The classical view of centrosome decentering and migration to the cell periphery during ciliogenesis is that it is pulled toward its final destination. Here, Pitaval et al. argue that microtubule stabilization in the early stages of ciliogenesis generates pushing forces that propel the centrosome toward the apical pole.
Abstract
Primary cilia are sensory organelles located at the cell surface. Their assembly is primed by centrosome migration to the apical surface, yet surprisingly little is known about this initiating step. To gain insight into the mechanisms driving centrosome migration, we exploited the reproducibility of cell architecture on adhesive micropatterns to investigate the cytoskeletal remodeling supporting it. Microtubule network densification and bundling, with the transient formation of an array of cold-stable microtubules, and actin cytoskeleton asymmetrical contraction participate in concert to drive apical centrosome migration. The distal appendage protein Cep164 appears to be a key actor involved in the cytoskeleton remodeling and centrosome migration, whereas intraflagellar transport 88’s role seems to be restricted to axoneme elongation. Together, our data elucidate the hitherto unexplored mechanism of centrosome migration and show that it is driven by the increase and clustering of mechanical forces to push the centrosome toward the cell apical pole.
Introduction
The centrosome, the major microtubule-organizing center of the cell, can transform to a primary cilium, a sensory organelle protruding from the cell surface, when the cell exits the cell cycle and enters a quiescent state (Bornens, 2012). Sensory function is endowed to the primary cilium by transmembrane receptors, which localize to and concentrate within the extracellular part of the primary cilium (Nigg and Raff, 2009). As a sensory organelle, the primary cilium plays important roles in embryonic development and cellular homeostasis. Genetic mutations that result in the failure to form a primary cilium cause developmental defects, including polydactyly, craniofacial defects, and heart malformation, and highlight the crucial role of this organelle in development (Goetz and Anderson, 2010).
Primary cilium assembly is a complex and highly coordinated process, which is reflected by the large number of primary ciliogenesis effectors and their diverse functions (Kim et al., 2010; Wheway et al., 2015). This multistep process begins with the assembly of a ciliary vesicle at the distal end of the mother centriole (Nachury et al., 2007; Knödler et al., 2010; Westlake et al., 2011; Lu et al., 2015). Recent work has shown that Cep164, a distal appendage protein (Graser et al., 2007), plays an important role in anchoring factors regulating centriole elongation (Cajánek and Nigg, 2014) and the formation of the primary ciliary vesicle (Schmidt et al., 2012). After ciliary vesicle formation, tubulin dimers are added to the minus ends of the centriolar microtubules of the mother centriole to form an axoneme, which protrudes from the cell surface and is ensheathed by ciliary membrane. This is dependent on the activity of a multisubunit complex, known as the intraflagellar transport (IFT) complex, interacting with kinesin and dynein molecular motors (Lechtreck, 2015). Finally, basal body anchoring to the cortex is mediated by the mother centriole’s distal appendages, and several of their components have been identified. Cep83 is a key distal appendage protein (Joo et al., 2013) that is responsible for anchoring four other components, Cep89 (Cep123/CCDC123; Sillibourne et al., 2013), SCLT1, Cep164 (Graser et al., 2007), and FBF1, to the distal appendages (Tanos et al., 2013). During or after the process of ciliary vesicle formation and axoneme extension, the mother centriole migrates to the cell surface, where it attaches to the cortex (Singla et al., 2010; Reiter et al., 2012). Despite much information regarding basal body maturation and anchoring and the players involved in the regulation of centrosome positioning (Barker et al., 2016), the physical mechanism powering centrosome displacement and migration to cell apical pole is poorly understood.
Microtubules regulate centrosome positioning at the cell center by exerting pushing and pulling forces (Burakov et al., 2003; Zhu et al., 2010; Kimura and Kimura, 2011). They have also been shown to support centrosome migration away from the cell center toward the cell surface by the production of pulling forces during immune synapse formation (Yi et al., 2013) or mitotic spindle positioning (Morin and Bellaïche, 2011). Interestingly, recent numerical simulations suggested that asymmetrical pushing forces could also efficiently promote centrosome off-centering (Letort et al., 2016). Whether pushing forces on the basal pole and/or pulling forces from the apical pole are involved in centrosome migration during primary ciliogenesis remains to be uncovered.
The actin cytoskeleton is also involved in the regulation of centrosome positioning and ciliogenesis (Dawe et al., 2009; Kim et al., 2010; Pitaval et al., 2010). Actomyosin contractility is required for basal body migration to the cell apical pole (Pitaval et al., 2010; Hong et al., 2015), but its hyperactivation impairs primary cilium formation and cilium elongation (Kim et al., 2010; Pitaval et al., 2010; Rao et al., 2014). In parallel, disruption of actin filament formation, either by depletion of Arp2/3 or treatment with a low dose of cytochalasin D, promotes primary ciliogenesis (Kim et al., 2010; Sharma et al., 2011). Indeed, branched actin filament formation impairs the recruitment of primary ciliogenesis effectors to a region surrounding the centrosome referred to as the pericentrosomal preciliary compartment (Kim et al., 2010; Rao et al., 2014). Recent work has demonstrated that the Arp2/3 complex is present at the centrosome, where it promotes the nucleation of actin filaments (Farina et al., 2015). This centrosomal network needs to be disassembled for the centrosome to detach from the nucleus and move to the periphery during immune synapse assembly (Obino et al., 2016). A similar regulation of the centrosome–nucleus link has been proposed to be required for centrosome migration during ciliogenesis (Dawe et al., 2009; Adams et al., 2012), although the physical mechanism powering centrosome motion has not yet been established. Thus, the exact mechanism by which actin network architecture and contractility regulate centrosome migration during ciliogenesis remains unclear and needs further characterization.
In this paper, we exploit the reproducibility of cell architecture on adhesive micropatterns to investigate the mechanisms driving apical centrosome migration during primary ciliogenesis. Dramatic remodeling of the actin and microtubule cytoskeletons was found to drive apical centrosome movement and relied on the activity of molecular motors. A candidate-based siRNA screen of primary ciliogenesis effectors identifies a role for the distal appendage protein Cep164 in centrosome movement. These data characterize in detail the previously understudied process of centrosome migration and identify unreported roles of known ciliogenesis effectors in this process.
Results
Centrosome migration in serum-starved cells
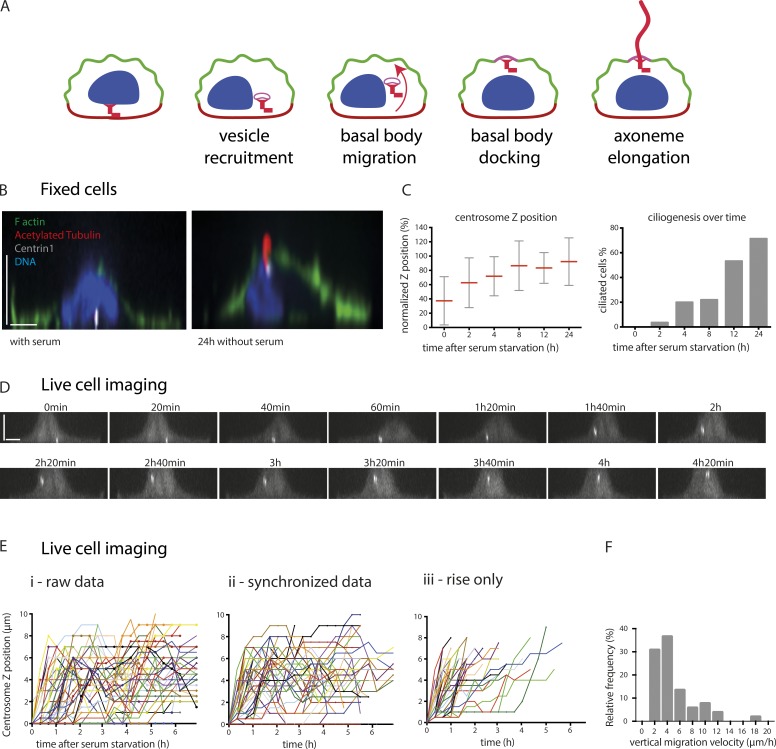
Previous studies have shown that the culture of cells on adhesive micropatterns promotes reproducible organelle positioning and results in a defined intracellular architecture (Théry et al., 2006; Pitaval et al., 2013). We have shown that primary ciliogenesis can be induced in isolated single retinal pigment epithelial 1 (RPE1) cells cultured on disk-shaped micropatterns and is influenced by cell confinement and contractility (Pitaval et al., 2010), as is the case in vivo (Blitzer et al., 2011). Highly confined cells exhibiting low contractility form primary cilia more frequently than less constrained cells exhibiting higher contractility. For this reason, we chose to monitor centrosome migration during primary ciliogenesis in RPE1 cells growing on small disk-shaped micropatterns with an area of 700 µm2. Furthermore, the increased cell height associated with cell confinement offered the possibility of monitoring basal body migration over a few micrometers (Fig. 1, A and B). Thereby, we could distinguish centrosome migration and axonemal elongation defects in cells where ciliogenesis was impaired.
Figure 1.
Adhesive micropatterns facilitate the study of centrosome migration during primary ciliogenesis. (A) Primary ciliogenesis is a multistep process that is proposed to begin with centrosome maturation and the formation of a ciliary vesicle at the distal end of the mother centriole, after which the centrosome migrates to the apical surface and attaches to the cortex. Full extension of the axoneme occurs once the mother centriole is anchored to the cortex. (B) Side view of micropatterned RPE1 cells expressing EGFP-centrin1 (white), cultured in the presence or absence of serum for 24 h, and stained with phalloidin to visualize F-actin, acetylated tubulin antibody to label the cilium, and DAPI to stain the DNA. (C) Measurement of centrosome z position as a percentage of nuclear height. Migration started within 2 h of serum starvation and appeared completed 6 h later. Measurement of the proportion of ciliated cells showed a delayed process compared with centrosome migration (one experiment, n = 60 cells per condition). Error bars represent standard deviation. (D) Side view of a representative time-lapse imaging of serum-starved RPE1 EGFP-centrin1 cells on micropatterns. Centrosome migration was engaged 1 h after starvation and completed 2 h later. (E) Representation of time-lapse centrosome movement in serum-starved RPE1 EGFP-centrin1 (data of three independent experiments, n = 53 cells). (i) The graph represents all the raw data. (ii) The centrosome trajectories are synchronized, i.e., they start when the centrosome leaves the basal pole. (iii) The plotting of the centrosome trajectories is limited to the maximal z position, i.e., they come to an end when centrosomes reach the apical pole. (F) Frequency distribution of centrosome migration velocity from basal pole to apical pole. Bars, 5 µm.
