Abstract
Background
Simple sequence repeats (SSR), also called microsatellites, have been widely used as genetic markers, and have been extensively studied in some model insects. At present, the genomes of more than 100 insect species are available. However, the features of SSRs in most insect genomes remain largely unknown.
Results
We identified 15.01 million SSRs across 136 insect genomes. The number of identified SSRs was positively associated with genome size in insects, but the frequency and density per megabase of genomes were not. Most insect SSRs (56.2−93.1%) were perfect (no mismatch). Imperfect (at least one mismatch) SSRs (average length 22−73 bp) were longer than perfect SSRs (16−30 bp). The most abundant insect SSRs were the di- and trinucleotide types, which accounted for 27.2% and 22.0% of all SSRs, respectively. On average, 59.1%, 36.8%, and 3.7% of insect SSRs were located in intergenic, intronic, and exonic regions, respectively. The percentages of various types of SSRs were similar among insects from the same family. However, they were dissimilar among insects from different families within orders. We carried out a phylogenetic analysis using the SSR frequencies. Species from the same family were generally clustered together in the evolutionary tree. However, insects from the same order but not in the same family did not cluster together. These results indicated that although SSRs undergo rapid expansions and contractions in different populations of the same species, the general genomic features of insect SSRs remain conserved at the family level.
Conclusion
Millions of insect SSRs were identified and their genome features were analyzed. Most insect SSRs were perfect and were located in intergenic regions. We presented evidence that the variance of insect SSRs accumulated after the differentiation of insect families.
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4234-0) contains supplementary material, which is available to authorized users.
Keywords: Insect, SSR, Genome features, Phylogenetic analysis, Taxon, Molecular marker
Background
Simple sequence repeats (SSR), also known as microsatellites, are tandem repetitions of 1–6 bp motifs that are found in all eukaryotic genomes [1]. SSRs are mainly distributed in noncoding regions, but are also found in coding regions [2]. Some studies have indicated that SSRs are preferentially associated with retrotransposons [3]. Due to replication slippage [4] and unequal crossing-over during meiosis [5–7], SSRs have undergone rapid expansions and contractions, leading to variation in SSR length among populations of a single species. Because of these characteristics, SSRs have been widely used as molecular markers for fingerprinting, parentage analysis, genetic mapping, and analysis of genome structure [8–12]. Moreover, numerous studies suggest that SSRs may have biological functions and evolve in a complex process under selective pressure [11, 13, 14]. For example, the expansion of a dinucleotide SSR (AC repeat) in the promoter region of CYP6CY3, a P450 gene, resulted in its overexpression, allowing a tobacco-adapted race of polyphagous aphid (Myzus persicae) to increase its ability to detoxify nicotine [15].
Insects are one of the most diverse animal classes on our planet. Microsatellite markers are highly polymorphic and selectively neutral [16, 17], and thus are powerful genetic tools to investigate the spatial and temporal population dynamics and evolutionary trends of insects. So far, SSR diversity has been extensively surveyed in more than 200 insects to validate their use as molecular markers to infer the demography and relationships of closely related populations or species [10, 18, 19]. A comparative analysis of SSRs occurring within protein-coding regions of 25 insect species suggested that these repeats represent characteristic features of insect genome diversity [13]. Investigation of imperfect microsatellites (at least one mismatch) in five taxonomic orders (Diptera, Hymenoptera, Lepidoptera, Coleoptera, and Hemiptera) revealed their evolutionary paths across genomes [20]. However, the diversity, features, and evolutionary significance of microsatellites in insects are far from being fully understood. Here, we investigated SSRs in 136 insect species, representing 16 taxonomic orders. Millions of insect SSRs were identified from these insects. Cluster and divergence analysis based on the frequencies of various SSRs were also performed.
Results
The number, density and relative abundance of SSRs in 136 insect genomes
We identified a total of 15.01 million SSRs from 136 insect genomes (Additional file 1: Table S1). Typically, SSRs represent only a very small proportion of insect genomes, ranging from 0.02% to 3.1% of the whole genome (Additional file 1: Table S1, Additional file 2: Fig. S1). Interestingly, we found that the body louse (Pediculus humanus) had a much higher percentage (10.53%) of SSRs. Some insects had very few SSRs, such as the cochineal insect Dactylopius coccus (225 SSRs), the small green stink bug Piezodorus guildinii (432 SSRs), and the beetle Priacma serrata (859 SSRs). In contrast, over 500,000 SSRs were identified in some insects, including the body louse Pediculus humanus, the German cockroach Blattella germanica, and the locust Locusta migratoria has the largest animal genome so far [21] and was also shown to have the largest number of SSRs (1.5 million in total). The number of SSRs is positively correlated with genome size (Spearman’s rho = 0.499, P < 0.001), indicating that the abundance of SSRs varies greatly with insect species (Fig. 1).
Fig. 1.
