Abstract
The kinase inhibitory domain of the cell cycle regulatory protein p27Kip1 (p27) was nuclear spin hyperpolarized using dissolution dynamic nuclear polarization (D-DNP). While intrinsically disordered in isolation, p27 adopts secondary structure, including α-helical structure, upon binding to cyclin-dependent kinase 2 (Cdk2)/cyclin A. The sensitivity gains obtained with hyperpolarization enable the real-time observation of 13C NMR signals during p27 folding upon binding to Cdk2/cyclin A on a time scale of several seconds. Time-dependent intensity changes are dependent on the extent of folding and binding, as manifested in differential spin relaxation. Analysis of signal decay rates suggests the existence of a partially folded p27 intermediate during the timescale of the D-DNP NMR experiment.
Keywords: hyperpolarization, intrinsically disordered proteins, protein folding, NMR spectroscopy
The cell cycle regulator p27Kip1 in isolation lacks secondary and tertiary structure and samples an ensemble of disordered conformations. It is thus classified as an intrinsically disordered protein (IDP).[1] However, in the presence of Cdk2/cyclin A, p27Kip1 sequentially folds into a conformation that blocks substrate binding to cyclin A and, independently, inhibits ATP binding to Cdk2 and substrate phosphorylation.[1, 2, 3] Many IDPs have been identified as hubs in protein interaction networks[4] and shown to play regulatory roles in a wide range of biological processes including cell division,[5, 6] signal transduction[7] and gene transcription.[8, 9] Due to their extensive functional interconnectivity, deregulation of hub IDPs is associated with deleterious cellular phenotypes[10] and numerous human diseases including neurodegenerative diseases (Alzheimers, Parkinsons and Huntingtons disease),[11] various cancers[12], infectious diseases[13], diabetes, and cardiovascular disease[14].
Characteriztion of protein–protein interactions is hindered due to the initially unfolded nature of p27Kip1. NMR is a potentially powerful technique for the characterization of transient changes in proteins due to the sensitivity of chemical shift to structure. Stopped-flow methods have long been proposed for the study of protein folding by NMR.[15] Signal enhancements by dissolution dynamic nuclear polarization (D-DNP)[16] after rapid mixing can facilitate real-time NMR of nuclei with lower sensitivity, such as 13C.[17] Direct 13C hyperpolarization of a protein allows making use of the large chemical shift dispersion of this nucleus to assess structural changes during folding with true real-time capability.[18, 19]
Here, we explore the versatility of D-DNP NMR for determining interactions of intrinsically disordered proteins. Hyperpolarization of a polypeptide such as p27 (the 10.4 kDa N-terminal kinase inhibitory domain of p27Kip1 was used and is referred hereafter as p27) by D-DNP can be achieved with rapid injection and mixing[20] in conjunction with deuterium isotope labeling. Both of these methods alleviate spin polarization loss due to short spin lattice relaxation times prior to data acquisition. Series of five spectra from single samples of thus hyperpolarized p27 were recorded with or without admixing of Cdk2/cyclin A. Signal acquisition started 770 ms after component injection and proceeded at 343 ms time intervals. Figure 1 shows the spectral regions corresponding to 13CO and 13Cα chemical shifts, while the full spectra are presented in Figure S1 (supporting information). Since the radio-frequency (rf) pulse length used for excitation in the DNP experiments was progressively increased, the depletion of hyperpolarization in later scans due to the effect of the pulses themselves was compensated.[21] Thus, changes observed in the signal should be attributed to spin relaxation and changes that occurred to the protein itself during the time of the experiment. In Figure 1, a reduction in signal intensity can be seen throughout the spectra due to these effects. In addition, differences in the spectra measured with or without admixing of Cdk2/cyclin A are evident.