RPE1 cells expressing EGFP-centrin1 were cultured either in the presence or absence of serum for 24 h and were then fixed and stained with phalloidin to label F-actin, DAPI to label DNA, and an antibody to acetylated tubulin to label cilia. This revealed that the centrosome was located at the basal surface in cells cultured in the presence of serum, whereas those cultured in the absence of serum had formed cilia, and the centrosome was at the apical surface (Fig. 1 B). To determine the timing of centrosome migration, RPE1 cells expressing EGFP-centrin1 were serum starved over a 24-h period and stained with an acetylated tubulin antibody, and the axial position of the centrosome was determined and expressed as a percentage of nuclear height. Ciliated cells were also enumerated. Surprisingly, centrosome migration was found to occur as soon as 2 h after serum starvation, and by 8 h, the centrosome was located at the apical surface (Fig. 1 C). As expected, primary cilium formation took longer to complete and reached a maximum after 24 h of serum starvation (Fig. 1 C). Monitoring of centrosome movement in serum-starved RPE1 EGFP-centrin1 cells by live imaging provided a more detailed picture of the 3D migration mechanism in individual cells (Fig. 1 D). The ignition of centrosome takeoff was preceded by a lag period that varied from a few minutes to a few hours. These variations confounded the analysis of centrosome apical motion (Fig. 1 E, i). To overcome this problem, centrosome trajectories were synchronized at the moment of centrosome takeoff from the basal surface (Fig. 1 E, ii). However, centrosome wandering after takeoff continued to complicate analysis. This problem was circumvented by plotting only the gain in elevation of each centrosome trajectory, which thus enabled the specific quantitation of centrosome movement (Fig. 1 E, iii). We found that the duration of centrosome motion from the basal to apical pole was quite variable, ranging from 30 to 300 min. The corresponding centrosome velocity was 2–20 µm/min with a median value close to 5 µm/min (Fig. 1 F). Interestingly, the vertical displacements from the basal to the apical pole was ∼100× slower than the reported velocities for centrosome migration toward immune synapse in T lymphocytes (Yi et al., 2013) or sperm aster centration in sea urchin eggs (Tanimoto et al., 2016), suggesting that the positioning mechanism was likely to differ from the pulling forces involved in these two examples. These data not only demonstrated the utility of micropatterns in quantifying centrosome migration during ciliogenesis, but also in characterizing the initial steps of migration and highlighting important differences between centrosome movement during ciliogenesis and immune synapse formation.
Specific implication of ciliogenesis effectors in centrosome migration or axoneme elongation
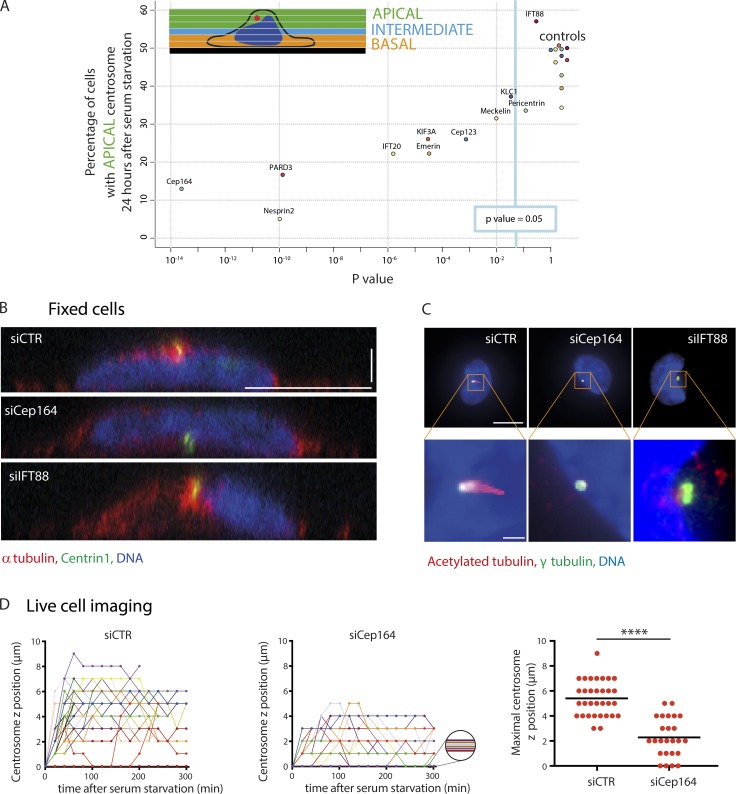
Next, we decided to investigate the role of known primary ciliogenesis effectors in centrosome migration by using siRNA to mediate their depletion. Candidates were chosen to reflect the diversity of ciliogenesis effectors and included Cep164 (Graser et al., 2007), Cep123 (Cep89/CCDC123; Sillibourne et al., 2013; Tanos et al., 2013), IFT20 (Follit et al., 2006), partitioning defective 3 (Pard3; Sfakianos et al., 2007), nesprin2, meckelin (Dawe et al., 2009), pericentrin (Jurczyk et al., 2004), KIF3A (Lin et al., 2003), and IFT88 (Pazour et al., 2000). The roles of kinesin light chain (KLC1) and emerin in the regulation of centrosome anchoring to the nucleus were also tested (Salpingidou et al., 2007; Roux et al., 2009; Schneider et al., 2011). siRNA-mediated protein depletion was used to assess the role of each candidate in centrosome migration (Table S1). RPE1 cells previously treated with siRNA were plated onto micropatterns, and after 24 h of serum starvation, the proportion of cells with a centrosome located near the apical pole (>3 µm above the glass substrate) was measured and compared with nontargeting control siRNA–treated cells (Fig. 2 A). The deletion of all tested proteins except pericentrin and IFT88 had a significant deleterious effect on centrosome migration, suggesting that these were specifically involved in axoneme elongation (Fig. 2 A). The finding that KIF3A depletion impacts centrosome migration agrees with previously published data showing that defective ciliogenesis was associated with basal body mispositioning in mouse hair cells (Sipe and Lu, 2011) and zebrafish photoreceptors (Pooranachandran and Malicki, 2016). The effect of IFT20, meckelin, and nesprin2 depletion on basal body positioning confirmed earlier observations in mouse kidney (Jonassen et al., 2008) and in cultured kidney cells (Dawe et al., 2009). To corroborate our results with those of others, we determined the frequency of primary cilium formation and found that it was significantly reduced after treatment with siRNA targeting the candidate mRNAs (Fig. S1 A). In addition, cilium length was measured and found to be reduced compared with the control siRNA, except where the cells were treated with siRNA to meckelin, KLC1, or IFT20 (Fig. S1 B). Together, these results suggested that depletion of the candidate proteins was successful.
Figure 2.
Implication of known ciliogenesis effectors in centrosome migration. (A) RPE1 EGFP-centrin1 cells were treated with siRNAs targeting known primary ciliogenesis effectors for 24 h to investigate their potential role in centrosome migration during primary cilium formation. The proportion of cells displaying a centrosome located >3 µm above the basal pole was determined and normalized to that of the nontargeting control siRNA for each condition. See Table S1 for the siRNA sequences and Fig. S1 (A and B) for the effect on the rate of ciliated cells and the length of primary cilia. (B) Side views of serum-starved RPE1 EGFP-centrin1 cells stained with DAPI to label the DNA and an antibody to α-tubulin to stain microtubules. Bars: (x and y) 10 µm; (z) 2.5 µm. (C) Staining of RPE1 cells with DAPI and with antibodies to acetylated tubulin and γ-tubulin. Images show maximal projection of z stacks. Bars: (top) 10 µm; (bottom) 1 µm. (D) Representation of synchronized time-lapse centrosome movement in serum-starved RPE1 EGFP-centrin1 treated with nontargeting control siRNA (one experiment, n = 32 cells) or siRNA against siCep164 (one experiment, n = 25 cells; left and middle). The graph represents the maximal centrosome z position for cells treated with nontargeting control siRNA and with siRNA against siCep164 (right). ****, P < 0.0001. Error bars represent standard deviation. Horizontal bars show mean values.
Two candidates, Cep164 and IFT88, were selected for further analysis, as their depletion (Fig. S1 C) resulted in opposing phenotypes, with the depletion of Cep164 blocking centrosome migration and IFT88 ablation having no discernable effect. Confocal imaging and side view reconstructions confirmed our initial results and showed that after serum starvation, the centrosome was at the basal surface in Cep164-depleted cells, whereas in IFT88-depleted cells, it was at the apical surface (Fig. 2 B), but neither possessed a cilium, in contrast to control siRNA–treated cells (Fig. 2 C). Cep164 is a core component of mother centriole distal appendages (Graser et al., 2007), and it is involved in the docking of the ciliary vesicle (Schmidt et al., 2012). Distal appendages are required for the mother centriole anchoring to lipid membranes and notably the apical pole (Tanos et al., 2013). This suggested that the defective positioning we observed might result from a lack of an anchor rather than a defective migration toward the apical pole. To distinguish between these two possibilities, we tracked centrosome 3D migration in EGFP-centrin1–expressing cells treated with either siRNA against Cep164 or nontargeting control siRNA. In Cep164 knocked-down cells, centrosome migration was severely defective. Few centrosomes took off from the basal pole, and the others could not reach the apical pole (Fig. 2 D), showing that the migration process itself, and not only the anchoring step afterward, was compromised. These results indicated that some primary ciliogenesis effectors participate in centrosome migration, notably Cep164, whereas others, such as IFT88, do not.
