Abstract
Fatty acid-binding proteins (FABP) are small molecular mass intracellular lipid chaperones that are expressed in a tissue-specific manner with some overlaps. FABP4 and FABP5 share ~55 % amino acid sequence homology and demonstrate synergistic effects in regulation of metabolic and inflammatory responses in adipocytes and macrophages. Recent studies have shown that FABP4 and FABP5 are also co-expressed in a subset of endothelial cells (EC). FABP4, which has a primarily microvascular distribution, enhances angiogenic responses of ECs, including proliferation, migration, and survival. However, the vascular expression of FABP5 has not been well characterized, and the role of FABP5 in regulation of angiogenic responses in ECs has not been studied to date. Herein we report that while FABP4 and FABP5 are co-expressed in microvascular ECs in several tissues, FABP5 expression is also detected in ECs of larger blood vessels. In contrast to FABP4, EC-FABP5 levels are not induced by VEGF-A or bFGF. FABP5 deficiency leads to a profound impairment in EC proliferation and chemotactic migration. These effects are recapitulated in an ex vivo assay of angiogenesis, the aortic ring assay. Interestingly, in contrast to FABP4-deficient ECs, FABP5-deficient ECs are significantly more resistant to apoptotic cell death. The effect of FABP5 on EC proliferation and survival is mediated, only in part, by PPARδ-dependent pathways. Collectively, these findings demonstrate that EC-FABP5, similar to EC-FABP4, promotes angiogenic responses under certain conditions, but it can also exert opposing effects on EC survival as compared to EC-FABP4. Thus, the balance between FABP4 and FABP5 in ECs may be important in regulation of angiogenic versus quiescent phenotypes in blood vessels.
Keywords: FABP4, FABP5, Angiogenesis, Endothelial cell, PPAR, Apoptosis, Cell survival, Aortic ring assay
Introduction
Fatty acid-binding proteins (FABPs) are a well-conserved family of intracellular lipid chaperones that are expressed in a tissue-specific manner with some overlaps [1–5]. Nine FABP genes (FABP1-9) have been identified in mammals. FABPs demonstrate an amino acid sequence homology of 20–70 % and bind long-chain fatty acids (FA) and other hydrophobic ligands with variable affinity and specificity [6]. Recent studies have suggested that FABPs have individual functions in different tissues despite the similarities in their tertiary structures and FA-binding profiles. However, the biologic functions of FABPs remain incompletely understood.
The endothelium is actively involved in lipid metabolism, and our recent studies have shown that FABP4 is abundantly expressed in microvascular endothelial cells (ECs) in several normal and pathologic tissues [7–10]. EC-FABP4 exhibits a pro-angiogenic role by promoting cell proliferation, survival, and migration [9–11]. The expression of EC-FABP4 is regulated by VEGF-A and mTORC1, and in turn, FABP4 regulates the activity of several mitogenic pathways, including stem cell factor/c-kit signaling, which plays an important role in its pro-angiogenic function [9, 10]. FABP4 also regulates the inflammatory activity of ECs by regulating the expression of genes that play key roles in EC activation, such as endothelial nitric oxide synthase (eNOS) and intercellular adhesion molecule 1 (ICAM-1).
FABP5, which shares 55 % amino acid sequence homology with FABP4, was also reported to have a primarily microvascular expression pattern in ECs [12, 13]. However, the function of FABP5 in ECs remains largely unknown. A recent study has suggested an essential role for FABP4 and FABP5 in FA uptake in the heart and skeletal muscle ECs [13]. FABP5 is expressed in several other cell types, including epidermal cells, adipocytes, macrophages, and alveolar epithelial cells [14–17]. Through its expression in adipocytes and macrophages, FABP5 contributes to regulation of inflammatory and metabolic responses [4, 18]. In animal models, combined deficiency of FABP4 and FABP5 provides a greater protection against diet-induced obesity, insulin resistance, and atherosclerosis than mice deficient for either FABP4 or FABP5 [19, 20]. In addition to long-chain FAs, FABP5 binds natural and synthetic peroxisome proliferator-activated receptor (PPAR)-δ ligands, including retinoic acid (RA), and, upon binding these ligands, mobilizes to the nucleus to activate PPARδ. Recent studies in a mouse model of breast cancer have demonstrated that the ratio of cellular retinoic acid-binding protein 2 (CRABP2) and FABP5 determines whether RA plays an inhibitory role in cell proliferation through the CRABP2/RA receptor signaling or a pro-survival role through the FABP5/PPARδ pathway [21, 22]. Thus, FABP5 may promote cell growth and differentiation by regulation of PPARδ signaling. Consistent with this notion, FABP5 up-regulation in certain cancers, such as breast cancer and squamous cell carcinoma, has been linked to poor prognosis and aggressive behavior [23–25].
Regulation of EC homeostasis and vascular integrity is critical to normal organ function as well as tissue repair, regeneration, and tumor growth. Based on the additive functions of FABP4 and FABP5 in other cell types and their co-expression in microvascular ECs, we investigated whether FABP5 also played a role in regulation of angiogenesis-related EC functions, such as proliferation, migration, and survival. We also examined the potential role of PPAR-δ as a downstream mediator of FABP5-related effects in ECs.
Methods and materials
Cell culture and reagents
Human umbilical vein ECs (HUVECs) were isolated as described previously [10, 26]. HUVECs from 3 to 4 cords were pooled for experiments, and passage 2–3 HUVECs were used for all experiments. HUVEC culture medium included M199 media (Invitrogen, Carlsbad, CA) supplemented with 10 % fetal bovine serum (FBS) (Invitrogen), 30 µg/ml endothelial cell growth factor supplement (ECGF) (BD Biosciences, San Jose, CA), 100 µg/ml heparin sodium (Sigma, St Louis, MO), 1 % sodium bicarbonate (Invitrogen), 1 % Glutamax™-1 (Invitrogen), 1 % penicillin/streptomycin (Invitrogen), 0.25 % fungizone (Invitrogen), and 0.25 % gentamicin (Invitrogen). Cells were cultured at 37 °C with 5 % CO2 and humidity. In some experiments, cells were treated with VEGF-A 165 (R&D Systems, Minneapolis, MN) or chemically defined lipid mixture 1 (Sigma). For induction of apoptosis, HUVECs were cultured in medium containing 5 % FBS and no ECGF, or treated with TNFα (40 ng/ml) for 24 h. PPARδ agonist GW0742 and PPARδ inhibitor GSK0660 were purchased from Cayman Chemicals (Ann Arbor, MI) and R&D Systems, respectively.
Treatment of HUVECs with VEGF-A, bFGF, and lipid mixture
1.5 × 105 HUVECs (passage 2) per well were seeded onto 1 % gelatin (Sigma)-coated 6-well plates and cultured in standard HUVEC medium as described above. Twenty-four hours later, standard culture medium was replaced with medium without ECGF and with VEGF-A (0, 1, 10, or 50 ng/ml), bFGF (1 or 10 ng/ml), or lipid mixture (0, 10, 20, or 100 ng/ml). Cells were harvested after 24 or 48 h and processed as described below for analysis by immunoblotting.