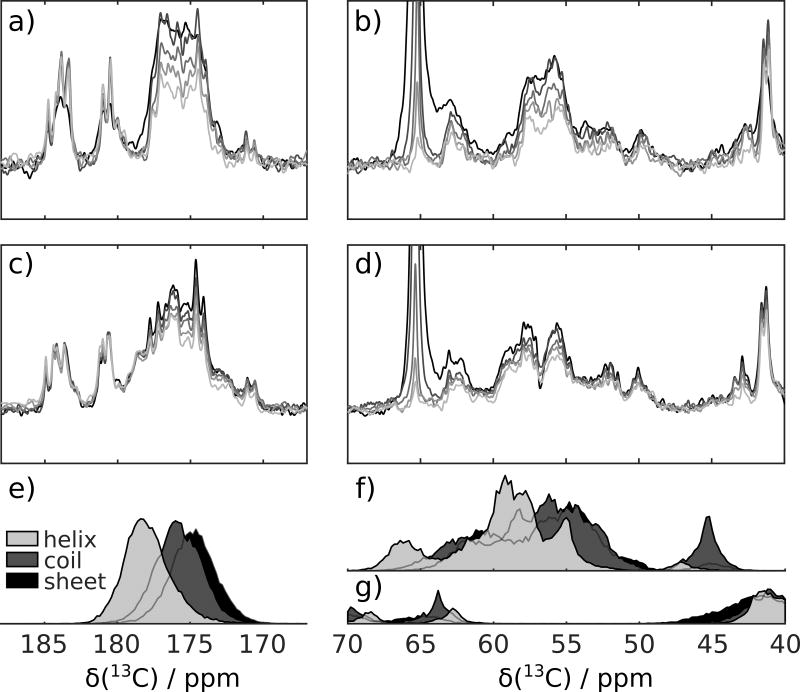
Figure 1.
a) and b) Series of spectra acquired following injection of hyperpolarized p27 into the NMR spectrometer, showing 13CO and 13C spectral regions. c) and d) Series of spectra as in (a) and (b), but with admixing of Cdk2/cyclin A complex in the NMR spectrometer. Final concentration of each protein after mixing was nominally 20 µM. The statistical distribution of chemical shifts in α-helix, random coil and β-sheet secondary structure based on those available from ref. [22] is plotted in (e) for 13CO, in (f) for 13Cα and in (g) for 13Cβ.
In the carbonyl region of the p27 DNP NMR spectra, additional signals between 177 and 180 ppm arise upon mixing with Cdk2/cyclin A. This spectral region is characteristic of α–helical secondary structure (Figure 1e).[22] For comparison, reference spectra of p27, p27/Cdk2, p27/cyclin A and p27/Cdk2/cyclin A were obtained without DNP hyperpolarization (Figures S2 and S3). The DNP and non-DNP NMR spectra are not expected to be exactly identical, foremost because of differences in spin-lattice (T1) relaxation throughout the spectra. Nevertheless, similar intensity in the region corresponding to α-helical secondary structure was observed in all of the samples of p27 in binary or ternary complexes. Likewise, in the region >56 ppm, where Cα in α-helical secondary structure is expected to shift, the DNP NMR spectrum of p27 mixed with Cdk2/cyclin A contains more intensity than p27 (Figure 1b,d,f,g).[22] The presence of 12 glutamate residues, which has a high propensity to form α–helix, likely contributes to the appearance of resonances around 58 ppm.[22]
A unique feature of the use of dissolution DNP is the ability to record spectra under non-equilibrium conditions and to analyze the evolution of signals from multiple spectra acquired in rapid succession. The isolated p27 polypeptide in solution is predominantly unstructured albeit with a propensity towards formation of α-helical secondary structure.[2] The above observations suggest that the DNP hyperpolarized p27 mixed with Cdk2/cyclin A is during the DNP measurement, at least, partially folded into an α-helical structure. However the appearance of the individual spectra within each data set does not change as much during the experiment.
