Abstract
Measuring the exposome remains a challenge due to the range and number of anthropogenic molecules that are encountered in our daily lives, as well as the complex systemic responses to these exposures. One option for improving the coverage, dynamic range and throughput of measurements is to incorporate ion mobility spectrometry (IMS) into current MS-based analytical methods. The implementation of IMS in exposomics studies will lead to more frequent observations of previously undetected chemicals and metabolites. LC-IMS-MS will provide increased overall measurement dynamic range, resulting in detections of lower abundance molecules. Alternatively, the throughput of IMS-MS alone will provide the opportunity to analyze many thousands of longitudinal samples over lifetimes of exposure, capturing evidence of transitory accumulations of chemicals or metabolites. The volume of data corresponding to these new chemical observations will almost certainly outpace the generation of reference data to enable their confident identification. In this perspective, we briefly review the state-of-the-art in measuring the exposome, and discuss the potential use for IMS-MS and the physico-chemical property of collisional cross section in both exposure assessment and molecular identification.
Keywords: : collision cross section, exposome, ion mobility spectrometry, mass spectrometry, metabolome
The majority of human disease (>90%) is attributable to a combination of individual genetic factors and nongenetic ‘environmental factors’ [1–3]. The former can be readily assessed using rapid genome sequencing technologies, whereas the latter can be assessed or estimated using a variety of analytical methods [3,4]. To represent these nongenetic environmental factors, the concept of the ‘exposome’ was defined by Christopher Wild as the sum of all exposures over a lifetime to chemical, social and biological agents that influence human health [1]. The exposome was proposed as the natural complement to the genome, with the recognition that the phenome (sum of individual traits) was a product of both the genome and an exposome – the environment within which genes are expressed.
The exposome embraces the totality of both external and internal exposure. External exposure is mainly to chemicals in the air, water, soil, diet or consumer products, and internal exposures correlate to molecular changes within body compartments. Furthermore, these internal exposures can be to products of normal metabolism or in response to external exposures, including chemical, infectious or social [5]. External exposure measurements are often more convenient and cost effective, particularly when the goal is to identify sources of exposure. Internal exposures are often preferred when the objective is to identify exposure–disease relationships because they better represent concentrations at or near the biological site of action. The extensive set of sample matrices – from blood, breath, hair and nails to soil and water – as well as the range of complexity or ‘space’ comprising xenobiotic chemicals and endogenous molecules, is a challenge for analytical chemists attempting to measure the exposome. As the exposome is the ‘totality’ of exposure, each measurement will provide a snapshot (or piece of the puzzle) that can be used to build the bigger picture.
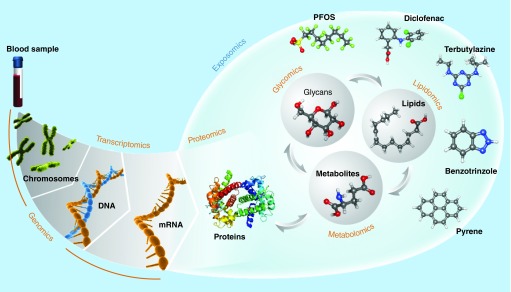
The use of ‘omics’ technologies to characterize the exposome [2,6–9], originally proposed by Wild [1], considers two distinct approaches: direct measurement of chemical exposures by measuring parent compounds and their transformation products in environmental samples or parent compounds and their metabolites in biofluids and tissues using similar analytical approaches as implemented in metabolomics, and inference of an exposure based on biological signatures (i.e., the ‘phenome’) obtained by one or more complementary approaches, such as transcriptomics or proteomics. A significant difference among these omics approaches is that genomics, transcriptomics and proteomics focus on a narrow chemistry – polymers of nucleotides and amino acids – whereas direct measurement of chemical exposure and of the metabolome requires dedicated or, better, complimentary approaches suitable for a very diverse chemical space [10–15] (Figure 1). Although the average molecular composition of a peptide [16] is not significantly different from that of a metabolite [17], the peptide chemistry is constrained to the typical peptide bonding between a fixed set of amino acids. In contrast, the metabolite chemistry is constrained only by the organism (for endogenous metabolites) and the boundaries of chemistry itself, while some xenobiotic compounds of anthropogenic origin even push the boundaries of chemistry.
Figure 1. . The ‘omes’ measured in exposome studies.
The genome and transcriptome are comprised of DNA and RNA, respectively, which are polymers of four defined nucleotides. Similarly, proteins are polymers composed of 20 defined amino acids. In contrast, the metabolome and related small molecule ‘omes’ are comprised of molecules with much greater chemical diversity.
In the search for causes of human disease, the genomics revolution brought unprecedented ability to obtain genetic information across individuals and populations, and a deeper appreciation for the importance of exposure as causative agent, yet tools for measuring the exposome were far less developed. For example, while advancements are continually being made in methods for broadly and accurately identifying metabolites in metabolomics studies [18–31], these developments are still relatively immature compared with the ease, robustness and throughput with which the proteomics community confidently identifies peptide sequences [32–34]. The expansion of more conventional analytical approaches to handle higher sample throughput, and to measure thousands of chemicals, will allow comprehensive characterization of chemical exposures across larger populations. Combined with emerging untargeted analytical methods (e.g., in metabolomics), there is the expectation that previously unknown or unexpected exposures will be identified, improving the search for environmental drivers of disease and providing exposure data to guide chemical selection for toxicity testing. Applied to both human (e.g., blood and urine) and environmental samples (e.g., water, air, dust and soil), and linked through geographical information systems, these new analytical approaches will also advance the search for sources of exposure, even when the identity of some chemicals involved remain unknown.
In this perspective paper, we will briefly review the state-of-the-art in measuring the small, organic molecular components of the exposome and discuss the potential use for ion mobility spectrometry-mass spectrometry (IMS-MS) and the physicochemical property of collisional cross section (CCS) in both exposure assessment and molecular identification in metabolomics studies.
State-of-the-art in measuring the exposome
Current exposure science leverages many disciplines to assess level(s) of environmental exposure and risk thereof, including epidemiology, toxicology and analytical chemistry, with the latter being used for environmental and biological monitoring [5]. In this section, state-of-the-art approaches used in environmental and biomonitoring are illustrated. Specifically, methods used to identify molecular signatures of exposure, whether evidence of parent compounds and their metabolites (i.e., direct measurement) or of pathogens, or the host's complex biological response to these molecules and organisms (i.e., indirect measurement) will be discussed. We further limit our discussion to direct measurements of chemical exposure and indirect inference of an exposure based on biological signatures contained in the metabolome. Macromolecules and metals will not be covered.
Regardless of whether direct or indirect measures of exposure are performed, the technical approaches used largely fall under one of two categories: targeted or untargeted. The next two sections will provide an overview of these two approaches, citing specific examples and covering benefits and caveats of each.
Targeted analysis
Methods for targeted analysis in assessment of the exposome focus on a single analyte, a class of chemically similar analytes or a set of analytes whose chemistries are sufficiently similar to allow their measurement in a single analysis, all of which aim to address specific questions of exposure. The analytical protocols for such methods, including sample preparation, have been optimized to provide accurate identification and quantification with high sensitivity (i.e., true positive rate) and specificity (i.e., true negative rate) in select matrices [35]. Select examples of targeted analysis include a multipanel LC-MS assay to assess exposure to pesticides, veterinary drugs and parabens [36], GC-MS analysis of flame retardants after flow-through air sampling [37] and the measurement of estrogens in wastewater samples using GC-MS [38]. The assays used for targeted analysis can be as simple as a glucose colorimetric [39] kit to determine the degree of hyperglycemia [40] associated with the response to exposure to streptozotocin [41], or certain drugs [42], to as complex as chromatography coupled with MS to quantify oxygenated polycyclic aromatic hydrocarbons [43]. Regardless of the degree of complexity of the assay, targeted analysis is rooted in the fundamentals of basic analytical chemistry: an analyte is selected for quantification in a given matrix; experiments are performed to optimize the detection of the analyte by the chosen measurement platform, based on analyses of authentic chemical standards; if necessary, experiments are performed to optimize the extraction of the chosen analyte from a given matrix, again using authentic chemical standards and with determination of extraction efficiency; finally, an approach for appropriate quantitative data analysis is established, usually based on calibration curves constructed through analyses of authentic chemical standards in the matrix of interest and, ideally using stable isotope-labeled internal standards. The benefits of targeted analysis include the accurate, quantification of the analyte(s) of interest, based on the optimization of all aspects of the analysis method; low limits of quantification (e.g., sufficient to quantify pM levels of chemicals in blood [44]), again due to the optimization of the method parameters; and usually absolute confidence in the identity of the analyte. Caveats of targeted approaches include the narrow snapshot of chemistry measured; the level of effort required to fully optimize and validate the analytical pipeline [43,45], and the relatively low analysis throughput, depending on the specifics of the method.