Immunohistochemistry and immunocytofluorescence
Immunohistochemistry was performed on formalin-fixed, paraffin-embedded wild-type (WT) C57BL/6 (Jackson Laboratories, Bar Harbor) and FABP5−/− murine (also on C57BL/6 background) [16] and human tissue sections as described previously [10, 27]. Discarded surgical or autopsy human tissue samples were obtained from Boston Children’s Hospital or Brigham and Women’s Hospitals and used with approval from the respective institutional review boards. For immunohistochemistry, human and murine sections were incubated with a rabbit polyclonal anti-FABP5 antibody (Abcam, 1:500) overnight at 4 °C. For immunocytofluorescence, HUVECs were grown on glass coverslips, fixed in 4 % paraformaldehyde for 10 min, washed in ice-cold PBS twice, and permeabilized by incubation with PBS containing 0.25 % Triton X-100 for 10 min. Following the blocking step with 1 % bovine serum albumin (Sigma) in PBS for 30 min, cells were incubated with rabbit polyclonal anti-FABP4 (Abcam, 1:50) and mouse monoclonal anti-FABP5 (R&D Systems, 1:200) antibodies for 1 h at room temperature. This was followed by incubations with 1:1000 dilutions of the secondary antibodies Alexa Fluor 488 goat anti-rabbit IgG and Alexa Fluor 594 goat anti-mouse IgG (Molecular Probes, Eugene, OR, USA) for 1 h at room temperature. After washing in PBS, coverslips were mounted in Vectashield (Vector Laboratories) and viewed under a Nikon Eclipse 80i microscope. Images were captured using NIS-Elements Basic Research® software.
RNA interference
Mission short-hairpin RNAs (shRNAs) targeting human FABP5 and shRNA control vector targeting firefly luciferase were purchased from Sigma. Lentiviral shRNA transfer vectors and four expression vectors encoding viral packaging proteins (provided by Dr. Richard Mulligan, Boston Children’s Hospital) were co-transfected into HEK293 cells, as described previously [9, 10]. Supernatants of HEK293T cells were collected and used to transduce HUVECs. Puromycin (2 ng/ml; Sigma) was added to the medium for 24 h for enrichment of transduced cells the day after transduction. Cells transduced with the control shRNA reached 70–80 % confluence 48–72 h after transduction.
Immunoblotting and densitometry
Cells were lysed in RIPA buffer (BioProducts, Ashland, MA) supplemented with phosphatase inhibitor cocktail set II (Calbiochem, San Diego, CA) and the protease inhibitor cocktail tablet (Roche, Indianapolis, IN). Immunoblot analysis was performed as described previously [28]. All primary antibody incubations were performed at 4 °C overnight at the following dilutions: rat monoclonal anti-FABP5 (R&D Systems), 1:1000; rabbit monoclonal anti-FABP4 (ab92501, Abcam, Cambridge, MA), 1:1000; rabbit polyclonal anti-caspase-3 (Cell Signaling, Danvers, MA), 1:1000; and mouse monoclonal anti-β-actin (Sigma), 1:10,000. Relative protein amounts were normalized to β-actin and quantified using NIH Image J software.
Proliferation assay and analysis of cell cycle
2 × 104 HUVECs per well were seeded onto 1 % gelatin-coated 96-well plates in quadruplicate and transduced with lentiviruses encoding shRNA for FABP5 or firefly luciferase. Cells were cultured in the starvation medium for 6 h and then in complete medium for 24 h. BrdU incorporation was measured using a chemiluminescence-based cell proliferation ELISA kit (Roche Diagnostics) following the manufacturer’s instructions. For cell cycle analysis, HUVECs were grown to 70–80 % confluency, treated with RNAase A, stained with propidium iodide, and then subjected to flow cytometry with a BD FACSCanto flow cytometer (BD Biosciences). Ten thousand events were collected from each sample. Data acquisition was carried out using the BD FACSDIVA software, and cell cycle distribution was calculated using the FlowJo software (Treestar, Ashland, OR).
Migration assays
Chemotaxis/directed migration assay was performed using polycarbonate filter wells (transwell, 8-µm pores; Coaster, Corning, NY) coated with 1 % gelatin. HUVECs were growth-arrested by addition of 2 mmol/L hydroxyurea to the medium and then plated in the upper chamber in 0.1 % FBS, ECGF-free medium at a density of 1 × 105 cells per well. Transwell migration of ECs was stimulated by addition of VEGF-A (50 ng/ml) or 10 % FBS to the culture medium in the lower well. After 6 h, the upper surface of the insert was swabbed to remove non-migrating cells. The cells that had migrated to the lower surface were fixed and stained by using Diff-Quik Stain Set (Dade Behring, Deerfield, IL). EC migration was quantified by counting the number of cells in three random fields per insert.
For cellular wound assay, cells were seeded in 6-well plates at a density of 5 × 105 cells per well to achieve 80–90 % confluency and 2 mmol/l hydroxyurea was added to the medium to induce growth arrest. Two hours later, three vertical scratches were made across each well with a flat-edge forceps. A horizontal reference line was drawn to denote the scratch field of view (FOV) at the scratch intersection. Images of the wounded cells were taken above and below the reference line at the scratch/cell interfaces at t = 0 and t = 8 h. The average scratch width was determined for each FOV, and the distance migrated was calculated [9, 29].
Adhesion assay
96-Well plates were coated with 10 ug/ml gelatin, and adhesion assay was performed as previously described [30, 31]. Briefly, HUVECs were seeded at a density of 5 × 104 cells per well and allowed to attach for 1 h. Non-adherent cells were removed by washing in PBS, and adherent cells were fixed and stained in 0.125 % Coomassie Blue [32]. After thorough destaining, the contents of the wells were solubilized in 2 % SDS, and optical density, indicative of adherent cells, was read at 620 nm.
Aortic ring assay
This study was approved by the Harvard Medical Area Standing Committee on Animals. FABP5−/− mice were backcrossed more than 10 generations onto C57BL/6J genetic background [16], and aortic ring assay was performed as previously described [9]. Abdominal aortas were harvested from 4- to 5-week-old FABP5−/− or WT mice. One-mm-long mouse aortic rings were embedded in Matrigel in 96-well plates and cultured in medium supplemented with bFGF for up to 7 days. Neovessel outgrowth was imaged using a Nikon D5-5M camera (Nikon, Tokyo, Japan) and quantified using NIH Image J software.
Flow cytometric analysis of apoptosis
Apoptosis was induced in HUVECs by serum deprivation (5 % FBS). After 24 h, floating and adherent cells were collected, stained with fluorescein isothiocyanate (FITC)-conjugated annexin V and propidium iodide (PI) (Biovision Inc, Mountain View, CA), and analyzed by flow cytometry.
Measurement of PPARδ-binding activity
Nuclear extracts were prepared from control and FABP5-KD HUVECs using a nuclear extraction kit (Chemicon, Temecula, CA) following the manufacturer’s instructions. PPARδ-binding activity in nuclear extracts was measured using a PPARδ transcription factor enzyme-linked immunosorbent assay (Cayman Chemicals).
