Abstract
The aim of this study was to identify virulence genes and antimicrobial resistance of Escherichia coli isolated from bovine clinical mastitis in dairy herds in Iran. Sampling was done from 86 inflamed quarters of dairy cows in 8 commercial farms of Alborz province, Iran in summer 2015. Shiga toxin-producing E. coli (STEC) virulence genes were detected by multiplex PCR and multi-drug resistance profiles were confirmed using disk diffusion method. Among 60 E. coli isolated from examined samples, 13 (21.6%) of them were STEC. The results of PCR assay showed that eaeA gene was carried by 4 (30.8%) of STEC isolates. Although stx1 in combination with eaeA gene was detected from 7 (53.8%) of STEC isolates, stx1 and stx2 genes were detected from only 1 (7.7%) of the examined samples. The result of the disk diffusion method showed that all E. coli isolates were resistant to penicillin, tylosin, oxytetracycline, erythromycin, ampicillin, streptomycin and neomycin. However all isolates were susceptible to enrofloxacin. Therefore, according to the results establishing a regular monitoring system for identification of cases with clinical mastitis and conducting antibiotic sensitivity tests are recommended.
Key Words: Antimicrobial resistance, Clinical mastitis, E. coli, STEC, Virulence factors
Introduction
There are several reports related to the incidence of bovine mastitis caused by Escherichia coli around the world (Bradley and Green, 2001 ▶). According to the previous studies Shiga toxin-producing E. coli (STEC) strains are an important group for mastitis (Kobori et al., 2004 ▶; Guler and Gündüz, 2007 ▶). Identification of virulence factors of E. coli isolated from bovine clinical mastitis has been conducted in numerous previous investigations (Wenz et al., 2006 ▶). These studies demonstrated that Shiga toxins (Stx1, Stx2) and eae (intimin) are the most significant virulence genes in E. coli strains isolated from bovine clinical mastitis (Momtaz et al., 2012 ▶). Antibiotic therapy is the common treatment for bovine clinical mastitis. Widespread use of antimicrobials in farm animals has resulted in a considerable rise of antimicrobial-resistant strains of bacteria which can increase treatment cost and period (Sawant et al., 2007 ▶).
Therefore, the aim of this study was to identify virulence genes and antimicrobial resistance of E. coli isolated from bovine clinical mastitis in dairy herds in Iran.
Materials and Methods
Sample collection
Sampling was performed from 86 inflamed quarters of dairy cows in eight commercial farms of Alborz province, Iran in summer 2015.
DNA extraction
Overnight cultures of the bacteria in 2 ml nutrient broth were centrifuged for 5 min at 5,000 rpm. The bacterial pellet was re-suspended in 200 μL of distilled water and boiled for 10 min. Tubes were centrifuged again, and the supernatant was used as template DNA (Pourtaghi et al., 2013 ▶; Pourtaghi and Sodagari, 2016 ▶).
STEC detection
PCR was performed on the samples to detect the presence of the stx1, stx2 and eaeA genes. The primer sets and related genes that encode virulence genes and PCR condition to amplification are described in Table 1. Amplification reactions were performed in a total volume of 25 μL containing 2.5 μL of 10 × PCR buffer, 0.5 mM Mgcl2, 250 μM dNTP, 1 μM of each primer and 0.5 U Taq DNA polymerase. To determine molecular weight, 100 bp DNA ladder (Fermentas) was used.
Table 1.
