ABSTRACT
The HIV Vaccine Trials Network (HVTN) 087 vaccine trial assessed the effect of increasing doses of pIL-12 (interleukin-12 delivered as plasmid DNA) adjuvant on the immunogenicity of an HIV-1 multiantigen (MAG) DNA vaccine delivered by electroporation and boosted with a vaccine comprising an attenuated vesicular stomatitis virus expressing HIV-1 Gag (VSV-Gag). We randomized 100 healthy adults to receive placebo or 3 mg HIV-MAG DNA vaccine (ProfectusVax HIV-1 gag/pol or ProfectusVax nef/tat/vif, env) coadministered with pIL-12 at 0, 250, 1,000, or 1,500 μg intramuscularly by electroporation at 0, 1, and 3 months followed by intramuscular inoculation with 3.4 × 107 PFU VSV-Gag vaccine at 6 months. Immune responses were assessed after the prime and boost and 6 months after the last vaccination. High-dose pIL-12 increased the magnitude of CD8+ T-cell responses postboost compared to no pIL-12 (P = 0.02), while CD4+ T-cell responses after the prime were higher in the absence of pIL-12 than with low- and medium-dose pIL-12 (P ≤ 0.05). The VSV boost increased Gag-specific CD4+ and CD8+ T-cell responses in all groups (P < 0.001 for CD4+ T cells), inducing a median of four Gag epitopes in responders. Six to 9 months after the boost, responses decreased in magnitude, but CD8+ T-cell response rates were maintained. The addition of a DNA prime dramatically improved responses to the VSV vaccine tested previously in the HVTN 090 trial, leading to broad epitope targeting and maintained CD8+ T-cell response rates at early memory. The addition of high-dose pIL-12 given with a DNA prime by electroporation and boosted with VSV-Gag increased the CD8+ T-cell responses but decreased the CD4+ responses. This approach may be advantageous in reshaping the T-cell responses to a variety of chronic infections or tumors. (This study has been registered at ClinicalTrials.gov under registration no. NCT01578889.)
KEYWORDS: DNA prime, HIV, IL-12, T cell, VSV vector, vaccine
INTRODUCTION
The need for a preventive HIV vaccine remains of utmost importance for global health. According to the 2014 UNAIDS Gap Report, of the 35 million people living with HIV, more than half (19 million) are unaware of their status (1) and therefore unlikely to take preventive measures to avoid transmission to others. Since HIV-specific antibodies were found to correlate with reduced HIV acquisition in the RV144 vaccine trial, which showed 31% efficacy (2), induction of humoral responses has been the primary outcome pursued in HIV vaccine trial design. Nevertheless, T-cell responses are considered likely to contribute to viral control (3) and to prevention of infection from becoming established (4, 5). Polyfunctional T cells have been associated with decreased risk of HIV infection in follow-up analyses of the RV144 trial, which identified highly polyfunctional CD4+ T cells expressing CD40L, interleukin-2 (IL-2), IL-4, gamma interferon (IFN-γ), and tumor necrosis factor alpha (TNF-α) as an additional inverse correlate of infection risk (6), as well as in the HIV Vaccine Trials Network (HVTN) 505 vaccine trial, which identified polyfunctional CD8+ T cells expressing IL-2, IFN-γ, TNF-α, and granzyme B as an additional inverse correlate of infection risk.
Replication-competent vectors, such as those based on vesicular stomatitis virus (VSV) or adenovirus, may increase the magnitude and durability of vaccine-induced responses compared to nonreplicating vectors (reviewed in reference 7). Recombinant replication-competent VSV (rVSV) vectors have been tested as an Ebola virus vaccine (8), with a ring vaccination randomized-efficacy trial supporting up to 100% efficacy at preventing Ebola virus disease with symptom onset at least 10 days after vaccination (9). We recently tested a vaccine comprising rVSV expressing HIV-1 Gag (VSV-Gag) in HVTN 090, where two doses of up to 3.4 × 107 PFU were found to be safe and well tolerated in humans, although Gag-specific immune responses were relatively modest and CD4+ T-cell biased (10).
DNA vaccines have been shown to efficiently prime T-cell responses when followed by various vectored boosts (11–13) and can be enhanced by cytokine adjuvants (14). IL-12 is crucial in the development of Th1 cells and enhances cytotoxic activity (15), representing an ideal molecular adjuvant for T-cell-based vaccines. Vaccines including pIL-12 (IL-12 delivered as a plasmid DNA [pDNA]) consistently induce increased cellular and humoral immunity in macaques (16), especially when delivered by electroporation (EP) (14, 17). The effect of IL-12 on immunogenicity to an HIV vaccine in humans is less clear; while response rates were somewhat increased in one study when combining IL-12 delivery with electroporation (18), no benefit of the adjuvant was observed in two other studies (19, 20). The adjuvant effect of IL-12 is dose dependent in mice, with no effect at low doses and immunosuppressive effects at high doses (21, 22); the optimal dose of IL-12 for human vaccine studies remains to be determined.
HVTN 087 (ClinicalTrials registration no. NCT01578889) is a randomized, double-blind, placebo-controlled phase 1a clinical trial that evaluated the effect of three doses of IL-12 delivered as plasmid DNA on the immunogenicity of an HIV-1 multiantigen plasmid DNA (HIV-MAG) vaccine delivered by intramuscular (i.m.) EP followed by an rVSV HIV-1 Gag (VSV-Gag) boost. The doses of pIL-12 chosen for this study included the 1,000-μg dose that showed an adjuvant effect in HVTN 080, as well as a lower dose (250 μg) and higher dose (1,500 μg). Cellular and humoral immune responses were measured after the prime and boost, including an early memory time point 6 months after the last vaccination, to determine the optimal dose of IL-12, the effect of the DNA prime on the immunogenicity of the VSV-Gag vaccine, and the longevity of immune responses induced by this vaccine regimen.
RESULTS
Participant accrual and vaccination schedule.
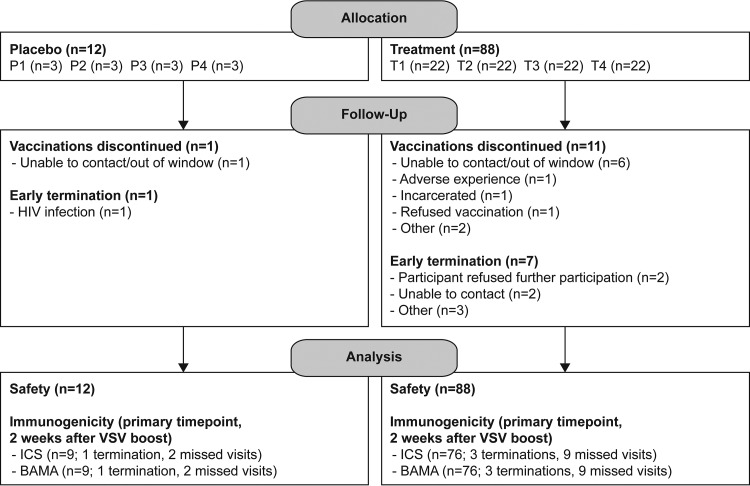
Participant accrual, demographic data, and vaccine safety and tolerability for HVTN 087 are presented in detail elsewhere (M. L. Elizaga, S. S. Li, N. Kochar, G. J. Wilson, M. Allen, H. V. Tieu, M. E. Sobieszczyk, K. W. Cohen, B. Sanchez, T. Latham, et al., submitted for publication); there were no serious adverse events believed to be related to the vaccine. Figure 1 shows the trial schema and CONSORT statement flow diagram.
FIG 1.
Trial schema and CONSORT statement 2010 flow diagram. Participants were randomized into one of four groups, each with a vaccine/placebo ratio of 22/3. The vaccine and adjuvant doses were mixed and divided between two injection sites on opposite deltoids. The numbers of participants enrolled, randomized, followed up, and analyzed are shown for placebo and treatment groups. pIL-12, plasmid IL-12 DNA.
Low Gag-specific T-cell responses after DNA prime are significantly boosted by VSV-Gag.