Statistical analysis
All results are presented as mean ± standard error of mean from a minimum of three independent experiments. Mann–Whitney U test was used to determine statistical significance, and p < 0.05 was considered significant.
Results
FABP5 expression in vascular endothelial cells
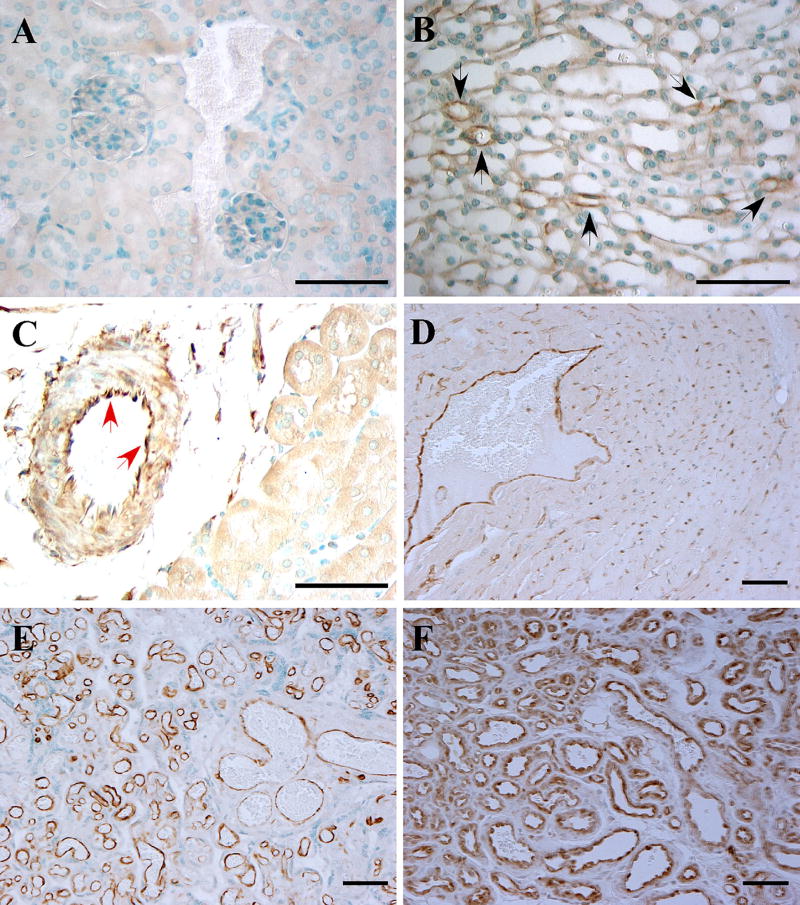
Previous studies have suggested that EC-FABP5 expression was restricted to the microvasculature in several tissues [12, 33]. We expanded these previous studies by characterizing the vascular expression of FABP5 in several mouse and human tissue samples using a polyclonal FABP5 antibody, the specificity of which was confirmed on FABP5−/− mouse tissues (Fig. 1a). In contrast to the previous reports, our analyses revealed FABP5 expression in ECs of some larger vessels, including arteries and veins, in addition to the microvasculature in several tissues, such as the kidney, heart, and placenta (Fig. 1b–e and Fig. S1). Notably, FABP5 expression was not detected in vascular ECs in the murine liver, spleen, and intestine (not shown). Abundant FABP5 expression was also detected in ECs in cutaneous hemangiomas, which are the most common EC-derived tumors (Fig. 1F).
Fig. 1.
FABP5 expression in vascular endothelial cells. FABP5 expression was analyzed by immunohistochemistry on a panel of paraffin-embedded mouse and human tissues. a FABP5−/− mouse kidney as a negative control; b WT murine kidney with occasional capillary EC immunoreactivity for FABP5 in the medulla (black arrows); c WT murine kidney with EC FABP5 immunoreactivity in an arcuate artery at the corticomedullary junction (red arrows); d WT murine heart with capillary and venous EC staining for FABP5; e human placenta with uniform vascular EC immunoreactivity for FABP5; f cutaneous hemangioma with uniform vascular EC immunoreactivity for FABP5. Representative images from a minimum of three different samples per tissue are shown. Scale bar 50 µm
FABP5 deficiency dramatically impairs endothelial cell proliferation
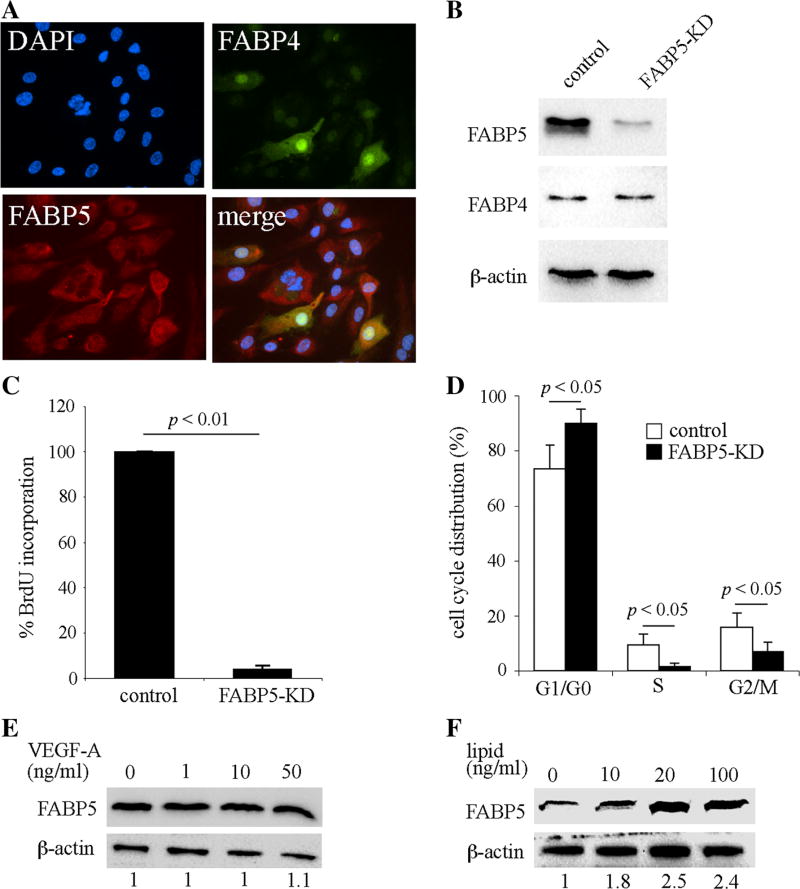
HUVECs are the best characterized and most commonly used in vitro culture system for primary ECs [26]. In previous studies, we have consistently observed similar cellular responses in HUVECs and primary microvascular ECs [10]. Therefore, in this study, we primarily employed HUVECs as an in vitro model of both microvascular and large vessel ECs. We found that, in contrast to the heterogenous expression pattern of FABP4, FABP5 was uniformly and abundantly expressed in HUVECs (Fig. 2a). To determine whether FABP5 is involved in regulation of EC proliferation, we generated FABP5-knockdown (FABP5-KD) HUVECs using lentivirus-mediated shRNA transduction, which consistently resulted in a greater than 80 % reduction in FABP5 protein levels without affecting FABP4 levels (Fig. 2b). Cell proliferation was measured using BrdU incorporation and found to be dramatically reduced in FABP5-KD HUVECs as compared to the control cells (Fig. 2c, p < 0.01). To further assess the effect of FABP5 on EC proliferation, cell cycle analysis was performed using flow cytometry. The number of FABP5-KD cells at the G0/G1 phase was significantly increased compared to control cells with subsequent decreases in S and G2/M phases (Fig. 2d). Thus, FABP5 deficiency resulted in cell cycle arrest in HUVECs at the G0/G1 phase.
