Abstract
A recent study led by Professor Rob Martienssen in Cell showed that 3′-tRNA-derived small RNAs can suppress long terminal repeat retrotransposon activity in mammalian cells by mechanisms independent of DNA-associated epigenetic marks, suggesting how the genome may defend itself from retrotransposon invasion during epigenetic reprogramming.
Transposon elements (TEs), known as the 'genomic parasites', are mobile genomic DNAs capable of autonomous or non-autonomous transposition. The transposition activities of TEs are potentially harmful to the host genome, and the transcription of TEs is normally suppressed by epigenetic marks such as DNA methylation and histone modifications1. However, it remains an open question how the genome defends itself during the window of epigenetic reprogramming, such as pre-implantation embryo development, when most of the epigenetic marks are wiped off and then reestablished2. In mammals such as mice, small RNA-mediated pathways centered by PIWI-interacting RNAs (piRNAs) are effective in controlling TE activities via post-transcriptional silencing or/and de novo methylation of TE loci3. However, piRNAs in mice are mostly expressed in the developing germ cells, but gradually depleted during sperm maturation and pre-implantation embryo development4, leaving the question whether other small RNA pathways can silence TEs during this period of vulnerability. In a recent study published in Cell, Andrea et al.5 discovered that 3′-tRNA-derived small RNAs (tsRNAs or tRFs) with different lengths (18 nt and 22 nt) can silence at least one class of mammalian TEs, the long terminal repeat (LTR)-retrotransposon, by blocking reverse transcription (RT; 18 nt) and post-transcriptional silencing (22 nt), respectively.
Retrotransposons, a major class of TEs, use RNA as an intermediate, which is reverse-transcribed into DNA and then inserted into host genome1. Retrotransposons have two major subclasses, LTR (also known as endogenous retrovirues (ERVs)) and non-LTR retrotransposons (e.g., LINEs, SINEs)1. Usually, Dnmt1-mediated DNA methylation inhibits most of the non-LTR LINEs, whereas a majority of the LTR-retrotransposons are silenced by Setdb1-mediated histone H3K9 trimethylation (H3K9me3). Andrea et al.5 set out the experiments by discovering two types of 3′-tsRNAs with different lengths, namely 18-nt-3′-tRF and 22-nt-3′-tRF, that were elevated in Setdb1−/−, but not in Dnmt1−/− mouse ESCs (mESCs). Both types of tsRNAs contain the 3′-CCA end of their tRNA precursor, indicating that they are cleaved from the mature tRNAs. They further found that these elevated 3′-tsRNAs from Setdb1−/− mESCs show sequence matches to ERVs (particularly young and active ones), thus suggesting a potential link between 3′-tsRNAs and ERV activity. They next examined the function of 3′-tsRNAs in TE control by using a well-defined retrotransposition assay in Hela cells, and found that transfection of synthesized 18-nt-3′-tRF or 22-nt-3′-tRF (tRF-Lys-AAA) with complementary sequence to ERVs can inhibit their transposition activity.
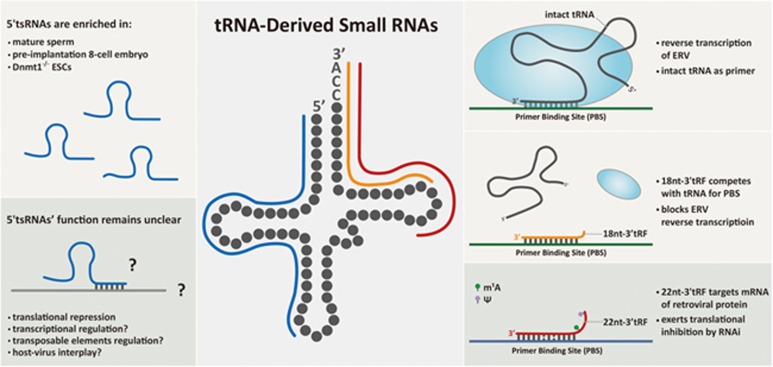
The authors next explored the mechanisms by which 18-nt-3′-tRFs inhibit ERV transposition. They first found that 3′-tsRNA-targeted ERV loci were not strongly associated with H3K9me3 elevation, suggesting an effect independent of H3K9me3-induced transcriptional suppression. They also excluded the possibility of post-transcriptional silencing because transfection of 18-nt-3′-tRFs did not induce RNAi-like mRNA degradation or translational inhibition. Crucially, the authors detected strong RT inhibition by 18-nt-3′-tRFs, the mechanism of which lays in the fact that many ERVs duplicate themselves by using 3′ terminus of intact mature tRNA as a primer for their RT, anchoring to the highly conserved primer binding sequence (PBS; Figure 1). 18-nt-3′-tRFs compete with mature tRNAs for the PBS of ERVs, leading to RT block and impeded retroviral cDNA synthesis (Figure 1). This RT blocking effect works efficiently when 18-nt-3′-tRF is completely complementary to the PBS, whereas 2-bp mismatch decreases the efficiency. On the other hand, the authors found that the 22-nt-3′-tRF has a different role in ERV inhibition, which is through inducing post-transcriptional silencing of protein-coding mRNA of autonomous ERV. The effect of 22-nt-3′-tRF also depends on the presence of PBS target site, but with a miRNA-like tolerance of 2-bp mismatch (Figure 1). Together, both 18-nt- and 22-nt-3′-tRFs contribute to the suppression of mammalian LTR-retrotransposon activity with distinct mechanisms.