Untargeted analysis
In contrast to targeted analysis, untargeted analysis does not focus on a specific analyte but instead seeks to comprehensively measure all analytes in a sample. Untargeted analysis of small molecules is increasingly popular in metabolomics and environmental studies [46–57]. For general metabolomics studies, NMR spectroscopy [58] and MS [59] have been the primary analytical techniques employed. NMR is inherently quantitative and offers the ability to elucidate molecular structures, but suffers from low measurement sensitivity and throughput [60,61]. The utility of NMR in exposome studies is limited to measuring relatively high-abundance xenobiotics and to assessing biological response to exposure. On the other hand, MS analyses when coupled with chromatography are highly sensitive, but not absolutely quantitative unless stable-isotope-labeled internal standards, matrix-matched external standards or the standard addition method are used. However, the high dynamic range and low of MS approaches tend to outweigh the typical limitation to semiquantitative analyses (if desired, full quantitation is possible using any of the approaches mentioned above) and offer the potential to measure both xenobiotic chemicals and the biological responses to exposure. Chromatography (e.g., LC; GC) coupled with MS is thus the most widely used analytical platform for exposome studies [46–52]. A typical workflow for untargeted analysis using chromatography coupled with MS involves SPE or liquid–liquid extraction of the sample, drying of the extracts and reconstitution in a suitable buffer or solvent, followed by analysis. If GC-MS is used, then chemical derivatization of extracted molecules may be required to increase their volatility and enhance their separation. For GC-MS, electron ionization (EI) is typically used to impart the required charge to the analyte for detection by the mass spectrometer, although chemical ionization can also be used. Molecular fragmentation occurs simultaneously with EI, and commercial and open access reference libraries of spectra can be used to identify detected molecules [18,22,62], often in conjunction with other information such as retention time and/or retention indices [22]. For LC-MS, ESI is most commonly used to introduce analyte ions to the instrument, and data are typically collected by repeatedly scanning over a wide mass range (e.g., 100–1000 m/z) to detect as many molecules as possible. In most LC-MS experiments, intact molecular ions are fragmented using collision induced dissociation to produce MS/MS spectra, which can be used to identify the structures of detected molecules in conjunction with libraries of reference spectra [20,28,63] or tentatively identify candidates using in silico fragmentation approaches combined with candidate look-up [26,64–66]. The benefit of untargeted analysis is that it is not biased by a priori assumptions and so offers the best opportunity to discover novel markers of exposure or to characterize the response to exposure by measuring a broad swath of chemical space. Caveats, however include possible artifacts in the data due to the lack of optimization of sample preparation procedures, difficulty detecting very low-abundance analytes in the presence of high-abundance analytes (particularly when using LC-MS due to ESI suppression) and an incomplete representation of chemical space in spectral reference libraries, leading to, in some cases, limited confidence in the identification of detected species where reference standards are not available.
Molecular identification confidence
As described above, the identification confidence of molecules measured in targeted analyses is generally very high, since methods are optimized for the chemistry of interest and the use of authentic standards ensures that the correct molecule is identified each time. Therefore, the discussion in this section will focus on the confidence in identification of metabolites and chemicals detected in untargeted, MS-based analyses.
The use of untargeted acquisition techniques brings a whole set of identification challenges with it as, unlike targeted acquisition, no preliminary hypothesis as to the number and identity of analytes of interest has been generated. It is possible (and increasingly common) to perform target analysis on data acquired with untargeted methods, and this is the best starting point for any untargeted investigation. This requires the availability of in-house reference standards (measured with the same technique) and, for quantitative results, internal standards and appropriate calibration curves [54,67]. Confident identification requires the match of two orthogonal pieces of experimental data between standard and sample, such as matching exact mass of the molecular ion and fragments as well as retention time. Typically, targets are ‘screened’ in data from untargeted analysis by searching for the exact mass in peak lists or via extracted ion chromatograms, hence the term ‘target screening’ [54].
In cases where investigations wish to pursue a substance of interest for which the reference standard is not available in house, a so-called ‘suspect screening’ can be performed. The exact masses of the ions relating to the substance(s) of interest can be used to search the data from untargeted analyses either via peak lists or extracted ion chromatograms, as is performed for target screening. Unlike target screening, however, additional steps need to be undertaken to prove (or disprove) the identity of the suspect, since a standard is not available. This is discussed further below. The lists used for suspect screening can vary widely and depend on the study question. One could consider screening the Human Metabolome Database (HMDB) [68] or other metabolite-specific databases as suspect screening for metabolomics. In environmental studies, one could use a list of registered pharmaceuticals [69] or pesticides [70], surfactants [53] or even all substances registered under the Registration, Evaluation, Authorization and Restriction of Chemicals regulation (contained, e.g., in STOFF-IDENT [71]). The lists used vary widely [54].
The final step in data analysis is nontarget screening – effectively all the remaining masses in the sample after target and suspect screening. Typically a peak picking step is performed to pick genuine peaks due to chromatographic or other separation, then a grouping step is highly recommended to find associated adduct and isotope peaks. This information provides confidence about the mass of the molecular ion and can help determine the molecular formula. To assist in the identification of the structure, fragmentation information is essential; identification can be performed via library searching or candidate look-up and comparison of predicted and measured fragments. Without a reference standard available, these identifications can be only tentative at best.
Communicating the confidence in chemical identification can be a challenge and several systems exist for small molecules, including the metabolomics standards initiative (MSI) [67], an LC-high resolution MS/MS specific set from Eawag [72] and many more. The essences of these are:
Confirmed identification with two orthogonal matching properties to an authentic reference standard measured in-house (MSI Level 1, Eawag Level 1).
Probable identification with all evidence indicating only one structure is possible, but authentic standard is not available for confirmation (Eawag Level 2a/b).
Putative annotation based on physicochemical properties and spectral matching (MSI Level 2, Eawag Level 2a).
Tentative identification/putative compound class – tentative identification using predictive techniques, multiple structures are possible, or insufficient evidence to eliminate other structures; substance class only is clear (MSI Level 3, Eawag Level 3).
Unknown compounds – molecular formula is unequivocal (Eawag Level 4) or exact mass only (Eawag Level 5; both MSI Level 4). These can be traced in samples and correspond to ‘detected features’ in the analyses, but the identity remains unclear.
To date, the evidence for coupled chromatographic-MS systems has been based on the mass spectral information (exact mass and fragments for MS/MS, EI-MS spectrum for GC-MS) plus retention time information (either as retention time or retention indices) as the orthogonal information. However, coupled chromatographic systems alone are not ideally suited to high-throughput studies due to the time required per analysis. Later in this article, the potential for IMS and CCS to provide orthogonal identification evidence together with (or in place of) the retention time in coupled chromatographic systems is discussed.
Introducing IMS as a new tool for the exposomics toolbox
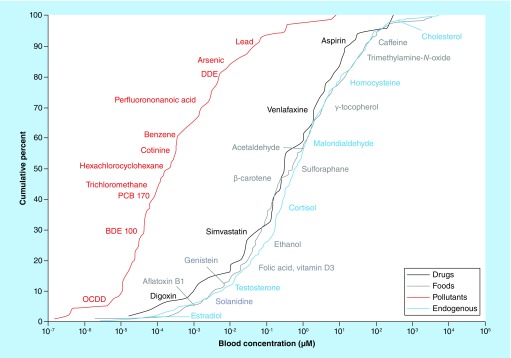
Comprehensive characterization of exposure is extremely challenging since individuals are generally subject to thousands of structurally and physicochemically diverse chemical agents per day (e.g., pharmaceuticals, pesticides, personal care products, industrial chemicals), with various systemic responses to these. Measuring these chemicals in clinical samples is very difficult since they typically occur from picomolar to millimolar concentrations in complex matrices (Figure 2). In addition, many of these molecules transform, either in the environment by biotic and abiotic processes, or in vivo due to xenobiotic metabolisms, such that the original chemical is not the final form that may accumulate to detectable levels in the sample of interest. Finally, the parent molecules or their transformation products may be transitory in regard to their detectability in the sample of interest, and therefore frequent sampling will be required in order to capture their presence. Comprehensive characterization of human exposure with high measurement dynamic range and with throughput sufficient for large, epidemiological studies requiring frequent sampling would transform the search for environmental causes of disease and revolutionize our understanding of the role of the environment and genome in health.
Figure 2. . Anthropogenic chemicals and endogenous metabolite concentrations in blood.
A wide dynamic range of concentrations of chemicals and metabolites are observed in human blood, making the comprehensive measurement of these in an exposomics approach extremely challenging.
Reproduced with permission from [44].
An appealing technique for enhancing current exposomic methods is the incorporation of IMS [73]. IMS allows ions to be conformationally separated based on the balance of two forces that impact ion movement, namely, the electric field and the drag force from collisions with buffer gas molecules. Currently there are many IMS-based platforms such as drift tube IMS (DTIMS), traveling wave IMS (TWIMS), trapped IMS [74], overtone IMS [75,76], differential IMS [77], field asymmetric IMS) [78–80] and transversal modulation IMS [81]. However, of these, DTIMS shows the greatest promise in small molecule measurements because it is able to directly determine molecular structural information without the external calibration approaches that are required by other IMS-based platforms [82]. In the following sections, we will focus the discussion on the benefits of DTIMS and how it can be used to address the challenges of exposomics.