Fig. 2.
FABP5 deficiency impairs endothelial cell proliferation. a Double immunocytofluorescence for FABP4 and FABP5 in HUVECs demonstrates uniform expression of FABP5 compared to heterogenous expression of FABP4. b FABP5-KD HUVECs were generated using lentivirus-mediated shRNA transduction. Cells were cultured in medium containing puromycin for enrichment for 24 h and then harvested 24–48 h later. FABP5 and FABP4 protein levels were analyzed by immunoblotting. β-actin was used as a loading control. c Cell proliferation of FABP5-KD and control HUVECs was measured by BrdU incorporation. Bar graph represents mean ± SEM from five independent experiments. d Cell cycle distribution of synchronized FABP5-KD and control HUVECs was analyzed by flow cytometry. Bar graph represents mean ± SEM from four independent experiments. e HUVECs were incubated with indicated doses of VEGF-A for 24 h. FABP5 and β-actin levels were assessed by immunoblotting and densitometry. Fold-FABP5 changes are shown at the bottom of the figure. f HUVECs were incubated with a chemically defined lipid mixture for 24 h. FABP5 and β-actin levels were assessed by immunoblotting and densitometry. The indicated lipid doses are based on the fatty acid concentration in the mixture. Results in e and f are representative of three independent experiments
Due to the potent regulation of EC proliferation by FABP5, we next investigated whether FABP5 was a downstream target of VEGF-A. However, FABP5 levels remained unchanged in response to VEGF-A (1, 10, and 50 ng/ml) treatment for 24 h (Fig. 2e) or 48 h (not shown). Similarly, bFGF (1 and 10 ng/ml) treatment for up to 48 h did not alter FABP5 protein expression levels in HUVECs (not shown). Since the assumed intracellular function of FABP5 is to bind fatty acids, we next assessed whether EC-FABP5 levels were regulated by lipids. Incubation of HUVECs with a chemically defined lipid mixture resulted in a dose-dependent increase in FABP5 protein levels (Fig. 2f). Thus, extracellular lipids, but not VEGF-A or bFGF, regulates FABP5 expression in ECs.
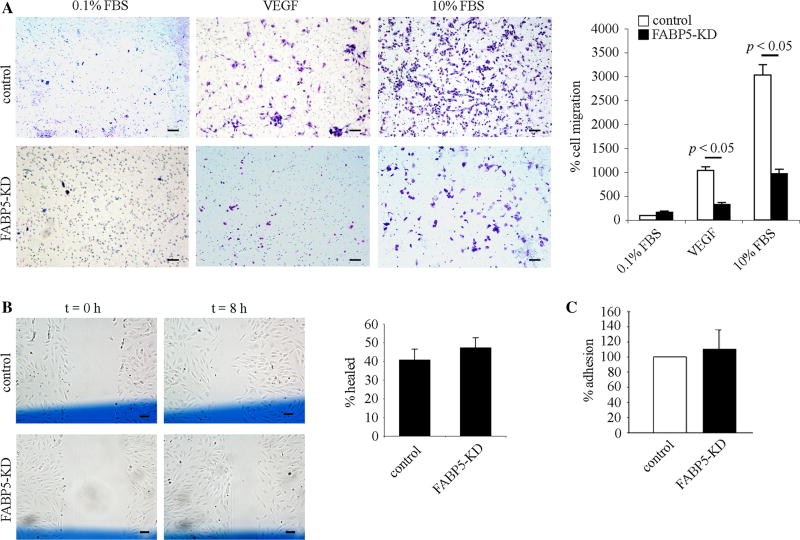
FABP5 deficiency attenuates chemotactic migration of endothelial cells
To determine whether FABP5 had an effect on EC motility, a critical process during angiogenesis, we first assessed the effect of FABP5 deficiency on chemotactic migration of HUVECs using the transwell migration assay (Fig. 3a). These experiments were performed using growth-arrested cells to ensure that the effect of FABP5 on cell proliferation would not be a confounding factor. The migration of FABP5-KD HUVECs was significantly reduced in comparison with control cells in response to both VEGF and 10 % FBS gradient across gelatin-coated membranes (p < 0.05). To determine whether FABP5 has an effect on random migration of ECs, the cellular wound assay was performed (Fig. 3b). There was no difference in chemokinesis of FABP5-KD HUVECs as compared to control cells. To determine whether FABP5 played a role in adhesive properties of HUVECs, adhesion of FABP5-KD and control HUVECs to gelatin was compared and was found to be similar (Fig. 3c).
Fig. 3.