Primers used for PCR and DNA sequencing
| Virulence factor | Primer sequence 5´-3´ | Position in open reading frame | Size of product | References |
|---|---|---|---|---|
| Stx1 | TTCGCTCTGCAATAGGTA | 125-142 of A subunit | 555* | Frank et al. 1998 |
| TTCCCCAGTTCAATGTAAGAT | 659-679 of A subunit | |||
| Stx2 | GTGCCTGTTACTGGTTTTTCTTC | 30-53 of A subunit | 118 | Frank et al. 1998 |
| AGGGGTCGATATCTCTGTCC | 128-147 of A subunit | |||
| eaeA | ATATCCGTTTTAATGGCTATCT | 992-1013 of eaeA | 425 | Frank et al. 1998 |
| AATCTTCTGCGTACTGTGTTCA | 1395-1416 of eaeA |
PCR condition: 35 × (94°C for 60 s, 50°C for 60 s, 72°C for 90 s)
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by using the disk diffusion method and interpreted according to the standards recommended by CLSI, for the following antimicrobial agents including ampicillin (10 μg), ceftifor (30 μg), colistin (10 μg), erythromycin (15 μg), florfenicol (30 μg), gentamicin (10 μg), lincospectin (15/200 μg), neomycin (30 μg), tetracyclin (30 μg), penicillin (10 IU), sulfamethoxazole/ trimethoprim (1.25/23.75 μg), streptomycin (10 μg), tylosin (30 μg), and enrofloxacin (5 μg). The results were interpreted in accordance with interpretive criteria provided by CLSI (2008). Escherichia coli ATCC 25922 was used as a quality control strain.
Results
As shown in Table 2 of the 60 E. coli isolated from examined samples, 13 (21.6%) of them were STEC. The results of PCR assay showed that eaeA gene was carried by 4 (30.8%) of the STEC isolates. Although stx1 in combination with eaeA gene were detected from 7 (53.8%) of STEC isolates, stx1 and stx2 genes were detected from only 1 (7.7%) of the examined samples. Furthermore, the present study revealed that there was only 1 (7.7%) STEC isolate that possessed all three identified virulence genes (Table 2). Figure 1 shows the virulence genes of some STEC isolates. The result of the disk diffusion method showed that all E. coli isolates were resistant to penicillin, tylosin, oxytetracyclin, erythromycin, ampicillin, streptomycin and neomycin. However, all isolates indicated susceptibility to enrofloxacin (Table 3). Additionally, multi-drug resistance was found among E. coli isolates (Table 3). Sixteen and eight multi-drug resistance patterns were observed for E. coli and STEC isolates respectively. According to the results, the pattern number 15 in Table 3 demonstrated the highest rate of multi-drug resistance.
Table 2.
Frequency of STEC virulence genes
| Virulence gene(s) | Number (%) |
|---|---|
| stx1 | 0 (0) |
| stx2 | 0 (0) |
| eaeA | 4 (30.8) |
| stx1 and stx2 | 1 (7.7) |
| stx1 and eaeA | 7 (53.8) |
| stx2 and eaeA | 0 (0) |
| stx1, stx2 and eaeA | 1 (7.7) |
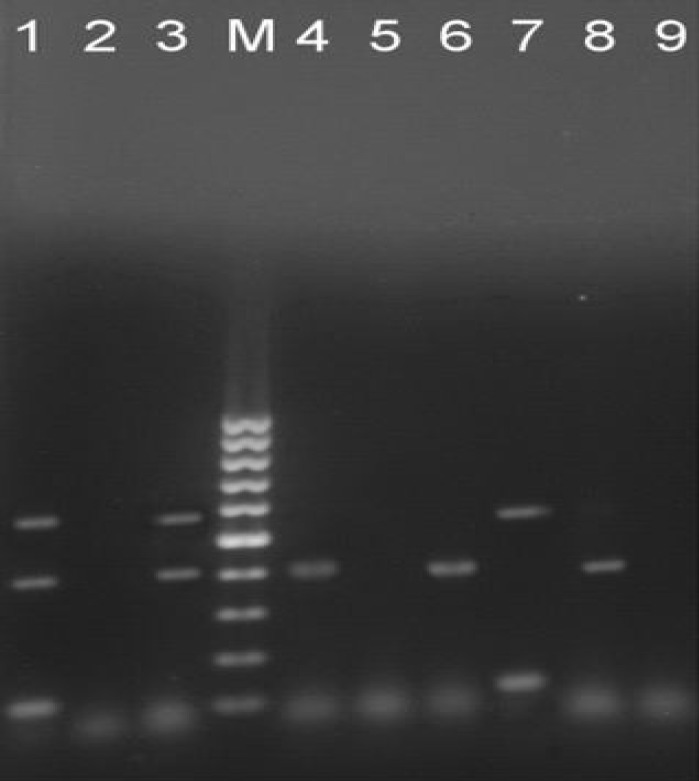
Fig. 1.