Previous kinetic measurements that were performed using surface plasmon resonance spectroscopy (SPR) indicated that the association of p27 with Cdk2/cyclin A complex occurs in two phases with rate constants of 1.57 × 106 M−1s−1 and 5.61 × 103 M−1s−1.[2] Notwithstanding the different observables in SPR and NMR, at the final concentration of 20 µM these rate constants seemingly place the time-scale of the DNP NMR experiments, extending from 770 ms to 2140 ms, between the rapid and the slow phase. It is therefore possible that the observed 13C NMR spectra stem from a folding intermediate of the intrinsically disordered protein that arises after the fast association phase. Under this assumption, the changes observed in the time series of spectra would predominantly be due to the effect of T1 relaxation. The T1 parameter can provide evidence for protein dynamics to the extent that peaks can be resolved in different regions of the spectrum. Rather than attempting to identify specific peaks in the one-dimensional spectra, the series of spectra were analyzed by fitting a single exponential decay to each chemical shift location; i.e., at the position of each data point in the spectrum obtained after smoothing. In the analysis, the first spectrum was omitted due to broadening that occurred during sample settling immediately following injection. The fitted intensity and decay rate constants resulting from the second to the fifth spectra are plotted in Figure 2. Intensity parameters are plotted as a continuous line over the entire chemical shift range, which as expected closely follows the intensity distribution of the original spectra (Figure 1). The decay parameters, shown as filled circles, are omitted in the regions where the intensity is less than 2% of the maximum to avoid the randomly fluctuating values in the spectral regions that only contain noise.
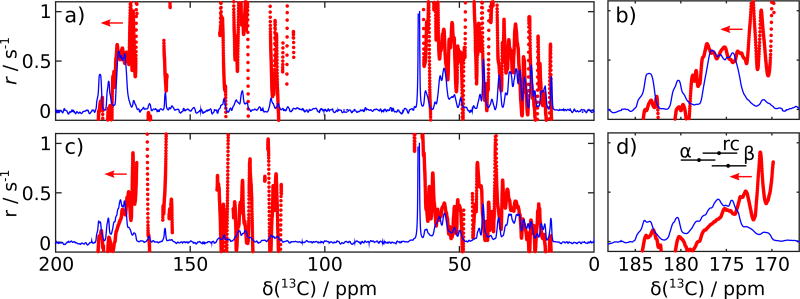
Figure 2.
Amplitudes, a, and decay rate constant parameters, r, obtained from fit of smoothed intensities of spectra 2 – 5, at each chemical shift, of a,b) p27 and c,d) p27 mixed with Cdk2/cyclin A. Fitted amplitudes, a, are shown as continuous lines (blue), and decay rate constants, r, are discrete points (red) plotted in all regions, where the amplitude is larger than 2% of the maximum. The arrows indicate that the scale on the left side corresponds to the decay rate constants. Ranges encompassing 80% of histogram values for helix (α), coil (rc) and sheet (β) chemical shifts from Figure 1e are indicated with horizontal bars for comparison with the data in (b) and (d).
Comparing Figures 2a and c indicates that signal decay rate constants in p27 with Cdk2/cyclin A are smaller at most chemical shift locations, including for aliphatic, aromatic and backbone carbonyl carbons. Exceptions are the 13CO side chain signals above 180 ppm. Some of these signals show a small increase in intensity, which gives rise to negative calculated rate constants. Increases in later scans may arise because of changes in line-shape due to sample settling after injection, or imperfections in radiofrequency pulses.
In the 13CO spectral region (Figures 2b and d), it is apparent that, while the decay rates from the predominantly random coil spectral region are similar between the two samples, the sample with Cdk2/cyclin A exhibits greatly reduced decay rate constants in the region from predominantly α-helical secondary structure (spectral regions are indicated with horizontal bars in the figure). This observation is consistent with the assumption that a bound α-helix experiences the longer rotational correlation time of the higher molecular weight complex leading to slower spin-lattice relaxation, combined with the possibility of forming some new α-helical secondary structure due to continued folding. Example decay curves from the predominantly α-helical and predominantly random-coil region are plotted in Figure 3a and b, respectively. Despite the use of only two fit parameters, the exponential decay closely models the data in this time regime. For comparison, Figure 3 also shows two example curves from the aliphatic region of the spectrum.
