Favorite’ and ‘Zaosu,’ four QTLs for red skin from ‘Clapp’s Favorite’ were on LG4, LG13 and LG16.50 The Red gene was located at 64.0 cM of LG4, whereas Pyb04_016, a QTL for red skin from ‘Clapp’s Favorite,’ was at 4.8 cM, indicating that they were far apart. The present study used the Asian pear cultivars ‘Mantianhong’ and ‘Hongxiangsu’ as parental lines. Their red skin trait was inherited from the ‘Huobali’ sand pear cultivar and the ‘Korla Xiangli’ Xinjiang pear cultivar. We located the R/G locus in a 582.5 -kb candidate interval at the bottom of LG5. This finding is not in agreement with the findings reported by Dondini et al.13 and Wu et al.50 These varying mapping results indicate that there are different mechanisms for the control and regulation of the red skin trait in Asian and European pears and different types of red-skinned phenotypes within European pears.
In the hybrid group of the red-skinned Asian pears, the coloration of the progeny showed the character of the alternative quality traits. Volz et al.29 analyzed the segregation of red characters in the progenies of 10 Asian pear hybrids and suggested that it could be controlled by two dominant gene loci, which is consistent with the findings in apples.68 In our study, the segregation ratios of red to green were 2:3 (Mantianhong×Hongxiangsu) and 8:9 (Yuluxiang×Mantianhong), which were lower than the minimum segregation ratio (9:7, with AaBb and AaBb genotypes) in the hypothesis assuming two independent dominant gene loci. The ratio of red to green is as low as 1:1 only when the two loci are closely linked or their Ab|aB alleles are cosegregated, the segregation type of these two loci is hk×hk and the homozygous and heterozygosity genotypes of progeny correspond to the green- and red-skinned phenotypes, respectively. However, in this study, the Asian pear red skin trait loci were in a continuous interval on the fifth chromosome, and the segregation types of three closely linked molecular markers with red-skinned traits were nn×np, not hk×hk.
Our findings support the hypothesis that the trait is controlled by a single dominant gene that tends toward green-skinned segregation. This same confusion was observed in apples. At present, only one locus has been found to control the red-skinned trait in apples.69 Deviation from Mendel's genetic separation in the hybrid progeny of the red-skinned Asian pear with bias toward green separation appears to be a common phenomenon 28,29,55 and may be a result of the linkage of red skin trait loci with lethal or semilethal gene loci.29 A higher distorted segregation ratio of molecular markers was also detected when we identified the hybrid progeny of ‘Mantianhong’ and ‘Hongxiangsu’.70 Additional work is needed to fully understand the genetic regularity of the red-skinned trait of Asian pears, including analysis of hybrid progeny segregation and genotypes, development and application of molecular markers, determination of the mechanism of distorted segregation and validation of gene function.
The color area and degree of redness in the skin of Asian pears are influenced by environmental conditions and show the characteristics of quantitative traits. The structure gene and transcription factor gene of the anthocyanin biosynthetic pathway play important regulatory roles in this process. However, it is unclear if the quality trait genes that control the coloration of the red-skinned pear are derived from the structural gene or transcription factor gene of the anthocyanin biosynthetic pathway, and no such genes were found in the known gene model within the candidate intervals in this study. Therefore, it is not possible to exclude the presence of a gene that acts as a switch in the coloration of red-skinned pears upstream of anthocyanin biosynthetic pathway-related genes or transcription factors. The most interesting candidate genes identified within the region containing the R/G locus were the NAC domain class transcription factor, the WRKY domain class transcription factor and the ABC transporter B family member 9-like gene, which have all been associated with anthocyanin synthesis and accumulation. The NAC transcription factors play important roles in plant stress responses, flowering and secondary cell-wall biosynthesis.71,72 An expression sequence tag encoding the transcription factor NAC domain protein was found to be selectively induced by cold in blood oranges but not in common oranges.73 A report on blood-fleshed peaches suggested that the heterodimer of two NAC transcription factors (BL and PpNAC1) can activate the transcription of PpMYB10.1, resulting in anthocyanin accumulation, and that BL is the key gene for the blood-flesh trait in peaches.74 Interestingly, there are two R2R3 MYB transcription factor 10 genes in scaffold NW_008988130.1 near the candidate region (listed in Supplementary Table 2). The relationship between the two R2R3 MYB genes and the NAC transcription factors in the candidate intervals merits additional investigation.