FABP5 deficiency attenuates chemotactic migration of endothelial cells. a Representative images of filters stained with Diff-Quick in a transwell cell migration assay. FABP5-KD or control HUVECs were plated in the upper chamber of a transwell at a density of 1 × 105 cells in 0.1 % FBS, ECGF-free medium, and transwell migration was stimulated by addition of VEGF (50 ng/ml) or 10 % FBS to the media in the lower well. EC migration was quantified by counting the number of cells in three random fields per insert at ×100 magnification. Bar graph represents mean ± SEM from three independent experiments. Scale bar 100 µm. b Representative images of a cellular wound assay at 0 and 8 h after wounding. Control and FABP5-KD HUVECs were grown to 70–90 % confluency in 6-well plates. Three vertical scratches were made across each well with a flat-edge forceps. A horizontal reference line (shown in blue color) was drawn to denote the scratch field of view (FOV) at the scratch intersection. Images of the wounded cells were taken above and below the reference line at the scratch/cell interfaces at t = 0 and t = 8 h. The average scratch width was determined for each FOV, and the distance migrated was calculated and expressed as a percentage of the initial scratch width. Bar graph represents mean ± SEM from four independent experiments. Scale bar 100 µm. c Control and FABP5-KD HUVECs were seeded in 96-well plates coated with gelatin at a density of 5 × 104 cells per well and allowed to attach for 1 h. Non-adherent cells were removed by washing in PBS, and adherent cells were fixed and stained in 0.125 % Coomassie Blue. After thorough destaining, the contents of the wells were solubilized in 2 % SDS, and optical density, indicative of adherent cells, was read at 620 nm. Bar graph represents mean ± - SEM from three independent experiments
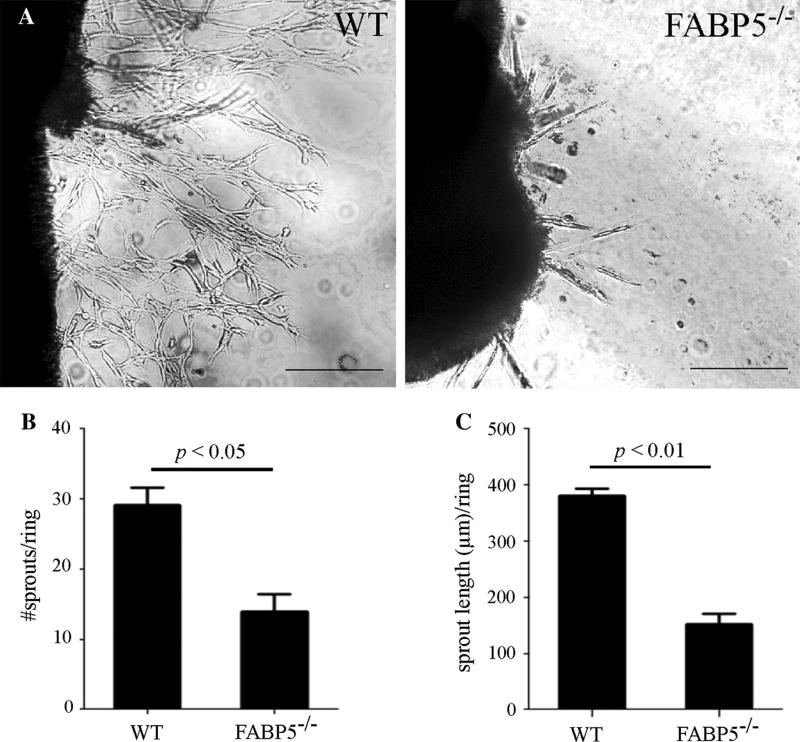
FABP5-knockout aortic rings exhibit reduced angiogenic sprouting
Since FABP5 promotes both EC proliferation and chemotactic migration, two critical processes that are essential in angiogenic responses, we next analyzed the role of FABP5 during blood vessel sprouting by using the aortic ring assay as an ex vivo model of angiogenesis. Aortic rings from 4-to 5-week-old, gender-matched FABP5−/− and WT mice were embedded in Matrigel and supplemented with bFGF for 7 days as previously described [9]. As expected, WT murine aortic rings demonstrated a robust sprouting angiogenesis, whereas the number and length of angiogenic sprouts were significantly reduced in FABP5−/− aortic rings (Fig. 4a–c). Thus, FABP5 promotes angiogenic responses of ECs both in vitro and ex vivo.
Fig. 4.
Angiogenic sprouting is impaired in FABP5−/− mouse aortic ring explants. a 1-mm long WT and FABP5−/− mouse aortic rings were embedded in Matrigel in 96-well plates and cultured in medium supplemented with bFGF. Representative images are shown. Scale bar 100 µm. b The number of angiogenic sprouts arising from the aortic rings was quantified. c The average length of angiogenic sprouts arising from the aortic rings was measured. Bar graphs represent mean ± SEM, n = 8–10 mice per group
FABP5 deficiency confers protection against endothelial cell death
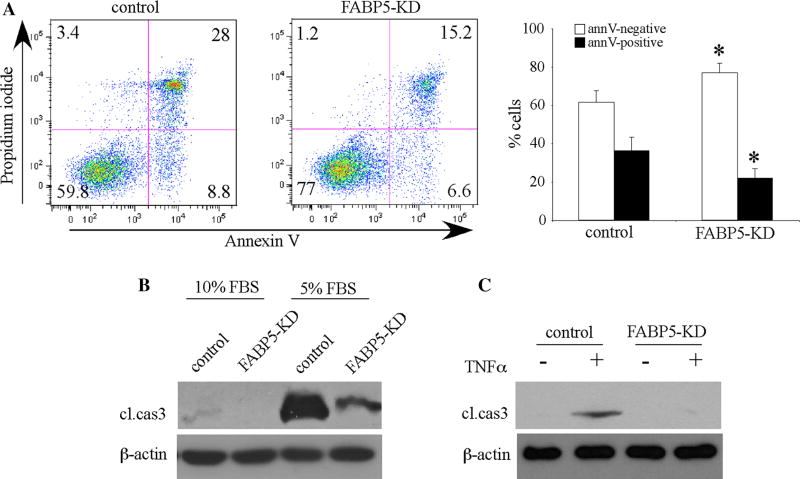
Our previous studies have demonstrated that EC-FABP4 has an anti-apoptotic function [9]. To determine whether FABP5 had a similar role in ECs, we first examined the response of FABP5-KD HUVECs to serum deprivation-induced cell death. Surprisingly, flow cytometry analysis demonstrated that approximately 22 % of FABP5-KD HUVECs were annexin V-positive (combined early and late apoptosis) compared to 37 % of control cells (p < 0.01, Fig. 5a). The positive effect of FABP5 deficiency on EC survival was verified by detection of cleaved caspase 3 as a marker of apoptosis by immunoblotting (Fig. 5b). As expected, caspase 3 cleavage was not detected or minimal in FABP5-KD or control HUVECs cultured in 10 % FBS, but was present in control HUVECs cultured in 5 % FBS. Consistent with the flow cytometry analysis, the cleaved caspase 3 signal was dramatically reduced in FABP5-KD HUVECs as compared to control HUVECs cultured in 5 % FBS. In addition, similar results were obtained using TNFα as the pro-apoptotic signal (Fig. 5c). Incubation of HUVECs with TNFα for 24 h resulted in apoptosis as evidenced by detection of cleaved caspase 3 in the control, but not in FABP5-KD cells. Thus, unexpectedly, FABP5 deficiency provided protection against EC apoptosis induced by two different methods, serum deprivation and TNF-α signaling.
Fig. 5.
FABP5 deficiency enhances endothelial cell survival. a FABP5-KD and control HUVECs were cultured in 5 % FBS-containing medium for 24 h. Cells were harvested, stained with FITC-conjugated annexin V and propidium iodide, and analyzed by flow cytometry. Representative plots and mean results ± SEM from five independent experiments are shown (*p < 0.01 vs control). b FABP5-KD and control HUVECs were cultured in 10 or 5 % FBS-containing media for 24 h. Cells were harvested, and immunoblotting for caspase 3 was performed. c FABP5-KD and control HUVECs were treated with TNFα (40 ng/ml) or the vehicle control for 24 h, and immunoblotting for caspase 3 was performed. All immunoblots are representative of a minimum of three independent experiments
Role of PPAR-δ activation on FABP5-induced responses in endothelial cells
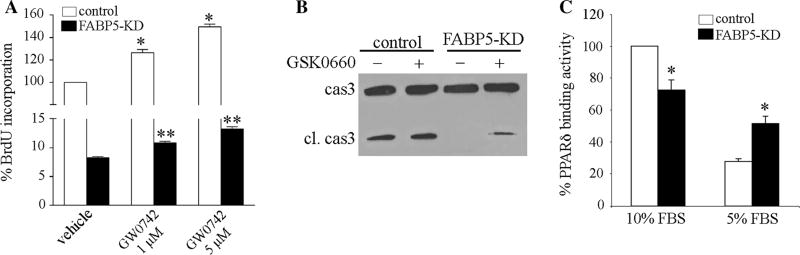
Previous studies have shown that PPARδ participates in regulation of EC proliferation and angiogenic responses [34–36]. FABP5 is known to regulate the activity of PPARδ in tumor cell lines [21, 22, 25], but whether this occurs in EC is not known. To determine whether PPARδ played a role on reduced EC proliferation in FABP5-KD HUVECs, we treated the cells with the selective PPARδ agonist GW0742 [37, 38] for 24 h and assessed its effect on cell proliferation using BrdU incorporation (Fig. 6a). As previously reported, PPARδ activation induced a significant increase in proliferation of control HUVECs in a dose-dependent manner. There was also a modest, but statistically significant increase in proliferation of FABP5-KD HUVECs treated with GW0742 as compared to the vehicle-treated cells. However, the cell proliferation rate of GW0742-treated FABP5-KD cells remained much lower than GW0742-treated control cells. These results suggested that both PPAR-δ-dependent and PPAR-δ-independent mechanisms were involved in inhibition of EC proliferation by FABP5 deficiency.