DTIMS increases the dynamic range of existing LC-MS methods
In DTIMS platforms, ions travel through a drift tube under the influence of a weak attractive electric field while colliding with a stationary buffer gas (Figure 3A). In this way, ions with small CCSs spend less time inside the drift tube than other equally charged ions with larger CCS values [83,84], as described in the following equation:
 |
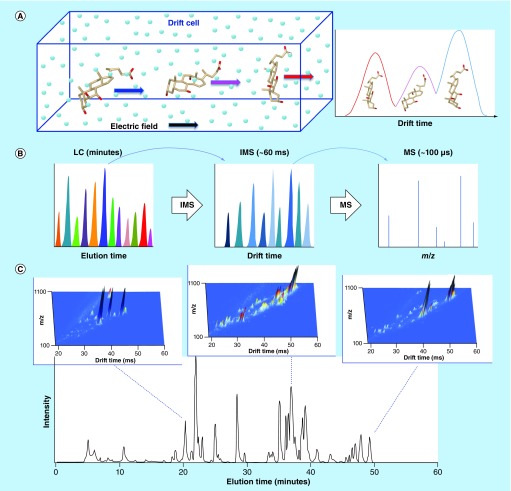
Figure 3. . Ion mobility spectrometry and its incorporation into LC-MS workflows.
(A) In ion mobility spectrometry (IMS), ions are separated based on their size and shape. In the example shown, the bile acids β-muricholic, urocholic and α-muricholic acids separate in order of fastest to slowest based on relative gas-phase ion shape. (B) The millisecond acquisitions of IMS are easily coupled between the minute and microsecond timescales of LC separations and TOF detection, since the time scale of each allows rapid sampling of the preceding separation. (C) Integration of IMS into LC-MS workflows provides an additional dimension of separation. Incorporating IMS provides greater coverage of the sample for the same LC separation length, or allows for decreasing the LC separation length while maintaining the same coverage. The example shows a human plasma total lipid extract analyzed by LC-IMS-MS. To indicate the increased separation capability obtained with the combination of LC and IMS, IMS-MS spectra were summed across three 10-s regions of the LC separation, creating three dimensional plots of m/z, drift time and intensity.
Figure 3C adapted with permission from [125] © Elsevier (2008).
where e is the elementary charge, z is the charge on the ion, N the gas density number (mol/m3), K is the mobility of the ion, μ is the reduced mass of the collision gas and ion, kb is the Boltzmann constant and T is absolute temperature. While the m/z of an ion strongly influences the time it spends in the drift tube, the CCS value directly reflects the mobility of each ion and correlates with the shape of the molecule based on its ion-neutral interaction potential [85]. As the m/z ratios of ions can be measured following the DTIMS separation using a mass spectrometer, this allows CCSs to be directly determined for each ion via the fundamental zero-field (i.e., Mason Schamp equation [86]). In general, this orthogonality between the separation mechanisms of IMS and MS is key to the utilization of this combination of techniques, particularly for the separation of isobaric compounds.
The ability to resolve isomers that are difficult to distinguish using LC-MS alone is an inherent strength of DTIMS, particularly in small molecule analysis since many metabolites and other chemicals have the same molecular formula but play very different roles in biological systems. Indeed, DTIMS has been used to separate some important classes of isomers [87–91]. Stephen et al. used library CCS values to confirm the identities of the isomeric tramadol and desvenlafaxine in wastewater samples [92], which have the same molecular formula and hence the same m/z and often co-elute in generic untargeted LC-MS methods [53]. DTIMS also provides separation of species based on their charge state and shape, which in turn, depends on the chemical makeup and spatial structure of the molecules. Because DTIMS instruments depend only on drift cell pressure, temperature and length, CCS values on a single instrument are extremely repeatable (<0.4% error) [92–95], allowing reliable molecular feature alignment (i.e., identity confirmation) across multiple samples. Furthermore, measurements from different instruments in different laboratories have also been compared and their values normally agree within <2% error [96], with recent DTIMS instruments yielding values with reproducibility precision of <1% [93,94]. The high reproducibility and speed of DTIMS allows it to be easily nested between LC and MS to provide additional separation power and dynamic range of detection in measuring the exposome (Figure 3B). The resulting LC-IMS-MS approach can either provide greater coverage of the sample for the same LC separation length (i.e., by increasing the dynamic range of the measurement) [97], or allows for decreasing the LC separation length while maintaining the same measurement coverage (Figure 3C). For example, incorporating DTIMS into a LC-MS-based proteomics method allowed shortening of the LC gradient from 100 to 15.5 min with no loss in coverage of 20 standard peptides spiked into a mouse plasma protein digest over a range of 1 ng/ml to 10 μ/ml [98]. Similarly, Stephen et al. incorporated DTIMS into 1D and 2D LC-MS methods, supporting the identity confirmation of 22 and 53 different compounds, respectively, in highly complex wastewater samples [92].
DTIMS separations are ultra-high throughput
While using chromatographic separations prior to the IMS stage will continue to play a role in exposome studies, DTIMS-MS alone lends itself extremely well to coping with the high-throughput demands of measuring large population cohorts. Interfacing DTIMS with high-resolution MS, such as TOF [99] permits simultaneous separation of sample constituents and acquisition of structural information and high-accuracy MS data, and when coupled with quadrupole-TOF MS, also allows for acquisition of tandem mass spectra in targeted, data-dependent or data-independent modes. IMS separations can also be coupled with trapping MS instruments such as Orbitrap and Fourier Transform Ion Cyclotron Resonance. However, in these cases the millisecond IMS separations would be faster than the second timescale sampling of the mass spectrometers, and so MS data acquisition manipulations would be required. Thus, TOF-based instruments that sample on the microsecond timescale are more practical to retain the IMS measurement throughput. In DTIMS-TOF MS analysis, a single separation typically occurs in 10–100 ms and the TOF MS pulser samples each peak approximately every 100 μs, so DTIMS separations can be reconstructed with sufficient points-per-(IMS)-peak to allow for reliable discrimination of features in 3D space (drift time, m/z, abundance). DTIMS transients are normally summed for at least 0.5 s to acquire enough signal for reproducible IMS peaks and repeatable determination of peak apices, which is essential for accurate determination of CCS.
DTIMS separations do not require derivatization and can be combined with different ionization sources for analysis of distinct compound types such as polar metabolites (ESI in positive or negative ionization modes) and as well as compounds for which other ionization modes are required (e.g., APCI in positive or negative modes for analysis of polyaromatic hydrocarbons), allowing for multiple analyses to obtain high coverage of the exposome. Without chromatographic or other preseparation prior to ESI, ionization suppression due to the presence of highly abundant chemicals has previously limited the utility of DTIMS-MS for analysis of complex mixtures. However, a front-end automated SPE system was recently coupled directly to DTIMS-MS and different cartridge chemistries were utilized to provide a means to reduce ion suppression in analysis of complex mixtures. This automated SPE-IMS-MS system provided a 10-s sample-to-sample duty cycle and a theoretical maximum throughput of >8000 injections per day [100,101]. This is 2–3 orders of magnitude higher throughput than conventional GC-MS or LC-MS methods and makes the study of large patient cohorts practical. When necessary, DTIMS-MS can also be coupled with GC or LC to address prohibitively complex samples and reduce front-end analysis times [98].
Incorporating CCS into molecular identification workflows
The implementation of IMS in exposomics studies will lead to increasingly more frequent observations of previously undetected chemicals and metabolites. LC-IMS-MS will provide increased overall measurement dynamic range, resulting in detections of lower abundance molecules, and the throughput of IMS-MS alone will provide the opportunity to analyze many thousands of longitudinal samples over lifetimes of exposure, capturing evidence of transitory accumulations of chemicals or metabolites. The volume of data corresponding to these new chemical observations will almost certainly outpace the development of reference data to enable their confident identification. Thus, the question remains as to how these molecules can then be identified in exposomics studies.
We here explore the possibility of using computationally predicted CCS to assist in the identifications of newly observed anthropogenic chemicals and metabolites, analogous to previous and ongoing efforts to predict retention times or tandem mass spectra during analysis of small molecules [29,30,102,103]. In this approach, CCSs are computationally predicted from the chemical structures of the molecules themselves, as captured using molecular modeling and quantum chemical calculations. In addition, unlike chromatographic retention times, the CCS parameter recorded under low drift field conditions is influenced by far fewer physicochemical parameters, meaning that excellent correspondence between measured values and library entries can be expected across different instruments.
To broadly predict CCS values, an automated computational pipeline is required as depicted in Figure 4. First, International Chemical Identifiers (InChIs; [104]) are converted into 2D structures, and then analyzed using the Marvin pKa plugin (Marvin 15.9.14, 2015, ChemAxon) to predict protonation/deprotonation states and adduct sites [105]. Initial geometry optimizations are performed using the Merck molecular force field [106] implemented in Avogadro (v.1.1.1) [107] and final geometry optimizations are performed using a density functional theory implemented in NWChem [108] at the B3LYP/6–31g* level [109–111]. Finally, the theoretical CCS values are calculated based on the geometry-optimized structures using the N2-optimized trajectory method implemented in the MOBCAL software [112–114]. In this way, the generation of accurate predicted CCS values can facilitate the broad identification of detected molecules in combination with accurate mass and MS/MS spectra, when available, and possibly with just accurate mass. The power of this approach is that metabolites can be annotated rapidly since only a chemical structure (e.g., in InChI format) obtained from either one of the many databases (e.g., HMDB, CHEBI, Chemspider, PubChem) or drawn by hand in chemical structure software is required for predicting theoretical CCS values to be matched with experimental numbers, together with other metrics, such as accurate mass and/or MS/MS spectra.