Although WRKY transcription factors play an important role in the plant stress response and hormone signaling,75 some WRKY family genes play an important role in regulation of anthocyanin accumulation. For example, the WRKY transcription factor AtTTG2 has been reported to regulate the accumulation of proanthocyanidins and anthocyanins in A. thaliana.76,77 The TTG2-like homolog protein VvWRKY26 is also involved in the regulation of vacuolar transport and flavonoid accumulation in grape berries.78 In addition, a study on A. thaliana photomorphogenesis and root development revealed that two ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, are required for normal accumulation of anthocyanins.79 The results of the present and previous studies clearly indicate the need for additional and more detailed work to identify candidate genes associated with anthocyanin synthesis and accumulation. Molecular analyses should be conducted on more taxa. Gene sequences and expression should also be compared between red- and green-skinned genotypes. These additional studies will elucidate the genetic control and regulatory processes responsible for the red-skinned trait in Asian pears.
Acknowledgments
We thank Dr Jialong Yao (Mount Albert Research Centre, The New Zealand Institute for Plant and Food Research Limited) and Dr Richard Volz (Hawke’s Bay Research Centre, The New Zealand Institute for Plant and Food Research Limited) for comments on this manuscript. This work was funded by the National Natural Science Foundation of China (31272140) and the Agricultural Science and Technology Innovation Program (ASTIP) (CAAS-ASTIP).
Footnotes
Supplemental Information for this article can be found on the Horticulture Research website (http://www.nature.com/hortres)
The authors declare no conflict of interest.
References
- Potter D, Eriksson T, Evans RC et al. Phylogeny and classification of Rosaceae. Plant Syst Evol 2007; 266: 5–43. [Google Scholar]
- Inoue E, Kasumi M, Sakuma F et al. Identification of RAPD marker linked to fruit skin color in Japanese pear (Pyrus pyrifolia Nakai). Sci Hortic 2006; 107: 254–258. [Google Scholar]
- Yang J, Li X. Research progress on the red-skinned pear in China [Chinese]. Chinese Agric Sci Bull 2002; 18: 87–89. [Google Scholar]
- Zhang D, Teng Y. Germplasm resources of red pears and research progress in pear coloring mechanism [Chinese]. J Fruit Sci 2011; 28: 485–492. [Google Scholar]
- Zhou J, Xin P, Xu Y et al. Current advance of red skin pear resource origin from China and molecular mechanism of red development in pear skin [Chinese]. Northern Hortic 2010; 6: 213–218. [Google Scholar]
- Gamble J, Jaeger SR, Harker FR. Preferences in pear appearance and response to novelty among Australian and New Zealand consumers. Postharvest Biol Technol 2006; 41: 38–47. [Google Scholar]
- Boyer J, Liu R. Apple phytochemicals and their health benefits. Nutr J 2004; 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang S, Qin G et al. Isolation and determination of major anthocyanin pigments in the pericarp of P. communis L. cv. Red Du Comices and their association with antioxidant activity. Afr J Agric Res 2012; 7: 3772–3780. [Google Scholar]
- Zhang Q, Yang J, Wang L et al. Advances in research of red-skinned pear coloration and anthocyanin synthesis [Chinese]. J Fruit Sci 2012; 29: 668–675. [Google Scholar]
- Huang C, Yu B, Su J et al. A study of the visual changes of the skin color and fruit transection structure of two Chinese red skinned pear cultivars during fruit coloring process [Chinese]. J Fruit Sci 2009; 26: 19–24. [Google Scholar]
- Feng S, Sun S, Chen X et al. PyMYB10 and PyMYB10.1 interact with bHLH to enhance anthocyanin accumulation in pears. PLoS ONE 2015; 10: e0142112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yao G, Zheng D et al. Expression differences of anthocyanin biosynthesis genes reveal regulation patterns for red pear coloration. Plant Cell Rep 2015; 34: 189–198. [DOI] [PubMed] [Google Scholar]
- Dondini L, Pierantoni L, Ancarani V et al. The inheritance of the red colour character in European pear (Pyrus communis) and its map position in the mutated cultivar ‘Max Red Bartlett’. Plant Breed 2008; 127: 524–526. [Google Scholar]
- Zhang D, Yu B, Bai J et al. Effects of high temperatures on UV-B/visible irradiation induced postharvest anthocyanin accumulation in ‘Yunhongli No. 1’ (Pyrus pyrifolia Nakai) pears. Sci Hortic 2012; 134: 53–59. [Google Scholar]
- Dussi MC, Sugar D, Wrolstad RE. Characterizing and quantifying anthocyanins in red pears and the effect of light quality on fruit color. J Am Soc Hortic Sci 1995; 120: 785–789. [Google Scholar]
- Steyn W, Holcroft D, Wand S et al. Anthocyanin degradation in detached pome fruit with reference to preharvest red color loss and pigmentation patterns of blushed and fully red pears. J Am Soc Hortic Sci 2004; 129: 13–19. [Google Scholar]
- Feng S, Chen X, Zhang C et al. A study of the relationship between anthocyanin biosynthesis and related enzymes activity in Pyrus pyrifolia ‘Mantianhong’and its bud sports ‘Aoguan’ [Chinese]. Sci Agric Sin 2008; 41: 3184–3190. [Google Scholar]