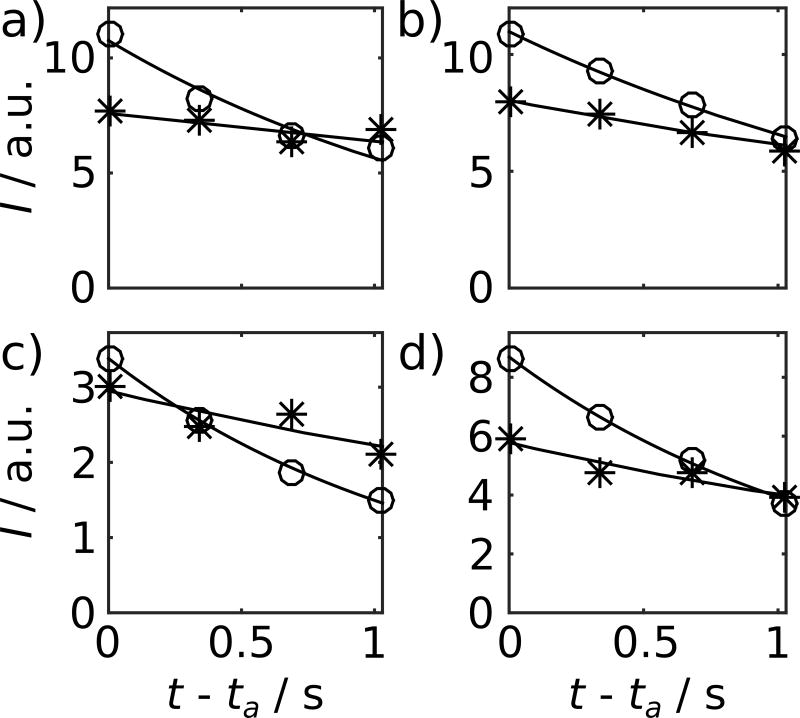
Figure 3.
Examples of fitted curves, comprising data points from the second to fifth spectra in the series of both the experiments with and without mixing with Cdk2/cyclin A. Curves are shown for the chemical shift positions of a) 177 ppm (r=0.64 s−1 and 0.15 s−1), b) 174 ppm (r=0.51 s−1 and 0.29 s−1), c) 62 ppm (r=0.82 s−1 and 0.28 s−1), and d) 56 ppm (r=0.79 s−1 and 0.36 s−1). ta is the time of the start of acquisition of the second spectrum in the series. Curves denoted by open circles are from p27 without mixing, and curves with (*) signs from p27 mixed with Cdk2/cyclin A.
The decay curves can be understood more fully by analyzing relaxation mechanisms for the 13C spins. Expected relaxation rates as a function of the rotational correlation time τc are calculated in Figure S4 based on average molecular parameters.[23, 24, 25] Based on these calculations, the expected decay rate constants are larger than the folding rate of the slow association phase suggested by the previous SPR measurements, lending support to the idea that the observed signal decay is dominated by relaxation. The expected R1 overall decreases when the rotational correlation time τc increases. Consistent with the observed decay rate constants for both 13CO and 13Cα would be an increase of the effective τc from on the order of a nanosecond for free p27, to on the order of 10 nanoseconds for p27 in the 75 kDa complex with Cdk2/cyclin A before completion of the slow phase of association, with some parts of the protein remaining more flexible.
Signal changes of hyperpolarized p27 upon association with Cdk2/cyclin A uniquely relate to dynamic and kinetic processes under non-equilibrium conditions. Additional resolution may in principle be available from rapid 2D NMR spectroscopy, which is possible through techniques ensuring rapid recovery of longitudinal magnetization[26] or pulsed field gradient based ultrafast acquisition.[27] The former has recently been used for measuring exchange kinetics from hyperpolarized water in an intrinsically disordered protein.[28] In the present work, 2D NMR would be interesting for further characterization of the directly hyperpolarized polypeptide conformation, since the overlap in the 13C NMR spectra prevents residue-specific assignments and structure determination. A tradeoff in the application of 2D NMR in this context may be the loss of the directly observable time dependence of signal intensities. Here, direct observation of hyperpolarized 13C allowed real-time NMR on the sub-second time scale. Additional signal gains in the spectra would further be possible through combination with cryogenically cooled NMR probes or higher field strength, both of which are compatible with D-DNP. These measures could reduce the achievable final protein concentration for 13C spectroscopy to the level of low single digit µM.