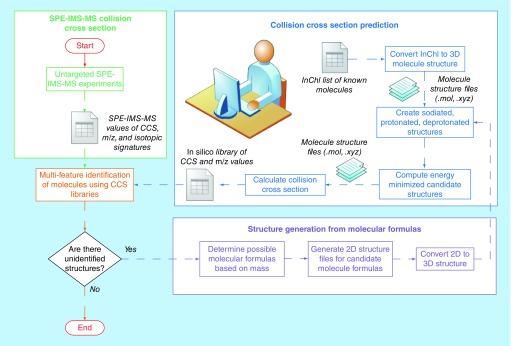
Figure 4. . Workflow for the identification of small molecules based on experimental and theoretical collision cross sections and accurate mass.
Untargeted SPE-ion mobility spectrometry-MS measurements (green box) provide experimental collision cross section (CCS) and accurate mass m/z values for sample molecules, as well as their isotopic signatures. Independently, theoretical CCS calculations (blue box) augment the contents of a CCS and accurate mass reference library. Separately, molecules that cannot be identified by matching to the CCS and accurate mass reference library or to MS/MS libraries are analyzed based on their masses to generate candidate molecular formulas, which are then converted into 3D structures for CCS prediction (purple box).
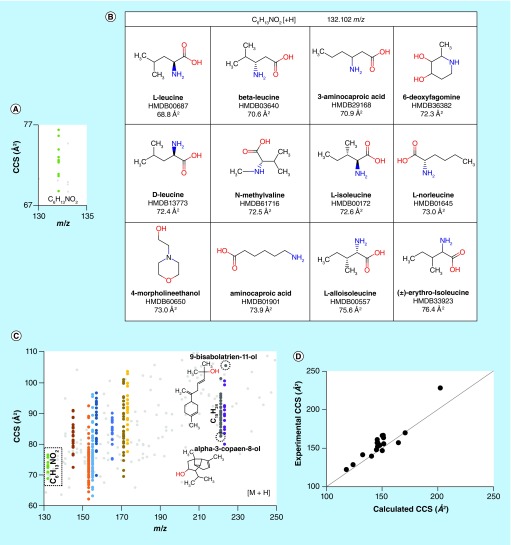
By utilizing a theoretical pipeline such as defined above, the feasibility of broadly predicting CCS for small molecules was determined by calculating theoretical CCSs for 5068 metabolites from the HMDB, in their protonated, deprotonated and sodiated forms based on pK a analysis, yielding predicted CCSs for a total of 11,046 unique ionized structures. To improve throughput for predicting so many structures, CCS was calculated using the IMPACT software [105,115], parameterized for helium as the drift gas. On average, it required approximately 0.5 s to calculate the CCS of a molecule on a windows desktop (with 16 GB RAM, Intel® Xeon® 1.6 GHz CPU); while the InChI to 2D/3D structure conversions, the pK a calculations and the construction of the ionized structures required about 3.3, 2.9 and 26.3 s per molecule, respectively, for 1465 molecules. The DFT calculations required approximately 1 h per molecule to run on a single computer node of a 3.4 petaflop linux cluster that has 1440 compute nodes and 16 cores per node. From these results, the chemical search space for possible molecules with a given accurate monoisotopic mass could be reduced by a minimum of 79% using CCS with a standard deviation (SD) of 2% (compared with a 0% reduction if using only m/z to find a match) within the HMDB library. For example, 76 different metabolites had the molecular formula C10H16O, highlighting the fact that annotation and probable identification based on a single property (i.e., m/z) is not sufficient. Figure 5A shows the predicted CCS values for 12 different isobaric small molecules with the formula C6H13NO2 (Figure 5B) plotted versus their corresponding accurate masses. As the searched databases grow in size (i.e., by including ChemSpider or others), and as novel undocumented molecules are considered, this problem will only increase in magnitude (Figure 5C).
Figure 5. . Performance of the molecular modeling and quantum chemical calculation pipeline.
(A) A plot of calculated collision cross section (CCS) versus m/z for the protonated forms of the molecules shown in (B), illustrating the degree of orthogonality of the two metrics; (B) metabolite structures from the Human Metabolome Database corresponding to the formula C6H13NO2; (C) a plot of CCS versus m/z for 200 metabolites, out of the >11 k solved structures obtained from the Human Metabolome Database. Different colors correspond to CCSs for different metabolites with identical molecular formulae. The data for C6H13NO2 from (B) are outlined in the dashed box, and two anthropogenic chemicals with the formula C15H24O, 9-bisabolatrien-11-ol and α-3-copaen-8-ol are shown to have very different CCS; and (D) the theoretical CCS values obtained for selected +H, -H and +Na forms of 11 small molecules show good agreement with experimental CCS values derived from an SPE-ion mobility spectrometry (IMS)-MS platform, with average error of appproximately 2%. All CCS values were predicted using He as the buffer gas.
In terms of the required agreement between predicted and measured CCS values, the combination of instrumental measurement precision and accuracy of CCS prediction is critical to the success of the described workflow. As discussed above, extremely good instrumental precision (<0.5%) is now readily possible on individual instruments, and interlaboratory exercises indicate that DTIMS-MS instrumentation can provide reproducible consensus CCS values with an RSD of less than 1% [93,94] for a wide range of biologically relevant molecules, which is a marked improvement over raw chromatographic retention time matching across different laboratories. This latter point is particularly critical to the success of the proposed approach, as a sound understanding of the uncertainty of CCS measurements must be ascertained in order to improve the identification potential. In an initial evaluation, experimental and theoretical CCS values of selected ionized structures (protonated, deprotonated and sodiated) of 11 metabolites were compared, resulting in an average error of approximately 2% (Figure 5D) and indicating the potential of predicted CCS as a metric for supporting chemical identification in exposomic analysis.
The approach of using IMS to compare experimental predicted CCS values is not new and dates back to the early 1990s [116]. More recently, the community has been moving in the direction of routine implementation of IMS and CCS in their methods for chemical analysis on a larger scale. Paglia et al. integrated TWIMS into an LC-MS method and established a retention time, CCS and accurate mass database of 125 common metabolites; CCSs were then measured for the same molecules and using identical instrumentation in three independent laboratories with RSD < 5% for 99% of molecules studied [117]. The authors also used computational methods to predict CCSs for the same 125 metabolites and found good agreement with the experimentally determined CCSs, with R2 of 0.93. Stephen et al. coupled DTIMS with 1D and 2D LC-MS in untargeted analysis of wastewater samples and used an in-house CCS reference database containing 500 entries together with accurate mass to identify 22 and 53 different compounds, respectively [92].
Conclusion
In summary, comprehensive characterization of exposure is challenging due to the thousands of structurally and physicochemically diverse chemicals one encounters per day, and which may be present at any given time in some amount in body compartments. Complicating this, when employing metabolomics approaches, is the presence of the endogenous metabolome and its localized or systemic response to exposure. Thus, the potential chemical space that can be measured is vast, and untargeted data acquisition approaches are best suited to comprehensively measure the exposome.
Advancing measurement technologies, including those based on or incorporating DTIMS, are enabling increasingly higher coverage of sample molecular compositions. The volume of data corresponding to these new chemical observations will require new informatics methodologies to confidently assign as many identifications as possible. To address this challenge, we propose that accurate, computationally predicted CCS values can facilitate the broad identification of detected molecules in combination with accurate mass and MS/MS spectra, when available, and possibly with just accurate mass
Future perspective
Unsurprisingly, some analytical and data processing challenges remain for full-scale implementation of IMS in standard exposomics workflows, including dealing with the annotation of adducts and dimers separated by DTIMS in untargeted analysis, which need to be reconciled with the principal monoisotopic ion for correct annotation. Similarly, the nature of the DTIMS separation necessitates an ion gating or trap-and-release mechanism that entails a loss of duty cycle and (in the latter case) the possibility of losses of ions or interactions between different ions in the trapping environment. As such, careful analytical method development must be pursued in practice to validate the robustness and minimize the false annotation potential for exposome studies.
The next generation of IMS instrumentation: structures for lossless ion manipulations
A current limitation of DTIMS technology is its limited resolution for a given fixed length drift tube, and since resolution is proportional to the length of the device for a given weak electric field [118], making the devices longer than several meters is not practical. Recently, ultra-high resolution IMS devices were constructed with Structures for Lossless Ion Manipulations (SLIM), utilizing traveling waves. In their present form, SLIM are ion conduits formed by confining electric fields using a pair of parallel printed circuit boards. Radiofrequency potentials prevent ion losses to the circuit boards and direct current potentials prevent lateral ion losses. SLIM has separated isomers from multiple molecular classes, including peptides, natural products, lipids and oligosaccharides that have not been fully resolved with currently available IMS systems. In initial evaluations, SLIM has separated compounds with differences in CCS < 0.5% (Figure 6). Recently, a 13-m length, serpentine path SLIM device was shown to have about fivefold higher resolution separations than present commercially available DTIMS or TWIMS platforms [119]. SLIM research at Pacific Northwest National Laboratory currently focuses on multipass separations where it should be possible to obtain resolutions at least fivefold higher than the current SLIM, with a reasonable number of passes (<50), and a future vision includes km-long path length devices. Though the separation resolution is increased, the sensitivity of these SLIM measurements is maintained. In addition, when coupled with TOF MS, SLIM separations are still carried out on a rapid (∼1s) time scale, providing the opportunity for ultra-high throughput studies.
