Abstract
Background:
Nontuberculous mycobacteria (NTM) are a group of opportunistic pathogens and these are widely dispersed in water and soil resources. Identification of mycobacteria isolates by conventional methods including biochemical tests, growth rates, colony pigmentation, and presence of acid-fast bacilli is widely used, but these methods are time-consuming, labor-intensive, and may sometimes remain inconclusive.
Materials and Methods:
The DNA was extracted from NTM cultures using CTAB, Chelex, Chelex + Nonidet P-40, FTA® Elute card, and boiling The quantity and quality of the DNA extracted via these methods were determined using UV-photometer at 260 and 280 nm, and polymerase chain reaction (PCR) amplification of the heat-shock protein 65 gene with serially diluted DNA samples.
Results:
The CTAB method showed more positive results at 1:10–1:100,000 at which the DNA amount was substantial. With the Chelex method of DNA extraction, PCR amplification was detected at 1:10 and 1:1000 dilutions.
Conclusions:
According to the electrophoresis results, the CTAB and Chelex DNA extraction methods were more successful in comparison with the others as regard producing suitable concentrations of DNA with the minimum use of PCR inhibitor.
Keywords: DNA extraction methods, heat-shock protein 65, nontuberculous mycobacteria, polymerase chain reaction
Introduction
Nontuberculous mycobacteria (NTM) are a group of environmental organisms commonly found in water and soil resources.[1,2,3,4] These opportunistic pathogens are a major cause of diseases in high-risk individuals such as immunocompromised or human immunodeficiency virus-infected patients, particularly in developed countries.[5,6,7] The reported incidence of nontuberculous mycobacterial disease appears to be increasing worldwide because of the spread of immunodeficiency diseases.[1,8,9,10,11] Identification of mycobacteria isolates by conventional methods including biochemical tests, growth rates, colony pigmentation, and presence of acid-fast bacilli is widely used, but these methods are time-consuming, labor-intensive, and may sometimes remain inconclusive.[5,8,9,12] Molecular methods have proved to be useful and more sensitive alternatives for species identification of mycobacteria.[9,12,13] DNA extraction is a routine step in many biological studies including molecular identification, phylogenetic inference, genetics, and genomics.[14,15] A variety of methods have been established to isolate DNA molecules from biological materials.[13,15] Different methods have various effects on DNA extraction.[16] For a successful polymerase chain reaction (PCR)-based identification, application of a suitable method for DNA extraction is necessary.[17] Due to the presence of lipid complex in the cell wall, disruption of the cell wall is an essential but difficult step in extracting DNA from mycobacteria.[13] Several methods for mycobacterial cell wall lysis and DNA extraction have been evaluated.[13] As the heat-shock protein 65 (hsp65) gene is expressed in all mycobacteria species, it would be a suitable target at the species level.[18] The purpose of this study was to compare five methods of DNA extraction including CTAB-Chloroform, FTA® Elute card, Chelex 100, Chelex 100+ Nonidet P-40, and boiling.
Materials and Methods
Bacteria
Four NTM strains consisting of one NTM clinical isolate, Mycobacterium gordonae, and three saprophytic isolates; Mycobacterium chelonae, M. gordonae, and Mycobacterium fortuitum (ATCC 49403) supplied by the Tuberculosis Center in Isfahan, Iran, were used as the model organisms with difficult-to-lyse cell walls in this study. A pure culture of NTMs was obtained using Middlebrook 7H10 agar plates, and a suspension of mycobacteria cells was prepared in sterile double-distilled water and adjusted to a 1.0 McFarland standard density. Five-fold serial dilutions were prepared in sterile phosphate-buffered saline, aliquoted, and stored at −20°C until use. Each dilution series also included a negative control, i.e., PBS without added bacteria. All were subjected to each of the five DNA extraction methods.