To assess the immunogenicity of the multiantigen (MAG) DNA prime plus VSV-Gag boost vaccine, we measured T-cell responses to Gag by intracellular cytokine staining (ICS) 2 weeks after the third DNA prime (month 3.5), 2 weeks after the VSV boost (month 6.5), and 6 months after the final vaccination (month 12). In addition to Gag, the HIV-1 MAG DNA used in this study encodes Pol in the same plasmid and Env, Nef, Tat, and Vif in a second plasmid; responses to the other proteins are discussed below. pIL-12, when given, was admixed with the HIV-MAG DNA prior to electroporation.
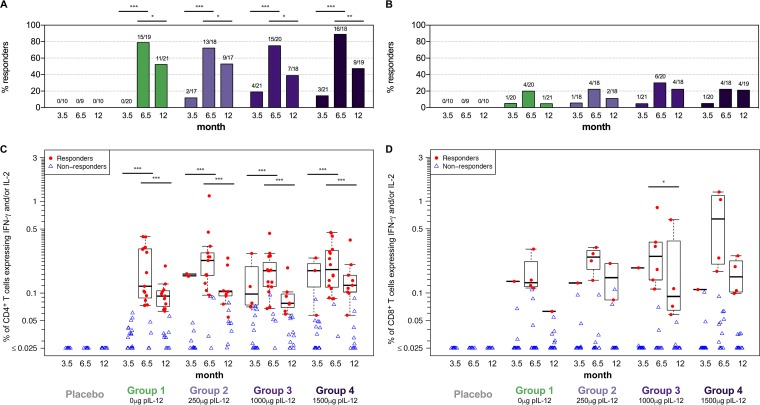
After the HIV-MAG DNA priming series, Gag-specific response rates were low for both CD4+ (0 to 19%) and CD8+ (5 to 6%) T cells (Fig. 2A and B, month 3.5). However, both response rates and magnitudes of Gag-specific CD4+ T cells were considerably boosted by VSV-Gag (Fig. 2A and C, month 6.5; P < 0.001 for all groups), leading to response rates of 72 to 89%. CD8+ T-cell responses were also increased after VSV-Gag boost, although not significantly so (Fig. 2B and D, month 6.5; B, P = 0.06 to 0.08; D, P = 0.11 to 0.13), resulting in 20 to 30% response rates. In line with previous studies using DNA, responses in this trial were mainly CD4+ T-cell mediated (Fig. 2A and C).
FIG 2.
Gag-specific T-cell responses. CD4+ (A and C) and CD8+ (B and D) T-cell responses to Gag were measured 2 weeks after the 3rd DNA prime, (prime), 2 weeks after the VSV boost (boost), and 6 months after the boost (memory) by ICS. Shown are response rates (percentage) (A and B) and response magnitudes (C and D) of CD4+ or CD8+ T cells producing IFN-γ and/or IL-2 for placebo recipients (combined for groups 1 to 4) and vaccinees in each treatment group. Positive responses are shown in filled red circles, and negative responses are shown in open blue triangles (C and D). Box plots represent the distribution for the positive responders only. Response rates were compared using Fisher's exact test (A and B); response magnitudes were compared using Wilcoxon's rank sum test for the comparisons between treatment groups among the responders and using Wilcoxon's signed-rank test for the comparisons between visits among the participants with a positive response at either or both visits (C and D). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. Significance bars represent longitudinal comparisons.
The VSV-Gag vaccine used in HVTN 087 was previously shown to be safe but only mildly immunogenic in a homologous prime-boost regimen in a small phase 1 dose escalation trial, HVTN 090 (10). A single vaccination with VSV-Gag at the same dose used in HVTN 087 (3.4 × 107 PFU, group 5 in HVTN 090) in that trial showed responses that were similar to those observed after the DNA prime in HVTN 087 (see Fig. S1 in the supplemental material), but contrary to the considerable boost observed in all groups in HVTN 087, a homologous VSV-Gag regimen did not lead to increased responses postboost in HVTN 090 (Fig. S1).
Addition of pIL-12 leads to increased magnitude of CD8+ T-cell responses.
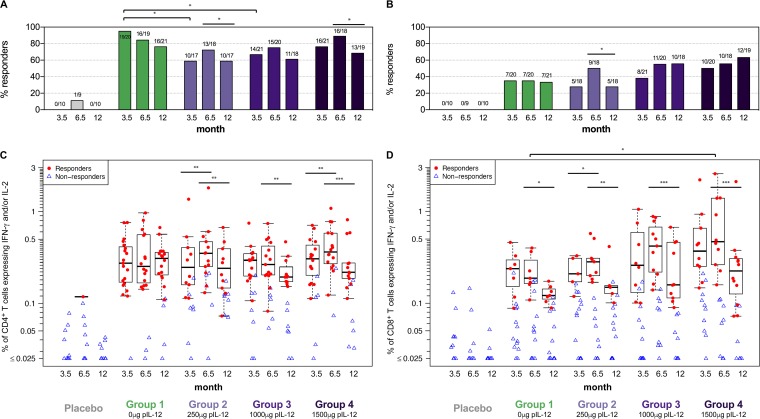
Adjuvanticity of pIL-12 was explored in doses ranging from 0 to 1,500 μg pIL-12 delivered by EP with the HIV-MAG DNA. For CD8+ T cells, pIL-12 had a generally positive effect. Response rates overall (Fig. 3B) and to individual antigens were higher in the medium- and high-dose pIL-12 groups (groups 3 and 4) after the prime as well as the boost, and the total magnitude of CD8+ T-cell responses was significantly increased in group 4 compared to group 1 after the final vaccination (Fig. 3D, month 6.5; P = 0.02).
FIG 3.
HIV-specific T-cell responses to any protein. CD4+ (A and C) and CD8+ (B and D) T-cell responses to Env, Pol, Gag, Nef, and Vif were measured 2 weeks after the 3rd DNA prime (prime), 2 weeks after the VSV boost (boost), and 6 months after the boost (memory) by ICS. Shown are response rates (percentage) (A and B) and response magnitudes (C and D) for the sum of individual antigen responses of CD4+ or CD8+ T cells producing IFN-γ and/or IL-2 for placebo recipients (combined for groups 1 to 4), and vaccinees in each treatment group. Positive responses are shown in filled red circles, and negative responses are shown in open blue triangles (C and D). Box plots represent the distribution for the positive responders only. Response rates were compared using Fisher's exact test (A and B); response magnitudes were compared using Wilcoxon's rank sum test for the comparisons between treatment groups among the responders, and using Wilcoxon's signed-rank test for the comparisons between visits among the participants with a positive response at either or both visits (C and D). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. Significance bars without tails represent the comparisons between visits within the same group, and those with tails represent the comparisons between treatment groups.
Unexpectedly, addition of pIL-12 led to significantly decreased CD4+ T-cell response rates to any antigen after the prime for the low- and medium-dose groups (Fig. 3A, month 3.5; P = 0.014 for group 2 versus 1 and P = 0.045 for group 3 versus 1). This effect was mainly driven by reduced Env-specific CD4+ T-cell responses (see Table S1 and Fig. S2 in the supplemental material [group 2 versus 1, P = 0.0007; group 3 versus 1, P = 0.004; and group 4 versus 1, P = 0.045]). In contrast, Gag-specific CD4+ T-cell response rates to the prime were somewhat improved with addition of pIL-12 (Fig. 2A). In addition to Env and Gag, Pol-specific CD4+ T-cell responses were observed in all groups (see Fig. S3 in the supplemental material). Addition of IL-12 had no effect on Pol-specific CD4+ T-cell responses after the DNA prime but significantly reduced the response after the VSV boost in group 2 compared to group 1 (P = 0.046).
Following the VSV boost, CD4+ T-cell response rates were not significantly different across groups, with the highest rate to any antigen (89% [16/18]) in the high-pIL-12 group (Fig. 3A, month 6.5). Similarly, no significant differences for CD4+ T-cell magnitudes were observed between treatment groups (Fig. 3C, month 6.5).
DNA with pIL-12 vaccination induces stable T-cell responses.
Considering that the VSV vaccine only expressed Gag, HVTN 087 allowed us to assess the stability of responses induced by three doses of DNA without further boosting for Env- and Pol-specific T-cell responses at two memory time points: 2 weeks after the VSV boost (i.e., 3.5 months after the last DNA [early memory]) and at the end of the study, 9 months after the last DNA (late memory). Gag-specific memory responses after DNA prime/VSV boost could only be assessed 6 months after the VSV boost, at the end of the study.