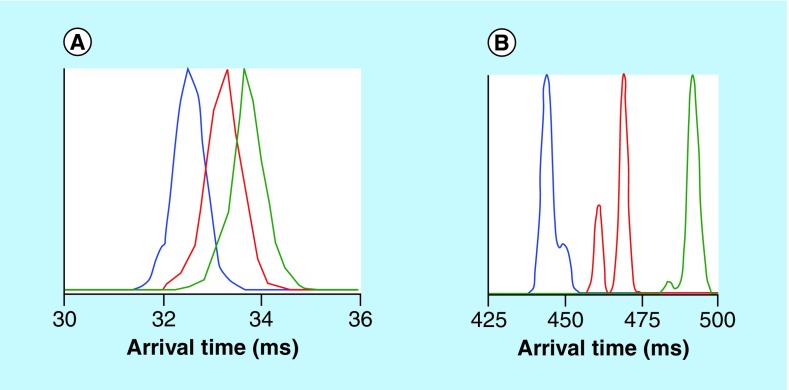
Figure 6. . Ion mobility spectrometry separations of a mixture of sodiated pentasaccharides.
The pentasacchsarides cellopentaose (blue, collision cross section [CCS] 250 Å2), maltopentaose (red, CCS 253 Å2) and mannopentaose (green, CCS 256 Å2) were measured as their sodiated forms using (A) an Agilent 6560 ion mobility spectrometry-quadrupole TOF and (B) a 31-m Structures for Lossless Ion Manipulations (SLIM)-TOF arrangement. The additional features (i.e., peak shoulders) can be putatively assigned to either different sodium cation binding locations or separation of α and β anomers. Estimating that roughly six peaks can fit in the SLIM-TOF separation space, the resolution of the measurement is at least less than half the difference between two of the sugars. The cellopentaose/maltopentaose and maltopentaose/mannopentaose pairs each differ by approximately 1% in CCS, and the cellopentaose/mannopentaose pair differs by approximately 2% in CCS. The additional features resolved in the SLIM separation show a resolution of at least less than 0.5% difference in CCS.
Traveling wave separations are the result of the abilities of ions to keep up with a direct current wave pushing them throughout the drift cell while undergoing collisions with the drift gas. Ions with larger CCS will be passed over by traveling waves more frequently than ions with smaller CCS. Due to this mechanism, the resulting separation is not linearly correlated to CCS, and the Mason Schamp equation does not hold. Thus, one caveat of TWIMS-based SLIM devices is that they do not allow for direct measurement of CCS. TWIMS does allow for calibration of CCS against standards that have been measured by DTIMS; however, calibrant ions should be from the same biomolecule class, should overlap in m/z space and should have the same charge states as the target ions, otherwise large errors in CCS (e.g., up to >5%) can result [82,120–123]. However, since accurate CCSs are measured from the DTIMS workflow, the identified compounds can be used as internal calibrants for SLIM measurements. Importantly, SLIM should allow CCS measurements to be made for many more mixture components and with greater precision due to the far greater resolution achieved [124], and potentially allowing CCS with much greater accuracy as better standards are developed. In this manner, small molecules that are not distinguishable by present DTIMS should be much better separated in SLIM-based IMS-MS and assigned with more accurate CCS values in a combined DTIMS/SLIM-MS pipeline, allowing more effective identification. Although the initial cost of devices and data management will be expensive, the time cost for measurement would be much lower than the traditional targeted/suspect screening using LC-MS/MS or GC-MS/MS analysis.
Executive summary.
Background
Human disease is a combination of individual genetic factors and nongenetic environmental factors.
The ‘exposome’ complements the genome and is the sum of all nongenetic exposures over a lifetime to chemical, social and biological agents.
Omics technologies can assist in characterizing the exposome by directly measuring chemical exposures and by inferring exposure based on biological signatures obtained by one or more complementary approaches.
As new analytical methodologies for measuring small molecules advance, there will be a need for computational approaches to rapidly and comprehensively identify chemicals and metabolites.
State-of-the-art in measuring the exposome
The technical approaches used to measure the exposome largely fall under one of two categories: targeted or untargeted.
Targeted analyses focus on a limited number of analytes, have high quantitative accuracy and low limits of quantification, but only measures a narrow snapshot of the sample molecular composition.
Untargeted analyses do not focus on a specific analyte but instead seek to comprehensively measure all analytes in a sample and offer the best opportunity to discover novel markers of exposure. Caveats include possible artifacts in the data due to the lack of optimization of sample preparation procedures, difficulty detecting very low-abundance analytes in the presence of high-abundance analytes and an incomplete representation of chemical space in spectral reference libraries.
NMR spectroscopy and LC-MS coupled with MS are typically employed in untargeted analysis of chemicals and metabolites.
Communicating the confidence in chemical identification can be a challenge and several systems exist for small molecules, including the Metabolomics Standards Initiative, an LC-high resolution MS/MS specific set from Eawag and many more. The essences of these are:
Confirmed identification with two orthogonal matching properties to an authentic reference standard measured in-house (MSI Level 1, Eawag Level 1).
Probable identification with all evidence indicating only one structure is possible, but authentic standard is not available for confirmation (Eawag Level 2a/b).
Putative annotation based on physicochemical properties and spectral matching (MSI Level 2, Eawag Level 2a).
Tentative identification/Putative compound class – tentative identification using predictive techniques, multiple structures are possible or insufficient evidence to eliminate other structures; substance class only is clear (MSI Level 3, Eawag Level 3).
Unknown compounds – molecular formula is unequivocal (Eawag Level 4) or exact mass only (Eawag Level 5; both MSI Level 4). These can be traced in samples and correspond to ‘detected features’ in the analyses, but the identity remains unclear.
Introducing ion mobility spectrometry as a new tool for the exposomics toolbox
Drift tube ion mobility spectrometry (DTIMS) shows great promise in small molecule measurements because it is able to directly determine molecular structural information.
The ability to resolve isomers that are difficult to distinguish using LC-MS alone is an inherent strength of DTIMS, particularly in small molecule analysis.
Because DTIMS instruments depend only on drift cell pressure, temperature and length, molecular collisional cross section (CCS) measurements are extremely reproducible. Furthermore, measurements from different instruments in different laboratories have also been compared and their values normally agree within <2% error, with recent DTIMS instruments yielding values with reproducibility precision of <1%.
The high reproducibility and speed of DTIMS allows it to be easily nested between LC and MS to provide additional separation power and dynamic range of detection in measuring the exposome.
When coupled with TOF time-of-flight MS, DTIMS-MS analysis can be ultra-high throughput, with a single ion mobility spectrometry (IMS) separation typically occurring in 10–100 ms.
A novel, automated SPE sample introduction system coupled with DTIMS-MS provides a 10-s sample-to-sample duty cycle and a theoretical maximum throughput of >8000 injections per day.
Accurate, computationally predicted CCS values can facilitate the broad identification of detected molecules in combination with accurate mass and MS/MS spectra, when available, and possibly with just accurate mass.
Future perspective
Analytical and data processing challenges remain for full-scale implementation of IMS in standard exposomics workflows.
Ultra-high resolution IMS devices constructed with Structures for Lossless Ion Manipulations are pushing the separation resolution possibilities of IMS.
Footnotes
Financial & competing interests disclosure
This work was supported by the Pacific Northwest National Laboratory (PNNL) Laboratory Directed Research and Development program and is a contribution of the Global Forensic Chemical Exposure Assessment for the Environmental Exposome project (JG Teeguarden and TO Metz) and the Microbiomes in Transition Initiative (RS Renslow and TO Metz), as well as the National Institutes of Health National Institute of Environmental Health Sciences grant R01ES022190) (ES Baker). EL Schymanski was supported in part by the SOLUTIONS project, funded by the European Union's Seventh Framework Programme for research, technological development and demonstration under Grant Agreement No. 603437. TJ Causon and S Hann would like to thank the Vienna Business Agency for funding, as well as EQ BOKU VIBT GmbH for providing mass spectrometry instrumentation. Research to develop Structures for Lossless Ion Manipulations (SLIM) was supported by the National Institutes of Health, National Institute of General Medical Sciences grant P41 GM103493 (RD Smith), and the US Department of Energy Office of Biological and Environmental Research via the Genome Science Program and is a contribution of the PNNL Pan-omics Program (RD Smith). The authors would like to thank Nathan Johnson for assistance in preparing figures. PNNL is a multiprogram national laboratory operated by Battelle for the DOE under Contract DE-AC05–76RLO 1830. Co-author RD Smith is a co-inventor on one of the patents licensed by Agilent for the 6560 IMS-QTOF MS instrument utilized for some of the work presented in this paper. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Attribution-NonCommercial-No Derivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0
References
Papers of special note have been highlighted as: •• of considerable interest
- 1.Wild CP. Complementing the genome with an ‘exposome’: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev. 2005;14(8):1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]; •• Introduces the concept of the exposome and the rationale for measuring it, citing the relative lack of expected progress from previous genome-based studies.