Extraction methods
Method 1: CTAB-chloroform
This method was performed according to De Almeida et al.[14] Briefly, a total of 500 μL of each dilution of the cell suspensions was transferred into lysozyme (10 mg/ml) and incubated at 37°C for 1 h. Then, proteinase K and sodium dodecyl sulfate 10% were added, and the suspension was incubated at 65°C for 10 min. Subsequently, a solution consisting of a mixture of NaCl and CTAB (NaCl 5 M and CTAB/NaCl) was added, and the suspension was incubated for 10 min at 65°C. DNA was then extracted with chloroform/isoamyl alcohol (24:1). The supernatant was transferred to a new tube and isopropanol was added. The suspension was kept at −20°C for 30 min and centrifuged for 15 min at 14,000 g. The pellet was washed with 1 mL of 70% ethanol and centrifuged for 5 min at 14,000 g, and 30 μL of TE buffer was added and used in PCR.
Method 2: FTA® Elute card
Five microliters of each dilution of mycobacteria suspension was added to a disc of FTA® Elute card (Cat. No. W B120410; Whatman® Inc., Florham Park, NJ, USA) measuring 5 mm in diameter which was then allowed to dry completely. The cards were washed rapidly with 500 μl of sterile water and immersed in a 0.2 ml PCR tube containing 30 μl of sterile water and incubated at 95°C for 20 min. The disc was removed, and the DNA was used in PCR.[19]
Method 3: Chelex 100+ Nonidet P-40
Five hundred microliters of each dilution of mycobacteria suspension was added to a solution containing 5% Chelex-100, 1% Nonidet P-40, 1% Tween 20, and 96 μL distilled water and incubated at 100°C for 30 min. Subsequently, the tubes were centrifuged for 10 min at 13,000 g, and the solution was transferred to a new tube as DNA and used in PCR.[14]
Method 4: Chelex 100
An aliquot of 500 μL of each dilution of mycobacteria suspension was added to tubes containing Chelex-100 and incubated at 100°C for 20 min. The samples were then centrifuged at 12,000 g for 15 min, and the supernatant was transferred to a new tube as DNA for application in PCR.[14]
Method 5: Boiling
Five hundred microliters of each dilution of mycobacteria suspension was added to TE buffer and boiled at 100°C for 15 min, centrifuged at 16,000 g for 5 min, and the supernatant was transferred to a new tube as DNA for application in PCR.[16]
Mycobacteria polymerase chain reaction
A 439 bp sequence of mycobacterial hsp65 was amplified with primers forward (Tb 11: 5’-ACCAACGATGGTGTGTCCAT-3’) and reverse (Tb12: 5’-CTTGTCGAACCGCATACCCT-3’) in a Biomerta gradient thermocycler and Eppendorf AG22331.[20] The PCR reaction contained 1 μl (10 pmol) of each primer, 12.5 μl of PCR master mix (Ampliqon RED, Denmark), 2 μl of extracted DNA, and 8.5 μl of RNase-free water in a final reaction volume of 25 μl. The thermal program was one cycle of initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min and extension at 72°C for 1 min, and a final extension at 72°C for 10 min. Tubes containing distilled water instead of DNA were included in each run. A 5 μl aliquot of PCR products was run in 1.5% agarose gel and electrophoresed into TBE buffer (Tris 90 mM, Boric acid 90 mM, and ethylenediaminetetraacetic acid 2 mM). The gel was stained with SYBR Green I and the DNA-dye-complex was photographed under blue light transilluminator.
Results
In this study, we described the use of five methods of DNA extraction including CTAB-chloroform, FTA® Elute card, Chelex 100, Chelex 100+ Nonidet P-40, and boiling procedure for mycobacterial DNA extraction for the purpose of standard DNA extraction and PCR. The comparison of PCR results revealed a 5.3-fold and 2.5-fold higher yield of bacterial DNA using CTAB and Chelex methods in juxtaposition with the mechanical lysis procedure. The DNA concentration resulting from all samples was determined spectrophotometrically (Biometra, Germany) (data not shown). The results showed that the CTAB and Chelex methods detected low concentrations of bacteria suspension. The CTAB method showed more positive results at 1:10–1:100,000 at which the DNA amount was substantial [Figure 1a]. In these DNA extraction methods, there was the least amount of RNA, proteins, or other PCR inhibitors that this was a significant factor for successful PCR. Method 2 (FTA® Elute card) was unsuccessful resulting in no detection of PCR amplification in any dilution. Method 3 (Chelex 100 + Nonidet P-40) was simple with PCR amplification detected at 1:10 dilution [Figure 1a]. DNA extraction with the Chelex method was completed within <1 h and PCR amplification was detected at 1:10 and 1:1000 dilutions [Figure 1b]. The result of the PCR amplification of boiling (method 5) showed that the use of this protocol enables detection at 1:10 dilution [Figure 1a].