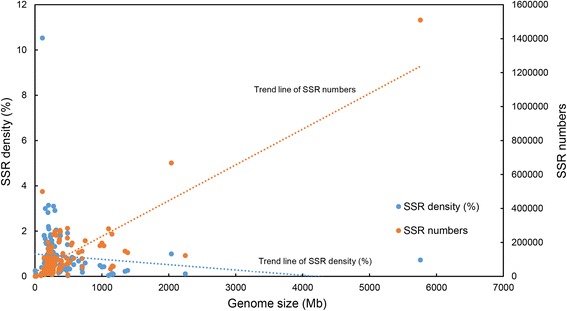
The relationship between SSR number, SSR density, and genome size. The number of identified SSRs was positively associated with genome size
SSR density (total bases of SSR (in bp) per Mb of genome) had a significantly negative relationship with genome size (Spearman’s rho = −0.228, P = 0.007) (Fig. 1). The density was significantly positively correlated with genome GC content, but the correlation was not strong (Spearman’s rho = 0.183, P = 0.033). Interestingly, the SSR frequency in various genomes (i.e., number of SSRs per Mb genome) varied tremendously in insects, ranging from 12 to 4509 (mean 251). The highest frequency (4509) was found in P. humanus, followed by the cactophilic fruitfly, Drosophila mojavensis (1038). SSR frequency was significantly negatively correlated with genome size (Spearman’s rho = −0.191, P = 0.026) and significantly positively correlated with genome GC content (Spearman’s rho = 0.178, P = 0.038), but none of the correlations was strong.
Most identified SSRs (56.2−93.1%) were perfect. The length of imperfect microsatellites (range 22−73 bp, average 34.8 bp) was significantly higher (t = −22.834, df = 175.875, P < 0.001) than that of perfect SSRs (range 16−30 bp, average 20.4 bp) in each species. To evaluate the relationship between SSR length and motif imperfection, we determined the frequency of mismatches in each locus. Imperfect SSRs contained 0.43−3.23% (1.85% on average) motif mismatches, which mainly appeared in the SSRs with a length of approximately 35 bp (Additional file 3: Table S2). Some closely related species had similar percentages of imperfect SSRs while other closely related species had very different numbers of SSRs. For example, the percentages of imperfect SSRs in 23 Drosophila species were very different, ranging from 23% to 44% [13, 20, 22]. In contrast, the imperfect SSR frequencies in two Nasonia species were the same at 21%. Similar phenomena were observed in three Papilio species (15–16%) and in three Batrocera species (22–24%).
Abundance of SSR motif types
When comparing the number of various classes of SSRs within genomes, we found that the percentages of di- and trinucleotide SSRs (27.2% and 22.0% on average, respectively) were significantly higher (P < 0.05, Tukey test) than those observed for mono- (11.6%), tetra- (17.9%), penta- (14.6%), and hexa-nucleotide repeat types (6.7%) (Additional file 4: Table S3, Fig. 2).
Fig. 2.
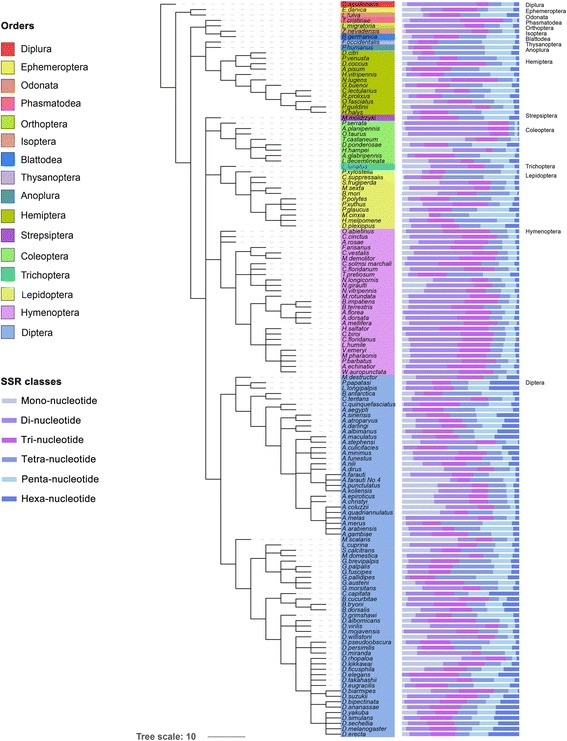
The relative percentages of six SSR motif types within 136 insect genomes. The percentages of mono-, di-, tri-, tetra-, penta- and hexanucleotide repeats are shown in different colors
Among mononucleotide repeats, the A/T type was predominant, accounting for 10.3% of the repeat motifs. AG/GA/CT/TC and AC/CA/GT/TG were the most frequent dinucleotide SSRs motifs, accounting for 10.2% and 10.0%, respectively. The next most abundant type was the sequences with AT/TA (6.6%). Among trinucleotide repeats, the AAT/ATA/TAA/ATT/TAT/TTA motif was most abundant (6.3%), and each of the other repeat types accounted for less than 3% (Additional file 5: Table S4).
Between the two types of monomer repeats, the A/T type was significantly more abundant (82.4%) than the G/C type (17.6%) (t = 22.962, df = 268, P < 0.001). Analysis of all dinucleotide repeats revealed that the GC/CG type accounted for only 0.9%, significantly lower (P < 0.05, Tukey test) than each of the other three types (AT/TA, AG/GA/CT/TC and AC/CA/GT/TG, each 30.5−36.4%). Among the trinucleotide SSR repeats, those containing two continuous Gs or Cs accounted for only 3.8−4.3%, which was significantly lower (P < 0.05, Tukey test) than other trinucleotide types (6.9−30.0%) (Additional file 6: Table S5). These data indicated that most SSRs in insect genomes consist of AT bases. AT-rich SSR containing motifs, such as AAT/AAAT/AAAAT/AAAAAT or ATA/ATAT/AATAAT/AAAATA, were very common (Fig. 3, Additional file 7: Table S6).
Fig. 3.