Gag-specific CD4+ T-cell response rates, which had been considerably boosted by the VSV-Gag vaccine, significantly declined in the 6 months following the VSV boost (Fig. 2A; P = 0.008 to 0.045). Similarly, the magnitude of these responses decreased significantly (Fig. 2C; P = <0.0001 to 0.0002). Gag-specific CD8+ T-cell responses also declined in the 6 months following the boost, but the only significant reduction was observed for the magnitude of responses in group 3 (Fig. 2D; P = 0.03).
Interestingly, DNA-induced Env-specific CD4+ T-cell response rates were fairly stable in the groups receiving pIL-12 (P > 0.08 for groups 2 to 4 between 2 weeks and 3 months after their last Env vaccination) but declined significantly in the absence of pIL-12 (P = 0.005, group 1) (Fig. S2A). No further decline in CD4+ T-cell response rates was observed from 3 to 9 months after the last DNA prime (P > 0.05 for all groups); of note, response rates were frequently higher at the late memory time point than at the early memory time point in groups 1 to 3. The response rate in group 1 remained significantly higher than that in group 2 at the late memory time point (P = 0.046).
Env-specific CD4+ T-cell magnitudes significantly declined from peak post-DNA to early memory 3 months later in groups 1 and 3 (P = 0.002 and 0.03, respectively) but did not change in the other groups or between 3 and 6 months post-last Env vaccination in all groups (Fig. S2C).
Env-specific CD8+ T-cell response rates and magnitudes remained stable throughout the trial (Fig. S2B and D), with no significant differences observed across groups or time points.
Pol-specific CD4+ T-cell response rates declined significantly in the first 3 months in groups 2 and 3 (P = 0.008 and P = 0.046, respectively) but not in groups 1 and 4 (P > 0.05) and stabilized thereafter for all groups (P > 0.05) (Fig. S3A).
Functionality of Gag-specific T-cell responses is significantly increased after VSV boost.
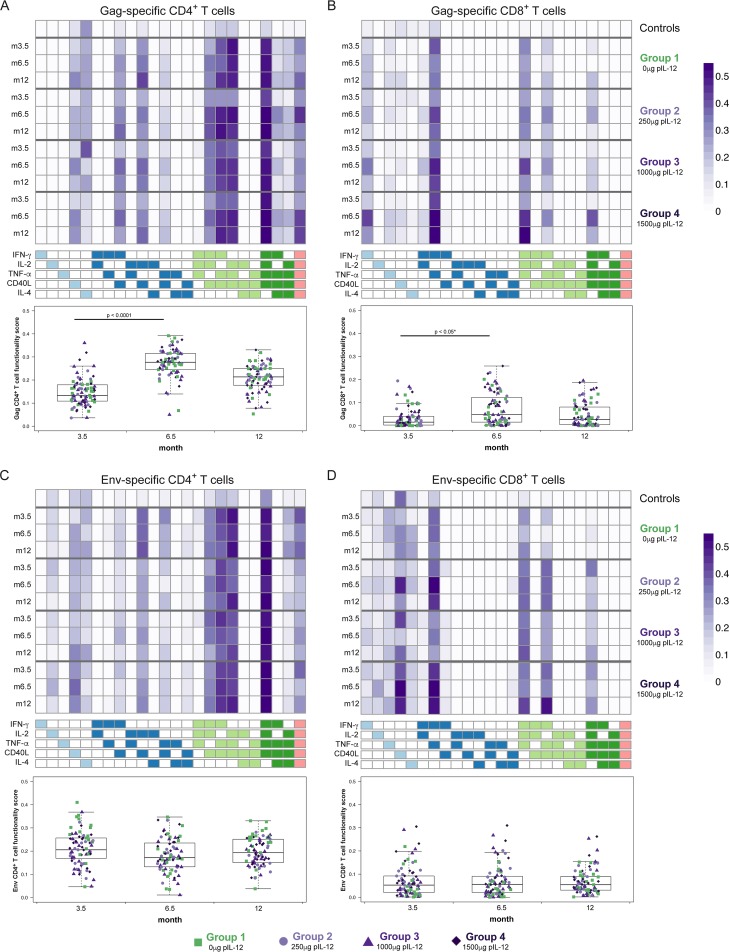
Combining DNA vaccination with pIL-12 and delivery by EP led to enhanced functionality of the induced T-cell response in macaques (17). We have recently developed a method based on a Bayesian hierarchical framework to model all observed cell subsets and select those most likely to have antigen-specific responses (6). This combinatorial polyfunctionality analysis of antigen-specific T-cell subsets (COMPASS) identified polyfunctional CD4+ T-cell responses as an inverse correlate of risk of infection in the RV44 Thai trial (6). We applied COMPASS to compute the functionality score for each individual, defined as the proportion of antigen-specific subsets detected among all 32 possible combinations of expression of IL-2, IL-4, IFN-γ, TNF-α, and CD40L.
The distributions of subsets expressing different combinations of functional markers were fairly consistent over time and were similar for Gag-specific (Fig. 4A and B) and Env-specific (Fig. 4C and D) T cells. The dominant CD4+ T-cell subsets were those expressing three or more markers (darker shades in the heat map), especially those expressing TNF-α, IL-2, IFN-γ, and CD40L (Fig. 4A and C). CD8+ T cells dominantly expressed TNF-α and IFN-γ (Fig. 4B and D). Functionality scores for Gag-specific CD4+ were significantly increased after the VSV boost (Fig. 4A, month 3.5 versus 6.5; P < 0.0001 for all groups), as were functionality scores for Gag-specific CD8+ T cells (Fig. 4B, P < 0.05 for all groups except group 2 [P = 0.1]). No significant differences in functionality scores for Gag-specific T cells were observed across treatment groups at any time point.
FIG 4.
Functionality analysis using COMPASS. Expression of different combinations of functional markers is represented as heat maps of COMPASS posterior probabilities for Gag-specific CD4+ (A) and CD8+ T cells (B), and Env-specific CD4+ (C) and CD8+ T cells (D) at months 3.5, 6.5, and 12. Columns correspond to the different cell subsets modeled by COMPASS, color coded by the cytokines they express (where white represents “off,” shaded represents “on,” and the color grouping represents the degree of functionality), and ordered by degree of functionality from one function on the left to five functions on the right. Subsets with maximum posterior probabilities of <0.005 were removed from the heat map. Rows correspond to the average within each treatment group at each time point; all time points are averaged for controls. Each cell shows the probability that the corresponding cell subset (column) exhibits an Ag-specific response in the corresponding treatment group (row), where the probability is color coded from white (zero) to purple (one). Subject-level posterior probabilities reflect the certainty from the COMPASS model that the subset exhibits an antigen-specific response in a subject (i.e., that the magnitude of the stimulated sample is above the magnitude of the [paired] nonstimulated sample). These subject-specific and category-specific posterior probabilities are summarized by their average across subjects within different treatment groups and time points, giving rise to the heat maps shown here. Below the heat maps, functionality scores provide a summary measure, defined as the proportion of antigen-specific subsets detected among all possible ones, and are not shown for controls. VSV-Gag boost led to significantly increased functionality scores for Gag-specific CD4+ T cells (P < 0.0001 for all groups) and for Gag-specific CD8+ T cells in groups 1 (P = 0.047), 3 (P = 0.032), and 4 (P = 0.032). *, does not apply to group 2. Group 1, green squares, no IL-12; group 2, lavender circles, 250 μg IL-12; group 3, plum triangles, 1,000 μg IL-12; group 4, purple diamonds, 1,500 μg IL-12.
Functionality scores were significantly higher for Env-specific CD4+ T cells in group 1 than for the combined groups receiving different doses of pIL-12 at peak (i.e., 2 weeks after the last DNA) (Fig. 4C, month 3.5; P = 0.004), but not at early memory 3 months later (Fig. 4C, month 6.5, green symbols versus purple symbols). Interestingly, Env-specific CD8+ T cells benefited from the inclusion of pIL-12, with higher functionality scores in the combined pIL-12 groups at peak (P = 0.051) and early memory (Fig. 4D, purple symbols versus green symbols; P = 0.03).
As expected, the functionality scores for Env- and Gag-specific T cells were highly correlated with the magnitude of these responses measured by expression of IFN-γ and/or IL-2 (Spearman ρ, 0.62 to 0.86; P < 10−10 for all subsets), as participants with more HIV-specific T cells are more likely to have increased functionality. However, the correlation is not perfect, which supports the inclusion of the functionality scores as measures of the quality of the response to vaccination.