In summary, dissolution DNP NMR spectra of the intrinsically disordered protein p27 provide evidence for structural change associated with the interaction with Cdk2/cyclin A. The time scale uniquely accessible with the DNP NMR experiments falls between the fast and slow phase of association with Cdk2/cyclin A, suggesting that the signals observed stem from a partially associated structure. From the spectra of this species, observed signal decay rate constants provide evidence for the presence of both rigid and flexible elements. In general, intrinsically disordered proteins are interesting targets for study by dissolution DNP due to their well-behaved nature in the freeze-thaw cycle and, at least in the case of p27, the ability to inject the sample into the NMR spectrometer using only an aqueous buffer. Measurement of transient NMR signals enabled by dissolution DNP, and their interpretation based on relaxation processes may be used to obtain insight into the association of this class of proteins with their binding partners.
Experimental Section
Uniformly 2H and 13C labeled p27 and unlabeled Cdk2/cyclin A complex was recombinantly expressed and purified as described previously.[2] An aliquot for DNP was prepared by mixing 8.8 µL protein stock (2.5 mM in 60%(v/v) ethylene glycol/water mixture) with 1.0 µL of 150 mM tris[8-carboxy-2,2,6,6-tetra(hydroxyethyl)-benzo-(1,2-d:4,5-d)-bis(1,3)-dithiole-4-yl] methyl sodium salt (OXO63, Oxford Instruments, Abingdon, UK) and 0.2 µL of 50 mM gadolinium diethylenetriaminepenta acetic acid (Gd-DTPA, Sigma-Aldrich, St. Louis, MO). The samples were hyperpolarized on 13C nuclei in a HyperSense DNP polarizer (Oxford Instruments). Hyperpolarization occurred for 4 h at a temperature of 1.4 K, using microwaves of 93.974 GHz frequency at a power of 60 mW. Subsequently, samples were dissolved using 4 mL of hot (vapor pressure of 10 bar) 20 mM 2-amino-2-(hydroxymethyl)-1,3-propanediol hydrochloride (TRIS-HCl) buffer, pH 7.2, containing 300 mM NaCl and 5 mM dithiothreitol (DTT). The leading 450 µL of the dissolved sample was injected into a 400 MHz NMR spectrometer (Bruker Biospin, Billerica, MA) using pressurized nitrogen gas in a previously described sample injection device.[20] For this purpose, a 5 mm NMR tube was pre-installed in a broadband observe (BBO, Bruker Biospin) probe head. In the experiments with Cdk2/cyclin A complex, the injected sample mixed with 50 µL of the Cdk2/cyclin A complex stock solution pre-loaded in the NMR tube. In the experiments without Cdk2/cyclin A, 50 µL of dissolution buffer was instead pre-loaded in the NMR tube. Following injection, NMR spectra were acquired with a repetition time of 343 ms at a temperature of 301 K. During acquisition, 1H and 2H decoupling were applied. Chemical shifts were referenced against the 13C resonance of ethylene glycol in the sample, which was determined using an external standard containing 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS). The overall dilution factor during sample dissolution and injection was determined in reference experiments, and above sample sizes were chosen to result in a nominal concentration of 20 µM for p27, Cdk2 and cyclin A complex. Spectra were Fourier transformed with 16384 points. Signal changes in the spectra from the same data set were quantified by smoothing the spectra with a moving average over 33 data points, followed by fitting of a single exponential with amplitude a and decay rate r to the obtained intensities from spectra 2 – 5 at each chemical shift position, using Topspin (Bruker) and Matlab (MathWorks, Natick, MA).