Figure 1.
Illustration of 3′-tsRNAs in suppressing LTR-retrotransposon (ERV), and the potential functions of 5′-tsRNAs. Right panel: 3′-tsRNAs with 18 nt in length block ERV reverse transcription by competing with intact tRNAs for PBS; 3′-tsRNAs with 22 nt in length exert post-transcriptional silencing of retroviral protein production by targeting its mRNA. Left panel: cell-specific distribution and potential functions of 5′-tsRNAs.
This work advanced our understanding of RNA-mediated retrotransposon control in mammals, although most evidence was obtained in vitro. It also inspires interesting questions, for example, with merely 4 extra nucleotides at the 5′ tail, why 22-nt-3′-tRF showed totally different working mechanisms compared to 18-nt-3′-tRF? Could the reasons go beyond simply their sequences but extend to the specific RNA modifications harbored by each? It is known that modifications in tRNAs can dramatically change their secondary structures6, and this may similarly apply to tsRNAs4. Indeed, the extra 4 nucleotides in 22-nt-3′-tRF harbor RNA modifications (Figure 1), which may contribute to altered RNA structure and interacting potential, possibly related to the recognition of ERV PBS and silencing complexes. Due to the concern of RNA modifications, one caveat of the findings is that the tsRNAs studied in vitro are modified with 2′-O-methylation in each nucleotide and might have different properties compared to the tsRNAs in vivo. The precise contribution of tsRNAs in retrotransposon control in vivo might be more complicated and deserves case-by-case study.
Nevertheless, the study is important and shows clear implications for many fundamental biological processes. For example, tsRNAs are the most abundant small RNA species in the mature sperm of mice4, 7. The sperm tsRNAs delivered at fertilization can change the transcriptome of pre-implantation embryo8, including LTR-associated gene expression9, and can generate long-standing effect that affect the phenotype of the offspring8. This is thought-inspiring, as the pre-implantation embryo undergoes genome-wide epigenetic reprogramming, leaving the genome unprotected by epigenetic marks when facing various TEs. Could the tsRNAs delivered by sperm interplay with genome protection/modulation during this period? Interestingly, recent small-RNA sequencing of different stages of mouse pre-implantation embryos has revealed a surge of tsRNAs at 8-cell stage10, with signatures similar to the sperm tsRNAs. This may suggest a self-loop induction of the initial sperm tsRNA input during early embryo development. However, one confounding fact is that the tsRNA signatures revealed by RNA-seq in both sperm and early embryos are mostly composed of 5′-tsRNAs (Figure 1), but not 3′-tsRNAs. This could be due to RNA-seq bias caused by RNA modifications in the sperm 3′-tsRNAs that interfered with RNA-seq library preparation11; or it may suggest that 5′-tsRNAs can also affect TE activity with unidentified mechanisms. Very intriguingly, 5′-tsRNAs are indeed abundantly upregulated in the Dnmt1−/−, but not in Setdb1−/− mESCs, as shown by Andrea et al. (Figure 1); their function under these circumstances apparently deserves future in-depth investigations.
Finally, since LTR-retrotransposons resemble the behavior of retroviruses from which they are derived, the function of tsRNAs in repressing TEs probably bears even wider implications in host-virus interplay. Interestingly, tsRNAs are highly enriched in the mammalian serum and elevated upon active infection12. The possibility of tsRNAs as an ancient part of innate immune system against virus infection is intriguing and awaits further evidence.
References
- Slotkin RK, Martienssen R. Nat Rev Genet 2007; 8:272–285. [DOI] [PubMed]
- Feng S, Jacobsen SE, Reik W. Science 2010; 330:622–627. [DOI] [PMC free article] [PubMed]
- Siomi MC, Sato K, Pezic D, et al. Nat Rev Mol Cell Biol 2011; 12:246–258. [DOI] [PubMed]
- Chen Q, Yan W, Duan E. Nat Rev Genet 2016; 17:733–743. [DOI] [PMC free article] [PubMed]
- Schorn AJ, Gutbrod MJ, LeBlanc C, et al. Cell 2017; 170:61–71. [DOI] [PMC free article] [PubMed]
- Motorin Y, Helm M. Biochemistry 2010; 49:4934–4944. [DOI] [PubMed]
- Peng H, Shi J, Zhang Y, et al. Cell Res 2012; 22:1609–1612. [DOI] [PMC free article] [PubMed]
- Chen Q, Yan M, Cao Z, et al. Science 2016; 351:397–400. [DOI] [PubMed]
- Sharma U, Conine CC, Shea JM, et al. Science 2016; 351:391–396. [DOI] [PMC free article] [PubMed]
- Yang Q, Lin J, Liu M, et al. Sci Adv 2016; 2:e1501482. [DOI] [PMC free article] [PubMed]
- Zhang X, Cozen AE, Liu Y, et al. Trends Mol Med 2016; 22:1025–1034. [DOI] [PMC free article] [PubMed]
- Zhang Y, Zhang Y, Shi J, et al. J Mol Cell Biol 2014; 6:172–174. [DOI] [PubMed]