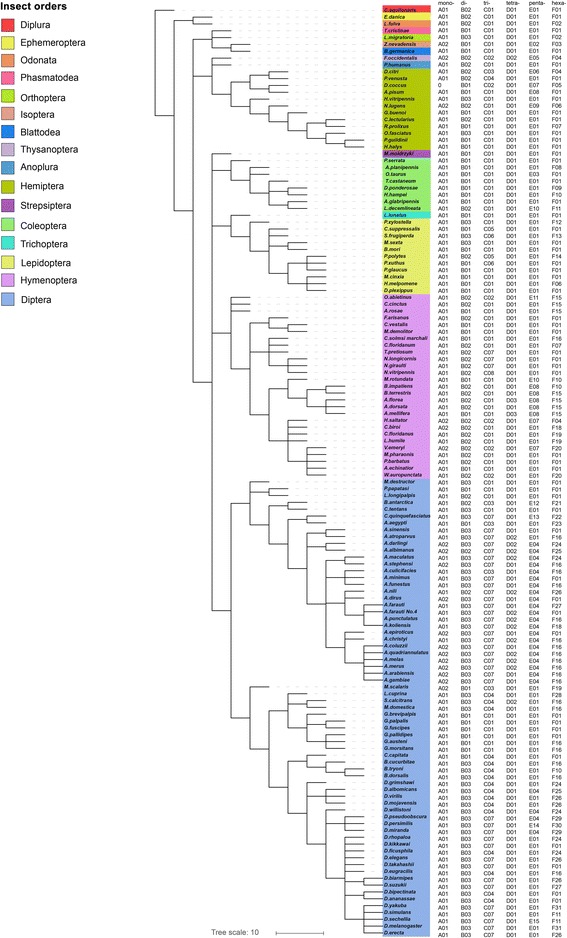
SSR abundance across insect genomes. Capital letters followed with number (A01 etc.) stand for different motif sequences as given in Additional file 3: Table S2. Zero means no SSRs of this class were identified
SSR diversity in different insect taxa
We analyzed the relative abundance of various types of SSRs in different insect taxa, and found that the frequencies of some SSR classes were different at the order level. For example, dinucleotide SSRs (the most abundant of the six types as revealed in Additional file 4: Table S3) accounted for 42.6% on average (range 13.2−71.5%) in the genomes of Hymenoptera, which was significantly higher than that observed in Hemiptera (18.2%), Lepidoptera (12.8%), and Coleoptera (9.2%) (P < 0.05, Tukey test) and also higher than that in Diptera (27.5%) (Fig. 2). At the family level, high conservation was observed in terms of the relative abundance of various types of SSRs. This was the most obvious for the families Cupedidae, Buprestidae, and Scarabaeinae that belong to the order Coleoptera. In addition, when viewed at the genus level, SSR frequency was also quite similar among genera within families (Fig. 2). The conservation of SSRs at the family level was also confirmed by analysis of the most abundant motifs. The results indicated that the most abundant motifs were conserved at the family level (Fig. 3).
At the species level, relative abundance of SSRs was very similar within some genera, such as Apis of Apidae, Nasonia of Pteromalidae, Anopheles of Culicidae, and Glossina of Glossinidae. However, interspecies differences in SSR frequency were also observed in some genera, such as Papilio, Drosophila, and Anopheles (Fig. 2). Taken together, the evolution of SSR diversity varied depending on the insect species, suggesting that insect SSRs face dissimilar selection pressures in different taxa.
Distribution of SSRs in different genomic regions
On average, 59.1% (range 49.6−62.0%) of SSRs were distributed in intergenic regions. Within genes, SSRs were mainly found in introns, which accounted for 36.8% (range 27.8−49.9%) of total SSRs, while only 3.7% (range 0.3−9.9%) were in exons (Table 1, Additional file 8: Table S7 and Additional file 9: Table S8). We compared the occurrence of SSRs in different genomic regions between Diptera and Hymenoptera genomes. In Diptera, the exonic SSRs reached 5.5% (range 1.2−9.95%), which was significantly higher than the 2.4% (range 0.3−5.8%) observed in Hymenoptera (t = 5.608, df = 36.312, P < 0.001). By contrast, the percentage of intronic SSRs were significantly lower in Diptera (average 33.2%) when compared to Hymenoptera (average 45.2%) (t = −2.296, df = 32.546, P = 0.028). The SSRs that occurred in mRNA regions were not significantly different (t = −1.758, df = 30.034, P = 0.089), accounting for 38.9% and 47.8% in Diptera and Hymenoptera, respectively.
Table 1.