Supplementary Material
Acknowledgments
Financial support from the Welch Foundation (Grant A-1658 to CH), the National Science Foundation (Grants CHE-0846402 and CHE-1362691 to CH), the National Institutes of Health (R01CA082491 to RWK and a National Cancer Institute Cancer Center Support Grant P30CA21765 to St. Jude Childrens Research Hospital), and ALSAC (to RWK).
References
- 1.Russo AA, Jeffrey PD, Patten AK, Massagu J, Pavletich NP. Crystal structure of the p27kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382(6589):325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 2.Lacy Eilyn R, Filippov Igor, Lewis William S, Otieno Steve, Xiao Limin, Weiss Sonja, Hengst Ludger, Kriwacki Richard W. p27 binds cyclinCDK complexes through a sequential mechanism involving binding-induced protein folding. Nature Structural & Molecular Biology. 2004;11(4):358–364. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]
- 3.Grimmler Matthias, Wang Yuefeng, Mund Thomas, Cilenšek Zoran, Keidel Eva-Maria, Waddell M Brett, Jäkel Heidelinde, Kullmann Michael, Kriwacki Richard W, Hengst Ludger. Cdk-inhibitory activity and stability of p27kip1 are directly regulated by oncogenic tyrosine kinases. Cell. 2007;128(2):269–280. doi: 10.1016/j.cell.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 4.Dunker A Keith, Cortese Marc S, Romero Pedro, Iakoucheva Lilia M, Uversky Vladimir N. Flexible nets. FEBS Journal. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 5.Dunker A Keith, et al. Intrinsically disordered protein. Journal of Molecular Graphics and Modelling. 2001;19(1):26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 6.Galea Charles A, Wang Yuefeng, Sivakolundu Sivashankar G, Kriwacki Richard W. Regulation of cell division by intrinsically unstructured proteins: Intrinsic flexibility, modularity, and signaling conduits. Biochemistry. 2008;47(29):7598–7609. doi: 10.1021/bi8006803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wright Peter E, Dyson H Jane. Intrinsically Disordered Proteins in Cellular Signaling and Regulation. Nature Reviews Molecular Cell Biology. 2015;16(1):18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garza Anna S, Ahmad Nihal, Kumar Raj. Role of intrinsically disordered protein regions/domains in transcriptional regulation. Life Sciences. 2009;84(78):189–193. doi: 10.1016/j.lfs.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Liu Jiangang, Perumal Narayanan B, Oldfield Christopher J, Su Eric W, Uversky Vladimir N, Dunker A Keith. Intrinsic disorder in transcription factors. Biochemistry. 2006;45(22):6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vavouri Tanya, Semple Jennifer I, Garcia-Verdugo Rosa, Lehner Ben. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138(1):198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 11.Uversky Vladimir N. The triple power of D: protein intrinsic disorder in degenerative diseases. Frontiers in Bioscience (Landmark Edition) 2014;19:181–258. doi: 10.2741/4204. [DOI] [PubMed] [Google Scholar]
- 12.Iakoucheva Lilia M, Brown Celeste J, Lawson J David, Obradovi Zoran, Dunker A Keith. Intrinsic disorder in cell-signaling and cancer-associated proteins. Journal of Molecular Biology. 2002;323(3):573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 13.Pushker Ravindra, Mooney Catherine, Davey Norman E, Jacqu Jean-Marc, Shields Denis C. Marked variability in the extent of protein disorder within and between viral families. PLOS ONE. 2013;8(4):1–13. doi: 10.1371/journal.pone.0060724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uversky Vladimir N. Wrecked regulation of intrinsically disordered proteins in diseases: pathogenicity of deregulated regulators. Frontiers in Molecular Biosciences. 2014;1:6. doi: 10.3389/fmolb.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimaldi John J, Sykes Brian D. Stopped flow fourier transform nuclear magnetic resonance spectroscopy. application to the .alpha.-chymotrypsin-catalyzed hydrolysis of tert-butyl-l-phenylalanine. Journal of the American Chemical Society. 1975;97(2):273–276. doi: 10.1021/ja00835a007. [DOI] [PubMed] [Google Scholar]