- 2.Wild CP. The exposome: from concept to utility. Int. J. Epidemiol. 2012;41(1):24–32. doi: 10.1093/ije/dyr236. [DOI] [PubMed] [Google Scholar]
- 3.Thomas D. Gene–environment-wide association studies: emerging approaches. Nat. Rev. Genet. 2010;11(4):259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norris SL, Nichols PJ, Caspersen CJ, et al. The effectiveness of disease and case management for people with diabetes: a systematic review. Am. J. Prev. Med. 2002;22(4):15–38. doi: 10.1016/s0749-3797(02)00423-3. [DOI] [PubMed] [Google Scholar]
- 5.Committee on Human and Environmental Exposure Science in the 21st Century. Exposure Science in the 21st Century: A Vision and a Strategy. The National Academies Press; Washington, DC: 2012. [PubMed] [Google Scholar]
- 6.Weis BK, Balshaw D, Barr JR, et al. Personalized exposure assessment: promising approaches for human environmental health research. Environ. Health Perspect. 2005;113(7):840–848. doi: 10.1289/ehp.7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson CH, Patterson AD, Idle JR, Gonzalez FJ. Xenobiotic metabolomics: major impact on the metabolome. Annu. Rev. Pharmacol. Toxicol. 2012;52:37–56. doi: 10.1146/annurev-pharmtox-010611-134748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bundy JG, Davey MP, Viant MR. Environmental metabolomics: a critical review and future perspectives. Metabolomics. 2008;5(1):3–21. [Google Scholar]
- 9.Vineis P, Khan AE, Vlaanderen J, Vermeulen R. The impact of new research technologies on our understanding of environmental causes of disease: the concept of clinical vulnerability. Environ. Health. 2009;8:54. doi: 10.1186/1476-069X-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Athersuch T. Metabolome analyses in exposome studies: profiling methods for a vast chemical space. Arch. Biochem. Biophys. 2016;589:177–186. doi: 10.1016/j.abb.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Ruddigkeit L, Awale M, Reymond JL. Expanding the fragrance chemical space for virtual screening. J. Cheminform. 2014;6:27. doi: 10.1186/1758-2946-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'hagan S, Kell DB. Understanding the foundations of the structural similarities between marketed drugs and endogenous human metabolites. Front. Pharmacol. 2015;6:105. doi: 10.3389/fphar.2015.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wishart DS. Advances in metabolite identification. Bioanalysis. 2011;3(15):1769–1782. doi: 10.4155/bio.11.155. [DOI] [PubMed] [Google Scholar]
- 14.Menikarachchi LC, Hill DW, Hamdalla MA, Mandoiu II, Grant DF. In silico enzymatic synthesis of a 400,000 compound biochemical database for nontargeted metabolomics. J. Chem. Inf. Model. 2013;53(9):2483–2492. doi: 10.1021/ci400368v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peironcely JE, Reijmers T, Coulier L, Bender A, Hankemeier T. Understanding and classifying metabolite space and metabolite-likeness. PLoS ONE. 2011;6(12):e28966. doi: 10.1371/journal.pone.0028966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senko MW, Beu SC, Mclaffertycor FW. Determination of monoisotopic masses and ion populations for large biomolecules from resolved isotopic distributions. J. Am. Soc. Mass Spectrom. 1995;6(4):229–233. doi: 10.1016/1044-0305(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 17.Metz TO, Zhang Q, Page JS, et al. The future of liquid chromatography–mass spectrometry (LC-MS) in metabolic profiling and metabolomic studies for biomarker discovery. Biomark. Med. 2007;1(1):159–185. doi: 10.2217/17520363.1.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein SE, Ausloos P, Lias SG. Comparative evaluations of mass spectral databases. J. Am. Soc. Mass Spectrom. 1991;2(5):441–443. doi: 10.1016/1044-0305(91)85012-U. [DOI] [PubMed] [Google Scholar]
- 19.Ausloos P, Clifton CL, Lias SG, et al. The critical evaluation of a comprehensive mass spectral library. J. Am. Soc. Mass Spectrom. 1999;10(4):287–299. doi: 10.1016/S1044-0305(98)00159-7. [DOI] [PubMed] [Google Scholar]
- 20.Smith CA, O'maille G, Want EJ, et al. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 2005;27(6):747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 21.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 2006;78(13):4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 22.Kind T, Wohlgemuth G, Lee DY, et al. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009;81(24):10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kind T, Liu KH, Lee Do Y, Defelice B, Meissen JK, Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods. 2013;10(8):755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bocker S, Rasche F. Towards de novo identification of metabolites by analyzing tandem mass spectra. Bioinformatics. 2008;24(16):i49–i55. doi: 10.1093/bioinformatics/btn270. [DOI] [PubMed] [Google Scholar]
- 25.Brouard C, Shen H, Duhrkop K, D'alche-Buc F, Bocker S, Rousu J. Fast metabolite identification with Input Output Kernel Regression. Bioinformatics. 2016;32(12):i28–i36. doi: 10.1093/bioinformatics/btw246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duhrkop K, Shen H, Meusel M, Rousu J, Bocker S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl Acad. Sci. USA. 2015;112(41):12580–12585. doi: 10.1073/pnas.1509788112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeffryes JG, Colastani RL, Elbadawi-Sidhu M, et al. MINEs: open access databases of computationally predicted enzyme promiscuity products for untargeted metabolomics. J. Cheminform. 2015;7:44. doi: 10.1186/s13321-015-0087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horai H, Arita M, Kanaya S, et al. MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010;45(7):703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 29.Wolf S, Schmidt S, Muller-Hannemann M, Neumann S. In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinformatics. 2010;11:148. doi: 10.1186/1471-2105-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerlich M, Neumann S. MetFusion: integration of compound identification strategies. J. Mass Spectrom. 2013;48(3):291–298. doi: 10.1002/jms.3123. [DOI] [PubMed] [Google Scholar]
- 31.Allen F, Pon A, Wilson M, Greiner R, Wishart D. CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res. 2014;42:W94–W99. doi: 10.1093/nar/gku436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eng JK, Mccormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 33.Craig R, Beavis RC. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 2003;17(20):2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J. Proteome Res. 2008;7(8):3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altman DG, Bland JM. Diagnostic tests. 1: sensitivity and specificity. BMJ. 1994;308(6943):1552. doi: 10.1136/bmj.308.6943.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cortejade A, Kiss A, Cren C, Vulliet E, Bulete A. Development of an analytical method for the targeted screening and multi-residue quantification of environmental contaminants in urine by liquid chromatography coupled to high resolution mass spectrometry for evaluation of human exposures. Talanta. 2016;146:694–706. doi: 10.1016/j.talanta.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 37.Davie-Martin CL, Hageman KJ, Chin YP, Nistor BJ, Hung H. Concentrations, gas-particle distributions, and sourceindicator analysis of brominated flame retardantsin air at Toolik Lake, Arctic Alaska. Environmental Science. Processes & Impacts. 2016;18:1274–1284. doi: 10.1039/c6em00395h. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez A, Avivar J, Cerda V. Estrogens determination in wastewater samples by automatic in-syringe dispersive liquid–liquid microextraction prior silylation and gas chromatography. J. Chromatogr. A. 2015;1413:1–8. doi: 10.1016/j.chroma.2015.08.031. [DOI] [PubMed] [Google Scholar]
- 39.Carroll JJ, Smith N, Babson AL. A colorimetric serum glucose determination using hexokinase and glucose-6-phosphate dehydrogenase. Biochem. Med. 1970;4(2):171–180. doi: 10.1016/0006-2944(70)90093-1. [DOI] [PubMed] [Google Scholar]
- 40.Rossini AA, Like AA, Chick WL, Appel MC, Cahill GF., Jr Studies of streptozotocin-induced insulitis and diabetes. Proc. Natl Acad. Sci. USA. 1977;74(6):2485–2489. doi: 10.1073/pnas.74.6.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vavra JJ, Deboer C, Dietz A, Hanka LJ, Sokolski WT. Streptozotocin, a new antibacterial antibiotic. Antibiot. Annu. 1959;7:230–235. [PubMed] [Google Scholar]
- 42.Luna B, Feinglos MN. Drug-induced hyperglycemia. JAMA. 2001;286(16):1945–1948. doi: 10.1001/jama.286.16.1945. [DOI] [PubMed] [Google Scholar]
- 43.O'connell SG, Haigh T, Wilson G, Anderson KA. An analytical investigation of 24 oxygenated-PAHs (OPAHs) using liquid and gas chromatography-mass spectrometry. Anal. Bioanal. Chem. 2013;405(27):8885–8896. doi: 10.1007/s00216-013-7319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rappaport SM, Barupal DK, Wishart D, Vineis P, Scalbert A. The blood exposome and its role in discovering causes of disease. Environ. Health Perspect. 2014;122(8):769–774. doi: 10.1289/ehp.1308015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dennis KK, Marder E, Balshaw DM, et al. Biomonitoring in the era of the exposome. Environ. Health Perspect. 2016 doi: 10.1289/EHP474. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ellis JK, Athersuch TJ, Thomas LD, et al. Metabolic profiling detects early effects of environmental and lifestyle exposure to cadmium in a human population. BMC Med. 2012;10:61. doi: 10.1186/1741-7015-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Go YM, Walker DI, Liang Y, et al. Reference standardization for mass spectrometry and high-resolution metabolomics applications to exposome research. Toxicol. Sci. 2015;148(2):531–543. doi: 10.1093/toxsci/kfv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Southam AD, Lange A, Al-Salhi R, Hill EM, Tyler CR, Viant MR. Distinguishing between the metabolome and xenobiotic exposome in environmental field samples analysed by direct-infusion mass spectrometry based metabolomics and lipidomics. Metabolomics. 2014;10(6):1050–1058. doi: 10.1007/s11306-014-0693-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics. 2013;9(Suppl. 1):S132–S143. doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson JM, Yu T, Strobel FH, Jones DP. A practical approach to detect unique metabolic patterns for personalized medicine. Analyst. 2010;135(11):2864–2870. doi: 10.1039/c0an00333f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao Y, Wang P, Shao G, Anzalota Del Toro LV, Codero J, Giese RW. Nontargeted analysis of the urine nonpolar sulfateome: a pathway to the nonpolar xenobiotic exposome. Rapid Commun. Mass Spectrom. 2016;30(21):2341–2350. doi: 10.1002/rcm.7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park YH, Lee K, Soltow QA, et al. High-performance metabolic profiling of plasma from seven mammalian species for simultaneous environmental chemical surveillance and bioeffect monitoring. Toxicology. 2012;295(1–3):47–55. doi: 10.1016/j.tox.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schymanski EL, Singer HP, Longree P, et al. Strategies to characterize polar organic contamination in wastewater: exploring the capability of high resolution mass spectrometry. Environ. Sci. Technol. 2014;48(3):1811–1818. doi: 10.1021/es4044374. [DOI] [PubMed] [Google Scholar]
- 54.Schymanski EL, Singer HP, Slobodnik J, et al. Non-target screening with high-resolution mass spectrometry: critical review using a collaborative trial on water analysis. Anal. Bioanal. Chem. 2015;407(21):6237–6255. doi: 10.1007/s00216-015-8681-7. [DOI] [PubMed] [Google Scholar]; •• Provides a ‘status quo’ of nontarget contaminant screening in Europe, and also provide perspectives for the future of environmental monitoring. Included are an overview of target, suspect and nontarget screening of data, a review of studies where these approaches were employed and a summary of the results of an interlab comparison of methods.