Figure 1.
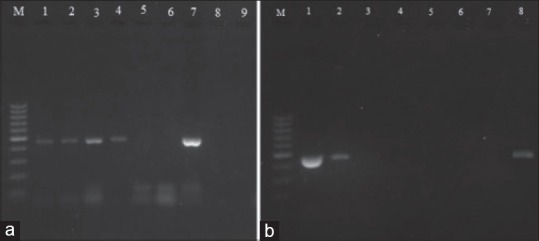
Gel electrophoresis of polymerase chain reaction. (a) M; markers (100-bp). 1–3; CTAB 1:10, 1:1000, 1:100000. 4–6; boiling 1:10, 1:1000, 1:100000. 7–9; Nonidet P-40, 1:10, 1:1000, 1:100000. (b) M; 100-bp size markers. 1–3, Chelex 100; 1:10, 1:1000, 1:100000. 4–6 FTA-card; 1:10, 1:1000, 1:100000. 7; Negative. 8; Positive control
Discussion
In the present work, we demonstrated that molecular detection of bacteria with resistant cell walls in clinical samples can be enhanced by the use of physical cell wall disintegration methods. Furthermore, the amplification of DNA derived from bacteria that are easily lysed is not compromised if the treatments are carefully optimized.
NTM causes different types of disease such as pulmonary disease, lymphadenitis, localized skin, soft tissue, and skeletal infection. Diagnosing NTM is more difficult than diagnosing other chronic pulmonary instances since the existing symptoms may harbor similarities.[21,22] Early identification of NTM is necessary in that it would help to procure the best plan for its treatment. Culture and biochemical tests have limitations for diagnosis including lack of accurate species identification.[9,12] DNA isolation is the first step in molecular methods and mycobacterial DNA is difficult to isolate; however, today there are various commercial kits for DNA extraction. DNA specimens extracted by each different kit may have different DNA purity which affects PCR sensitivity, and each commercial extraction kit has its advantages and disadvantages.[16,23,24]
Therefore, five DNA isolation methods including CTAB, Chelex 100, Chelex + Nonidet P-40, FTA® Elute card, and boiling were compared in this study. DNA extraction techniques can be divided into three groups, namely, physical, chemical, and a combination of physical and chemical methods. The methods used in this study are a combination of these approaches.
Methods 3 and 4 are a combination of chemical and physical methods using Chelex 100 and Nonidet P-40 and incubation at high temperature. Chelex is a resin that can help remove cell wall compounds at high temperatures; also, nuclei are inactivated and DNA is protected. We used Chelex as a DNA extraction method and detected PCR amplification at 1:10 and 1:1000 dilutions [Figure 1b]. Some studies have used Chelex for DNA extraction from different samples such as cerebrospinal fluid (CSF), blood, and tissue concluding that this method can produce suitable amounts DNA.[25,26,27] Nagdev et al. compared six Chelex methods with the PC method in CSF spiked and concluded that among the six Chelex methods, the extraction method using 70% ethanol produced a higher yield of DNA compared to the PC method. These studies show that in PCR assay with the Chelex method, sensitivity and specificity are better compared to the PC method.[25] DNA extraction using the Chelex method results in higher quality DNA in comparison with boiling. The Chelex method can be very quick and cheap. Care must be taken not to transfer any beads in the PCR because they might act as inhibitors.
de Almeida et al. used Nonidet P-40-Chelex 100 for DNA extraction from Mycobacterium tuberculosis, in which specificity and sensitivity of the PCR were 100%, and 52.3%, respectively.[28] This protocol is fast as compared to enzymatic methods which are used to extract DNA.[14] Based on our results, this protocol showed the lowest degree of DNA purity (absorbance at 260 and 280 nm) and thus unsuitability as regard PCR amplification.