VSV boost leads to significant broadening of the Gag-specific T-cell response.
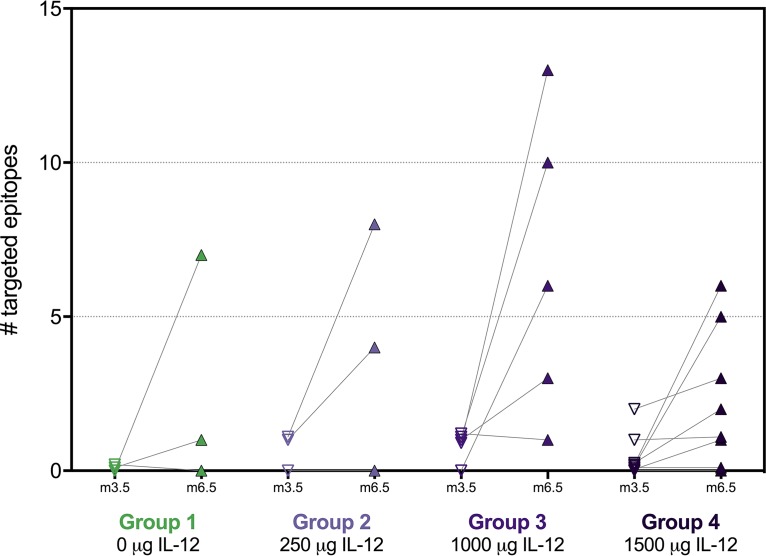
Since boosting with VSV led to significantly increased Gag-specific T-cell responses measured by ICS, we next assessed whether this increase was mediated by targeting few epitopes with high magnitudes or whether it was due to a broadening of the response to several epitopes. Epitope mapping was performed by IFN-γ enzyme-linked immunosorbent spot (ELISpot) assay 2 weeks after the last DNA prime and 2 weeks after the VSV-Gag boost for 19 vaccinees who had a detectable Gag-specific CD4+ and/or CD8+ T-cell response after the boost, as measured by ICS, and for whom samples were available, as well as for five placebo recipients.
The median number of targeted epitopes increased significantly following the VSV-Gag boost, from a median of 0 after DNA prime to a median of 4 after VSV boost (Fig. 5; P < 0.0001 for all groups combined). Increases were observed in all groups regardless of the dose of pIL-12 received; the numbers tested in each group were too low to allow for statistical testing across groups. No epitopes were identified in placebo recipients (data not shown).
FIG 5.
Number of targeted epitopes in Gag. The number of epitopes in Gag was determined 2 weeks after the last DNA prime (V10, month 3.5) and 2 weeks after the VSV boost (V15, month 6.5) by IFN-γ ELISpot using a matrix approach. Responses to two overlapping 15-mer peptides were counted as one epitope. Each data point represents the number of targeted epitopes per vaccinee; lines connect longitudinal data for the same individual. Epitope mapping was restricted to vaccinees who had a detectable CD4+ and/or CD8+ T-cell response at V15 as measured by ICS and for whom samples were available.
DNA prime/VSV boost does not induce significant binding antibody responses.
DNA vaccines, whether delivered with or without EP, have been shown to be poorly immunogenic for antibody responses in previous clinical trials (18, 19). We assessed whether adding a VSV boost to the DNA prime would improve induction of binding antibodies.
As shown in Table 1, the MAG DNA delivered by EP did not lead to significant induction of binding antibody responses to Env or Gag, regardless of the dose of IL-12 plasmid DNA (pDNA). Gag p24-specific IgG response rates were slightly higher after the VSV HIV gag boost, but these increases were not significant, possibly in part due to the high frequency of antibodies occurring at baseline that cross-react with Gag p24.
TABLE 1.
Response rates for IgG and IgA to Env and Gag measured 2 weeks post-DNA prime and 2 weeks post-VSV boost
| Treatment group | Response rate, % (no. positive/total) |
|||
|---|---|---|---|---|
| 6101 Env gp140 |
Gag p24 |
|||
| Prime | Boost | Prime | Boost | |
| IgG | ||||
| Placebo | 0 (0/10) | 0 (0/9) | 0 (0/10) | 11.1 (1/9) |
| Group 1 | 5 (1/20) | 5.3 (1/19) | 5 (1/20) | 15.8 (3/19) |
| Group 2 | 0 (0/17) | 0 (0/17) | 5.9 (1/17) | 5.9 (1/17) |
| Group 3 | 0 (0/20) | 0 (0/18) | 10.5 (2/19) | 15.8 (3/19) |
| Group 4 | 5.6 (1/18) | 0 (0/15) | 0 (0/18) | 13.3 (2/15) |
| IgA | ||||
| Placebo | 0 (0/10) | 0 (0/9) | 0 (0/10) | 0 (0/9) |
| Group 1 | 5 (1/20) | 0 (0/19) | 0 (0/20) | 0 (0/19) |
| Group 2 | 0 (0/18) | 0 (0/18) | 5.6 (1/18) | 0 (0/18) |
| Group 3 | 0 (0/21) | 0 (0/20) | 4.8 (1/21) | 5 (1/20) |
| Group 4 | 5.3 (1/19) | 6.3 (1/16) | 0 (0/19) | 6.3 (1/16) |
These low-antibody responses are also reflected in the low induction of vaccine-induced HIV seroreactivity (VISR): only one participant in group 1 and two in group 4 showed reactivity in the Abbott Prism enzyme immunoassay (EIA), while none of the other tested EIAs in the HVTN end-of-study algorithm detected positive responses (data not shown).
DISCUSSION
HVTN 087, the first clinical HIV vaccine trial combining a DNA prime, with and without IL-12, with a replication-competent VSV boost, demonstrated the safety of this heterologous prime-boost regimen as well as its ability to induce CD4+ and CD8+ T-cell responses in a majority of vaccine recipients. This trial was designed to answer two primary questions: (i) whether preceding a modestly immunogenic VSV-Gag vaccine with a DNA prime would lead to increased immune responses and (ii) whether we could identify an optimal dose of IL-12 as a molecular adjuvant to a DNA prime delivered by electroporation.
The VSV-Gag vaccine previously tested in HVTN 090 induced fairly unremarkable HIV Gag-specific immunity, even though vaccine take was evident by induction of VSV-specific neutralizing antibodies (10). However, preceding it with a DNA prime given by electroporation strikingly enhanced Gag-specific immunity, as shown by CD4+ T-cell response rates up to 89% in HVTN 087 after the VSV-Gag boost compared to 11% after administration of VSV-Gag alone in HVTN 090 when using the same assay (Fig. S1). This increase is especially noteworthy since the frequency of Gag-specific CD4+ T-cell responses induced by the DNA in HVTN 087 only ranged from 0 to 19%; these were significantly boosted to 72 to 89% by VSV-Gag. CD8+ T-cell responses increased 4- to 6-fold from postprime to postboost, even though a single vaccination with VSV-Gag at the highest dose did not induce any detectable CD8+ T cells in HVTN 090. These data clearly demonstrate that Gag-specific responses were primed by the DNA, even though they were too low to detect in our assays.
CD4+ T cells induced by vaccination were highly polyfunctional, with the majority expressing three or more functional markers; polyfunctionality was already apparent post-DNA prime, and further enhanced post-VSV boost for Gag-specific CD4+ T cells. The patterns observed for Env-specific and Gag-specific CD4+ T cells were fairly similar, even though functionality scores were higher after the VSV boost; the dominant populations in both cases were T cells expressing IL-2, IFN-γ, TNF-α, and CD40L. Of note, vaccination in HVTN 087 led to significant induction of 5-functional CD4+ T cells (including IL-4 in addition to the markers described above), the subset that was most dominantly associated with reduced infection risk in RV144 and which was also identified in HVTN 100, lending support to the initiation of HVTN 702 (23). Additionally, T-cell responses were quite stable over time, remaining detectable in the majority of participants for most treatment groups.