- 55.Ibanez M, Sancho JV, Mcmillan D, Rao R, Hernandez F. Rapid non-target screening of organic pollutants in water by ultraperformance liquid chromatography coupled to time-of-light mass spectrometry. Trends Analyt. Chem. 2008;27(5):481–489. [Google Scholar]
- 56.Hug C, Ulrich N, Schulze T, Brack W, Krauss M. Identification of novel micropollutants in wastewater by a combination of suspect and nontarget screening. Environ. Pollut. 2014;184:25–32. doi: 10.1016/j.envpol.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 57.Chiaia-Hernandez AC, Schymanski EL, Kumar P, Singer HP, Hollender J. Suspect and nontarget screening approaches to identify organic contaminant records in lake sediments. Anal. Bioanal. Chem. 2014;406(28):7323–7335. doi: 10.1007/s00216-014-8166-0. [DOI] [PubMed] [Google Scholar]
- 58.Larive CK, Barding GA, Jr, Dinges MM. NMR spectroscopy for metabolomics and metabolic profiling. Anal. Chem. 2014;87(1):133–146. doi: 10.1021/ac504075g. [DOI] [PubMed] [Google Scholar]
- 59.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007;26(1):51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagana Gowda GA, Gowda YN, Raftery D. Expanding the limits of human blood metabolite quantitation using NMR spectroscopy. Anal. Chem. 2014;87(1):706–715. doi: 10.1021/ac503651e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 2006;78(13):4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 62.Kopka J, Schauer N, Krueger S, et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21(8):1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- 63.Vinaixa M, Schymanski EL, Neumann S, Navarro M, Salek RM, Yanes O. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: state of the field and future prospects. Trends Analyt. Chem. 2016;78:23–35. [Google Scholar]
- 64.Hufsky F, Scheubert K, Bocker S. Computational mass spectrometry for small-molecule fragmentation. Trends Analyt. Chem. 2014;53:41–48. doi: 10.1186/1758-2946-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vaniya A, Fiehn O. Using fragmentation trees and mass spectral trees for identifying unknown compounds in metabolomics. Trends Analyt. Chem. 2015;69:52–61. doi: 10.1016/j.trac.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruttkies C, Schymanski EL, Wolf S, Hollender J, Neumann S. MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J. Cheminform. 2016;8:3. doi: 10.1186/s13321-016-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sumner LW, Amberg A, Barrett D, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3(3):211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wishart DS, Jewison T, Guo AC, et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singer HP, Wossner AE, Mcardell CS, Fenner K. Rapid screening for exposure to “non-target” pharmaceuticals from wastewater effluents by combining HRMS-based suspect screening and exposure modeling. Environ. Sci. Technol. 2016;50(13):6698–6707. doi: 10.1021/acs.est.5b03332. [DOI] [PubMed] [Google Scholar]
- 70.Moschet C, Piazzoli A, Singer H, Hollender J. Alleviating the reference standard dilemma using a systematic exact mass suspect screening approach with liquid chromatography-high resolution mass spectrometry. Anal. Chem. 2013;85(21):10312–10320. doi: 10.1021/ac4021598. [DOI] [PubMed] [Google Scholar]
- 71.NORMAN Suspect List Exchange. www.norman-network.com/?q=node/236
- 72.Schymanski EL, Jeon J, Gulde R, et al. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ. Sci. Technol. 2014;48(4):2097–2098. doi: 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- 73.Suhr H. Plasma Chromatography. Plenum Press; NY, USA: 1984. [Google Scholar]
- 74.Michelmann K, Silveira JA, Ridgeway ME, Park MA. Fundamentals of trapped ion mobility spectrometry. J. Am. Soc. Mass Spectrom. 2015;26(1):14–24. doi: 10.1007/s13361-014-0999-4. [DOI] [PubMed] [Google Scholar]
- 75.Ewing MA, Conant CRP, Zucker SM, Griffith KJ, Clemmer DE. Selected overtone mobility spectrometry. Anal. Chem. 2015;87(10):5132–5138. doi: 10.1021/ac504555u. [DOI] [PubMed] [Google Scholar]
- 76.Zucker SM, Ewing MA, Clemmer DE. Gridless overtone mobility spectrometry. Anal. Chem. 2013;85(21):10174–10179. doi: 10.1021/ac401568r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rus J, Moro D, Sillero JA, et al. IMS–MS studies based on coupling a differential mobility analyzer (DMA) to commercial API–MS systems. Int. J. Mass Spectrom. 2010;298(1–3):30–40. [Google Scholar]
- 78.Brown LJ, Creaser CS. Field asymmetric waveform ion mobility spectrometry analysis of proteins and peptides: a review. Curr. Anal. Chem. 2013;9(2):192–198. [Google Scholar]
- 79.Guevremont R. High-field asymmetric waveform ion mobility spectrometry: a new tool for mass spectrometry. J. Chromatogr. 2004;1058(1–2):3–19. [PubMed] [Google Scholar]
- 80.Kolakowski BM, Mester Z. Review of applications of high-field asymmetric waveform ion mobility spectrometry (FAIMS) and differential mobility spectrometry (DMS) Analyst. 2007;132(9):842–864. doi: 10.1039/b706039d. [DOI] [PubMed] [Google Scholar]
- 81.Vidal-De-Miguel G, Macía M, Cuevas J. Transversal modulation ion mobility spectrometry (TM-IMS), a new mobility filter overcoming turbulence related limitations. Anal. Chem. 2012;84(18):7831–7837. doi: 10.1021/ac301127u. [DOI] [PubMed] [Google Scholar]
- 82.Hines KM, May JC, Mclean JA, Xu L. Evaluation of collision cross section calibrants for structural analysis of lipids by traveling wave ion mobility-mass spectrometry. Anal. Chem. 2016;88(14):7329–7336. doi: 10.1021/acs.analchem.6b01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suhr H. Plasma chromatography. In: Carr TW, editor. Berichte der Bunsengesellschaft für physikalische Chemie. Plenum Press; NY, USA: 1984. p. 924. [Google Scholar]
- 84.Mason EA, Mcdaniel EW. Transport Properties of Ions in Gases. John Wiley and Sons; NY, USA: 1988. p. 35. [Google Scholar]
- 85.Revercomb HE, Mason EA. Theory of plasma chromatography gaseous electrophoresis – review. Anal. Chem. 1975;47(7):970–983. [Google Scholar]
- 86.Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH., Jr Ion mobility-mass spectrometry. J. Mass Spectrom. 2008;43(1):1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 87.May JC, Mclean JA. Ion mobility-mass spectrometry: time-dispersive instrumentation. Anal. Chem. 2015;87(3):1422–1436. doi: 10.1021/ac504720m. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Provides an overview of the historical developments in ion mobility spectrometry (IMS) and reviews basic concepts and characteristics of IMS (e.g., resolving power), and gives details on different types of IMS approaches.