Although the boiling method is easy and economical, it is not recommendable relative to the other protocols, perhaps because of the presence of lipids in mycobacterial cell walls. Thus, this method might not be appropriate for clinical samples.[10] Some studies suggest that boiling samples at 100°C in a buffer could extract enough mycobacterial DNA to perform PCR.[17]
FTA® Elute card is one of the simplest protocols that can be used for sample collection (such as the M. leprae), transport, and DNA preparation. FTA® Elute card can also be used directly in PCR with the advantage that samples may be stored for a long time at room temperature.[19,29,30,31] Furthermore, DNA remains detectable using FTA® Elute card more than 4 years after sample collection.[19]
CTAB is one of the oldest enzymatic extraction methods used to separate DNA from polysaccharides and has been accredited with producing the greatest amount of DNA.[15,22,32] This protocol uses proteinase K and lysozyme which damage cell membranes and release the cytoplasmic components.[14] Salgado et al. used three DNA extraction methods, UACH, QIAamp, and CTAB, for real-time PCR with results suggesting that the UACH method is cheaper and more useful than the other methods.[33] Our results showed that the CTAB method was the best method and can produce suitable DNA with a minimum amount of PCR inhibitor. The least expensive methods were 5 (boiling) and 4 (Chelex), followed by 2 (FTA® Elute card). Finally, 3 (Nonidet P-40) and 1 (CTAB) were the most expensive. DNA extraction methods FTA® Elute card, Nonidet P-40, Chelex, and Boiling had the same labor intensity, whereas the CTAB method was the most time-consuming.
Conclusions
We reported here five rapid and simple methods for the extraction of DNA from a mycobacterial colony that can be used in the laboratory. Two methods, namely, CTAB and Chelex appeared to be the suitable choices to extract DNA from mycobacteria. We hope that these methods will be used to extract mycobacterial DNA from tissue and sputum. A quantitative real-time PCR is recommended for a more accurate comparison of these methods.
Financial support and sponsorship
This study was supported by grant No. 394262 from Isfahan University of Medical Sciences.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Crago B, Ferrato C, Drews SJ, Louie T, Ceri H, Turner RJ, et al. Surveillance and molecular characterization of non-tuberculous mycobacteria in a hospital water distribution system over a three-year period. J Hosp Infect. 2014;87:59–62. doi: 10.1016/j.jhin.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Tortoli E. Microbiological features and clinical relevance of new species of the genus Mycobacterium. Clin Microbiol Rev. 2014;27:727–52. doi: 10.1128/CMR.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Werf MJ, Ködmön C, Katalinic-Jankovic V, Kummik T, Soini H, Richter E, et al. Inventory study of non-tuberculous mycobacteria in the European Union. BMC Infect Dis. 2014;14:62. doi: 10.1186/1471-2334-14-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katoch VM. Infections due to non-tuberculous mycobacteria (NTM) Indian J Med Res. 2004;120:290–304. [PubMed] [Google Scholar]
- 5.Mokaddas E, Ahmad S. Development and evaluation of a multiplex PCR for rapid detection and differentiation of Mycobacterium tuberculosis complex members from non-tuberculous mycobacteria. Jpn J Infect Dis. 2007;60:140–4. [PubMed] [Google Scholar]
- 6.Nasr-Esfahani B, Sarikhani E, Moghim S, Faghri J, Fazeli H, Hoseini N, et al. Molecular characterization of environmental non-tuberculous mycobacteria using PCR – RFLP analysis of 441 Bp heat shock protein 65 fragments. Iran J Public Health. 2012;41:108–14. [PMC free article] [PubMed] [Google Scholar]