In line with a significant increase in the rate and magnitude of Gag-specific responses, we also observed significant broadening of the response after the VSV-Gag boost by mapping individual epitopes. After the boost, a median of four Gag epitopes was targeted in responders across all treatment groups; this finding is of particular interest based on the inverse association of the number of vaccine-induced Gag epitopes and viral load in vaccine recipients who acquired HIV infection in the Step study (24). HVTN 087 was designed to determine whether there was a discernible dose response to adjuvanting the DNA prime with IL-12 pDNA; unfortunately, the data obtained in this study are inconclusive. The only significant IL-12-mediated benefits we observed were the increased postboost magnitude of vaccine-induced CD8+ T cells in group 4 receiving the highest dose of IL-12, as well as increased functionality scores for Env-specific CD8+ T cells at peak in the groups receiving pIL-12. CD8+ T-cell response rates were lowest in the absence of IL-12 (group 1), but there was no clear dose response among groups 2 to 4. On the other hand, CD4+ T-cell response rates were significantly higher in the absence of IL-12 than in the low- and intermediate-dose groups, but there was a weak IL-12 dose response for groups 2 to 4 both after the DNA prime and after the VSV boost. The magnitude of CD4+ T cells seemed to be unaffected by IL-12; similarly, the functionalities of T-cell responses were comparable for all doses. Antibody responses to p24, which were low overall, were highest after the boost in the intermediate-dose group (group 3). This dichotomy is reflected in the current literature about the effects of IL-12 in HIV clinical trials: one study showed no or possibly detrimental effects (20), another detected some benefit of IL-12 at intermediate dosing (19), and yet another described a substantial though not statistically significant increase in response rates with IL-12 (18). Additional investigations of IL-12, especially in the context of immunogens designed to induce Env-specific antibodies, are clearly warranted to determine whether pIL-12 should be considered a general adjuvant for DNA vaccines.
Contrary to our observations for T-cell responses, priming with DNA only minimally enhanced the poor p24-specific antibody responses observed in HVTN 090 following VSV-Gag (10); response rates ranged from 6 to 16% after the boost with no significant influence of IL-12. Env-specific antibodies induced by the HIV-MAG DNA were detected in only two of 74 vaccinees (3%), in line with the generally poor induction of antibodies by DNA vaccines (18).
There were some shortcomings of the present study due to limitations in the size of the trial. Group sizes were fairly small, and some differences, though apparent in the graphical representations, did not reach statistical significance. Additionally, current assays may not be ideal to assess immunogenicity of DNA vaccines, since responses after the DNA prime were frequently below the level of detection, becoming apparent only after the VSV boost.
Overall, HVTN 087 contributes to the growing evidence that priming with DNA enhances vaccine-induced cellular immune responses, independently of the boosting modality. Therefore, in view of the relative ease of DNA vaccine design and production, vaccines aimed at inducing HIV-specific T cells would benefit from including a DNA prime. The safety and immunogenicity of the combination tested in this trial, including the induction of Gag-specific polyfunctional T cells with enhanced epitope breadth and improved durability of the induced responses, lend support to the continued use of the VSV platform as part of an HIV vaccine solution, especially in the context of a VSV-Env vector to enhance induction of protective antibodies.
MATERIALS AND METHODS
Study participants.
One hundred participants were enrolled between May 2012 and July 2013 and followed up for a year after the first vaccination. After written informed consent, volunteers were screened for eligibility and willingness to participate at four HVTN sites in the United States: Nashville, TN, New York, NY, Rochester, NY, and Philadelphia, PA. Participants were eligible if they were between 18 and 50 years of age, in good general health based on history, clinical examination, and laboratory investigations, practiced behaviors that placed them at low risk for HIV acquisition, and had no history of receiving investigational products, immunosuppressive medications, blood products, immunoglobulins, or vaccines within study-defined periods prior to enrollment. Female participants of childbearing potential were eligible if they were not pregnant or planning to become pregnant and agreed to consistently use contraception for 21 days prior to their first vaccination until 6 months after their final vaccination. All participants were counseled about HIV risk reduction and assessed for potential social impacts of study participation at each study visit.
Vaccine products and schedule. (i) HIV-MAG.
The HIV-MAG vaccine consists of two plasmid DNA (pDNA) expression vectors: ProfectusVax HIV-1 gag/pol and ProfectusVax HIV-1 nef/tat/vif, env. HIV-1 gag/pol expresses an HIV-1 clade B Gag-Pol fusion under the control of a human cytomegalovirus (hCMV) promoter and bovine growth hormone (BGH) polyadenylation signal. HIV-1 nef/tat/vif, env expresses (i) an HIV-1 clade B Nef-Tat-Vif fusion under the control of an hCMV promoter and a simian virus 40 (SV40) polyadenylation signal and (ii) Env gp160 from an HIV-1 clade B primary isolate, 6101, under the control of a simian cytomegalovirus (sCMV) promoter and BGH polyadenylation signal.
(ii) pIL-12 adjuvant.
The HIV-MAG vaccine was administered with or without a third expression plasmid encoding the p35 and p40 subunits of human IL-12 as previously described, also provided by Profectus Biosciences, Inc. (18, 19).
(iii) Administration of DNA study products with electroporation.
The HIV-MAG vaccine, pIL-12 adjuvant, and placebo were mixed together and immediately delivered by intramuscular (i.m.) injection followed by EP using the Ichor Medical Systems TriGrid Delivery System (TDS) EP device. Each dose was delivered as two injections, one into each deltoid.
(iv) rVSVIN N4CT1gag1 (VSV-Gag) vaccine.
The VSV-Gag vaccine candidate is an attenuated recombinant Indiana serotype vesicular stomatitis virus (rVSVIN) vector containing the HIV-1 gag gene, delivered at 3.4 × 107 PFU as 2× 1-ml injections by needle and syringe i.m. at 1 ml into each deltoid.
(v) Placebo for HIV-MAG vaccine, pIL-12 adjuvant, and VSV-Gag.
The placebo used was sodium chloride for injection (USP, 0.9%).
Sample size calculation.
The sample size of 22 vaccine recipients per group provided a 90% chance of observing at least one serious adverse experience if the true rate of such an event were at least 10%; there was a 90% chance that we would not observe at least one serious adverse experience if the true rate was no more than 0.4%. The precision to estimate immunogenicity was somewhat limited and therefore not a primary objective of the study. For example, the two-sided 95% confidence interval for the true response rate was approximately (43 to 82%) if the observed response rate was 65%, assuming 10% loss of data.
Randomization.
Groups 1 to 3 were opened to enrollment simultaneously. The participants who consented for more frequent visits for the evaluation of innate immune responses were randomized into one of groups 1 and 3 to receive the vaccine or the placebo in a vaccine/placebo ratio of 22:3. Group 2 was randomized separately, with 22 participants given vaccine and 3 given placebo. After the 24 participants in groups 1 and 3 and all 25 participants in group 2 reached 14 days and their safety and tolerability data were reviewed by the Protocol Safety Review Team (PSRT), group 4 was opened for enrollment, with 22 participants being randomized to receive the vaccine or 3 to receive the placebo.
Blinding.
Participants and site staff (except for site pharmacists) were blind to participant treatment arm assignments (i.e., active vaccine or control) but not to group assignment. Study product assignments were accessible to those HVTN clinical research site (CRS) pharmacists, Division of AIDS (DAIDS) protocol pharmacists and contract monitors, and Statistical and Data Management Center (SDMC) staff who were required to know this information in order to ensure proper trial conduct. The HVTN Safety Monitoring Board members were not blind to treatment assignment in order to conduct review of trial safety. Laboratory personnel remained blind to treatment and group assignments throughout data generation.
Sample processing.
Serum for humoral assays was obtained from serum-separating tubes (SSTs) and frozen at −80°C until use. Peripheral blood mononuclear cells (PBMCs) for cellular assays were isolated and cryopreserved from acid citrate dextrose (ACD)-anticoagulated whole blood within 6 h of venipuncture, as described previously (25). PBMCs were thawed and cultured overnight at 37°C in 5% CO2 in R10 (RPMI 1640 [GibcoBRL, Carlsbad, CA], 10% fetal bovine serum [FBS; Gemini Bioproducts, West Sacramento, CA], 2 mM l-glutamine [Gibco], 100 μg/ml streptomycin sulfate [Gibco], 100 U/ml penicillin G [Gibco]). The cells were then counted using the Guava ViaCount kit (Millipore, Bedford, MA) according to the manufacturer's instructions prior to stimulation.
ICS assay.