Remodeling of the microtubule network during centrosome migration
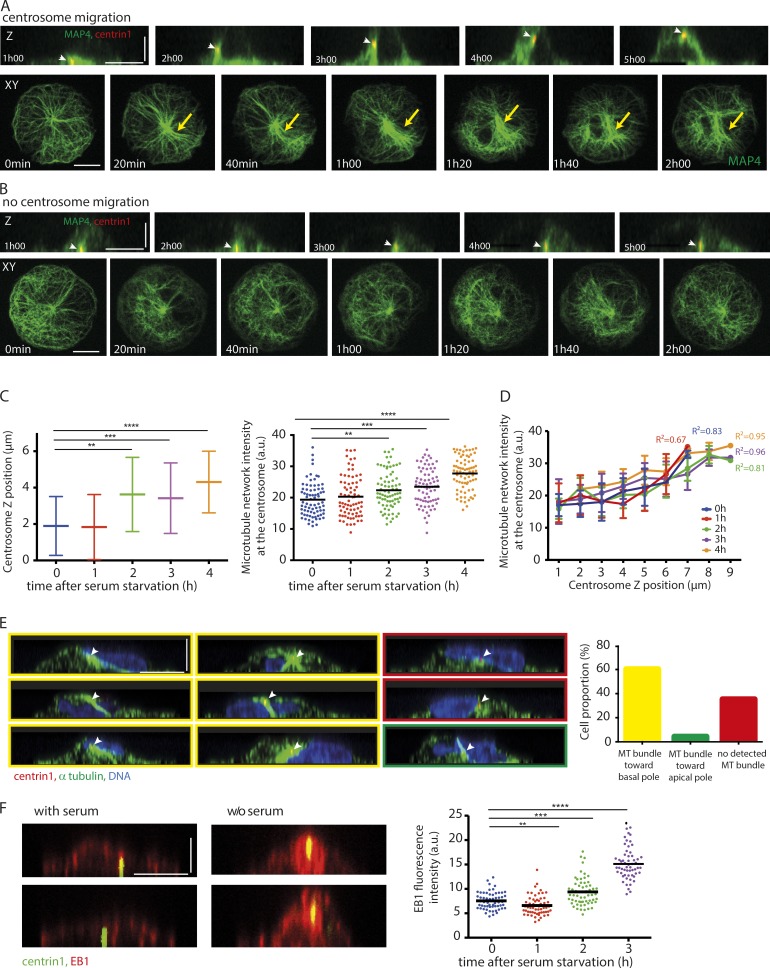
We then investigated whether microtubules were involved in the regulation of centrosome migration upon starvation. The addition of nocodazole to depolymerize microtubules immediately after serum starvation blocked centrosome migration and ciliogenesis (Fig. S2). When added 5 h after starvation, i.e., after the completion of centrosome migration, it had no detectable effect on centrosome position and cilia elongation (Fig. S2). This confirmed the specific implication of microtubules in centrosome migration. To investigate the role of microtubules in centrosome migration in more detail, live imaging of micropatterned RPE1 EGFP-centrin1 cells transduced with MAP4-RFP (Ganguly et al., 2013) to label the microtubules was performed (Fig. 3, A and B; and Video 1). Imaging of cells cultured in the absence of serum revealed that centrosome migration typically occurred 2–4 h after serum starvation, and this coincided with a dramatic increase in the number of microtubules surrounding the centrosome (Fig. 3 A). In cases where centrosome migration did not occur, no increase in microtubule density surrounding the centrosome was observed (Fig. 3 B). MAP4, being a known microtubule bundler, could be responsible for this effect. We controlled this by measuring the changes in microtubule density in nontransfected cells by fixing and staining serum-starved, micropatterned RPE1 cells with an antibody to α-tubulin (Fig. 3, C and D). Plotting of these measurements against the axial position of the centrosome showed that microtubule density increased significantly after serum starvation, and there was a positive correlation between centrosome position and microtubule density (correlation coefficient of 0.9; Fig. 3 D). The monitoring of microtubule network reorganization during centrosome 3D migration showed that microtubule network densification was associated with the clustering of microtubules in a large bundle (Video 1). The orientation of this bundle between the centrosome and the basal pole suggested that it was exerting pushing rather than pulling forces. Indeed, pulling forces would rather be associated with a bundle connecting the centrosome to the apical pole, as in the case of centrosome motion toward immune synapse (Yi et al., 2013). To quantify the occurrence of such pushing bundles, we fixed cells during the centrosome migration process, i.e., 80 min after serum starvation, and measured the frequency of bundle orientation toward the basal or apical pole whenever such a bundle was discernable (Fig. 3 E). Most centrosomes were associated with a microtubule bundle pointing toward the basal pole, suggesting that pushing forces had a major contribution to centrosome propulsion toward the apical pole.
Figure 3.
Microtubule network remodeling during centrosome migration. (A and B) Microtubule network organization was studied by time-lapse imaging of RPE1 cells expressing EGFP-centrin1 and MAP4-RFP. Two examples are shown: one where the centrosome (indicated by white arrowheads) migrated to the apical pole (A) and another where it did not (B). Orthogonal and top views are shown. Microtubule network symmetry breaking and densification are shown with yellow arrows. (C) Measurement of centrosome z position (left) and α-tubulin fluorescence intensity in a 5-µm box surrounding the centrosome (right) in thymidine-synchronized serum-starved RPE1 cells expressing EGFP-centrin1 for various delays after serum removal (n = 75 cells per condition). Horizontal bars show mean values. Error bars represent standard deviation. (D) The graph shows the microtubule network density at the centrosome against centrosome z position at various time points after serum starvation in thymidine-synchronized RPE1 cells expressing EGFP-centrin1. In all conditions, the two parameters were correlated. (E) RPE1 cells expressing EGFP-centrin1 (centrosome indicated by white arrowheads) were fixed 120 min after serum withdrawal and stained for α-tubulin and DAPI. The side views facilitated the visualization of microtubule bundle orientation and quantification of cell proportion exhibiting either a microtubule bundle toward the basal pole (images and bar graph with yellow outlines), a microtubule bundle toward the apical pole (images and bar graph with green outlines), or no detected microtubule bundle (images and bar graph with red outlines; results of four independent experiments, n = 116 cells). (F) Staining of serum-starved RPE1 cells expressing EGFP-centrin1 with an antibody to EB1. The graph shows EB1 fluorescence intensity measurements in a 5-µm box surrounding the centrosome (one experiment, n = 60 cells per condition). Bars: (x and y) 10 µm; (z) 5 µm. a.u., arbitrary units. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Combined, these data suggested that increased microtubule nucleation and/or stabilization was responsible for the densification of the microtubule network, which further generated pushing forces required to move the centrosome to the apical surface. To confirm such a model, serum-starved RPE1 cells were stained with an antibody to the microtubule end-binding protein EB1, and its fluorescence intensity was measured over time (Fig. 3 F). Strikingly, EB1 levels were found to be nearly twofold higher at the centrosome after 3 h of serum starvation, indicating that increased microtubule nucleation was likely responsible for driving centrosome migration during primary cilium formation.
Microtubule stabilization drives centrosome movement
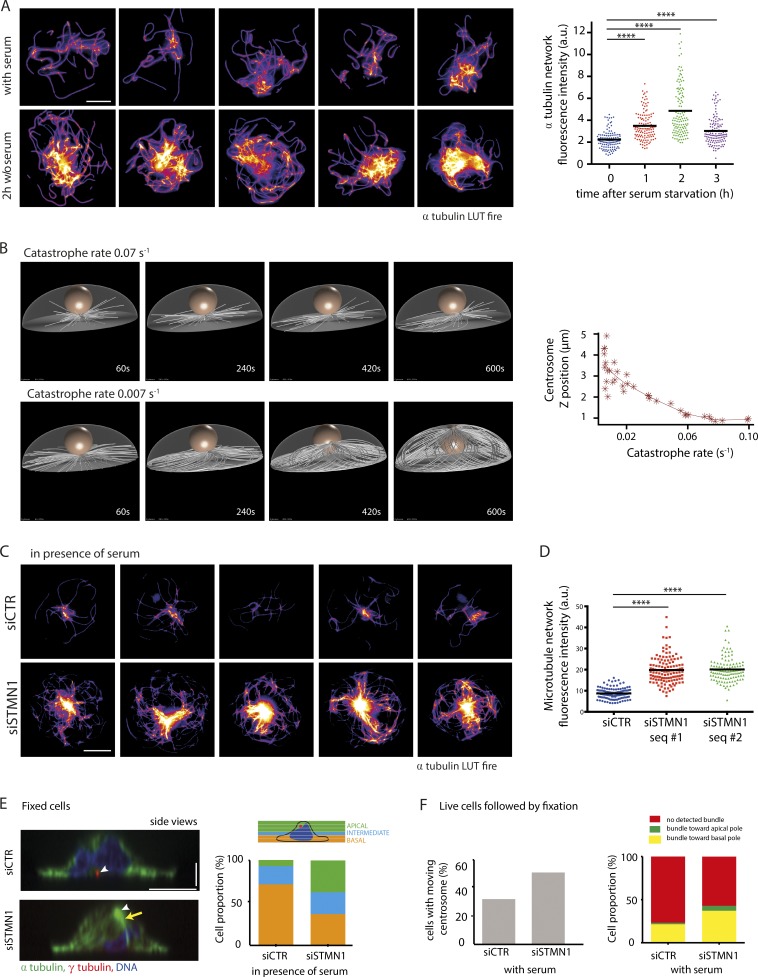
Microtubule stabilization could also participate in network densification and centrosome migration. Interestingly, serum starvation has been shown to reduce microtubule dynamics (Danowski, 1998) and increase characteristic posttranslational modifications of stabilized microtubules during primary ciliogenesis (Berbari et al., 2013). To investigate whether such changes could specifically contribute to centrosome migration, we subjected micropatterned RPE1 cells (serum starved for 1, 2, or 3 h) to a brief cold shock for 12 min and stained them with an antibody to α-tubulin (Fig. 4 A). Quantification of the amount of polymerized tubulin present in the cell showed that there was a remarkable twofold increase in the number of cold-stable microtubules 2 h after serum starvation. This increase in microtubule stability appeared to be transient, with tubulin levels decreasing after 3 h of serum starvation but remaining significantly higher than those of serum-fed cells. These data support a model whereby increased microtubule nucleation and microtubule stabilization work synergistically to generate a dense network of stable microtubules upon which the centrosome can migrate.
Figure 4.