Total number of SSRs in different genome regions
| Order | Species | Exon | Intron | Intergenic regions | Spanning exon-intron | Spanning intergenic-genetic | total number |
|---|---|---|---|---|---|---|---|
| Anoplura | P. humanus | 3797 | 82,101 | 377,312 | 340 | 287 | 463,837 |
| Coleoptera | D. ponderosae | 275 | 1726 | 6182 | 23 | 10 | 8216 |
| T. castaneum | 734 | 7623 | 9493 | 43 | 21 | 17,914 | |
| Diptera | A. aegypti | 4510 | 272,342 | 91,215 | 537 | 50 | 368,654 |
| A. coluzzii | 3896 | 22,894 | 52,062 | 127 | 95 | 79,074 | |
| A. darlingi | 27,936 | 45,779 | 213,703 | 2749 | 586 | 290,753 | |
| A. gambiae | 4781 | 28,746 | 74,093 | 108 | 106 | 107,834 | |
| A. sinensis | 3219 | 5862 | 23,306 | 75 | 30 | 32,492 | |
| A. stephensi | 4125 | 11,654 | 54,969 | 112 | 488 | 71,348 | |
| B. cucurbitae | 2388 | 42,930 | 40,223 | 46 | 161 | 85,748 | |
| B. dorsalis | 1903 | 27,991 | 29,792 | 37 | 77 | 59,800 | |
| C. capitata | 3862 | 113,867 | 100,518 | 73 | 288 | 218,608 | |
| C. quinquefasciatus | 10,339 | 53,060 | 221,906 | 1283 | 396 | 286,984 | |
| D. ananassae | 3823 | 15,074 | 32,900 | 73 | 26 | 51,896 | |
| D. erecta | 3810 | 10,660 | 23,757 | 49 | 27 | 38,303 | |
| D. grimshawi | 8444 | 47,707 | 95,949 | 172 | 70 | 152,342 | |
| D. melanogaster | 2201 | 30,075 | 15,571 | 56 | 95 | 47,998 | |
| D. mojavensis | 8222 | 56,598 | 124,059 | 203 | 69 | 189,151 | |
| D. persimilis | 5613 | 34,289 | 62,326 | 172 | 76 | 102,476 | |
| D. pseudoobscura | 6227 | 32,734 | 67,334 | 137 | 65 | 106,497 | |
| D. sechellia | 2527 | 9763 | 20,328 | 63 | 36 | 32,717 | |
| D. simulans | 2253 | 9234 | 19,388 | 57 | 24 | 30,956 | |
| D. virilis | 8114 | 39,513 | 88,738 | 140 | 60 | 136,565 | |
| D. willistoni | 6731 | 45,312 | 97,124 | 142 | 59 | 149,368 | |
| D. yakuba | 3571 | 13,101 | 28,165 | 79 | 35 | 44,951 | |
| M. destructor | 3883 | 17,407 | 59,110 | 519 | 415 | 81,334 | |
| M. scalaris | 268 | 770 | 2581 | 25 | 7 | 3651 | |
| M. domestica | 2298 | 58,145 | 64,685 | 45 | 227 | 125,400 | |
| Hemiptera | A. pisum | 2349 | 93,793 | 71,989 | 164 | 511 | 168,806 |
| D. citri | 459 | 78,562 | 78,172 | 115 | 179 | 157,487 | |
| N. lugens | 955 | 9002 | 25,103 | 73 | 35 | 35,168 | |
| R. prolixus | 505 | 24,038 | 87,771 | 42 | 45 | 112,401 | |
| Hymenoptera | A. dorsata | 1083 | 83,173 | 59,480 | 119 | 334 | 144,189 |
| A. florea | 1484 | 81,374 | 60,290 | 123 | 368 | 143,639 | |
| A. mellifera | 4455 | 82,325 | 133,229 | 724 | 397 | 221,130 | |
| A. rosae | 973 | 54,968 | 41,550 | 60 | 240 | 97,791 | |
| B. impatiens | 890 | 27,439 | 14,460 | 43 | 260 | 43,092 | |
| B. terrestris | 892 | 25,178 | 13,437 | 27 | 267 | 39,801 | |
| C. floridanus | 3493 | 22,024 | 92,188 | 682 | 227 | 118,614 | |
| C. biroi | 1446 | 30,066 | 14,794 | 57 | 179 | 46,542 | |
| C. solmsi marchali | 1579 | 115,235 | 126,743 | 424 | 985 | 244,966 | |
| F. arisanus | 744 | 8932 | 4937 | 32 | 54 | 14,699 | |
| H. saltator | 12,331 | 79,960 | 388,788 | 2777 | 818 | 484,674 | |
| L. humile | 1635 | 6724 | 41,722 | 67 | 61 | 50,209 | |
| M. rotundata | 1018 | 13,356 | 7637 | 35 | 84 | 22,130 | |
| M. demolitor | 2343 | 60,307 | 44,657 | 186 | 553 | 108,046 | |
| N. vitripennis | 1897 | 39,230 | 70,786 | 175 | 24 | 112,112 | |
| P. barbatus | 1860 | 13,131 | 78,291 | 119 | 97 | 93,498 | |
| V. emeryi | 1787 | 33,971 | 22,043 | 59 | 304 | 58,164 | |
| W. auropunctata | 1724 | 51,892 | 36,798 | 64 | 345 | 90,823 | |
| A. echinatior | 4443 | 35,426 | 115,736 | 790 | 203 | 156,598 | |
| Isoptera | Z. nevadensis | 879 | 16,054 | 48,801 | 61 | 27 | 65,822 |
| Lepidoptera | B. mori | 501 | 12,750 | 76,795 | 41 | 21 | 90,108 |
| C. suppressalis | 861 | 5346 | 8584 | 157 | 6 | 14,954 | |
| D. plexippus | 553 | 11,184 | 24,726 | 40 | 18 | 36,521 | |
| H. melpomene | 571 | 13,315 | 23,606 | 45 | 45 | 37,582 | |
| M. sexta | 864 | 17,324 | 24,138 | 40 | 67 | 42,433 | |
| P. xylostella | 9700 | 74,901 | 144,119 | 1862 | 291 | 230,873 |
Evolution analysis of insect SSR
Clustering analysis showed that the frequencies of various SSRs were largely similar within different insect orders (Fig. 4). A symmetrized Kullback-Leibler divergence analysis, based on the percentage of dinucleotide combinations, could almost perfectly separate Hymenoptera and Diptera from other insects (Additional file 10: Fig. S2). For Diptera, 54 species (80.6% of the total) clustered into two branches: 1) Diptera-I, which contained only flies of most families; and 2) Diptera-II, which was comprised solely of mosquitoes. Most Dipteran families can be readily separated from others, except for several Drosophilidae species (Additional file 11: Fig. S3). All hymenopteran species, except for Cotesia vestalis and Microplitisde molitor, clustered together (Additional file 10: Fig. S2). Similar results were obtained when the analysis was carried out using the tri-, tetra-, and penta-nucleotide motif information (Additional file 12: Fig. S4, Additional file 13: Fig. S5, Additional file 14: Fig. S6). In general, most insects were clearly divided using SSR frequencies at the family level, but not at the order level.