Agarose (1%) gel electrophoresis of STEC PCR products of virulence factors genes. Lane M: 100 bp DNA marker. Lane 1: Positive isolate for stx1, eaeA and stx2 (555, 425 and 118 bp), Lanes 2, 5 and 9: Negative control and negative isolates, Lane 3: Positive isolate for stx1 and eaeA, Lanes 4, 6 and 8: Positive isolates for eaeA, and Lane 7: Positive isolate for eaeA and stx2
Table 3.
Multi-drug resistance patterns in 60 E. coli isolated from clinical mastitis
| Number | Antibiotics resistance patterns | Number (%) of multi-resistant isolates |
|
|---|---|---|---|
| All E. coli isolates | STEC isolates | ||
| 1 | P, Ty, Ot, E, Am, S, N, Sxt | 2 (3.3%) | 0 (0%) |
| 2 | P, Ty, Ot, E, Am, S, N, Sxt, Ls | 1 (1.7%) | 0 (0%) |
| 3 | P, Ty, Ot, E, Am, S, N, FF, Ls | 3 (5%) | 2 (15.4%) |
| 4 | P, Ty, Ot, E, Am, S, N, Gm, Ls | 2 (3.4%) | 0 (0%) |
| 5 | P, Ty, Ot, E, Am, S, N, Sxt, Cf, Cl | 4 (6.7%) | 1 (7.7%) |
| 6 | P, Ty, Ot, E, Am, S, N, FF, Ls, Cl | 2 (3.3%) | 1 (7.7%) |
| 7 | P, Ty, Ot, E, Am, S, N, FF, Ls, Cf | 5 (8.3%) | 0 (0%) |
| 8 | P, Ty, Ot, E, Am, S, N, Gm, FF, Cf | 2 (3.3%) | 0 (0%) |
| 9 | P, Ty, Ot, E, Am, S, N, Sxt, FF, Ls, Cf | 3 (5%) | 1 (7.7%) |
| 10 | P, Ty, Ot, E, Am, S, N, Sxt, Gm, FF, Cf | 5 (8.3%) | 1 (7.7%) |
| 11 | P, Ty, Ot, E, Am, S, N, Sxt, Gm, FF, Ls | 3 (5%) | 0 (0%) |
| 12 | P, Ty, Ot, E, Am, S, N, Sxt, Gm, Cf, Cl | 4 (6.7%) | 1 (7.7%) |
| 13 | P, Ty, Ot, E, Am, S, N, Gm, FF, Ls, Cl | 2 (3.3%) | 0 (0%) |
| 14 | P, Ty, Ot, E, Am, S, N, Sxt, Gm, FF, Ls, Cf | 4 (6.7%) | 0 (0%) |
| 15 | P, Ty, Ot, E, Am, S, N, Sxt, Gm, FF, Cf, Cl | 16 (26.7%) | 5 (38.4%) |
| 16 | P, Ty, Ot, E, Am, S, N, Gm, FF, Ls, Cf, Cl | 2 (3.3%) | 1 (7.7%) |
P: Penicillin, Ty: Tylosin, Ot: Oxytetracyclin, E: Erythromycin, Am: Ampicillin, S: Streptomycin, N: Neomycin, Sxt: Sulfamethoxazole-trimethoprim, Gm: Gentamicin, FF: Florfenicol, Ls: Lincospectin, Cf: Ceftifor, and Cl: Colistin
Discussion
In the present study, 13 (21.6%) of the E. coli isolates were STEC which was higher than the previous report in Iran (Mansouri-Najand and Khalili, 2007 ▶) but lower than those found in Belgium (Vivegnis et al., 1999 ▶) and another investigation in Iran (Momtaz et al., 2012 ▶). According to the PCR assay the eaeA gene was carried by 4 (30.8%) of the STEC isolates. Higher result (33.3%) was reported in previous study in Iran (Momtaz et al., 2010 ▶). Numerous previous investigations have proved that there is a direct