A validated intracellular cytokine staining (ICS) assay (26) was performed on cryopreserved PBMCs by flow cytometry to examine HIV-1-specific vaccine-induced CD4+ and CD8+ T-cell responses at 2 weeks after the third DNA vaccination (completion of the prime, visit 10 [V10]) and fourth (boost, V15) vaccination, as well as 6 months after the boost (memory, V17). Cytokine production was assessed after stimulation with global potential T-cell epitope (PTEg) peptide pools (27) representing 15-mer peptides from Gag, Pol, Env, Nef, and Vif (Bio-Synthesis) at 1 μg/ml as previously described (26, 28). Briefly, the 6-h stimulation included brefeldin A (10 μg/ml; Sigma-Aldrich) and anti-CD28/anti-CD49d (each at 1 μg/ml; BD Biosciences). Phytohemagglutinin (PHA [Remel]) was used as a positive control, and peptide diluent (dimethyl sulfoxide [DMSO] at a final concentration of 1%) was used as a negative control. Cells were stained with Aqua Live/Dead fixable dead cell stain (Invitrogen) and then fixed, permeabilized, and stained intracellularly with fluorescently labeled antibodies to CD14 (exclusion marker), CD3, CD4, CD8, IFN-γ, IL-2, IL-4, TNF-α, CD40L, and granzyme B (28). Data were acquired on an LSRII and analyzed using FlowJo. Positivity of the ICS responses of individual cytokines or cytokine combination was determined by a one-sided Fisher's exact test applied to each peptide pool-specific response versus the negative-control response with a discrete Bonferroni adjustment for the multiple comparisons due to testing against multiple peptide pools. Peptide pools with adjusted P values of less than α = 0.00001 were considered positive. If at least one peptide pool for a specific HIV-1 protein is positive, then the overall response to the protein is considered positive (26). Data were filtered if background responses (DMSO control) were >0.1% cytokine secretion or if <5,000 events were acquired within the CD4+ or CD8+ T-cell subpopulations. Data were available for 89 participants after the DNA prime, 84 participants after the VSV boost, and 85 participants at the memory time point for CD4+ T-cell responses; for CD8+ T-cell responses, data were available for 89, 85, and 86 participants at those time points, respectively. Of note, the statistical nature of our positivity test—while extensively vetted and leading to false-positive rates under 5%—makes low-level responses (around 0.1% cytokine-producing T cells) prone to fluctuations in and out of positivity, as exemplified by the longitudinal Env-specific T-cell responses (Fig. S3). Even though the magnitude of responses is fairly stable, responses may be called negative at some time points and positive at others. This inherent variability in low-level responses leads to sometimes increasing response rates in the absence of stimulation of the immune system through vaccination.
Epitope mapping.
Cryopreserved PBMC samples from HVTN 087 were obtained from 24 study subjects from two visits (V10 and V15). Cryopreserved PBMCs were thawed at 37°C for 1.5 min, washed once with complete culture medium (RPMI 1640 containing 10% FBS and 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin sulfate, 1 mM sodium pyruvate, 10 mM HEPES, 100 μM nonessential amino acids) with 10 U/ml Benzonase (Novagon catalog no. 70664-3), and recovered overnight in a CO2 incubator at 37°C. PBMCs were then collected, counted, and resuspended to a concentration of 2 × 106 cells/ml in R10 for use in the ELISpot assay.
Design of HIV Gag peptide matrices.
One hundred twenty-two 15-mer peptides overlapping by 11 amino acids spanning HIV-1 HXBc2 Gag were synthesized by New England Peptide, Inc. (Gardner, MA), with FlashPure peptide array technology. All 122 overlapping peptides were pooled into a combined peptide matrix. Within the matrix, each peptide was represented in two different peptide pools, allowing for the identification of the respective peptide by responses in the two corresponding pools (29). Twenty-two matrix peptide pools were prepared (named from A to V), with 11 peptides per pool, except pools L and S, which comprised 12 peptides. The final concentration of each peptide within a peptide pool was 100 μM.
ELISpot assays.
The human IFN-γ ELISpot kit (Cellular Technology, Ltd., catalog no. HIFNG-10) was used in this assay per the manufacturer's instructions, and the resulting spots were counted using an Immunospot reader (CTL, Inc., Cleveland, OH). Peptide-specific IFN-γ ELISpot responses were considered positive if the response (minus medium background) was >3-fold above the medium response and ≥50 spot-forming cells (SFCs)/106 PBMCs.
Data analysis.
The identification of the individual 15-mer peptides being recognized was accomplished by conducting two sequential IFN-γ ELISpot assays on the same participant sample. The first IFN-γ ELISpot assay was done with PBMCs stimulated by each of the 22 matrix peptide pools. The intersections of all matrix peptide pools giving a positive response identified a potential positive-response peptide or peptides, which were selected for additional testing. A second IFN-γ ELISpot assay was run against those individual 15-mer peptides. Individual 15-mer peptides giving an IFN-γ ELISpot response of >55 SFCs/106 PBMCs and >4-fold above the wells containing medium alone were considered positive.
Binding antibody responses.
Serum HIV-1 specific IgG and IgA responses (1/50 dilution) against four HIV proteins (ConS gp140 and 6101 gp140, gp41, and p24) were measured at 2 weeks after the fourth vaccination by a validated HIV-1 binding antibody multiplex assay (BAMA) as previously described (30, 31). Antibody measurements were acquired on a Bio-Plex instrument (Bio-Rad), and the readout was expressed as mean fluorescent intensity (MFI). The positive control in each assay was HIV-positive serum, along with IgG- and IgA-specific controls. The negative control was HIV-negative human serum and blank beads. Samples with a blank bead MFI of >5,000 after one retesting were excluded. Samples were determined to be positive if both the MFI and blank-subtracted MFI were greater than 3-fold over the baseline MFI and blank-subtracted MFI, respectively, and if the blank-subtracted MFI values were above an antigen-specific cutoff that equals the antigen-specific average MFI plus 3 standard deviations from 60 seronegative plasma samples. Binding antibody data were available for 84 participants after the prime and 78 participants after the VSV boost.
VISR.
Vaccine-induced seroreactivity (VISR) was assessed at the end of the study (at month 12 [i.e., 1 year following enrollment]) using a diagnostic algorithm that includes four different enzyme immunoassays (EIAs): Abbott Architect HIV Ag/Ab Combo, Abbot Prism, Bio-Rad Genetic Systems HIV Combo Ag/Ab EIA, and Bio-Rad Multispot HIV-1/HIV-2 Rapid test. For participants with a positive result in any of these assays, RNA PCR (Abbott m2000 HIV-1 real-time PCR) was performed to distinguish vaccine-induced responses from actual infection. VISR data were available for 86 participants.
Statistical analyses.
All data from the participants who received at least one injection were analyzed according to the initial randomization assignment (modified intention-to-treat [mITT] analysis).
Box plots were used to summarize the distribution of various immune responses among the positive responders, where the midline of the box denotes the median and the ends of the box denote the 25th and 75th percentiles, with whiskers extended to the extreme data points that are no more than 1.5 times the interquartile range or, if no value meets this criterion, to the data extremes. Positive responses are determined by Fisher's exact test. The box plots are overlaid with the scatter plots of the immune responses of both positive responders (in red circles) and negative responders (in blue triangles).
T-cell response rates at each visit were compared between treatment group 1 (without IL-12 pDNA) and each of treatment groups 2 to 4 (with increasing pIL-12 doses) using Fisher's exact test. The comparisons were done for responses to any (i.e., at least one) antigen (as a summary measure of the overall HIV-specific response) as well as for responses to individual HIV antigens. The Cochran-Armitage test was used to assess trends in the pIL-12 dose response in groups 2 to 4. McNemar's test was used to compare response rates between visits for individual treatment groups.
Similarly, T-cell response magnitudes of positive responders were compared using Wilcoxon's rank sum tests between group 1 and each of groups 2 to 4 at each visit, using the Jonckheere-Terpstra test for trend in the pIL-12 dose response in groups 2 to 4, and using Wilcoxon's signed rank tests between different visits within each treatment group.
T-cell functionality analyses.