- 7.Serkani JE, Isfahani BN, Safaei HG, Kermanshahi RK, Asghari G. Evaluation of the effect of Humulus lupulus alcoholic extract on rifampin-sensitive and resistant isolates of Mycobacterium tuberculosis. Res Pharm Sci. 2012;7:235–42. [PMC free article] [PubMed] [Google Scholar]
- 8.Covert TC, Rodgers MR, Reyes AL, Stelma GN., Jr Occurrence of nontuberculous mycobacteria in environmental samples. Appl Environ Microbiol. 1999;65:2492–6. doi: 10.1128/aem.65.6.2492-2496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soini H, Musser JM. Molecular diagnosis of mycobacteria. Clin Chem. 2001;47:809–14. [PubMed] [Google Scholar]
- 10.Timms VJ, Mitchell HM, Neilan BA. Optimisation of DNA extraction and validation of PCR assays to detect Mycobacterium avium subsp. paratuberculosis. J Microbiol Methods. 2015;112:99–103. doi: 10.1016/j.mimet.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Hadifar S, Moghim S, Fazeli H, GhasemianSafaei H, Havaei SA, Farid F, et al. Molecular typing of Iranian mycobacteria isolates by polymerase chain reaction-restriction fragment length polymorphism analysis of 360-bp rpoB gene. Adv Biomed Res. 2015;4:152. doi: 10.4103/2277-9175.161579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nasr Esfahani B, Rezaei Yazdi H, Moghim S, Ghasemian Safaei H, Zarkesh Esfahani H. Rapid and accurate identification of Mycobacterium tuberculosis complex and common non-tuberculous mycobacteria by multiplex real-time PCR targeting different housekeeping genes. Curr Microbiol. 2012;65:493–9. doi: 10.1007/s00284-012-0188-2. [DOI] [PubMed] [Google Scholar]
- 13.Hosek J, Svastova P, Moravkova M, Pavlik I, Bartos M. Methods of mycobacterial DNA isolation from different biological material: A review. Vet Med. 2006;51:180–92. [Google Scholar]
- 14.De Almeida IN, Da Silva Carvalho W, Rossetti ML, Costa ER, De Miranda SS. Evaluation of six different DNA extraction methods for detection of Mycobacterium tuberculosis by means of PCR-IS6110: Preliminary study. BMC Res Notes. 2013;6:561. doi: 10.1186/1756-0500-6-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amaro A, Duarte E, Amado A, Ferronha H, Botelho A. Comparison of three DNA extraction methods for Mycobacterium bovis, Mycobacterium tuberculosis and Mycobacterium avium subsp. avium. Lett Appl Microbiol. 2008;47:8–11. doi: 10.1111/j.1472-765X.2008.02372.x. [DOI] [PubMed] [Google Scholar]
- 16.Aldous WK, Pounder JI, Cloud JL, Woods GL. Comparison of six methods of extracting Mycobacterium tuberculosis DNA from processed sputum for testing by quantitative real-time PCR. J Clin Microbiol. 2005;43:2471–3. doi: 10.1128/JCM.43.5.2471-2473.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amita J, Vandana T, Guleria R, Verma R. Qualitative evaluation of mycobacterial DNA extraction protocols for polymerase chain reaction. Mol Biol Today. 2002;3:43–9. [Google Scholar]
- 18.Rastogi N, Goh KS, Berchel M. Species-specific identification of Mycobacterium leprae by PCR-restriction fragment length polymorphism analysis of the hsp65 gene. J Clin Microbiol. 1999;37:2016–9. doi: 10.1128/jcm.37.6.2016-2019.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aye KS, Matsuoka M, Kai M, Kyaw K, Win AA, Shwe MM, et al. FTA card utility for PCR detection of Mycobacterium leprae. Jpn J Infect Dis. 2011;64:246–8. [PubMed] [Google Scholar]