Fig. 4.
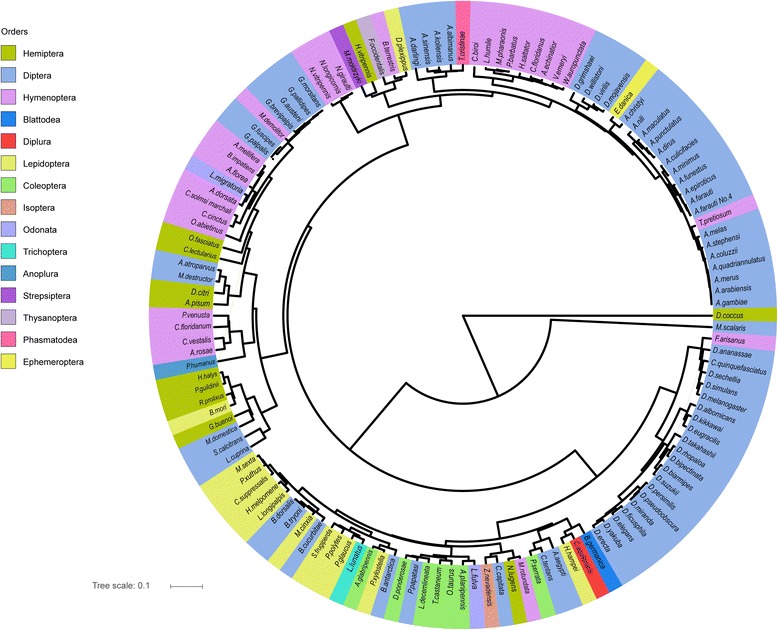
Phylogenetic analysis of insect SSRs. Symmetrized Kullback-Leibler divergence analysis was used and the evolutionary tree was constructed using the UPGMA method with MEGA6 software, and viewed with ITOL software
Discussion
Here, we identified millions of SSRs in 136 insect genomes and analyzed their features. The abundance and densities of insect SSRs were correlated with genome sizes. However, no strong correlation was found between the SSR density and GC content, nor was there a strong correlation between SSR frequency and genome size. This pattern was also observed in Tombusviridae [23], potexvirus [24] and carlaviruses [25]. In contrast, SSR frequency was shown to be inversely related to genome sizes in plants [26], Bovid species [27], fungi [28], and maize [29]. Both SSR frequency and density were negatively correlated with GC-content in these species.
SSRs only account for a very small proportion of insect genomes, and they generally do not have clear functions. Therefore, SSRs are believed to face relatively low selection pressures and accumulate mutations faster than coding genes [11]. SSRs have been widely used as genetic markers to distinguish individual insects from geographically distinct populations [30, 31]. Phylogenetic analysis of 136 insect SSRs indicated that the evolutionary tree constructed with SSR genome features was largely inconsistent with species trees, especially at the family level. This suggested that too many mutations have accumulated in insect SSRs since the division of the insect families. However, we found that insect species belonging to the same family tended to cluster together in the evolutionary tree, suggesting that selection pressures of SSRs were maintained at the family level. Thus, insect SSRs are good molecular markers to distinguish closely related insect species.
We found that perfect SSRs were significantly more abundant than imperfect SSRs in insects. Overall, perfect SSRs accounted for 56−93% of the identified microsatellites, whereas imperfect SSRs made up only 7−44%. This is consistent with similar results in plants, such as the Triticeae species [32], and previous reports in insects [13, 20]. The frequency of forming mismatch motifs varied with the length of the motif. For mono-, penta-, and hexanucleiotide SSRs, <19% were imperfect. In contrast, in di- and trinucleotide SSRs, ~31% of motifs were imperfect. This pattern was conserved in almost all tested insects, suggesting that this is a conserved feature in insect microsatellites [20]. A large fraction of the trinucleotide SSRs were derived from codon repeats, and the occurrence of these trinucleotide mismatches contributed to codon bias in the insect genomes [13, 20].
In insects, nearly three fifths of SSRs were found in intergenic regions, consistent with previous reports. Only 3.7% (0.3−9.9%) of SSRs occurred in exonic regions, which can be attributed to negative selection against frameshift mutations in coding regions [33]. In contrast, intronic SSRs accounted for 36.8%, which is 10-fold higher than exonic SSRs. It has been reported that intronic SSRs may affect gene expression [34], suggesting that the functions of intronic SSR will require more exploration.
Conclusions
In this study, we carried out a comprehensive analysis of SSRs in 136 insects. This is the first large-scale analysis of insect SSRs, and included more than 100 insect species. The results confirmed some previous conclusions about insect SSRs. The numbers of insect SSRs were positively associated with the genome sizes whereas the frequency and density were not. Both phylogenetic analysis and most abundant motif analysis showed that the insect SSRs were generally evolutionary conserved at the family level but not at the order level.
Methods
Genome sequences
At present, the genomes of 136 insect species are publically available, including 67 Diptera species, 27 Hymenoptera species, 12 Hemiptera species, 11 Lepidoptera species, eight Coleoptera species, and one species each of Diplura, Ephemeroptera, Odonata, Strepsiptera, Trichoptera, Thysanoptera, Anoplura, Phasmatodea, Orthoptera, Isoptera, and Blattodea (Additional file 1: Table S1). The genome sequences of all insects were downloaded from InsectBase 1.0 [35]. Ambiguous nucleotides were removed from the genomes prior to analysis.