COMPASS (combinatorial polyfunctionality analysis of single cells) is a computational framework for unbiased polyfunctionality analysis of antigen-specific T-cell subsets (6). COMPASS uses a Bayesian hierarchical framework to model all observed functional cell subsets and select those most likely to exhibit antigen-specific responses. Cell subset responses are quantified by posterior probabilities, while subject-level responses are quantified by a summary statistic (functionality score) that can be correlated directly with outcomes of interest and describe the quality of an individual's functional response. The functionality score is defined as the proportion of antigen-specific subsets detected among all possible ones. For this analysis, expression of IFN-γ, IL-2, TNF-α, IL-4, and CD40L were included. Functionality scores were compared between group 1 and groups 2, 3, and 4 combined using Wilcoxon rank sum test statistics.
Study approval.
The study was conducted at five clinical research centers in the United States and was approved by all relevant local and governmental ethics and regulatory bodies for each clinical research center. Written informed consent was obtained from each participant prior to inclusion in the study. The CONSORT statement 2010 flow diagram is presented in Fig. 1.
Supplementary Material
ACKNOWLEDGMENTS
This trial was conducted by the HIV Vaccine Trials Network. We gratefully acknowledge the participation and support of many colleagues and staff on the protocol team and at the sites and are particularly grateful for the participation of the 100 study participants. We thank Stephen Voght for technical editing of the manuscript. We thank Terri Stewart, Kevin Hawkins, Jane Vasilyeva, Paul Newling, Vicki Ashley, Judith Lucas, and Yong Lin for technical assistance and Marcella Sarzotti-Kelsoe and Michael Stirewalt for quality assurance oversight. We are grateful to David Clarke for his VSV expertise, Julie Foster for her role in protocol development, Jenny Tseng for supporting safety reviews, and Shelly Ramirez for protocol operations management. We thank the James B. Pendleton Charitable Trust for their generous equipment donation.
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) U.S. Public Health Service grants UM1 AI068614 (LOC: HIV Vaccine Trials Network), UM1 AI068635 (SDMC: HIV Vaccine Trials Network), UM1 AI068618 (LC: HIV Vaccine Trials Network), UM1 AI069470 (Columbia Partnership for Prevention and Control of HIV/AIDS Clinical Trials Unit), UM1 AI069534 and P30 AI 045008 (Philadelphia HIV Therapeutics and Prevention Clinical Trials Unit), UM1 AI069511 (University of Rochester HIV/AIDS Clinical Trials Unit), and UM1 AI069439 (Vanderbilt HIV Clinical Trials Unit). The content of the manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
R.X., A.O.S., D.K.C., M.A.E., and J.H.E. are employees of Profectus Biosciences, Inc., and own stock in the company. All other authors have declared that no conflict of interest exists.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/CVI.00263-17.
REFERENCES
- 1.UNAIDS. 2014. The gap report. UNAIDS, Geneva, Switzerland: http://www.unaids.org/sites/default/files/media_asset/UNAIDS_Gap_report_en.pdf. [Google Scholar]
- 2.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robb ML, Ananworanich J. 2016. Lessons from acute HIV infection. Curr Opin HIV AIDS 11:555–560. doi: 10.1097/COH.0000000000000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M Jr, Lifson JD, Picker LJ. 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen SG, Piatak M Jr, Ventura AB, Hughes CM, Gilbride RM, Ford JC, Oswald K, Shoemaker R, Li Y, Lewis MS, Gilliam AN, Xu G, Whizin N, Burwitz BJ, Planer SL, Turner JM, Legasse AW, Axthelm MK, Nelson JA, Fruh K, Sacha JB, Estes JD, Keele BF, Edlefsen PT, Lifson JD, Picker LJ. 2013. Immune clearance of highly pathogenic SIV infection. Nature 502:100–104. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin L, Finak G, Ushey K, Seshadri C, Hawn TR, Frahm N, Scriba TJ, Mahomed H, Hanekom W, Bart PA, Pantaleo G, Tomaras GD, Rerks-Ngarm S, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Michael NL, Kim JH, Robb ML, O'Connell RJ, Karasavvas N, Gilbert P, DeRosa S, McElrath MJ, Gottardo R. 2015. COMPASS identifies T-cell subsets correlated with clinical outcomes. Nat Biotechnol 33:610–616. doi: 10.1038/nbt.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robert-Guroff M. 2007. Replicating and non-replicating viral vectors for vaccine development. Curr Opin Biotechnol 18:546–556. doi: 10.1016/j.copbio.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agnandji ST, Huttner A, Zinser ME, Njuguna P, Dahlke C, Fernandes JF, Yerly S, Dayer JA, Kraehling V, Kasonta R, Adegnika AA, Altfeld M, Auderset F, Bache EB, Biedenkopf N, Borregaard S, Brosnahan JS, Burrow R, Combescure C, Desmeules J, Eickmann M, Fehling SK, Finckh A, Goncalves AR, Grobusch MP, Hooper J, Jambrecina A, Kabwende AL, Kaya G, Kimani D, Lell B, Lemaitre B, Lohse AW, Massinga-Loembe M, Matthey A, Mordmuller B, Nolting A, Ogwang C, Ramharter M, Schmidt-Chanasit J, Schmiedel S, Silvera P, Stahl FR, Staines HM, Strecker T, Stubbe HC, Tsofa B, Zaki S, Fast P, Moorthy V, et al. 2015. Phase 1 trials of rVSV Ebola vaccine in Africa and Europe—preliminary report. N Engl J Med 374:1647–1660. doi: 10.1056/NEJMoa1502924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henao-Restrepo AM, Longini IM, Egger M, Dean NE, Edmunds WJ, Camacho A, Carroll MW, Doumbia M, Draguez B, Duraffour S, Enwere G, Grais R, Gunther S, Hossmann S, Konde MK, Kone S, Kuisma E, Levine MM, Mandal S, Norheim G, Riveros X, Soumah A, Trelle S, Vicari AS, Watson CH, Keita S, Kieny MP, Rottingen JA. 2015. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: interim results from the Guinea ring vaccination cluster-randomised trial. Lancet 386:857–866. doi: 10.1016/S0140-6736(15)61117-5. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs JD, Frank I, Elizaga ML, Allen M, Frahm N, Kochar N, Li S, Edupuganti S, Kalams SA, Tomaras GD, Sheets R, Pensiero M, Tremblay MA, Higgins TJ, Latham T, Egan MA, Clarke DK, Eldridge JH, HVTN Study Group and the National Institutes of Allergy and Infectious Diseases HIV Vaccine Trials Network, Mulligan M, Rouphael N, Estep S, Rybczyk K, Dunbar D, Buchbinder S, Wagner T, Isbell R, Chinnell V, Bae J, Escamilla G, Tseng J, Fair R, Ramirez S, Broder G, Briesemeister L, Ferrara A. 2015. First-in-human evaluation of the safety and immunogenicity of a recombinant vesicular stomatitis virus human immunodeficiency virus-1 gag vaccine (HVTN 090). Open Forum Infect Dis 2:ofv082. doi: 10.1093/ofid/ofv082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Churchyard GJ, Morgan C, Adams E, Hural J, Graham BS, Moodie Z, Grove D, Gray G, Bekker LG, McElrath MJ, Tomaras GD, Goepfert P, Kalams S, Baden LR, Lally M, Dolin R, Blattner W, Kalichman A, Figueroa JP, Pape J, Schechter M, Defawe O, De Rosa SC, Montefiori DC, Nabel GJ, Corey L, Keefer MC, NIAID HIV Vaccine Trials Network . 2011. A phase IIA randomized clinical trial of a multiclade HIV-1 DNA prime followed by a multiclade rAd5 HIV-1 vaccine boost in healthy adults (HVTN204). PLoS One 6:e21225. doi: 10.1371/journal.pone.0021225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goepfert PA, Elizaga ML, Sato A, Qin L, Cardinali M, Hay CM, Hural J, DeRosa SC, DeFawe OD, Tomaras GD, Montefiori DC, Xu Y, Lai L, Kalams SA, Baden LR, Frey SE, Blattner WA, Wyatt LS, Moss B, Robinson HL, National Institute of Allergy and Infectious Diseases HIV Vaccine Trials Network . 