Microtubule stabilization after serum starvation promotes centrosome migration. (A) Identification of cold-resistant microtubules. Serum-starved RPE1 cells were subjected to cold shock (on ice for 12 min) and fixed and stained with an antibody to α-tubulin (lookup table [LUT] fire [ImageJ, National Institutes of Health]). Images show five examples of serum-starved and five examples of serum-fed cells (LUT fire). The graph shows measurements of α-tubulin fluorescence intensity after cold shock for various delays after serum removal (results of three independent experiments, n = 125 cells per condition). (B) 3D numerical simulations of microtubule growth from the centrosome at the basal pole. They showed that longer microtubules, assembled by reducing the catastrophe rate, induced a symmetry break in the network architecture that was capable of pushing the centrosome to the apical surface. The graph shows the centrosome z position according to the catastrophe rate. (C and D) Identification of cold-resistant microtubules in serum-fed cells treated with either control siRNA or siRNA against the tubulin sequestering protein stathmin 1. The same conditions as in A. Images show five examples of serum-fed cells treated with control siRNA and five examples of serum-fed cells treated with stathmin 1 siRNA (C). (D) Graph shows measurements of α-tubulin fluorescence intensity after cold shock (results of two independent experiments, control siRNA, n = 125 cells; stathmin 1 siRNA, two siRNA sequences, n = 125 cells each). Horizontal bars show mean values. (E) Stathmin 1 was depleted by siRNA from RPE1 cells cultured in the presence of serum to promote microtubule growth and observe its effect on centrosome position. Cells were fixed and stained for α-tubulin, γ-tubulin, and DNA. Centrosomes are indicated by white arrowheads, and the microtubule bundle is shown with a yellow arrow (left side view images). The graph shows cell percentage displaying basal centrosome (located from 0 to 2 µm above the glass substrate), intermediate centrosome (between 2 and 3 µm above the glass substrate), and apical centrosome (located >3 µm above the glass substrate; results of three independent experiments, control siRNA, n = 100 cells; stathmin 1 siRNA, two siRNA sequences, n = 150 cells each). (F) Time-lapse imaging for 80 min of serum-fed RPE1 cells expressing EGFP-centrin1 treated with control siRNA or siRNA against stathmin 1, then fixation/immunostaining for α-tubulin. The left graph shows the percentage of cells exhibiting a moving centrosome toward the apical pole in each condition (siCTR vs. siStathmin 1). The right graph shows microtubule bundle orientation frequency for each condition (one experiment, control siRNA, n = 63 cells; siRNA against stathmin 1, n = 45 cells). Bars: (x and y) 10 µm; (z) 2.5 µm. a.u., arbitrary units. ****, P < 0.0001.
We tested this model using 3D numerical simulations (Foethke et al., 2009) to investigate how microtubule network density could impact on the axial position of the centrosome (Fig. 4 B). We built upon our previous simulations showing centrosome decentering upon microtubule lengthening in 2D (Letort et al., 2016; Burute et al., 2017) by taking into account cell shape in 3D and the centrosome’s interaction with the nucleus. Catastrophe rate variations were used to modulate microtubule length and network density. Increasing microtubule length by reducing their catastrophe rate from 0.06 to 0.02 events/s (range estimated from Janson et al., 2003) resulted in the formation of a dense network of stable microtubules capable of generating sufficient force to push the centrosome to the apical surface (Fig. 4 B and Video 2). No minus end–directed motors capable of exerting pulling forces from the cell cortex were added to these simulations. These results indicated that a reduction in the catastrophe rate that resulted in the formation of a network of stable microtubules, as observed experimentally in serum-starved cells, could reorganize microtubule network architecture and the net orientation of pushing forces to destabilize centrosome basal position and push it toward the apical pole.
To gather evidence to support the numerical simulation data, the tubulin-sequestering protein stathmin 1 (Belmont and Mitchison, 1996) was depleted from cells to increase the pool of free tubulin available and promote microtubule polymerization (Fig. 4 C, Fig. S1 D, and Table S1). Imaging of the microtubule network in RPE1 cells treated with siRNA against stathmin 1 in the presence of serum indeed showed a network of cold-stable microtubules reminiscent of those observed in serum-starved cells. The quantification revealed that tubulin levels were twofold higher after stathmin 1 depletion compared with siRNA control (Fig. 4 D). siRNA-treated RPE1 cells were cultured in the presence of serum and fixed and stained with γ-tubulin and α-tubulin antibodies (Fig. 4 E). Confocal imaging and measurement of the axial position of the centrosome showed that it was significantly closer to the apical surface in the stathmin 1–depleted cells than the control siRNA–treated cells, although serum had not been withdrawn in these experiments (Fig. 4 E). Furthermore, live imaging of EGFP-centrin1–labeled centrosomes followed by fixation and immunolabeling of microtubules revealed an increased proportion of centrosomes moving toward the apical pole and an increased frequency of microtubule bundles pointing toward the basal pole in stathmin 1–knocked down cells compared with control cells in the presence of serum (Fig. 4 F). These results support the proposal that increased microtubule polymerization and stabilization are sufficient to generate a microtubule network capable of pushing the centrosome toward the cell apical pole.
Actin network contraction and symmetry breaking promote apical centrosome motion
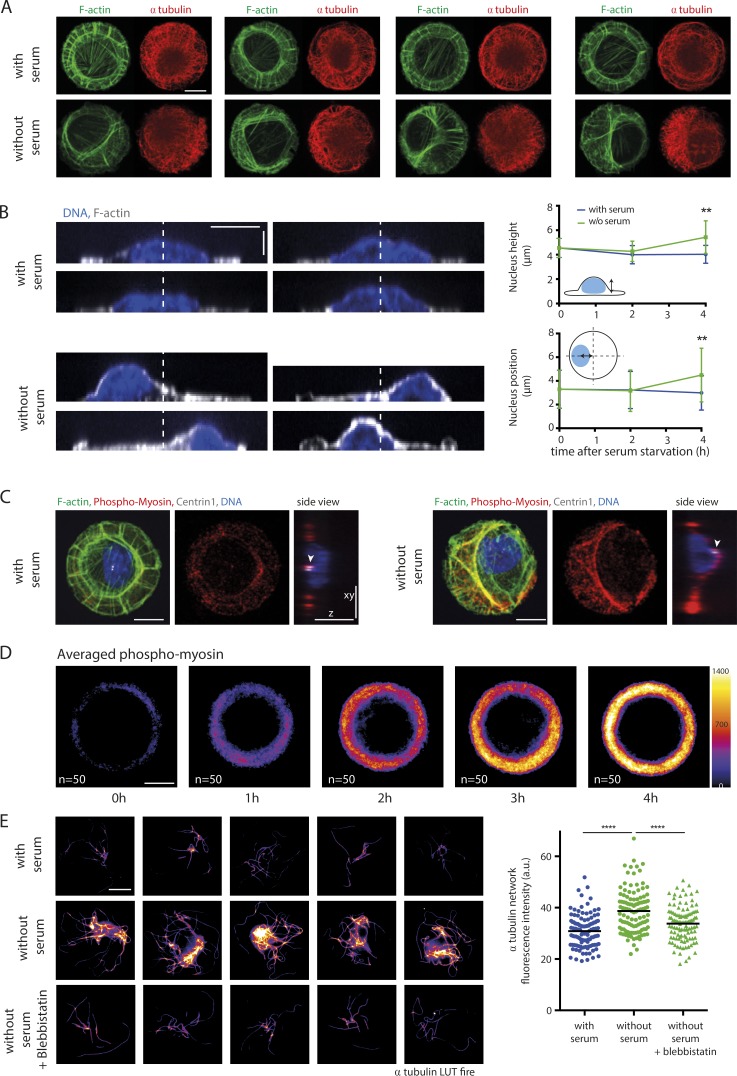
Numerous studies have implicated actin remodeling as part of the process of primary cilium formation (Dawe et al., 2009; Kim et al., 2010; Pitaval et al., 2010). We sought to characterize changes in the actin cytoskeleton during primary ciliogenesis by staining RPE1 cells with phalloidin to label filamentous actin and α-tubulin antibody to label the microtubules (Fig. 5 A). Before serum starvation, the actin cytoskeleton of micropatterned RPE1 cells was observed to be radially symmetrical in agreement with previously published data (Tee et al., 2015). However, after 4 h of serum starvation, the symmetry was broken, and actin filaments appeared preferentially clustered to one side of the cell. Transverse arcs forming a ring of bundled filaments tended to contract toward an off-centered position (Fig. 5 A). This not only impacted the microtubule cytoskeleton, resulting in the asymmetrical copartitioning of microtubules with F-actin; it also compressed the nucleus, which became higher and less spread out. The symmetry break in the actin network forced the nucleus to be displaced from the center (Fig. 5 B).
Figure 5.
Contractility increase breaks actin cytoskeleton symmetry and promotes microtubule stabilization. (A) Reorganization of the actin and microtubule cytoskeletons upon serum starvation. RPE1 cells were fixed 4 h after serum withdrawal, stained with phalloidin to visualize F-actin, immunostained with antibodies against α-tubulin, and compared with serum-fed cells. Images show four examples of serum-starved and serum-fed cells. (B) Nucleus positioning upon serum starvation. RPE1 cells were fixed 4 h after serum starvation, stained with phalloidin and DAPI and compared with serum-fed cells. Images show four examples of serum-starved and serum-fed cells. White dotted lines indicate the cell symmetry axis. The top graph shows measurements of nucleus height, and the bottom graph shows measurements of nucleus position in the x–y plane in fed and starved cells at various time points after serum starvation (results of two experiments, n = 75 cells). Error bars represent standard deviation. (C) Immunostaining against phosphomyosin showed an intense staining along the noncircular actin bundle (green) in serum-starved cells. White arrowheads point at centrosomes detected with anticentrin1 antibodies (white). RPE1 cells were fixed 4 h after serum withdrawal. (D) Averaging of phosphomyosin fluorescence intensity levels (LUT fire), obtained by stacking and averaging 50 images per condition, showed that the myosin phosphorylation increased after serum starvation. (E) Identification of cold-resistant microtubules in serum-fed and serum-starved cells in the presence or absence of the myosin II ATPase inhibitor blebbistatin. RPE1 cells were fixed 2 h after serum withdrawal. Images show five examples of serum-starved cells, five examples of serum-fed cells, and five examples of serum-starved cells treated with blebbistatin for 2 h (LUT fire). The graph shows measurements of α-tubulin fluorescence intensity after cold shock (results of two independent experiments, n = 110 cells for each condition). Bars: (x and y) 10 µm; (z) 5 µm. **, P < 0.01; ****, P < 0.0001. Horizontal bars show mean values. a.u., arbitrary units.