relationship between the presence of eaeA gene and the capacity of STEC to cause severe human disease, especially HUS (Beutin et al., 2004 ▶). Furthermore, the results indicated that stx1 and stx2 genes were detected from only 1 (7.7%) of the examined samples which was much lower than that found in the study conducted by Momtaz (2010) ▶. Another investigation on mastitis milk samples demonstrated that stx1 gene was the predominant virulence factor with a prevalence of 31% and 35%, respectively (Seyda et al., 2014 ▶), while according to our results this virulence gene was carried by none of the STEC isolates. The significant difference between the reports was associated with the origin of the strains (Seyda et al., 2014 ▶). Moreover, the result of the disk diffusion method showed that all E. coli isolates were resistant to penicillin, tylosin, oxytetracyclin, erythromycin, ampicillin, streptomycin and neomycin. However in contrast with Seyda et al.’s study (2014) ▶, all isolates indicated susceptibility to enrofloxacin. It reveals that enrofloxacin is less applied in farm animal medicine in Iran and can be more effective for treatment of clinical mastitis caused by E. coli. Similar to our study, resistance to some of the above antibiotics has also been frequently reported in several previous investigations. However, the results of these studies showed lower resistance rate than our findings (Stephan et al., 2008 ▶; Momtaz et al., 2012 ▶; Dubravka et al., 2015 ▶). Multi-drug resistance was found among all E. coli isolates (Table 3). In addition to the present study, multiple antibiotic resistances have been indicated in numerous previous researches around the world (Rangel and Marin, 2009 ▶; Spnu et al., 2012 ▶). The high rate of antimicrobial resistance identified in this study can be explained by the widespread use of common antimicrobials in dairy farms for treatment of clinical mastitis. Our findings, similar to previous investigations indicated that cattle with clinical mastitis are recognized as the main reservoir of STEC. Therefore, establishing a regular monitoring system for identification of cases with clinical mastitis, restriction of widespread use of common antibiotics as well as conducting antibiotic sensitivity tests is recommended for reducing the prevalence of resistant strains of STEC in industrial dairy herds.
Conflict of interest
No conflict of interests is declared.