- 20.Steingrube VA, Gibson JL, Brown BA, Zhang Y, Wilson RW, Rajagopalan M, et al. PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J Clin Microbiol. 1995;33:149–53. doi: 10.1128/jcm.33.1.149-153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mirsaeidi M, Farnia P, Sadikot R, Hsueh PR, Aliberti S. Nontuberculous mycobacteria: Epidemiologic, mycobacteriologic, and clinical aspects. Biomed Res Int. 2015;2015:523697. doi: 10.1155/2015/523697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinha P, Gupta A, Prakash P, Anupurba S, Tripathi R, Srivastava GN. Differentiation of Mycobacterium tuberculosis complex from non-tubercular mycobacteria by nested multiplex PCR targeting IS6110, MTP40 and 32kD alpha antigen encoding gene fragments. BMC Infect Dis. 2016;16:123. doi: 10.1186/s12879-016-1450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leite FL, Stokes KD, Robbe-Austerman S, Stabel JR. Comparison of fecal DNA extraction kits for the detection of Mycobacterium avium subsp. paratuberculosis by polymerase chain reaction. J Vet Diagn Invest. 2013;25:27–34. doi: 10.1177/1040638712466395. [DOI] [PubMed] [Google Scholar]
- 24.Mita A, Mori Y, Nakagawa T, Tasaki T, Utiyama K, Mori H. Comparison of fecal pooling methods and DNA extraction kits for the detection of Mycobacterium avium subspecies paratuberculosis. Microbiologyopen. 2016;5:134–42. doi: 10.1002/mbo3.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagdev KJ, Kashyap RS, Deshpande PS, Purohit HJ, Taori GM, Daginawala HF. Determination of polymerase chain reaction efficiency for diagnosis of tuberculous meningitis in Chelex-100 extracted DNA samples. Int J Tuberc Lung Dis. 2010;14:1032–8. [PubMed] [Google Scholar]
- 26.Santos A, Cremades R, Rodríguez JC, García-Pachón E, Ruiz M, Royo G. Comparison of methods of DNA extraction for real-time PCR in a model of pleural tuberculosis. APMIS. 2010;118:60–5. doi: 10.1111/j.1600-0463.2009.02558.x. [DOI] [PubMed] [Google Scholar]
- 27.Polski JM, Kimzey S, Percival RW, Grosso LE. Rapid and effective processing of blood specimens for diagnostic PCR using filter paper and Chelex-100. Mol Pathol. 1998;51:215–7. doi: 10.1136/mp.51.4.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Almeida IN, Aleixo AV, Carvalho Wda S, de Miranda SS. In-house PCR with DNA extracted directly from positive slides to confirm or exclude the diagnosis of tuberculosis: Focus on biosafety. Rev Argent Microbiol. 2015;47:47–9. doi: 10.1016/j.ram.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Jaravata CV, Smith WL, Rensen GJ, Ruzante J, Cullor JS. Survey of ground beef for the detection of Mycobacterium avium paratuberculosis. Foodborne Pathog Dis. 2007;4:103–6. doi: 10.1089/fpd.2006.54. [DOI] [PubMed] [Google Scholar]
- 30.Gustavsson I, Lindell M, Wilander E, Strand A, Gyllensten U. Use of FTA card for dry collection, transportation and storage of cervical cell specimen to detect high-risk HPV. J Clin Virol. 2009;46:112–6. doi: 10.1016/j.jcv.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 31.Milne E, van Bockxmeer FM, Robertson L, Brisbane JM, Ashton LJ, Scott RJ, et al. Buccal DNA collection: Comparison of buccal swabs with FTA cards. Cancer Epidemiol Biomarkers Prev. 2006;15:816–9. doi: 10.1158/1055-9965.EPI-05-0753. [DOI] [PubMed] [Google Scholar]
- 32.Hill EB, Wayne LG, Gross WM. Purification of mycobacterial deoxyribonucleic acid. J Bacteriol. 1972;112:1033–9. doi: 10.1128/jb.112.3.1033-1039.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salgado M, Verdugo C, Heuer C, Castillo P, Zamorano P. A novel low-cost method for Mycobacterium avium subsp. paratuberculosis DNA extraction from an automated broth culture system for real-time PCR analysis. J Vet Sci. 2014;15:233–9. doi: 10.4142/jvs.2014.15.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]