- 88.Williams JP, Bugarcic T, Habtemariam A, et al. Isomer separation and gas-phase configurations of organoruthenium anticancer complexes: ion mobility mass spectrometry and modeling. J. Am. Soc. Mass Spectrom. 2009;20(6):1119–1122. doi: 10.1016/j.jasms.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 89.Wu C, Siems WF, Klasmeier J, Hill HH. Separation of isomeric peptides using electrospray ionization/high-resolution ion mobility spectrometry. Anal. Chem. 2000;72(2):391–395. doi: 10.1021/ac990601c. [DOI] [PubMed] [Google Scholar]
- 90.Kyle JE, Zhang X, Weitz KK, et al. Uncovering biologically significant lipid isomers with liquid chromatography, ion mobility spectrometry and mass spectrometry. Analyst. 2016;141(5):1649–1659. doi: 10.1039/c5an02062j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng X, Zhang X, Schocker N, et al. Enhancing glycan isomer separations with metal ions and positive and negative polarity ion mobility spectrometry-mass spectrometry. Anal. Bioanal. Chem. 2016 doi: 10.1007/s00216-016-9866-4. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stephan S, Hippler J, Kohler T, Deeb AA, Schmidt TC, Schmitz OJ. Contaminant screening of wastewater with HPLC-IM-qTOF-MS and LC+LC-IM-qTOF-MS using a CCS database. Anal. Bioanal. Chem. 2016;408(24):6545–6555. doi: 10.1007/s00216-016-9820-5. [DOI] [PubMed] [Google Scholar]; •• The first comprehensive application of collisional cross section (CCS), as measured using IMS, in contaminant monitoring in wastewaters. The authors show the power of using an in-house library of 500 experimentally determined CCS values from analysis of authentic standards.
- 93.Fjeldsted J, Kurulugama RT, Mordehai A, et al. 64th Annual Meeting of the American Society for Mass Spectrometry. San Antonio, TX, USA: June 5–9, 2016. Highly accurate collision cross section measurements for comprehensive high throughput applications. Presented at. [Google Scholar]
- 94.Causon TJ, Mairinger T, Hung LS, et al. 12th Annual Conference of the Metabolomics Society. Dublin, Ireland: June 27–30, 2016. Addition of drift-tube ion mobility to liquid chromatography-mass spectrometry workflows: examining the potential for cellular metabolomics. Presented at. [Google Scholar]
- 95.Wyttenbach T, Bowers MT. Gas-phase conformations: the ion mobility/ion chromatography method. Mod. Mass Spectrom. 2003;225:207–232. [Google Scholar]
- 96.Baker ES, Clowers BH, Li F, et al. Ion mobility spectrometry-mass spectrometry performance using electrodynamic ion funnels and elevated drift gas pressures. J. Am. Soc. Mass Spectrom. 2007;18(7):1176–1187. doi: 10.1016/j.jasms.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu X, Valentine SJ, Plasencia MD, Trimpin S, Naylor S, Clemmer DE. Mapping the human plasma proteome by SCX-LC-IMS-MS. J. Am. Soc. Mass Spectrom. 2007;18(7):1249–1264. doi: 10.1016/j.jasms.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baker ES, Livesay EA, Orton DJ, et al. An LC-IMS-MS platform providing increased dynamic range for high-throughput proteomic studies. J. Proteome Res. 2010;9(2):997–1006. doi: 10.1021/pr900888b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Three-dimensional ion mobility/TOFMS analysis of electrosprayed biomolecules. Anal. Chem. 1998;70(11):2236–2242. doi: 10.1021/ac980059c. [DOI] [PubMed] [Google Scholar]
- 100.Baker ES, Zhang X, Burnum-Johnson KE, et al. The Pittsburgh Conference and Exposition. Atlanta, GA, USA: March 6–10, 2016. Enhancing ion mobility-mass spectrometry metabolomic analyses with high throughput front end separations. Presented at. [Google Scholar]
- 101.Baker ES, Zhang X, Zheng X, et al. 64th Annual Meeting of the American Mass Spectrometry Society. San Antonio, TX, USA: June 5–9, 2016. Automated solid phase extractions coupled with ion mobility-mass spectrometry analyses allow rapid metabolomic screening of complex plasma and urine samples. Presented at. [Google Scholar]
- 102.Kangas LJ, Metz TO, Isaac G, et al. In silico identification software (ISIS): a machine learning approach to tandem mass spectral identification of lipids. Bioinformatics. 2012;28(13):1705–1713. doi: 10.1093/bioinformatics/bts194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bauer CA, Grimme S. First principles calculation of electron ionization mass spectra for selected organic drug molecules. Org. Biomol. Chem. 2014;12(43):8737–8744. doi: 10.1039/c4ob01668h. [DOI] [PubMed] [Google Scholar]
- 104.Heller S, Mcnaught A, Stein S, Tchekhovskoi D, Pletnev I. InChI – the worldwide chemical structure identifier standard. J. Cheminform. 2013;5(1):7. doi: 10.1186/1758-2946-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.O'boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J. Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Halgren TA. Merck molecular force field. 1. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996;17(5–6):490–519. [Google Scholar]
- 107.Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012;4(1):17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Valiev M, Bylaska EJ, Govind N, et al. NWChem: a comprehensive and scalable open-source solution for large scale molecular simulations. Comput. Phys. Commun. 2010;181(9):1477–1489. [Google Scholar]
- 109.Hehre WJ, Ditchfie R, Pople JA. Self-consistent molecular-orbital methods. 12. Further extensions of gaussian-type basis sets for use in molecular-orbital studies of organic-molecules. J. Chem. Phys. 1972;56(5):2257. [Google Scholar]
- 110.Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ. Ab-initio calculation of vibrational absorption and circular-dichroism spectra using density-functional force-fields. J. Phys. Chem. 1994;98(45):11623–11627. [Google Scholar]
- 111.Becke AD. Density-functional thermochemistry. 3. The role of exact exchange. J. Chem. Phys. 1993;98(7):5648–5652. [Google Scholar]
- 112.Shvartsburg AA, Jarrold MF. An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chem. Phys. Lett. 1996;261(1–2):86–91. [Google Scholar]
- 113.Campuzano I, Bush MF, Robinson CV, et al. Structural characterization of drug-like compounds by ion mobility mass spectrometry: comparison of theoretical and experimentally derived nitrogen collision cross sections. Anal. Chem. 2012;84(2):1026–1033. doi: 10.1021/ac202625t. [DOI] [PubMed] [Google Scholar]
- 114.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. Structural information from ion mobility measurements: effects of the long-range potential. J. Phys. Chem. 1996;100(40):16082–16086. [Google Scholar]
- 115.Marklund EG, Degiacomi MT, Robinson CV, Baldwin AJ, Benesch JL. Collision cross sections for structural proteomics. Structure. 2015;23(4):791–799. doi: 10.1016/j.str.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 116.Von Helden G, Kemper PR, Gotts NG, Bowers MT. Isomers of small carbon cluster anions: linear chains with up to 20 atoms. Science. 1993;259(5099):1300–1302. doi: 10.1126/science.259.5099.1300. [DOI] [PubMed] [Google Scholar]
- 117.Paglia G, Williams JP, Menikarachchi L, et al. Ion mobility derived collision cross sections to support metabolomics applications. Anal. Chem. 2014;86(8):3985–3993. doi: 10.1021/ac500405x. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Demonstrates the utility of CCS in characterization of small molecules, and report the first comparison of experimental and theoretical CCS in a metabolomics workflow.
- 118.Rokushika S, Hatano H, Baim MA, Hill HH. Resolution measurement for ion mobility spectrometry. Anal. Chem. 1985;57(9):1902–1907. [Google Scholar]
- 119.Deng L, Ibrahim YM, Hamid AM, et al. Ultra-high resolution ion mobility separations utilizing traveling waves in a 13-m serpentine path length structures for lossless ion manipulations module. Anal. Chem. 2016;88(18):8957–8964. doi: 10.1021/acs.analchem.6b01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Knapman TW, Berryman JT, Campuzano I, Harris SA, Ashcroft AE. Considerations in experimental and theoretical collision cross-section measurements of small molecules using travelling wave ion mobility spectrometry-mass spectrometry. Int. J. Mass spectrom. 2010;298(1–3):17–23. [Google Scholar]
- 121.Ridenour WB, Kliman M, Mclean JA, Caprioli RM. Structural characterization of phospholipids and peptides directly from tissue sections by MALDI traveling-wave ion mobility-mass spectrometry. Anal. Chem. 2010;82(5):1881–1889. doi: 10.1021/ac9026115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal. Chem. 2010;82(22):9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 123.Gelb AS, Jarratt RE, Huang Y, Dodds ED. A study of calibrant selection in measurement of carbohydrate and peptide ion-neutral collision cross sections by traveling wave ion mobility spectrometry. Anal. Chem. 2014;86(22):11396–11402. doi: 10.1021/ac503379e. [DOI] [PubMed] [Google Scholar]
- 124.Deng L, Ibrahim YM, Hamid AM, et al. Ultra-high resolution ion mobility separations utilizing traveling waves in a 13-m serpentine path length structures for lossless ion manipulations module. Anal. Chem. 2016;88(18):8957–8964. doi: 10.1021/acs.analchem.6b01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Metz TO, Page JS, Baker ES, et al. High resolution separations and improved ion production and transmission in metabolomics. Trends Analyt. Chem. 2008;27(3):205–214. doi: 10.1016/j.trac.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]