References
- Beutin, L, Krause, G, Zimmermann, S, Kaulfuss, S, Gleier, K. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 2004;42:1099–1108. doi: 10.1128/JCM.42.3.1099-1108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, AJ, Green, MJ. Etiology of clinical mastitis in six somerset dairy herds. Vet. Rec. 2001;148:683–686. doi: 10.1136/vr.148.22.683. [DOI] [PubMed] [Google Scholar]
- CLSI . Performance and standards for antimicrobial disk diffusion and dilution susceptibility tests for bacteria isolated from animals; approved standards. 3rd Ed. Pennsylvania, USA: M31-A3. Clinical and Laboratory Standard Institute; 2008. pp. M31–A3. [Google Scholar]
- Dubravka M, Bojana P, Maja V, Dalibor T, Vladimir P. Investigation of biofilm formation and phylogenetic typing of Escherichia coli stains isolated from milk of cows with mastitis. Acta Vet. Beograd. 2015;65:202–216. [Google Scholar]
- Frank SM, Bosworth BT, Moon HW. Multiplex PCR for enterotoxigenic, attaching and effacing, and Shiga toxin-producing Escherichia coli strains from calves. J. Clin. Microbiol. 1998;36:1795–1797. doi: 10.1128/jcm.36.6.1795-1797.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler L, Gündüz K. Virulence properties of Escherichia coli isolated from clinical bovine mastitis. Turk. J. Vet. Anim. Sci. 2007;31:361–365. [Google Scholar]
- Kobori D, Rigobelo EC, Macedo C, Marin JM, Avila FA. Virulence properties of Shiga toxin-producing Escherichia coli isolated from cases of bovine mastitis in Brazil. Rev. Elev. Med. Vet. Pays. Trop. 2004;57:15–20. [Google Scholar]
- Mansouri-Najand L, Khalili M. Detection of Shiga-like toxigenic Escherichia coli from raw milk cheeses produced in Kerman-Iran. Veterinarski. Arhiv. 2007;77:515–522. [Google Scholar]
- Momtaz H. Investigation of virulence factors in Escherichia coli isolated from clinical and subclinical bovine mastitis. Bulg. J. Vet. Med. 2010;13:122–126. [Google Scholar]
- Momtaz H, Dehkordi SF, Taktaz T, Rezvani A, Yarali S. Shiga toxin-producing Escherichia coli isolated from bovine mastitic milk: serogroups, virulence factors, and antibiotic resistance properties. Sci. World J. 2012;2012:618709. doi: 10.1100/2012/618709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourtaghi H, Dahpahlavan V, Momtaz H. Virulence genes in Escherichia coli isolated from calves with diarrhoea in Iran. Com. Clin. Pathol. 2013;12:513–515. [Google Scholar]
- Pourtaghi H, Sodagari HR. Antimicrobial resistance of entrotoxigenic and non-entrotoxigenic Escherichia coli isolated from diarrheic calves in Iran. Int. J. Enteric. Pathog. 2016;4:e34557. [Google Scholar]
- Rangel P, Marin JM. Analysis of Escherichia coli isolated from bovine mastitic milk. Pesq. Vet. Bras. 2009;29:363–368. [Google Scholar]
- Sawant AA, Hegde NV, Straley BA, Donaldson SC, Love BC, Knabel SJ, Jayarao BM. Antimicrobial-resistant enteric bacteria from dairy cattle. Appl. Environ. Microbiol. 2007;73:156–163. doi: 10.1128/AEM.01551-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyda C, Gökçen D, Ünlü SM. Detection of several virulence properties, antibiotic resistance and phylogenetic relationship in E coli isolated from cow mastitis. Acta Vet. Beograd. 2014;64:413–425. [Google Scholar]
- Spnu M, Kobolkuti L, Cadar D, Niculae M, Bianu G, Popescu S, Lukacs L. Changes in antibiotic resistance indices of animal Escherichia coli strains with number of isolates. Ann. Rom. Soc. Cell. Biol. 2012;17:361–366. [Google Scholar]
- Stephan R, Schumacher S, Corti S, Krause G, Danuser J, Beutin L. Prevalence and characteristics of Shiga toxin producing Escherichia coli in Swiss raw milk cheeses collected at producer level. J. Dairy. Sci. 2008;91:2561–2565. doi: 10.3168/jds.2008-1055. [DOI] [PubMed] [Google Scholar]
- Vivegnis J, Lioui EM, Leclercq A, Lambert B, Decallonne J. Detection of Shiga-like toxin producing Escherichia coli from raw milk cheeses produced in Wallonia. Biotechnol. Agron. Soc. Environ. 1993;3:159–164. [Google Scholar]
- Wenz JR, Barrington GM, Garry FB, Ellis RP, Magnuson RJ. Escherichia coli isolates’ serotypes, genotypes, and virulence genes and clinical coliform mastitis severity. J. Dairy. Sci. 2006;89:3408–3412. doi: 10.3168/jds.S0022-0302(06)72377-3. [DOI] [PubMed] [Google Scholar]