Identification of insect SSRs
SSRs were identified using the SciRoKo 3.4 using default parameters [36]. According to the motifs, the repeat sequences were divided into six classes: mono-, di-, tri-, tetra-, penta-, and hexa-nucleotide SSRs [20]. For each class, only sequences with a length of ≥15 nucleotides were considered as SSRs. Briefly, SSRs with no mismatch in the motif were defined as perfect SSRs, while SSRs with at least one mismatch in the motif were defined as imperfect SSRs. The criteria used for defining imperfect SSRs was as following: ≥ 30 bp SSRs with 1–3 mismatches and ≤30 bp SSRs with ≥3 mismatches.
Analysis of SSRs
We calculated the frequency and density of SSRs in each of the available insect genomes. The frequency was determined as the percentage of the total number of SSRs per megabase (Mb) of genome sequence. The relative density was determined as the length (in bp) of SSRs sequences in the total Mb of genomic sequence analyzed. The relative abundances of perfect and imperfect repeat classes were calculated within each class of SSR, and their size distribution range and mean lengths were calculated. Associations between SSR number, frequency, and density with the genome sizes and GC contents were tested using Spearman rank correlation to determine whether there was significant correlation between the two variables (IBM SPSS Statistics, 2011).
Microsatellite distribution in insect genomes
Among the 136 insect species, 58 genomes were annotated with protein-coding genes accompanied by gff3 annotation files containing the positional information on exons and introns. The distribution of SSRs in these different regions was determined by mapping the SSRs to the genome using a Perl script.
Evolutionary analysis of insect SSRs
We constructed phylogenetic trees using insect SSRs with symmetrized Kullback-Leibler divergence analysis [37, 38]. The differences between two species were measured quantitatively with the percentages of SSRs ([p(x) and q(x)] in two species respectively, where x represents the class of SSR (di-, tri-, tetra-, penta-, and hexa-nucleotide repeat types). All pairwise comparisons among the 136 insect species were performed. Cluster analysis was performed using the UPGMA method [39] with the MEGA6 software package. Phylogenetic trees were visualized with ITOL software (http://itol.embl.de/) [40].
Additional files
Table S1. The genome sizes, GC content, SSR numbers and densities of 136 insect genomes. (DOCX 40 kb)
Figure S1. The insect genome sizes and SSR densities of 136 insects, showing that SSR densities have no relationship with genome size. (TIFF 2488 kb)
Table S2. Perfect and imperfect SSRs in insect genomes. (DOCX 36 kb)
Table S3. Percentage (%) of different types of SSRs. (DOCX 14 kb)
Table S4. Percentage of different types of SSRs in all SSRs (DOCX 15 kb)
Table S5. Percentage of different types of SSRs calculated within classes. (DOCX 15 kb)
Table S6. The sequences of SSR motifs. (DOCX 21 kb)
Table S7. Relative abundance of perfect SSRs in different genomic regions. (DOCX 24 kb)
Table S8. Relative abundance of imperfect SSRs in different genomic regions. (DOCX 26 kb)
Figure S2. Phylogenetic analysis of 136 insect genomes using the relative abundance information from di-nucleotide SSRs. (TIFF 2484 kb)
Figure S3. Phylogenetic analysis of Diptera using the relative abundance information from six types of SSRs, showing that dipteran insects can be clearly classified at the family level. (TIFF 1521 kb)
Figure S4. Phylogenetic analysis of 136 insect genomes using the relative abundance information from tri-nucleotide SSRs (TIFF 1694 kb)
Figure S5. Phylogenetic analysis of 136 insects using the relative abundance information from tetra-nucleotide SSR. (TIFF 2340 kb)
Figure S6. Phylogenetic analysis of 136 insects using the relative abundance information from penta-nucleotide SSR. (TIFF 2093 kb)
Acknowledgements
We thank Chuanlin Yin, Kun Lang, Dianhao Guo, and Ruizheng Tian for helpful discussions.
Funding
F.L. was funded by the National Key Research and Development Program [2017YFC1200602, 2016YFC1200600, 2017YFD0200900], National Science Foundation of China [31,772,238, 31,672,033].
Availability of data and materials
The gene data used in this study can be retrieved from the InsectBase (http://genome.zju.edu.cn/) [35]. The data of phylogenetic trees are deposited in the TreeBASE (Accession Number: 21,658).
Abbreviations
- Bp
base pair
- di-
dinucleotide
- gff3
generic Feature Format3
- hexa-
hexanucleotide
- mb
megabase
- mono-
mononucleotide
- penta-
pentanucleotide
- SSR
simple sequence repeats
- tetra-
tetranucleotide
- tri-
trinucleotide
Authors’ contributions
F.L. and M.J. designed the project; S.D. carried out SSR identification and analysis; S.W. distinguished fruit fries; K.H. did the PCR validation experiment; S.D. made the figures; S.D. drafted the manuscript; M.J. and S. D. analyzed the data; F.L., M.J. and S.W. wrote the paper. All authors read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12864-017-4234-0) contains supplementary material, which is available to authorized users.
Contributor Information
Simin Ding, Email: dingsm@zju.edu.cn.
Shuping Wang, Email: wangsp@shciq.gov.cn.
Kang He, Email: hekang@zju.edu.cn.
Mingxing Jiang, Email: mxjiang@zju.edu.cn.