To ascertain whether myosin II was involved in actin remodeling during primary ciliogenesis, RPE1 cells were stained with phalloidin and phospho–MLC II antibody (Fig. 5 C). After serum starvation, actin filaments were found to be decorated with phospho–MLC II antibody, suggesting that remodeling of the actin cytoskeleton was caused by myosin II activity. Staining of RPE1 cells with phosphomyosin antibody followed by averaging of the fluorescent signal indicated that the level of phosphorylated myosin II increased with time after the induction of primary ciliogenesis (Fig. 5 D). Though these data suggested that myosin II activity was required for actin remodeling, they did not provide direct evidence of a role for myosin II–dependent contractility in centrosome migration. To test for such a role, RPE1 cells were treated with the myosin II inhibitor blebbistatin, and microtubule cold-resistance assays were performed. A dramatic reduction in the number of cold-stable microtubules, compared with controls, was observed after blebbistatin treatment and indicated that myosin II–dependent contractility was involved in microtubule reorganization and stabilization (Fig. 5 E).
Apical pole maturation follows centrosome migration
These results suggested the implication of an internal symmetry break in cytoskeleton organization that contrasted with the more classical view of the centrosome being off-centered by the action of localized pulling forces from a defined portion of the cell cortex (Tang and Marshall, 2012; Barker et al., 2016). We challenged our interpretation by looking at classical markers of the apical pole that could be involved in the local activation of pulling forces (ERM [ezrin, radixin, and moesin], NuMA, and p150Glued). Increased phosphorylation of ERM was observed after serum starvation (Fig. S3 A). However, this increase could only be detected after the centrosome migration process, suggesting that it was a more downstream event. Increased recruitment of the dynein-interacting proteins p150Glued and NuMA to the apical cortex was also observed after serum starvation but was initiated only 4 h after serum withdrawal when most centrosomes had already reached their apical position (Fig. S3, B and C), further confirming that their local accumulation at the apical pole followed rather than promoted centrosome migration.
Investigation of the role of ciliogenesis effectors in centrosome migration
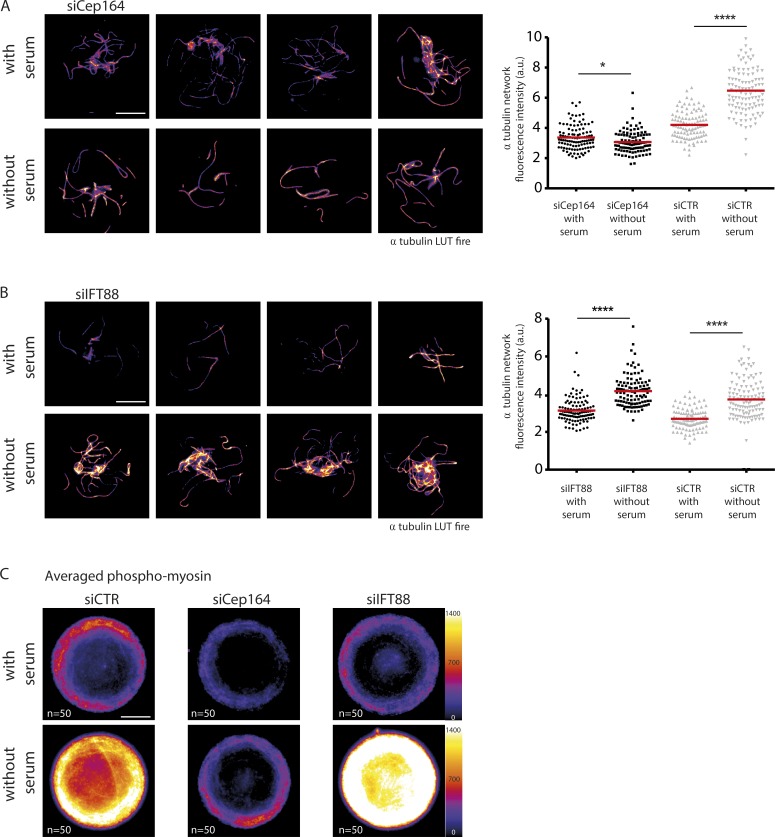
We concluded that centrosome movement was driven by concomitant and likely related (Joo and Yamada, 2014; Rao et al., 2014) stabilization of microtubules and contraction of the actin network, allowing symmetry breaks in both networks and the efficient production of microtubule-based pushing forces on the basal pole. Within this context, we decided to revisit the effect of the depletion of ciliogenesis effectors and test whether they affected centrosome migration via the mechanism we hypothesized. To that end, we investigated in more detail cytoskeleton organization resulting from the depletion of Cep164 or IFT88 because their ablation had opposing effects upon centrosome migration. In a microtubule cold-resistance assay, Cep164-depleted cells had fewer cold-resistant microtubules upon serum starvation (Fig. 6 A), whereas control and IFT88-depleted cells had more (Fig. 6, A and B). In the presence of serum, Cep164 knockdown decreased the amount of microtubules at the centrosome, whereas IFT88 tended to increase it (Fig. S4 A). Although the absence of Cep164 may affect centrosome position via its numerous partners (notably Rabin8, Rab8, and TTBK2; Westlake et al., 2011; Schmidt et al., 2012; Cajánek and Nigg, 2014), these observations suggested that the centrosome migration defect in Cep164-depleted cells could actually be caused by a failure to stabilize microtubules and remodel the network architecture accordingly. The proper centrosome migration in IFT88-depleted cells was consistent with proper microtubule reorganization in those cells. In parallel, phosphomyosin density appeared reduced in Cep164-depleted cells compared with control or IFT88-depleted cells (Fig. 6 C), further confirming the specific implication of the mechanism we discovered in the control of centrosome migration. Finally, the level of NuMA at the apical pole of serum-starved cells, which was seen to rise a few hours after centrosome migration, was lower in Cep164-depleted cells and higher in IFT88-depleted cells compared with controls (Fig. S4 B). As Cep164 is a centrosome regulator with no described effect in cortical actin, the absence of accumulation of NuMA at the apical pole is likely to be a consequence of the defective centrosome migration. Altogether, these results supported our conclusion that apical pole maturation in these conditions was a consequence rather than the cause of centrosome migration.
Figure 6.
The ciliogenesis effector Cep164 affects microtubule stabilization and actomyosin contractility upon serum starvation. (A) Identification of cold-resistant microtubules in serum-starved and serum-fed cells treated with siRNA against Cep164. Images show four representative examples of Cep164-depleted serum-starved and Cep164-depleted serum-fed cells (experiments obtained with two distinct Cep164 siRNA sequences). Cells fixed after a brief cold shock of 12 min after 3 h of serum starvation. The graph shows measurements of α-tubulin fluorescence intensity after cold shock (LUT fire; results of two independent experiments, n = 110 cells per condition). (B) Same as in A with siRNA against IFT88 (results of two independent experiments, n = 110 cells per condition). (C) Averaging of phosphomyosin fluorescence intensity levels (LUT fire), obtained by stacking and averaging 50 images per condition. Cells were fixed 4 h after serum starvation. Averaged images showed that myosin phosphorylation increased in control and IFT88-depleted cells, but not in Cep164-depleted cells. Bars, 10 µm. *, P < 0.05; ****, P < 0.0001. a.u., arbitrary units.
Discussion
In this paper, we have exploited the technique of cell micropatterning to characterize in detail the previously poorly defined step of centrosome migration during primary ciliogenesis. Microtubules in cells undergoing primary ciliogenesis were found to be more resistant to cold treatment, suggesting that they are more stable. In addition, increased levels of EB1 and tubulin at the centrosome were also observed. Elevated EB1 levels at the centrosome could represent an increase in microtubule nucleation or anchoring, as EB1 is involved in both (Yan et al., 2005). Numerical simulations in 3D showed an interesting consequence of this remodeling of the microtubule network. Increasing microtubule stability appeared sufficient to force network reorganization, leading to a vortex-like conformation that destabilized the basal position of the centrosome and pushed it up toward the cell apical pole. This mechanism was further confirmed experimentally, using the depletion of the tubulin-sequestering protein stathmin 1 to increase the level of free tubulin available for incorporation into polymers to generate an array of long microtubules. This resulted in the formation of an array capable of transmitting sufficient force to push the centrosome to the apical surface in the absence of any of the other compounding effects associated with serum starvation. Furthermore, ciliogenesis effectors involved in the regulation of centrosome migration, such as Cep164, also impacted microtubule stabilization and cell contractility, further supporting the implication of cytoskeleton remodeling in centrosome migration. This mechanism, relying on a symmetry break in the spatial organization of pushing forces, contrasts with the previously described mechanisms of centrosome off-centering in which unbalanced forces result from the asymmetrical distribution cortical pulling forces (Morin and Bellaïche, 2011; Tang and Marshall, 2012).