2011. Phase 1 safety and immunogenicity testing of DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J Infect Dis 203:610–619. doi: 10.1093/infdis/jiq105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perreau M, Welles HC, Harari A, Hall O, Martin R, Maillard M, Dorta G, Bart PA, Kremer EJ, Tartaglia J, Wagner R, Esteban M, Levy Y, Pantaleo G. 2011. DNA/NYVAC vaccine regimen induces HIV-specific CD4 and CD8 T-cell responses in intestinal mucosa. J Virol 85:9854–9862. doi: 10.1128/JVI.00788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jalah R, Patel V, Kulkarni V, Rosati M, Alicea C, Ganneru B, von Gegerfelt A, Huang W, Guan Y, Broderick KE, Sardesai NY, LaBranche C, Montefiori DC, Pavlakis GN, Felber BK. 2012. IL-12 DNA as molecular vaccine adjuvant increases the cytotoxic T cell responses and breadth of humoral immune responses in SIV DNA vaccinated macaques. Hum Vaccin Immunother 8:1620–1629. doi: 10.4161/hv.21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trinchieri G, Wysocka M, D'Andrea A, Rengaraju M, Aste-Amezaga M, Kubin M, Valiante NM, Chehimi J. 1992. Natural killer cell stimulatory factor (NKSF) or interleukin-12 is a key regulator of immune response and inflammation. Prog Growth Factor Res 4:355–368. doi: 10.1016/0955-2235(92)90016-B. [DOI] [PubMed] [Google Scholar]
- 16.Chong SY, Egan MA, Kutzler MA, Megati S, Masood A, Roopchard V, Garcia-Hand D, Montefiori DC, Quiroz J, Rosati M, Schadeck EB, Boyer JD, Pavlakis GN, Weiner DB, Sidhu M, Eldridge JH, Israel ZR. 2007. Comparative ability of plasmid IL-12 and IL-15 to enhance cellular and humoral immune responses elicited by a SIVgag plasmid DNA vaccine and alter disease progression following SHIV(89.6P) challenge in rhesus macaques. Vaccine 25:4967–4982. doi: 10.1016/j.vaccine.2006.11.070. [DOI] [PubMed] [Google Scholar]
- 17.Hirao LA, Wu L, Khan AS, Hokey DA, Yan J, Dai A, Betts MR, Draghia-Akli R, Weiner DB. 2008. Combined effects of IL-12 and electroporation enhances the potency of DNA vaccination in macaques. Vaccine 26:3112–3120. doi: 10.1016/j.vaccine.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalams SA, Parker SD, Elizaga M, Metch B, Edupuganti S, Hural J, De Rosa S, Carter DK, Rybczyk K, Frank I, Fuchs J, Koblin B, Kim DH, Joseph P, Keefer MC, Baden LR, Eldridge J, Boyer J, Sherwat A, Cardinali M, Allen M, Pensiero M, Butler C, Khan AS, Yan J, Sardesai NY, Kublin JG, Weiner DB, NIAID HIV Vaccine Trials Network . 2013. Safety and comparative immunogenicity of an HIV-1 DNA vaccine in combination with plasmid interleukin 12 and impact of intramuscular electroporation for delivery. J Infect Dis 208:818–829. doi: 10.1093/infdis/jit236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalams SA, Parker S, Jin X, Elizaga M, Metch B, Wang M, Hural J, Lubeck M, Eldridge J, Cardinali M, Blattner WA, Sobieszczyk M, Suriyanon V, Kalichman A, Weiner DB, Baden LR, NIAID HIV Vaccine Trials Network . 2012. Safety and immunogenicity of an HIV-1 gag DNA vaccine with or without IL-12 and/or IL-15 plasmid cytokine adjuvant in healthy, HIV-1 uninfected adults. PLoS One 7:e29231. doi: 10.1371/journal.pone.0029231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mpendo J, Mutua G, Nyombayire J, Ingabire R, Nanvubya A, Anzala O, Karita E, Hayes P, Kopycinski J, Dally L, Hannaman D, Egan MA, Eldridge JH, Syvertsen K, Lehrman J, Rasmussen B, Gilmour J, Cox JH, Fast PE, Schmidt C. 2015. A phase I double blind, placebo-controlled, randomized study of the safety and immunogenicity of electroporated HIV DNA with or without interleukin 12 in prime-boost combinations with an Ad35 HIV vaccine in healthy HIV-seronegative African adults. PLoS One 10:e0134287. doi: 10.1371/journal.pone.0134287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gherardi MM, Ramirez JC, Esteban M. 2000. Interleukin-12 (IL-12) enhancement of the cellular immune response against human immunodeficiency virus type 1 env antigen in a DNA prime/vaccinia virus boost vaccine regimen is time and dose dependent: suppressive effects of IL-12 boost are mediated by nitric oxide. J Virol 74:6278–6286. doi: 10.1128/JVI.74.14.6278-6286.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee K, Overwijk WW, O'Toole M, Swiniarski H, Restifo NP, Dorner AJ, Wolf SF, Sturmhoefel K. 2000. Dose-dependent and schedule-dependent effects of interleukin-12 on antigen-specific CD8 responses. J Interferon Cytokine Res 20:589–596. doi: 10.1089/10799900050044787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bekker LG, Laher F, Moodie Z, Tomaras G, Grunenberg N, Allen M, Daniels B, Innes C, Mngadi K, Malahleha M, Gilbert P, Michael N, Phogat S, Diaz Granados C, Kanesa-Thasan N, Corey L, Gray G, McElrath JM, HVTN100 Study Team . 2016. Meeting the “Go” criteria: immunogenicity from HVTN100, a phase 1/2 randomized, double blind, placebo-controlled trial of clade C ALVAC (vCP2438) and bivalent subtype C gp120/MF59 in HIV-uninfected South African adults, abstr TUAX0102LB. Abstr AIDS 2016, Durban, South Africa. [Google Scholar]
- 24.Janes H, Friedrich DP, Krambrink A, Smith RJ, Kallas EG, Horton H, Casimiro DR, Carrington M, Geraghty DE, Gilbert PB, McElrath MJ, Frahm N. 2013. Vaccine-induced gag-specific T cells are associated with reduced viremia after HIV-1 infection. J Infect Dis 208:1231–1239. doi: 10.1093/infdis/jit322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bull M, Lee D, Stucky J, Chiu YL, Rubin A, Horton H, McElrath MJ. 2007. Defining blood processing parameters for optimal detection of cryopreserved antigen-specific responses for HIV vaccine trials. J Immunol Methods 322:57–69. doi: 10.1016/j.jim.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, Huang Y, Chiu YL, McElrath MJ, De Rosa SC. 2007. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods 323:39–54. doi: 10.1016/j.jim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F, Malhotra U, Gilbert PB, Hawkins NR, Duerr AC, McElrath JM, Corey L, Self SG. 2006. Peptide selection for human immunodeficiency virus type 1 CTL-based vaccine evaluation. Vaccine 24:6893–6904. doi: 10.1016/j.vaccine.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 28.De Rosa SC, Carter DK, McElrath MJ. 2012. OMIP-014: validated multifunctional characterization of antigen-specific human T cells by intracellular cytokine staining. Cytometry A 81:1019–1021. doi: 10.1002/cyto.a.22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, Johnston MN, Corcoran C, Wurcel AG, Fitzpatrick CA, Feeney ME, Rodriguez WR, Basgoz N, Draenert R, Stone DR, Brander C, Goulder PJR, Rosenberg ES, Altfeld M, Walker BD. 2003. Comprehensive epitope analysis of HIV-1-specific T cell responses directed against the entire expressed HIV-1 genome demonstrates broadly directed responses, but no correlation to viral load. J Virol 77:2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, Decamp AC, Parks RJ, Ashley VC, Lucas JT, Cohen M, Eron J, Hicks CB, Liao HX, Self SG, Landucci G, Forthal DN, Weinhold KJ, Keele BF, Hahn BH, Greenberg ML, Morris L, Karim SS, Blattner WA, Montefiori DC, Shaw GM, Perelson AS, Haynes BF. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol 82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yates NL, Liao HX, Fong Y, deCamp A, Vandergrift NA, Williams WT, Alam SM, Ferrari G, Yang ZY, Seaton KE, Berman PW, Alpert MD, Evans DT, O'Connell RJ, Francis D, Sinangil F, Lee C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Pinter A, Zolla-Pazner S, Gilbert PB, Nabel GJ, Michael NL, Kim JH, Montefiori DC, Haynes BF, Tomaras GD. 2014. Vaccine-induced Env V1-V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Sci Transl Med 6:228ra239. doi: 10.1126/scitranslmed.3007730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.