Fei Li, Email: lifei18@zju.edu.cn.
References
- 1.Tautz D, Trick M, Dover GA. Cryptic simplicity in DNA is a major source of genetic-variation. Nature. 1986;322(6080):652–656. doi: 10.1038/322652a0. [DOI] [PubMed] [Google Scholar]
- 2.Mayer C, Leese F, Tollrian R. Genome-wide analysis of tandem repeats in Daphnia Pulex - a comparative approach. BMC Genomics. 2010;11 [DOI] [PMC free article] [PubMed]
- 3.Tay WT, Behere GT, Batterham P, Heckel DG. Generation of microsatellite repeat families by RTE retrotransposons in lepidopteran genomes. BMC Evol Biol. 2010;10 [DOI] [PMC free article] [PubMed]
- 4.Huntley MA, Golding GB. Selection and slippage creating serine homopolymers. Mol Biol Evol. 2006;23(11):2017–2025. doi: 10.1093/molbev/msl073. [DOI] [PubMed] [Google Scholar]
- 5.Richard GF, Paques F. Mini- and microsatellite expansions: the recombination connection. EMBO Rep. 2000;1(2):122–126. doi: 10.1093/embo-reports/kvd031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlotterer C, Tautz D. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 1992;20(2):211–215. doi: 10.1093/nar/20.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levinson G, Gutman GA. Slipped-strand Mispairing - a major mechanism for DNA-sequence evolution. Mol Biol Evol. 1987;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 8.Jones AG, Small CM, Paczolt KA, Ratterman NL. A practical guide to methods of parentage analysis. Mol Ecol Resour. 2010;10(1):6–30. doi: 10.1111/j.1755-0998.2009.02778.x. [DOI] [PubMed] [Google Scholar]
- 9.Selkoe KA, Toonen RJ. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol Lett. 2006;9(5):615–629. doi: 10.1111/j.1461-0248.2006.00889.x. [DOI] [PubMed] [Google Scholar]
- 10.Behura SK. Molecular marker systems in insects: current trends and future avenues. Mol Ecol. 2006;15(11):3087–3113. doi: 10.1111/j.1365-294X.2006.03014.x. [DOI] [PubMed] [Google Scholar]
- 11.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5(6):435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 12.Cipriani G, Marrazzo MT, Marconi R, Cimato A, Testolin R. Microsatellite markers isolated in olive (Olea Europaea L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theor Appl Genet. 2002;104(2–3):223–228. doi: 10.1007/s001220100685. [DOI] [PubMed] [Google Scholar]
- 13.Behura SK, Severson DW. Genome-wide comparative analysis of simple sequence coding repeats among 25 insect species. Gene. 2012;504(2):226–232. doi: 10.1016/j.gene.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huntley M, Golding GB. Evolution of simple sequence in proteins. J Mol Evol. 2000;51(2):131–140. doi: 10.1007/s002390010073. [DOI] [PubMed] [Google Scholar]
- 15.Bass C, Zimmer CT, Riveron JM, Wilding CS, Wondji CS, Kaussmann M, Field LM, Williamson MS, Nauen R. Gene amplification and microsatellite polymorphism underlie a recent insect host shift. P Natl Acad Sci USA. 2013;110(48):19460–19465. doi: 10.1073/pnas.1314122110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyle JM, Siegmund G, Ruhl JD, Eo SH, Hale MC, Marra NJ, Waser PM, DeWoody JA. Microsatellite analyses across three diverse vertebrate transcriptomes (Acipenser Fulvescens, Ambystoma Tigrinum, and Dipodomys Spectabilis) Genome. 2013;56(7):407–414. doi: 10.1139/gen-2013-0056. [DOI] [PubMed] [Google Scholar]
- 17.Leo SST, Millien V. Microsatellite markers reveal low frequency of natural hybridization between the white-footed mouse (Peromyscus Leucopus) and deer mouse (Peromyscus Maniculatus) in southern Quebec, Canada. Genome. 2017;60(5):454–463. doi: 10.1139/gen-2016-0163. [DOI] [PubMed] [Google Scholar]
- 18.Chapuis MP, Streiff R, Sword GA. Long microsatellites and unusually high levels of genetic diversity in the Orthoptera. Insect Mol Biol. 2012;21(2):181–186. doi: 10.1111/j.1365-2583.2011.01124.x. [DOI] [PubMed] [Google Scholar]
- 19.Sharma PC, Grover A, Kahl G. Mining microsatellites in eukaryotic genomes. Trends Biotechnol. 2007;25(11):490–498. doi: 10.1016/j.tibtech.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Behura SK, Severson DW. Motif mismatches in microsatellites: insights from genome-wide investigation among 20 insect species. DNA Res. 2015;22(1):29–38. doi: 10.1093/dnares/dsu036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang XH, Fang XD, Yang PC, Jiang XT, Jiang F, Zhao DJ, Li BL, Cui F, Wei JN, Ma CA, et al. The locust genome provides insight into swarm formation and long-distance flight. Nat Commun. 2014;5:1–9. doi: 10.1038/ncomms3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross CL, Dyer KA, Erez T, Miller SJ, Jaenike J, Markow TA. Rapid divergence of microsatellite abundance among species of drosophila. Mol Biol Evol. 2003;20(7):1143–1157. doi: 10.1093/molbev/msg137. [DOI] [PubMed] [Google Scholar]
- 23.George B, George B, Awasthi M, Singh RN. Genome wide survey and analysis of microsatellites in Tombusviridae family. Genes Genom. 2015;37(8):657–667. doi: 10.1007/s13258-015-0295-0. [DOI] [Google Scholar]