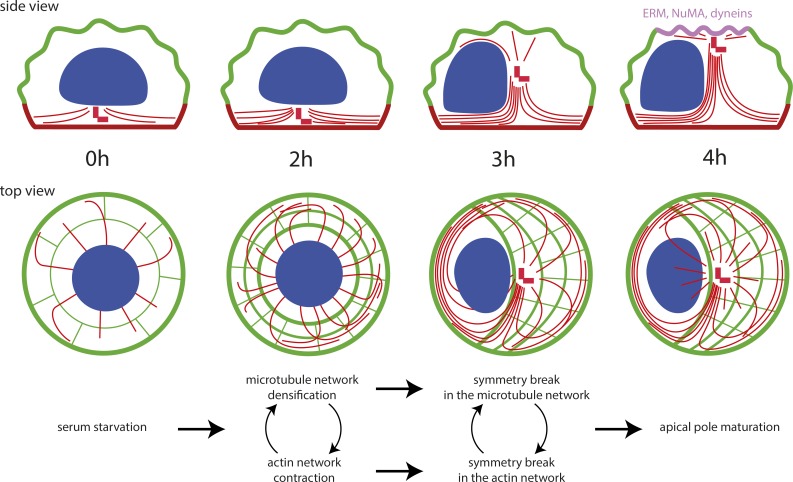
These observations led us to propose the following speculative scenario for the induction of centrosome migration when cells enter quiescence (Fig. 7). Centrosome maturation is initiated in the first 2 h after serum starvation and primes ciliogenesis (Westlake et al., 2011; Lu et al., 2015). Although we have no evidence that this maturation directly impacts microtubule dynamics, the lack of microtubule stabilization in response to Cep164 knockdown suggests that the Cep164-dependent recruitment of the ciliary vesicle and associated components (Westlake et al., 2011; Schmidt et al., 2012; Cajánek and Nigg, 2014) contributes to microtubule nucleation and stabilization. Microtubule lengthening appeared sufficient to induce a symmetry break in the spatial arrangement of microtubules. In parallel, microtubule stabilization is likely to feed back to actomyosin contractility via specific kinases and phosphatases coregulating the two pathways, such as myosin phosphatase (Joo and Yamada, 2014; Rao et al., 2014). The increase in actomyosin activity is sufficient to break the symmetry of the contracting network, as previously observed in several contractile systems (Yam and Theriot, 2004; Paluch et al., 2005; Sedzinski et al., 2011). This asymmetrical actin flow can move the nucleus away from the cell center (Gomes et al., 2005) and thereby facilitates centrosome apical movement from its initial central position and further contributes to the asymmetrical reconfiguration of the microtubule network that was initiated by microtubule stabilization. In addition, actomyosin contractility further contributes to microtubule stabilization. The increase in pushing forces associated with microtubule polymerization (Laan et al., 2008) and the imbalance in microtubule distribution destabilize the centrosome’s position at the basal pole (Pinot et al., 2009; Letort et al., 2016) and push it toward the apical pole, allowing microtubule elongation and the release of elastic stress that accumulated as a result of their bent conformation. When the centrosome reaches the apical membrane, it brings along minus end–directed motors, such as dyneins, and their associated proteins like NuMA (Merdes et al., 1996), which can then interact with the plasma membrane (Kotak et al., 2014). This local accumulation of microtubule-interacting proteins further contributes to the anchoring of the centrosome at the apical pole and the consequential accumulation of centrosome-associated proteins, as well as cargoes transported along the microtubules, which contribute to later stages of ciliogenesis (Reiter et al., 2012). Although our observations do not exclude a possible contribution of pulling forces exerted on centrosomal microtubules by minus end–directed motors anchored to a nascent apical pole, they provide compelling evidence for a major role played by microtubule pushing on the basal pole.
Figure 7.
Proposed sequence of events driving centrosome migration to the cell apical pole. These schemes show a side and top view of cytoskeleton rearrangements after serum withdrawal. Microtubule network densification and actin network contraction break the symmetry of both networks, which results in the production of pushing forces moving the centrosome to the dorsal surface. Upon contact, the centrosome promotes local surface maturation into an apical pole and centriole anchoring and elongation to form the primary cilium.
The key step in the mechanism we described is the transient stabilization of microtubules upon serum starvation. Serum starvation or activation of Rac1 or Cdc42 is known to result in microtubule end stabilization and life span increase (Danowski, 1998; Grigoriev et al., 2006), but the underlying mechanism still remains to be uncovered. Interestingly, microtubule stabilization and bundling upon entry into quiescence has been described in other systems. Recent work in fission yeast has shown that upon entry into quiescence, Schizosaccharomyces pombe assembles its microtubules into a single bundle that is attached to the spindle pole body, the yeast equivalent of the centrosome (Laporte et al., 2015). The assembly of this single microtubule bundle from the three to five bundles of microtubules that are normally present during interphase is Ase I and Mto I dependent. Interestingly, homologues of these proteins exist in humans, and they are PRC1 and CDK5RAP2 (Cep215), respectively. PRC1 is a microtubule-bundling protein that binds to the microtubules of the central spindle that forms in late mitosis (Mollinari et al., 2002), whereas CDK5RAP2 is a pericentriolar material protein involved in the nucleation of microtubules through the stimulation of γ-tubulin ring complex activity (Choi et al., 2010). It would be interesting to determine whether CDK5RAP2 participates in the increased nucleation of microtubules during centrosome migration and whether PRC1 is involved in reorganizing the microtubule network during primary cilium formation. The exact role of other centrosomal proteins, such as Cep164, and the associated recruitment of the ciliary vesicle in microtubule network reorganization during early ciliogenesis is still obscure. Interestingly, epithelial to mesenchymal transition was recently shown to be associated with a reduction in microtubule number and stability that was required for cell migration (Burute et al., 2017). In parallel, the simple knockdown of Cep164 was shown to be sufficient to induce such a transition and to foster the migration of epithelial kidney cells (Slaats et al., 2014). These two observations further support a probable role for Cep164 in the regulation of microtubule nucleation and stabilization that definitely deserves further investigation.
Our data also suggest a role for myosin-based contractility in reorganizing the actin cytoskeleton, which seems to facilitate centrosome migration by moving the nucleus and force microtubule network asymmetry. Reorganization of the actin cytoskeleton from a radially symmetrical to an asymmetrical array occurred within 3 h of the induction of primary ciliogenesis and was abolished by treatment with the myosin II inhibitor blebbistatin (unpublished data). Myosin II and ROCK inhibition were shown previously to impair centrosome migration (Pitaval et al., 2010). However, actin could have additional and independent effects on the regulation of centrosome migration. Here, we confirm earlier observations that the knockdown of meckelin, emerin, and nesprin2, which are actin-binding proteins ensuring the centrosome–nucleus connection, perturbs centrosome migration (Dawe et al., 2009). Interestingly, this suggests that the centrosome–nucleus link needs to be maintained during migration. Nucleus deformation or rotation upon actomyosin contraction may help apical centrosome displacement. The nucleus could also act as a guide to orient the pushing forces produced by the microtubule network. Our observations add to the increasing knowledge about the implication of actomyosin contractility in ciliogenesis, but the exact mechanism remains to be established.
Altogether, our data demonstrate that centrosome migration to the apical surface is orchestrated by coordinated changes in the actin and microtubule cytoskeletons, with an increase in microtubule stability playing an important part in the process. Identifying the factors responsible for mediating microtubule stability in response to serum starvation and the connection with the actin cytoskeleton remodeling should allow us to further understand the major intracellular reorganization that occurs when cells enter quiescence.
Materials and methods
Cell culture
Human telomerase-immortalized RPE1 cells (Clontech) and RPE1 cells stably expressing EGFP-centrin1 (a gift from A. Khodjakov, Wadsworth Center, Albany, NY) or Lifeact-GFP were cultured in a humidified incubator at 37°C in DMEM/F12 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (all from Life Technologies).
Cell plating on micropattern slides
Disk-shaped micropatterned coverslips were obtained from CYTOO or produced in-house according to previously established protocols (Azioune et al., 2009).
Inhibitors
RPE1 cells were treated with 50 µM blebbistatin for 2 h in the absence of serum. Synchronization of RPE1 cells was performed using a double thymidine block, culturing the cells in medium containing 2 mM thymidine (Sigma-Aldrich) for 16 h, releasing for 10 h, and culturing again in thymidine-containing medium for a further 16 h. Cells were released from the block by removing the thymidine-containing medium, and, after 10 h, when the cells were in early G1, they were plated onto micropatterns.
Viral transduction
RPE1 cells were transduced with BacMam MAP4-RFP virus (Life Technologies) according to the manufacturer’s instructions.
siRNA treatment
RPE1 cells were transfected with siRNAs (Qiagen and Dharmacon) using Lipofectamine RNAi Max transfection reagent (Life Technologies) at a final concentration of 10 nM according to the manufacturer’s instructions. At least two independent siRNAs were tested for each target and two or three independent experiments.
Antibodies and cytoskeletal labeling agents
Primary antibodies used in this study were used at the following dilutions and obtained from the following sources: mouse antiacetylated tubulin (1:10,000 for immunofluorescence [IF]; clone 6-11B-1; Sigma-Aldrich), rabbit anti–α-tubulin (1:3,000 for IF; AbD MCA77G; Serotec), rabbit anti-Cep164 (1:2,000 for Western blot [WB]; provided by E. Nigg, University of Basel, Basel, Switzerland), mouse anti-EB1 (1:500 for IF; 610535; BD Biosciences), rabbit anti–γ-tubulin (1:1,000 for IF; ab11317; Abcam), rabbit anti-GAPDH (1:2,000 for WB; 25778; Santa Cruz), rabbit anti-IFT88 (1:200 for WB; 13967; Proteintech), mouse anti–lamin A/C (1:5,000 for WB; clone 4C11; Sigma-Aldrich), rabbit anti-NuMA (1:100 for IF; 48773; Santa Cruz), mouse anti-p150Glued (1:100 for IF; 612709; BD Biosciences), anti-pERM (1:800 for IF; 3141; Cell Signaling Technology), rabbit anti–stathmin 1 (1:50,000 for WB; 52630; Abcam), and rabbit antiphosphomyosin light chain 2 (Ser19; 1:50 for IF; 3671; Cell Signaling Technology). Alexa fluorophore–conjugated secondary antibodies (Molecular Probes) were diluted 1:1,000. Alexa fluorophore–conjugated phalloidin (Molecular Probes) was resuspended in methanol and diluted 1:500 in PBS.
IF staining
Different fixation protocols were used and depended on the antigen-binding characteristics of the antibody. For the siRNA screening experiments, in which cells were stained with γ-tubulin and acetylated tubulin antibodies, fixation was performed using cold methanol/acetone (50:50) on ice for 5 min. EB1 staining required the cells to be fixed with cold methanol for 5 min. Phosphorylated myosin light chain 2 antibody staining required prepermeabilization of the cells with 0.1% Triton X-100 (Sigma-Aldrich) in MTBS buffer (60 mM Pipes, 25 mM Hepes, 5 mM EGTA, and 1 mM MgCl, pH 7) before fixation with 4% PFA for 15 min at ambient temperature. Stainings for phosphorylated ERM proteins, p150Glued, and NuMA were performed after fixation with 4% PFA for 15 min, followed by permeabilization with 0.1% Triton X-100 in PBS for 3 min and incubation with the antibody overnight at 4°C for pERM. Where cells were stained with α-tubulin antibody and phalloidin, fixation was performed with 0.5% glutaraldehyde in MTBS buffer containing 0.1% Triton X-100 for 15 min, followed by quenching with NaBH4 for 10 min, both at ambient temperature.
For all conditions, after fixation, the cells were washed and then blocked with PBSA (PBS containing 1.5% bovine serum albumin; Sigma-Aldrich) for 30 min. The cells were stained with primary antibodies diluted in PBSA for 1 h, with the exception of the pERM, where the cells were stained overnight at 4°C, followed by extensive washing with PBSA and staining with secondary antibodies diluted in PBSA for 30 min. The cells were washed with PBS, and the DNA was labeled with 0.2 µg/ml DAPI (Sigma-Aldrich) for 1 min. After washing the cells with water, the coverslips were air dried and mounted onto slides using Mowiol (Sigma-Aldrich).