- Huang C, Yu B, Su J et al. A study on coloration physiology of fruit in two red Chinese sand pear cultivars 'Meirensu' and 'Yunhongli No.1' [Chinese]. Sci Agric Sin 2010; 43: 1433–1440. [Google Scholar]
- Steyn W, Holcroft D, Wand S et al. Regulation of pear color development in relation to activity of flavonoid enzymes. J Am Soc Hortic Sci 2004; 129: 6–12. [Google Scholar]
- Fischer TC, Gosch C, Pfeiffer J et al. Flavonoid genes of pear (Pyrus communis). Trees 2007; 21: 521–529. [Google Scholar]
- Feng S, Wang Y, Yang S et al. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010; 232: 245–255. [DOI] [PubMed] [Google Scholar]
- Li L, Ban Z, Li X et al. Differential expression of anthocyanin biosynthetic genes and transcription factor PcMYB10 in pears (Pyrus communis L.). PLoS ONE 2012; 7: e46070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian M, Yu B, Li X et al. Isolation and expression analysis of anthocyanin biosynthesis genes from the red Chinese sand pear, Pyrus pyrifolia Nakai cv. Mantianhong, in response to methyl jasmonate treatment and UV-B/VIS conditions. Plant Mol Biol Rep 2014; 32: 428–437. [Google Scholar]
- Pierantoni L, Dondini L, De Franceschi P et al. Mapping of an anthocyanin-regulating MYB transcription factor and its expression in red and green pear Pyrus communis. Plant Physiol Biochem 2010; 48: 1020–1026. [DOI] [PubMed] [Google Scholar]
- Wang Z, Meng D, Wang A et al. The methylation of the PcMYB10 promoter Is associated with green-skinned sport in Max Red Bartlett Pear. Plant Physiol 2013; 162: 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski Q, Reimer F, Quackenbush V. Breeding behavior of fruit characteristics in pears, Pyrus communis. Proc Am Soc Hort Sci 1965; 86: 81–87. [Google Scholar]
- Booi S, Van Dyk M, Du Preez M et al. Molecular typing of red and green phenotypes of 'Bon Rouge' pear trees, with the use of microsatellites. Acta Hortic 2005; 671: 293–297. [Google Scholar]
- Wang Y, Allan W, Lester B et al. Report on breeding red-skinned Chinese pear varieties [Chinese]. J Fruit Sci 1997; 14: 71–76. [Google Scholar]
- Volz RK, White AG, Brewer LR. Breeding for red skin colour in interspecific pears. Acta Hortic 2008; 800: 469–474. [Google Scholar]
- Michelmore RW, Paran I, Kesseli R. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 1991; 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Terauchi R, McCouch SR. Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. PLoS Biol 2014; 12: e1001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JT, Demarest BL, Bisgrove BW et al. MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome Res 2013; 23: 687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier FD, Valentim CL, LoVerde PT et al. Efficient linkage mapping using exome capture and extreme QTL in schistosome parasites. BMC Genomics 2014; 15: 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Bloom J, Torabi N et al. Genetic architecture of highly complex chemical resistance traits across four yeast strains. PLoS Genet 2012; 8: e1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe A, Kosugi S, Yoshida K et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 2012; 30: 174–178. [DOI] [PubMed] [Google Scholar]
- Takagi H, Abe A, Yoshida K et al. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 2013; 74: 174–183. [DOI] [PubMed] [Google Scholar]
- Fekih R, Takagi H, Tamiru M et al. MutMap+: genetic mapping and mutant identification without crossing in rice. PLoS ONE 2013; 8: e68529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Takagi H, Mukainari Y et al. Genetic analysis of NEKODE1 gene involved in panicle branching of foxtail millet, Setaria italica (L.) P. Beauv., and mapping by using QTL-seq. Mol Breed 2016; 36: 1–8. [Google Scholar]
- Lu H, Lin T, Klein J et al. QTL-seq identifies an early flowering QTL located near flowering lLocus T in cucumber. Theor Appl Genet 2014; 127: 1491–1499. [DOI] [PubMed] [Google Scholar]
- Illa-Berenguer E, Van Houten J, Huang Z et al. Rapid and reliable identification of tomato fruit weight and locule number loci by QTL-seq. Theor Appl Genet 2015; 128: 1329–1342. [DOI] [PubMed] [Google Scholar]
- Hiremath PJ, Kumar A, Penmetsa RV et al. Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol J 2012; 10: 716–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh VK, Khan AW, Jaganathan D et al. QTL-seq for rapid identification of candidate genes for 100-seed weight and root/total plant dry weight ratio under rainfed conditions in chickpea. Plant Biotechnol J 2016; 14: 2110–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Upadhyaya HD, Bajaj D et al. Deploying QTL-seq for rapid delineation of a potential candidate gene underlying major trait-associated QTL in chickpea. DNA Res 2015; 22: 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Niu L, Chagné D et al. Fine mapping of the temperature-sensitive semi-dwarf (Tssd) locus regulating the internode length in peach (Prunus persica). Mol Breed 2016; 36: 1–11. [Google Scholar]