- 16.Ardenkjær-Larsen Jan H, Fridlund Bjrn, Gram Andreas, Hansson Georg, Hansson Lennart, Lerche Mathilde H, Servin Rolf, Thaning Mikkel, Golman Klaes. Increase in signal-to-noise ratio of >10,000 times in liquid-state nmr. Proceedings of the National Academy of Sciences. 2003;100(18):10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowen Sean, Hilty Christian. Time-resolved dynamic nuclear polarization enhanced nmr spectroscopy. Angewandte Chemie International Edition. 2008;47(28):5235–5237. doi: 10.1002/anie.200801492. [DOI] [PubMed] [Google Scholar]
- 18.Ragavan Mukundan, Chen Hsueh-Ying, Sekar Giridhar, Hilty Christian. Solution nmr of polypeptides hyperpolarized by dynamic nuclear polarization. Analytical Chemistry. 2011;83(15):6054–6059. doi: 10.1021/ac201122k. [DOI] [PubMed] [Google Scholar]
- 19.Chen Hsueh-Ying, Ragavan Mukundan, Hilty Christian. Protein folding studied by dissolution dynamic nuclear polarization. Angewandte Chemie International Edition. 2013;52(35):9192–9195. doi: 10.1002/anie.201301851. [DOI] [PubMed] [Google Scholar]
- 20.Bowen Sean, Hilty Christian. Rapid sample injection for hyperpolarized nmr spectroscopy. Physical Chemistry Chemical Physics. 2010;12:5766–5770. doi: 10.1039/c002316g. [DOI] [PubMed] [Google Scholar]
- 21.Zeng Haifeng, Bowen Sean, Hilty Christian. Sequentially acquired two-dimensional {NMR} spectra from hyperpolarized sample. Journal of Magnetic Resonance. 2009;199(2):159–165. doi: 10.1016/j.jmr.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Haiyan, Neal Stephen, Wishart David S. Refdb: A database of uniformly referenced protein chemical shifts. Journal of Biomolecular NMR. 2003;25(3):173–195. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 23.Engelke Jan, Rüterjans Heinz. Backbone dynamics of proteins derived from carbonyl carbon relaxation times at 500, 600 and 800 mhz: Application to ribonuclease t1. Journal of Biomolecular NMR. 1997;9(1):63–78. doi: 10.1023/A:1018675618785. [DOI] [PubMed] [Google Scholar]
- 24.Farmer Bennett T, Venters Ronald A. NMR of Perdeuterated Large Proteins. Springer US; Boston, MA: 2002. pp. 75–120. [Google Scholar]
- 25.Case David A, et al. Amber 2015 Reference Manual. University of California; San Francisco: 2015. [Google Scholar]
- 26.Schanda Paul, Forge Vincent, Brutscher Bernhard. Protein folding and unfolding studied at atomic resolution by fast two-dimensional nmr spectroscopy. Proceedings of the National Academy of Sciences. 2007;104(27):11257–11262. doi: 10.1073/pnas.0702069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gal Maayan, Schanda Paul, Brutscher Bernhard, Frydman Lucio. Ultrasofast hmqc nmr and the repetitive acquisition of 2d protein spectra at hz rates. Journal of the American Chemical Society. 2007;129(5):1372–1377. doi: 10.1021/ja066915g. [DOI] [PubMed] [Google Scholar]
- 28.Kurzbach Dennis, Canet Estel, Flamm Andrea G, Jhajharia Aditya, Weber Emmanuelle MM, Konrat Robert, Bodenhausen Geoffrey. Investigation of intrinsically disordered proteins through exchange with hyperpolarized water. Angewandte Chemie International Edition. 2017;56(1):389–392. doi: 10.1002/anie.201608903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.