Cold resistance assay
Cells were subjected to a cold shock on ice for 12 min and then prepermeabilized with MTBS buffer containing 0.1% Triton X-100 for 20 s and fixed with 0.2% glutaraldehyde in the same buffer for 15 min.
Live and fixed cell acquisition and analysis
Live-cell imaging was performed on a spinning-disk microscope (Nikon) equipped with a 60×, 1.4 NA objective lens and a camera (HQ2 CoolSnap; Photometrics; images were taken every 20 min). Images of fixed cells were captured on a microscope (BX61; Olympus) equipped with a 100×, 1.4 NA objective lens, and a camera (HQ2 CoolSnap). Some fluorescent images shown are maximal projections of z stacks acquired with oil immersion objectives at 100× (NA = 1.4) mounted on a piezo ceramic (Physics Instruments). Both microscopes were controlled with Metamorph software (MDS Analytical Technologies). Fluorescence images were also taken using confocal z stacks acquired with a confocal microscope (TCS-SP2; Leica) through a 63× objective (NA = 1.4).
Automated image acquisition and analysis were performed as previously described (Pitaval et al., 2013). For the siRNA screen, the centrosome z position, percentage of ciliated cells, and cilia length were quantified for each treated cell.
Western blotting
Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membrane using a semidry Western blotting apparatus (Bio-Rad). The membranes were blocked with PBS containing 5% nonfat milk for 1 h at ambient temperature. After blocking, the membranes were probed with primary antibodies overnight at 4°C. The membranes were washed four times with blocking buffer before adding HRP-conjugated secondary antibodies (Life Technologies), diluting as recommended in blocking buffer, and incubating for 30 min at ambient temperature. After washing three times with PBS containing 0.1% Tween 20 (Sigma-Aldrich), the membranes were developed using ECL reagent (Life Technologies) and imaged on the ChemiDoc system (Bio-Rad) or by exposing to scientific imaging film (Kodak).
Numerical simulations
Numerical simulations were performed using Cytosim software (Nedelec and Foethke, 2007).
Statistical tests
A Fisher exact test was performed to analyze contingency tables comparing the number of cells in two conditions for both control and siRNA, using R version 3.3.2. All the other data are presented as mean ± standard deviation. Results were analyzed using the Mann-Whitney test (Prism; GraphPad). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Online supplemental material
Fig. S1 shows the validation of siRNA effect on ciliogenesis and primary cilium length. Fig. S2 shows the effect of nocodazole on centrosome migration. Fig. S3 shows apical marker immunostainings at early and late stages of centrosome migration. Fig. S4 shows the effect of siRNA against Cep164 and IFT88 on NuMA recruitment at the cell apical pole. Video 1 shows microtubule network reorganization and bundle formation during centrosome migration upon serum starvation. Video 2 shows numerical simulation of microtubule network reconfiguration and centrosome motion in 3D at high (0.06 s−1) and low (0.007 s−1) microtubule catastrophe rates. Table S1 displays the sequences used in the candidate-based siRNA screen.
Supplementary Material
Acknowledgments
We thank Gregory Pazour (University of Massachusetts Medical School, Worcester, MA) for sharing ift20-null and Cep290-null cells, which we unfortunately did not have time to study in great detail, and Laurent Blanchoin and Maxence Nachury for helpful discussions.
This work was supported by a European Research Council Starting Grant to M. Théry (grant SpiCy 310472).
The authors declare no competing financial interests.
Author contributions: A. Pitaval performed most experiments with the help of F. Senger and J. Sillibourne. G. Letort performed numerical simulations. L. Guyon performed statistical analyses. X. Gidrol contributed expertise on the use of siRNA. M. Théry and J. Sillibourne supervised the project, analyzed the data, and wrote the manuscript.
Footnotes
Abbreviations used:
- ERM
- ezrin, radixin, and moesin
- IF
- immunofluorescence
- IFT
- intraflagellar transport
- LUT
- lookup table
- Pard3
- partitioning defective 3
- RPE1
- retinal pigment epithelial 1
- WB
- Western blot
References
- Adams M., Simms R.J., Abdelhamed Z., Dawe H.R., Szymanska K., Logan C.V., Wheway G., Pitt E., Gull K., Knowles M.A., et al. . 2012. A meckelin-filamin A interaction mediates ciliogenesis. Hum. Mol. Genet. 21:1272–1286. 10.1093/hmg/ddr557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azioune A., Storch M., Bornens M., Théry M., and Piel M.. 2009. Simple and rapid process for single cell micro-patterning. Lab Chip. 9:1640–1642. 10.1039/b821581m [DOI] [PubMed] [Google Scholar]
- Barker A.R., McIntosh K.V., and Dawe H.R.. 2016. Centrosome positioning in non-dividing cells. Protoplasma. 253:1007–1021. 10.1007/s00709-015-0883-5 [DOI] [PubMed] [Google Scholar]
- Belmont L.D., and Mitchison T.J.. 1996. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 84:623–631. 10.1016/S0092-8674(00)81037-5 [DOI] [PubMed] [Google Scholar]
- Berbari N.F., Sharma N., Malarkey E.B., Pieczynski J.N., Boddu R., Gaertig J., Guay-Woodford L., and Yoder B.K.. 2013. Microtubule modifications and stability are altered by cilia perturbation and in cystic kidney disease. Cytoskeleton. 70:24–31. 10.1002/cm.21088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitzer A.L., Panagis L., Gusella G.L., Danias J., Mlodzik M., and Iomini C.. 2011. Primary cilia dynamics instruct tissue patterning and repair of corneal endothelium. Proc. Natl. Acad. Sci. USA. 108:2819–2824. 10.1073/pnas.1016702108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. 2012. The centrosome in cells and organisms. Science. 335:422–426. 10.1126/science.1209037 [DOI] [PubMed] [Google Scholar]
- Burakov A., Nadezhdina E., Slepchenko B., and Rodionov V.. 2003. Centrosome positioning in interphase cells. J. Cell Biol. 162:963–969. 10.1083/jcb.200305082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burute M., Prioux M., Blin G., Truchet S., Letort G., Tseng Q., Bessy T., Lowell S., Young J., Filhol O., and Théry M.. 2017. Polarity Reversal by Centrosome Repositioning Primes Cell Scattering during Epithelial-to-Mesenchymal Transition. Dev. Cell. 40:168–184. 10.1016/j.devcel.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajánek L., and Nigg E.A.. 2014. Cep164 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proc. Natl. Acad. Sci. USA. 111:E2841–E2850. 10.1073/pnas.1401777111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.-K., Liu P., Sze S.K., Dai C., and Qi R.Z.. 2010. CDK5RAP2 stimulates microtubule nucleation by the γ-tubulin ring complex. J. Cell Biol. 191:1089–1095. 10.1083/jcb.201007030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danowski B.A. 1998. Microtubule dynamics in serum-starved and serum-stimulated Swiss 3T3 mouse fibroblasts: implications for the relationship between serum-induced contractility and microtubules. Cell Motil. Cytoskeleton. 40:1–12. [DOI] [PubMed] [Google Scholar]
- Dawe H.R., Adams M., Wheway G., Szymanska K., Logan C.V., Noegel A.A., Gull K., and Johnson C.A.. 2009. Nesprin-2 interacts with meckelin and mediates ciliogenesis via remodelling of the actin cytoskeleton. J. Cell Sci. 122:2716–2726. 10.1242/jcs.043794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina F., Gaillard J., Guérin C., Couté Y., Sillibourne J., Blanchoin L., and Théry M.. 2015. The centrosome is an actin-organizing centre. Nat. Cell Biol. 18:65–75. 10.1038/ncb3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foethke D., Makushok T., Brunner D., and Nédélec F.. 2009. Force- and length-dependent catastrophe activities explain interphase microtubule organization in fission yeast. Mol. Syst. Biol. 5:241 10.1038/msb.2008.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J.A., Tuft R.A., Fogarty K.E., and Pazour G.J.. 2006. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 17:3781–3792. 10.1091/mbc.E06-02-0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A., Yang H., and Cabral F.. 2013. Detection and quantification of microtubule detachment from centrosomes and spindle poles. Methods Cell Biol. 115:49–62. 10.1016/B978-0-12-407757-7.00004-9 [DOI] [PubMed] [Google Scholar]
- Goetz S.C., and Anderson K.V.. 2010. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 11:331–344. 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E.R., Jani S., and Gundersen G.G.. 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 121:451–463. 10.1016/j.cell.2005.02.022 [DOI] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.-D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., and Nigg E.A.. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179:321–330. 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I., Borisy G., and Vorobjev I.. 2006. Regulation of microtubule dynamics in 3T3 fibroblasts by Rho family GTPases. Cell Motil. Cytoskeleton. 63:29–40. 10.1002/cm.20107 [DOI] [PubMed] [Google Scholar]
- Hong H., Kim J., and Kim J.. 2015. Myosin heavy chain 10 (MYH10) is required for centriole migration during the biogenesis of primary cilia. Biochem. Biophys. Res. Commun. 461:180–185. 10.1016/j.bbrc.2015.04.028 [DOI] [PubMed] [Google Scholar]