- Doyle JJ. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 1987; 19: 11–15. [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang Z, Shi Z et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 2013; 23: 396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li L, Li M et al. High-density genetic linkage map construction and identification of fruit-related QTLs in pear using SNP and SSR markers. J Exp Bot 2014; 65: 5771–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007; 23: 1289–1291. [DOI] [PubMed] [Google Scholar]
- Van Ooijen J. Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genome Res 2011; 93: 343–349. [DOI] [PubMed] [Google Scholar]
- Zuryn S, Le Gras S, Jamet K et al. A strategy for direct mapping and identification of mutations by whole-genome sequencing. Genetics 2010; 186: 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsidou M, Jarriault S, Poole RJ. Next-generation sequencing-based approaches for mutation mapping and identification in Caenorhabditis elegans. Genetics 2016; 204: 451–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Kirk C, Deng C et al. Genotyping-by-sequencing of pear (Pyrus spp.) accessions unravels novel patterns of genetic diversity and selection footprints. Hort Res 2017; 4: 17015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Fresnedo-Ramírez J, Wang M et al. A next-generation marker genotyping platform (AmpSeq) in heterozygous crops: a case study for marker-assisted selection in grapevine. Hort Res 2016; 3: 16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Orozco-terWengel P, De Maio N et al. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS ONE 2011; 6: e15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Pandey RV, Schlötterer C. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 2011; 27: 3435–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin RS, Vidaurre D, Stamatiou G et al. Next-generation mapping of Arabidopsis genes. Plant J 2011; 67: 715–725. [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C et al. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 2009; 6: 550–551. [DOI] [PubMed] [Google Scholar]
- Doitsidou M, Poole RJ, Sarin S et al. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS ONE 2010; 5: e15435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minevich G, Zhang F, Carrera I et al. An update on mutant identification by combined SNP mapping/whole genome sequencing and CloudMap data analysis. Worm Breeder's Gazette 2014; 20: 5–6. [Google Scholar]
- Obholzer N, Swinburne IA, Schwab E et al. Rapid positional cloning of zebrafish mutations by linkage and homozygosity mapping using whole-genome sequencing. Development 2012; 139: 4280–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshchiner I, Alexa K, Kelsey P et al. Mutation mapping and identification by whole-genome sequencing. Genome Res 2012; 22: 1541–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voz ML, Coppieters W, Manfroid I et al. Fast homozygosity mapping and identification of a zebrafish ENU-induced mutation by whole-genome sequencing. PLoS ONE 2012; 7: e34671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Sarkar A, Yram MA et al. Sox2+ adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 2011; 9: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Fu W, Wang Y et al. Rapid identification of fruit length loci in cucumber (Cucumis sativus L.) using next-generation sequencing (NGS)-based QTL analysis. Sci Rep 2016; 6: 27496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AG, Lespinasse Y. The inheritance of fruit colour in apple (Malus pumila Mill.). Agronomie 1986; 6: 105–108. [Google Scholar]
- Chagné D, Kirk C, How N et al. A functional genetic marker for apple red skin coloration across different environments. Tree Genet Genom 2016; 12: 1–9. [Google Scholar]
- Xue H, Wang F, Wang L et al. Identification of the hybrids and analysis of genetic variation of a pear progeny derived from crossing between 'Mantianhong' and 'Hongxiangsu' by Genic-SSR [Chinese]. J Fruit Sci 2017; 34: 925–934. [Google Scholar]
- Ooka H, Satoh K, Nagata T et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 2003; 10: 239–247. [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL et al. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 2005; 10: 79–87. [DOI] [PubMed] [Google Scholar]
- Crifò T, Petrone G, Lo Cicero L et al. Short cold storage enhances the anthocyanin contents and level of transcripts related to their biosynthesis in blood oranges. J Agric Food Chem 2011; 60: 476–481. [DOI] [PubMed] [Google Scholar]
- Zhou H, Lin-Wang K, Wang H et al. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J 2015; 82: 105–121. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P et al. WRKY transcription factors. Trends Plant Sci 2010; 15: 247–258. [DOI] [PubMed] [Google Scholar]
- Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 2002; 14: 1359–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Hattori S, Sano R et al. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 2007; 19: 2531–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato A, Cavallini E, Zenoni S et al. A grapevine TTG2-like WRKY transcription factor is involved in regulating vacuolar transport and flavonoid biosynthesis. Front Plant Sci 2017; 7: 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Wang H. Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol 2005; 138: 949–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.