Fig. 6.
Role of PPARδ on FABP5-induced responses in ECs. a HUVECs were transduced with control shRNA or FABP5-shRNA. Cells were treated with DMSO or GW0742 at the indicated doses for 24 h. Cell proliferation was analyzed by BrdU incorporation using an ELISA kit. *p < 0.05 versus control/vehicle; **p < 0.05 versus FABP5-KD/vehicle. b FABP5-KD and control HUVECs were cultured in 5 % FBS-containing medium with or without PPARδ specific inhibitor GSK0660 (100 nM) for 24 h. Cells were harvested, and immunoblotting for caspase 3 was performed. c FABP5-KD and control HUVECs were cultured in 10 or 5 % FBS-containing medium for 24 h. DNA-binding activity of PPARδ in nuclear extracts was measured using an ELISA kit. *p < 0.05 versus control
We next examined the effect of PPARδ activity on survival of FABP5-KD HUVECs. We reasoned that if increased resistance of FABP5-KD cells to serum deprivation-induced apoptosis was mediated via PPARδ, then inhibition of PPARδ activity would increase their susceptibility to apoptotic cell death. Indeed, treatment of FABP5-KD HUVECs cultured in 5 % FBS with the PPARδ specific inhibitor GSK0660 [39] led to the detection of a weak cleaved caspase 3 band, which was not evident in vehicle-treated FABP5-KD cells (Fig. 6b). However, GSK0660 did not cause a significant increase in caspase 3 activation in control cells. Thus, PPARδ activation appeared to mediate, only in part, the resistance of FABP5-KD cells to serum deprivation-induced apoptosis.
Next we examined whether differential regulation of PPARδ activity, in a nutrient-dependent manner, might be responsible for the opposing effects of FABP5 on EC proliferation and survival. To examine this hypothesis, we examined the DNA-binding activity of PPARδ using nuclear extracts from 10 to 5 % FBS-exposed control and FABP5-KD HUVECs (Fig. 6c). FABP5-KD HUVECs grown in 10 % FBS demonstrated an approximately 25 % decrease in PPARδ-binding activity as compared to control HUVECs (p < 0.05). Serum deprivation led to a dramatic 75 % reduction in PPARδ activity in control cells, whereas FABP5-KD cells grown in 5 % FBS demonstrated a significantly higher PPARδ activity as compared to control cells (p < 0.05). These results suggest that while EC-FABP5 functions as a positive regulator of PPARδ under normal culture conditions, it inhibits the activity of PPARδ in serum deprivation.
Discussion
FABP4 and FABP5 are co-expressed in macrophages, adipocytes, and a subset of ECs. Previous studies have shown that these two FABPs have complementary functions in macrophages and adipocytes [2, 20]. In ECs, FABP4 exerts a pro-angiogenic effect [9–11], but the role of EC-FABP5 has not been investigated previously. Our study demonstrates that in contrast to FABP4, the expression of FABP5 is not restricted to microvascular ECs, but also extends to larger vascular ECs. Furthermore, while EC-FABP5 has some overlapping pro-angiogenic functions with FABP4, i.e., promotion of proliferation and chemotactic migration, contrary to EC-FABP4, it can also enhance apoptotic EC death under certain conditions. These functions of EC-FABP5 are mediated by both PPARδ-dependent and PPAR-δ-independent pathways.
FABP5-KD ECs had a profound impairment in cell proliferation. The magnitude of this effect (>90 % reduction) was greater than that observed in FABP4-KD cells (~70–80 %). Double FABP4/5 knockdown ECs demonstrated similar proliferation rates compared to those of FABP5-KD ECs (data not shown). Cell cycle analysis demonstrated that the percentage of FABP5-KD cells were higher in the G0/G1 phase and lower in the S and M phases compared to control cells. Taken together, these results demonstrate that EC-FABP5 is a potent positive regulator of cell cycle progression under normal conditions. In addition, similar to FABP4-KD ECs, FABP5-KD ECs demonstrated decreased motility in response to chemotactic factors such as VEGF and 10 % FBS. Furthermore, FABP5-deficient aortic rings exhibited reduced sprouting ex vivo, thus indicating that reduced EC proliferation and migration secondary to FABP5 deficiency result in impaired angiogenic responses. Interestingly, in contrast to FABP4, EC-FABP5 levels are not induced by VEGF-A or bFGF, but are induced by a lipid mixture in a dose-dependent manner. In this study, we did not identify the specific fatty acids in the lipid mixture that are responsible for this effect, but it will be important to do so in future studies to clarify whether FABP5 can be a link between “good lipids” or “bad lipids,” and angiogenic responses in ECs. Thus, it will be important to identify both inducers and inhibitors of EC-FABP5 expression as they might have potential therapeutic applications. Of note, although stem cell factor/c-kit pathway was identified to have a key role in mediating the angiogenic effects of FABP4 in ECs, we did not find any significant alterations in this pathway in FABP5-KD HUVECs by Q-RT-PCR (data not shown).
An unexpected finding in our study was the effect of FABP5 deficiency on EC survival. Although FABP4 and FABP5 had similar effects on EC proliferation and migration, FABP5 had a distinctly diverse effect on EC survival than FABP4. In previous studies, we have shown that EC-FABP4 provides significant protection against apoptotic cell death, whereas in the studies described here, we found that FABP5 rendered ECs more susceptible to apoptotic cell death induced by two different stimuli: serum starvation and TNF-α exposure. How can the opposing effects of FABP5 on EC proliferation versus survival be reconciled? One potential explanation is that EC-FABP5 functions as a sensor that links the extracellular conditions to cellular responses. Thus, under optimal conditions (as modeled by culturing cells in 10 % FBS), FABP5 promotes EC proliferation and migration, whereas in the presence of nutrient deprivation or other stressors, FABP5 facilitates EC death likely by activating apoptotic pathways. In this context, FABP5 appears to play a similar role to that of autophagy and further studies will be needed to investigate whether FABP5 could in fact be linked to autophagic responses in ECs. From a functional standpoint, positive regulation of EC proliferation and migration by FABP5 may be critical in promotion of angiogenic responses of microvascular ECs, where FABP4 and FABP5 are co-expressed. However, FABP5 may also contribute to vascular regression by promotion of apoptosis during conditions associated with nutrient deprivation, such as those occurring during tissue ischemia or starvation. These hypotheses will need to be addressed in future studies.