- Janson M.E., de Dood M.E., and Dogterom M.. 2003. Dynamic instability of microtubules is regulated by force. J. Cell Biol. 161:1029–1034. 10.1083/jcb.200301147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen J.A., San Agustin J., Follit J.A., and Pazour G.J.. 2008. Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J. Cell Biol. 183:377–384. 10.1083/jcb.200808137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo E.E., and Yamada K.M.. 2014. MYPT1 regulates contractility and microtubule acetylation to modulate integrin adhesions and matrix assembly. Nat. Commun. 5:3510 10.1038/ncomms4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo K., Kim C.G., Lee M.-S., Moon H.-Y., Lee S.-H., Kim M.J., Kweon H.-S., Park W.-Y., Kim C.-H., Gleeson J.G., and Kim J.. 2013. CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc. Natl. Acad. Sci. USA. 110:5987–5992. 10.1073/pnas.1220927110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyk A., Gromley A., Redick S., San Agustin J., Witman G., Pazour G.J., Peters D.J.M., and Doxsey S.. 2004. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J. Cell Biol. 166:637–643. 10.1083/jcb.200405023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee J.E., Heynen-Genel S., Suyama E., Ono K., Lee K., Ideker T., Aza-Blanc P., and Gleeson J.G.. 2010. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 464:1048–1051. 10.1038/nature08895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., and Kimura A.. 2011. A novel mechanism of microtubule length-dependent force to pull centrosomes toward the cell center. BioArchitecture. 1:74–79. 10.4161/bioa.1.2.15549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knödler A., Feng S., Zhang J., Zhang X., Das A., Peränen J., and Guo W.. 2010. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA. 107:6346–6351. 10.1073/pnas.1002401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Busso C., and Gönczy P.. 2014. NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane. EMBO J. 33:1815–1830. 10.15252/embj.201488147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L., Husson J., Munteanu E.L., Kerssemakers J.W.J., and Dogterom M.. 2008. Force-generation and dynamic instability of microtubule bundles. Proc. Natl. Acad. Sci. USA. 105:8920–8925. 10.1073/pnas.0710311105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D., Courtout F., Pinson B., Dompierre J., Salin B., Brocard L., and Sagot I.. 2015. A stable microtubule array drives fission yeast polarity reestablishment upon quiescence exit. J. Cell Biol. 210:99–113. 10.1083/jcb.201502025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.F. 2015. IFT-Cargo Interactions and Protein Transport in Cilia. Trends Biochem. Sci. 40:765–778. 10.1016/j.tibs.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letort G., Nedelec F., Blanchoin L., and Théry M.. 2016. Centrosome centering and decentering by microtubule network rearrangement. Mol. Biol. Cell. 27:2833–2843. 10.1091/mbc.E16-06-0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Hiesberger T., Cordes K., Sinclair A.M., Goldstein L.S.B., Somlo S., and Igarashi P.. 2003. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc. Natl. Acad. Sci. USA. 100:5286–5291. 10.1073/pnas.0836980100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Insinna C., Ott C., Stauffer J., Pintado P.A., Rahajeng J., Baxa U., Walia V., Cuenca A., Hwang Y.-S., et al. . 2015. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat. Cell Biol. 17:228–240. 10.1038/ncb3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J.D., and Cleveland D.W.. 1996. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 87:447–458. 10.1016/S0092-8674(00)81365-3 [DOI] [PubMed] [Google Scholar]
- Mollinari C., Kleman J.-P., Jiang W., Schoehn G., Hunter T., and Margolis R.L.. 2002. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 157:1175–1186. 10.1083/jcb.200111052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X., and Bellaïche Y.. 2011. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell. 21:102–119. 10.1016/j.devcel.2011.06.012 [DOI] [PubMed] [Google Scholar]
- Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peränen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., and Jackson P.K.. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 129:1201–1213. 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- Nedelec F., and Foethke D.. 2007. Collective Langevin dynamics of flexible cytoskeletal fibers. New J. Phys. 9:427 10.1088/1367-2630/9/11/427 [DOI] [Google Scholar]
- Nigg E.A., and Raff J.W.. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678. 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Obino D., Farina F., Malbec O., Sáez P.J., Maurin M., Gaillard J., Dingli F., Loew D., Gautreau A., Yuseff M.-I., et al. . 2016. Actin nucleation at the centrosome controls lymphocyte polarity. Nat. Commun. 7:10969 10.1038/ncomms10969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch E., Piel M., Prost J., Bornens M., and Sykes C.. 2005. Cortical actomyosin breakage triggers shape oscillations in cells and cell fragments. Biophys. J. 89:724–733. 10.1529/biophysj.105.060590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Dickert B.L., Vucica Y., Seeley E.S., Rosenbaum J.L., Witman G.B., and Cole D.G.. 2000. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151:709–718. 10.1083/jcb.151.3.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinot M., Chesnel F., Kubiak J.Z., Arnal I., Nedelec F.J., and Gueroui Z.. 2009. Effects of confinement on the self-organization of microtubules and motors. Curr. Biol. 19:954–960. 10.1016/j.cub.2009.04.027 [DOI] [PubMed] [Google Scholar]
- Pitaval A., Tseng Q., Bornens M., and Théry M.. 2010. Cell shape and contractility regulate ciliogenesis in cell cycle-arrested cells. J. Cell Biol. 191:303–312. 10.1083/jcb.201004003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitaval A., Christ A., Curtet A., Tseng Q., and Théry M.. 2013. Probing ciliogenesis using micropatterned substrates. Methods Enzymol. 525:109–130. 10.1016/B978-0-12-397944-5.00006-7 [DOI] [PubMed] [Google Scholar]
- Pooranachandran N., and Malicki J.J.. 2016. Unexpected Roles for Ciliary Kinesins and Intraflagellar Transport Proteins. Genetics. 203:771–785. 10.1534/genetics.115.180943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Y., Hao R., Wang B., and Yao T.-P.. 2014. A Mec17-Myosin II Effector Axis Coordinates Microtubule Acetylation and Actin Dynamics to Control Primary Cilium Biogenesis. PLoS One. 9:e114087 10.1371/journal.pone.0114087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J.F., Blacque O.E., and Leroux M.R.. 2012. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 13:608–618. 10.1038/embor.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Crisp M.L., Liu Q., Kim D., Kozlov S., Stewart C.L., and Burke B.. 2009. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl. Acad. Sci. USA. 106:2194–2199. 10.1073/pnas.0808602106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salpingidou G., Smertenko A., Hausmanowa-Petrucewicz I., Hussey P.J., and Hutchison C.J.. 2007. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J. Cell Biol. 178:897–904. 10.1083/jcb.200702026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K.N., Kuhns S., Neuner A., Hub B., Zentgraf H., and Pereira G.. 2012. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 199:1083–1101. 10.1083/jcb.201202126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M., Lu W., Neumann S., Brachner A., Gotzmann J., Noegel A.A., and Karakesisoglou I.. 2011. Molecular mechanisms of centrosome and cytoskeleton anchorage at the nuclear envelope. Cell. Mol. Life Sci. 68:1593–1610. 10.1007/s00018-010-0535-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedzinski J., Biro M., Oswald A., Tinevez J.-Y., Salbreux G., and Paluch E.. 2011. Polar actomyosin contractility destabilizes the position of the cytokinetic furrow. Nature. 476:462–466. 10.1038/nature10286 [DOI] [PubMed] [Google Scholar]
- Sfakianos J., Togawa A., Maday S., Hull M., Pypaert M., Cantley L., Toomre D., and Mellman I.. 2007. Par3 functions in the biogenesis of the primary cilium in polarized epithelial cells. J. Cell Biol. 179:1133–1140. 10.1083/jcb.200709111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N., Kosan Z.A., Stallworth J.E., Berbari N.F., and Yoder B.K.. 2011. Soluble levels of cytosolic tubulin regulate ciliary length control. Mol. Biol. Cell. 22:806–816. 10.1091/mbc.E10-03-0269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne J.E., Hurbain I., Grand-Perret T., Goud B., Tran P., and Bornens M.. 2013. Primary ciliogenesis requires the distal appendage component Cep123. Biol. Open. 2:535–545. 10.1242/bio.20134457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla V., Romaguera-Ros M., Garcia-Verdugo J.M., and Reiter J.F.. 2010. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev. Cell. 18:410–424. 10.1016/j.devcel.2009.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe C.W., and Lu X.. 2011. Kif3a regulates planar polarization of auditory hair cells through both ciliary and non-ciliary mechanisms. Development. 138:3441–3449. 10.1242/dev.065961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaats G.G., Ghosh A.K., Falke L.L., Le Corre S., Shaltiel I.A., van de Hoek G., Klasson T.D., Stokman M.F., Logister I., Verhaar M.C., et al. . 2014. Nephronophthisis-associated CEP164 regulates cell cycle progression, apoptosis and epithelial-to-mesenchymal transition. PLoS Genet. 10:e1004594 10.1371/journal.pgen.1004594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N., and Marshall W.F.. 2012. Centrosome positioning in vertebrate development. J. Cell Sci. 125:4951–4961. 10.1242/jcs.038083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto H., Kimura A., and Minc N.. 2016. Shape-motion relationships of centering microtubule asters. J. Cell Biol. 212:777–787. 10.1083/jcb.201510064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos B.E., Yang H.-J., Soni R., Wang W.-J., Macaluso F.P., Asara J.M., and Tsou M.-F.B.. 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27:163–168. 10.1101/gad.207043.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee Y.H., Shemesh T., Thiagarajan V., Hariadi R.F., Anderson K.L., Page C., Volkmann N., Hanein D., Sivaramakrishnan S., Kozlov M.M., and Bershadsky A.D.. 2015. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat. Cell Biol. 17:445–457. 10.1038/ncb3137 [DOI] [PubMed] [Google Scholar]
- Théry M., Racine V., Piel M., Pépin A., Dimitrov A., Chen Y., Sibarita J.-B., and Bornens M.. 2006. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. USA. 103:19771–19776. 10.1073/pnas.0609267103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake C.J., Baye L.M., Nachury M.V., Wright K.J., Ervin K.E., Phu L., Chalouni C., Beck J.S., Kirkpatrick D.S., Slusarski D.C., et al. . 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA. 108:2759–2764. 10.1073/pnas.1018823108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheway G., Schmidts M., Mans D.A., Szymanska K., Nguyen T.T., Racher H., Phelps I.G., Toedt G., Kennedy J., Wunderlich K.A., et al. University of Washington Center for Mendelian Genomics . 2015. An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat. Cell Biol. 17:1074–1087. 10.1038/ncb3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam P.T., and Theriot J.A.. 2004. Repeated cycles of rapid actin assembly and disassembly on epithelial cell phagosomes. Mol. Biol. Cell. 15:5647–5658. 10.1091/mbc.E04-06-0509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Habedanck R., and Nigg E.A.. 2005. A complex of two centrosomal proteins, CAP350 and FOP, cooperates with EB1 in microtubule anchoring. Mol. Biol. Cell. 17:634–644. 10.1091/mbc.E05-08-0810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J., Wu X., Chung A.H., Chen J.K., Kapoor T.M., and Hammer J.A.. 2013. Centrosome repositioning in T cells is biphasic and driven by microtubule end-on capture-shrinkage. J. Cell Biol. 202:779–792. 10.1083/jcb.201301004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Burakov A., Rodionov V., and Mogilner A.. 2010. Finding the cell center by a balance of dynein and myosin pulling and microtubule pushing: a computational study. Mol. Biol. Cell. 21:4418–4427. 10.1091/mbc.E10-07-0627 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.