- 24.Alam CM, Singh AK, Sharfuddin C, Ali S. Incidence, complexity and diversity of simple sequence repeats across potexvirus genomes. Gene. 2014;537(2):189–196. doi: 10.1016/j.gene.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Alam CM, Singh AK, Sharfuddin C, Ali S. Genome-wide scan for analysis of simple and imperfect microsatellites in diverse carlaviruses. Infect Genet Evol. 2014;21:287–294. doi: 10.1016/j.meegid.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 26.Morgante M, Hanafey M, Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet. 2002;30(2):194–200. doi: 10.1038/ng822. [DOI] [PubMed] [Google Scholar]
- 27.Qi WH, Jiang XM, LM D, Xiao GS, TZ H, Yue BS, Quan QM. Genome-wide survey and analysis of microsatellite sequences in bovid species. PLoS One. 2015;10(7) [DOI] [PMC free article] [PubMed]
- 28.Lim S, Notley-McRobb L, Lim M, Carter DA: a comparison of the nature and abundance of microsatellites in 14 fungal genomes. Fungal Genet Biol 2004, 41(11):1025–1036. [DOI] [PubMed]
- 29.Tian XJ, Strassmann JE, Queller DC. Genome nucleotide composition shapes variation in simple sequence repeats. Mol Biol Evol. 2011;28(2):899–909. doi: 10.1093/molbev/msq266. [DOI] [PubMed] [Google Scholar]
- 30.He L, Wang SB, Miao XX, Wu H, Huang YP. Identification of necrophagous fly species using ISSR and SCAR markers. Forensic Sci Int. 2007;168(2–3):148–153. doi: 10.1016/j.forsciint.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 31.Miao XX, SJ X, Li MH, Li MW, Huang JH, Dai FY, Marino SW, Mills DR, Zeng PY, Mita K, et al. Simple sequence repeat-based consensus linkage map of Bombyx Mori. P Natl Acad Sci USA. 2005;102(45):16303–16308. doi: 10.1073/pnas.0507794102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng PC, Wang M, Feng KW, Cui LC, Tong W, Song WN, Nie XJ. Genome-wide characterization of microsatellites in Triticeae species: abundance, distribution and evolution. Sci Rep-Uk. 2016;6 [DOI] [PMC free article] [PubMed]
- 33.Metzgar D, Bytof J, Wills C. Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 2000;10(1):72–80. [PMC free article] [PubMed] [Google Scholar]
- 34.Li YC, Korol AB, Fahima T, Nevo E. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol. 2004;21(6):991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- 35.Yin CL, Shen GY, Guo DH, Wang SP, Ma XZ, Xiao HM, Liu JD, Zhang Z, Liu Y, Zhang YQ, et al. InsectBase: a resource for insect genomes and transcriptomes. Nucleic Acids Res. 2016;44(D1):D801–D807. doi: 10.1093/nar/gkv1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kofler R, Schlotterer C, Lelley T. SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics. 2007;23(13):1683–1685. doi: 10.1093/bioinformatics/btm157. [DOI] [PubMed] [Google Scholar]
- 37.Xie C, Zhang S, Li M, Li X, Hao Z, Bai L, Zhang D, Liang Y. Inferring genome ancestry and estimating molecular relatedness among 187 Chinese maize inbred lines. Journal of Genetics & Genomics. 2007;34(8):738. doi: 10.1016/S1673-8527(07)60083-6. [DOI] [PubMed] [Google Scholar]
- 38.Kullback S, Leibler RA. On information and sufficiency. Ann Math Stat. 1951;22(1):79–86. doi: 10.1214/aoms/1177729694. [DOI] [Google Scholar]
- 39.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The genome sizes, GC content, SSR numbers and densities of 136 insect genomes. (DOCX 40 kb)
Figure S1. The insect genome sizes and SSR densities of 136 insects, showing that SSR densities have no relationship with genome size. (TIFF 2488 kb)
Table S2. Perfect and imperfect SSRs in insect genomes. (DOCX 36 kb)
Table S3. Percentage (%) of different types of SSRs. (DOCX 14 kb)
Table S4. Percentage of different types of SSRs in all SSRs (DOCX 15 kb)
Table S5. Percentage of different types of SSRs calculated within classes. (DOCX 15 kb)
Table S6. The sequences of SSR motifs. (DOCX 21 kb)
Table S7. Relative abundance of perfect SSRs in different genomic regions. (DOCX 24 kb)
Table S8. Relative abundance of imperfect SSRs in different genomic regions. (DOCX 26 kb)
Figure S2. Phylogenetic analysis of 136 insect genomes using the relative abundance information from di-nucleotide SSRs. (TIFF 2484 kb)
Figure S3. Phylogenetic analysis of Diptera using the relative abundance information from six types of SSRs, showing that dipteran insects can be clearly classified at the family level. (TIFF 1521 kb)
Figure S4. Phylogenetic analysis of 136 insect genomes using the relative abundance information from tri-nucleotide SSRs (TIFF 1694 kb)
Figure S5. Phylogenetic analysis of 136 insects using the relative abundance information from tetra-nucleotide SSR. (TIFF 2340 kb)
Figure S6. Phylogenetic analysis of 136 insects using the relative abundance information from penta-nucleotide SSR. (TIFF 2093 kb)
Data Availability Statement
The gene data used in this study can be retrieved from the InsectBase (http://genome.zju.edu.cn/) [35]. The data of phylogenetic trees are deposited in the TreeBASE (Accession Number: 21,658).