In previous studies, FABP5 was shown to deliver ligands from the cytosol and enhance the transcriptional activity of PPARδ in other cell types [21, 25, 40, 41], but to our knowledge, this relationship has not been examined in primary ECs previously. PPARδ is known to promote cell proliferation and angiogenic sprouting and protect ECs against apoptosis [34, 35]. Based on these previous data, we focused on PPARδ as a potential mediator of FABP5-related effects on EC responses. Consistent with previous studies, we found that a highly specific agonist of PPARδ, GW0742, enhanced proliferation of HUVECs and also modestly improved the proliferation of FABP5-KD ECs. We also found that while a PPARδ inhibitor did not have an obvious effect on serum deprivation-induced apoptosis of control cells, it induced some apoptosis in FABP5-KD cells. Our studies also suggested that PPARδ activity is differentially regulated under conditions that promote EC proliferation versus apoptosis. This notion was further supported by the opposite effects of FABP5-KD on PPARδ-binding activity in baseline conditions versus serum deprivation. Thus, taken together, our studies show that PPARδ activity is, only in part, responsible for mediating the divergent effects of FABP5 on EC proliferation and survival.
In summary, FABP5 is co-expressed with FABP4 in ECs in microvascular beds of several tissues, but the expression of FABP5 also extends to some larger vessel ECs, where FABP4 is not detected. Similar to FABP4, FABP5 significantly enhances EC proliferation, chemotactic migration, and angiogenic sprouting. These findings indicate that FABP4 and FABP5 play similar roles in regulation of angiogenic responses in microvascular ECs. However, an important difference is that, in contrast to the anti-apoptotic effects of FABP4, FABP5 promotes apoptotic EC death under certain conditions. This effect of FABP5 may be particularly relevant in larger vessel ECs, which lack FABP4 expression. The effect of FABP5 on EC proliferation and survival is mediated, at least in part, by PPARδ. Collectively, our findings suggest that the balance between FABP4 and FABP5 in ECs may be critical in regulation of quiescent versus angiogenic phenotypes of blood vessels. Further studies will be required to fully characterize the biologic significance of the interplay between FABP4 and FABP5 in vascular ECs in both physiologic and pathologic conditions.
Supplementary Material
Acknowledgments
This study was supported by grants to SC from the American Heart Association (11GRNT4900002) and BWH Biomedical Research Institute, and Clinical Translational Science Award (UL1RR025758) to Harvard University and Brigham, and Women’s Hospital from the National Center for Research Resources.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10456-015-9491-4) contains supplementary material, which is available to authorized users.
References
- 1.Hertzel AV, Bernlohr DA. The mammalian fatty acid-binding protein multigene family: molecular and genetic insights into function. Trends Endocrinol Metab. 2000;11:175–180. doi: 10.1016/s1043-2760(00)00257-5. [DOI] [PubMed] [Google Scholar]
- 2.Makowski L, Hotamisligil GS. The role of fatty acid binding proteins in metabolic syndrome and atherosclerosis. Curr Opin Lipidol. 2005;16:543–548. doi: 10.1097/01.mol.0000180166.08196.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smathers RL, Petersen DR. The human fatty acid-binding protein family: evolutionary divergences and functions. Hum Genomics. 2011;5:170–191. doi: 10.1186/1479-7364-5-3-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furuhashi M, Ishimura S, Ota H, Miura T. Lipid chaperones and metabolic inflammation. Int J Inflam. 2011;2011:642612. doi: 10.4061/2011/642612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Storch J, Thumser AE. Tissue-specific functions in the fatty acid-binding protein family. J Biol Chem. 2010;285:32679–32683. doi: 10.1074/jbc.R110.135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richieri GV, Ogata RT, Zimmerman AW, Veerkamp JH, Kleinfeld AM. Fatty acid binding proteins from different tissues show distinct patterns of fatty acid interactions. Biochemistry. 2000;39:7197–7204. doi: 10.1021/bi000314z. [DOI] [PubMed] [Google Scholar]
- 7.Cataltepe O, Arikan MC, Ghelfi E, Karaaslan C, Ozsurekci Y, et al. Fatty acid binding protein 4 is expressed in distinct endothelial and non-endothelial cell populations in glioblastoma. Neuropathol Appl Neurobiol. 2012;38:400–410. doi: 10.1111/j.1365-2990.2011.01237.x. [DOI] [PubMed] [Google Scholar]
- 8.Cataltepe S, Arikan MC, Liang X, Smith TW, Cataltepe O. Fatty acid binding protein 4 expression in cerebral vascular malformations: implications for vascular remodelling. Neuropathol Appl Neurobiol. 2015;41:646–656. doi: 10.1111/nan.12159. [DOI] [PubMed] [Google Scholar]
- 9.Elmasri H, Ghelfi E, Yu CW, Traphagen S, Cernadas M, et al. Endothelial cell-fatty acid binding protein 4 promotes angiogenesis: role of stem cell factor/c-kit pathway. Angiogenesis. 2012;15:457–468. doi: 10.1007/s10456-012-9274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elmasri H, Karaaslan C, Teper Y, Ghelfi E, Weng M, et al. Fatty acid binding protein 4 is a target of VEGF and a regulator of cell proliferation in endothelial cells. FASEB J. 2009;23:3865–3873. doi: 10.1096/fj.09-134882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghelfi E, Yu CW, Elmasri H, Terwelp M, Lee CG, et al. Fatty acid binding protein 4 regulates VEGF-induced airway angiogenesis and inflammation in a transgenic mouse model: implications for asthma. Am J Pathol. 2013;182:1425–1433. doi: 10.1016/j.ajpath.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masouye I, Hagens G, Van Kuppevelt TH, Madsen P, Saurat JH, et al. Endothelial cells of the human microvasculature express epidermal fatty acid-binding protein. Circ Res. 1997;81:297–303. doi: 10.1161/01.res.81.3.297. [DOI] [PubMed] [Google Scholar]
- 13.Iso T, Maeda K, Hanaoka H, Suga T, Goto K, et al. Capillary endothelial fatty acid binding proteins 4 and 5 play a critical role in fatty acid uptake in heart and skeletal muscle. Arterioscler Thromb Vasc Biol. 2013;33:2549–2557. doi: 10.1161/ATVBAHA.113.301588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madsen P, Rasmussen HH, Leffers H, Honore B, Celis JE. Molecular cloning and expression of a novel keratinocyte protein (psoriasis-associated fatty acid-binding protein [PA-FABP]) that is highly up-regulated in psoriatic skin and that shares similarity to fatty acid-binding proteins. J Invest Dermatol. 1992;99:299–305. doi: 10.1111/1523-1747.ep12616641. [DOI] [PubMed] [Google Scholar]
- 15.Ghelfi E, Karaaslan C, Berkelhamer S, Akar S, Kozakewich H, et al. Fatty acid-binding proteins and peribronchial angiogenesis in bronchopulmonary dysplasia. Am J Respir Cell Mol Biol. 2011;45:550–556. doi: 10.1165/rcmb.2010-0376OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda K, Uysal KT, Makowski L, Gorgun CZ, Atsumi G, et al. Role of the fatty acid binding protein mal1 in obesity and insulin resistance. Diabetes. 2003;52:300–307. doi: 10.2337/diabetes.52.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siegenthaler G, Hotz R, Chatellard-Gruaz D, Didierjean L, Hellman U, et al. Purification and characterization of the human epidermal fatty acid-binding protein: localization during epidermal cell differentiation in vivo and in vitro. Biochem J. 1994;302(Pt 2):363–371. doi: 10.1042/bj3020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furuhashi M, Fucho R, Gorgun CZ, Tuncman G, Cao H, et al. Adipocyte/macrophage fatty acid-binding proteins contribute to metabolic deterioration through actions in both macrophages and adipocytes in mice. J Clin Invest. 2008;118:2640–2650. doi: 10.1172/JCI34750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boord JB, Maeda K, Makowski L, Babaev VR, Fazio S, et al. Combined adipocyte-macrophage fatty acid-binding protein deficiency improves metabolism, atherosclerosis, and survival in apolipoprotein E-deficient mice. Circulation. 2004;110:1492–1498. doi: 10.1161/01.CIR.0000141735.13202.B6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda K, Cao H, Kono K, Gorgun CZ, Furuhashi M, et al. Adipocyte/macrophage fatty acid binding proteins control integrated metabolic responses in obesity and diabetes. Cell Metab. 2005;1:107–119. doi: 10.1016/j.cmet.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Schug TT, Berry DC, Toshkov IA, Cheng L, Nikitin AY, et al. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/delta to RAR. Proc Natl Acad Sci USA. 2008;105:7546–7551. doi: 10.1073/pnas.0709981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu RZ, Graham K, Glubrecht DD, Germain DR, Mackey JR, et al. Association of FABP5 expression with poor survival in triple-negative breast cancer: implication for retinoic acid therapy. Am J Pathol. 2011;178:997–1008. doi: 10.1016/j.ajpath.2010.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang LY, Wong TY, Chiang WF, Chen YL. Fatty-acid-binding protein 5 promotes cell proliferation and invasion in oral squamous cell carcinoma. J Oral Pathol Med. 2010;39:342–348. doi: 10.1111/j.1600-0714.2009.00836.x. [DOI] [PubMed] [Google Scholar]
- 25.Kannan-Thulasiraman P, Seachrist DD, Mahabeleshwar GH, Jain MK, Noy N. Fatty acid-binding protein 5 and PPARbeta/delta are critical mediators of epidermal growth factor receptor-induced carcinoma cell growth. J Biol Chem. 2010;285:19106–19115. doi: 10.1074/jbc.M109.099770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaffe EA, Nachman RL, Becker CG, Minick CR. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J Clin Invest. 1973;52:2745–2756. doi: 10.1172/JCI107470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altiok O, Yasumatsu R, Bingol-Karakoc G, Riese RJ, Stahlman MT, et al. Imbalance between cysteine proteases and inhibitors in a baboon model of bronchopulmonary dysplasia. Am J Respir Crit Care Med. 2006;173:318–326. doi: 10.1164/rccm.200503-425OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yasumatsu R, Altiok O, Benarafa C, Yasumatsu C, Bingol-Karakoc G, et al. SERPINB1 upregulation is associated with in vivo complex formation with neutrophil elastase and cathepsin G in a baboon model of bronchopulmonary dysplasia. Am J Physiol Lung Cell Mol Physiol. 2006;291:L619–L627. doi: 10.1152/ajplung.00507.2005. [DOI] [PubMed] [Google Scholar]
- 29.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 30.Humphries MJ. Cell adhesion assays. Mol Biotechnol. 2001;18:57–61. doi: 10.1385/MB:18:1:57. [DOI] [PubMed] [Google Scholar]
- 31.Dumartin L, Quemener C, Laklai H, Herbert J, Bicknell R, et al. Netrin-1 mediates early events in pancreatic adenocarcinoma progression, acting on tumor and endothelial cells. Gastroenterology. 2010;138:1595–1606. 1606, e1591–e1598. doi: 10.1053/j.gastro.2009.12.061. [DOI] [PubMed] [Google Scholar]
- 32.Talbot NC, Blomberg LA, Mahmood A, Caperna TJ, Garrett WM. Isolation and characterization of porcine visceral endoderm cell lines derived from in vivo 11-day blastocysts. In Vitro Cell Dev Biol Anim. 2007;43:72–86. doi: 10.1007/s11626-007-9014-8. [DOI] [PubMed] [Google Scholar]
- 33.Ha MK, Chung KY, Lee JH, Bang D, Park YK, et al. Expression of psoriasis-associated fatty acid-binding protein in senescent human dermal microvascular endothelial cells. Exp Dermatol. 2004;13:543–550. doi: 10.1111/j.0906-6705.2004.00196.x. [DOI] [PubMed] [Google Scholar]
- 34.Jiang B, Liang P, Zhang B, Huang X, Xiao X. Enhancement of PPAR-beta activity by repetitive low-grade H(2)O(2) stress protects human umbilical vein endothelial cells from subsequent oxidative stress-induced apoptosis. Free Radic Biol Med. 2009;46:555–563. doi: 10.1016/j.freeradbiomed.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 35.Piqueras L, Reynolds AR, Hodivala-Dilke KM, Alfranca A, Redondo JM, et al. Activation of PPARbeta/delta induces endothelial cell proliferation and angiogenesis. Arterioscler Thromb Vasc Biol. 2007;27:63–69. doi: 10.1161/01.ATV.0000250972.83623.61. [DOI] [PubMed] [Google Scholar]
- 36.Muller-Brusselbach S, Komhoff M, Rieck M, Meissner W, Kaddatz K, et al. Deregulation of tumor angiogenesis and blockade of tumor growth in PPARbeta-deficient mice. EMBO J. 2007;26:3686–3698. doi: 10.1038/sj.emboj.7601803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graham TL, Mookherjee C, Suckling KE, Palmer CN, Patel L. The PPARdelta agonist GW0742X reduces atherosclerosis in LDLR(−/−) mice. Atherosclerosis. 2005;181:29–37. doi: 10.1016/j.atherosclerosis.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 38.Berry DC, Noy N. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation- activated receptor beta/delta and retinoic acid receptor. Mol Cell Biol. 2009;29:3286–3296. doi: 10.1128/MCB.01742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shearer BG, Steger DJ, Way JM, Stanley TB, Lobe DC, et al. Identification and characterization of a selective peroxisome proliferator-activated receptor beta/delta (NR1C2) antagonist. Mol Endocrinol. 2008;22:523–529. doi: 10.1210/me.2007-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan NS, Shaw NS, Vinckenbosch N, Liu P, Yasmin R, et al. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol Cell Biol. 2002;22:5114–5127. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgan E, Kannan-Thulasiraman P, Noy N. Involvement of fatty acid binding protein 5 and PPARbeta/delta in prostate cancer cell growth. PPAR Res. 2010 doi: 10.1155/2010/234629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.