Adherens junction remodeling allows changes in cell shape and position. Zilberman et al. show through live imaging that CDC-42 is dispensable for epithelial cell polarization, but its RhoGAP-regulated activity is needed to control junctional actin organization during embryo elongation.
Abstract
During morphogenesis, adherens junctions (AJs) remodel to allow changes in cell shape and position while preserving adhesion. Here, we examine the function of Rho guanosine triphosphatase CDC-42 in AJ formation and regulation during Caenorhabditis elegans embryo elongation, a process driven by asymmetric epidermal cell shape changes. cdc-42 mutant embryos arrest during elongation with epidermal ruptures. Unexpectedly, we find using time-lapse fluorescence imaging that cdc-42 is not required for epidermal cell polarization or junction assembly, but rather is needed for proper junctional actin regulation during elongation. We show that the RhoGAP PAC-1/ARHGAP21 inhibits CDC-42 activity at AJs, and loss of PAC-1 or the interacting linker protein PICC-1/CCDC85A-C blocks elongation in embryos with compromised AJ function. pac-1 embryos exhibit dynamic accumulations of junctional F-actin and an increase in AJ protein levels. Our findings identify a previously unrecognized molecular mechanism for inhibiting junctional CDC-42 to control actin organization and AJ protein levels during epithelial morphogenesis.
Introduction
Polarized cell shape changes provide forces that alter the morphology of tissues, organs, and embryos. For example, changes in the shapes of Caenorhabditis elegans epidermal cells transform the embryo from an ellipse into an elongated worm-shaped cylinder in the absence of cell division. Epidermal cells are born on the dorsal surface of the embryo, then migrate ventrally and form new junctions with contralateral epidermal cells to wrap the embryo in skin (“ventral enclosure”; Chisholm and Hardin, 2005; Vuong-Brender et al., 2016). After completing ventral enclosure, epidermal cells begin to lengthen along their anterior-posterior axis and simultaneously shrink along their dorsal-ventral axis (“elongation”; Fig. 1 A). Actomyosin contractions in lateral epidermal cells provide the forces that alter epidermal cell shape during the early stage of elongation (Armenti and Nance, 2012; Cram, 2014; Vuong-Brender et al., 2016). Subsequently, the contraction of underlying muscles attached to epidermal cells provides forces that allow elongation to continue up to the fourfold stage (Armenti and Nance, 2012; Cram, 2014; Vuong-Brender et al., 2016). It is unclear how epidermal cells regulate adherens junctions (AJs) and their associated microfilaments during elongation to allow the remodeling needed for these asymmetric cell shape changes while still preserving cell adhesion. This problem is common to all types of epithelial cells that alter their shapes or change positions relative to neighbors during morphogenesis (Collinet and Lecuit, 2013; Röper, 2015).
Figure 1.
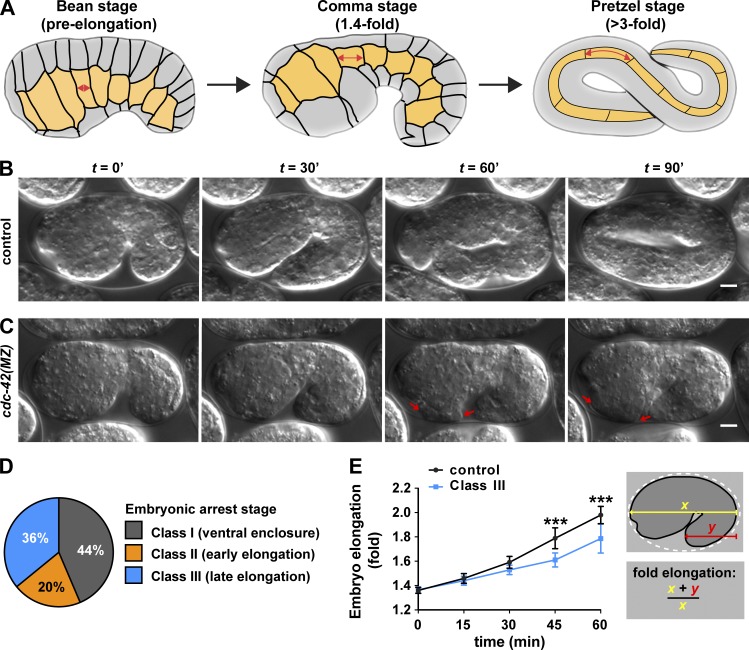
cdc-42 embryos have defects in ventral enclosure and elongation. (A) Stages of embryo elongation: bean stage (pre-elongation), comma stage (1.4-fold), and pretzel stage (>3-fold). Junctions between epidermal cells are indicated with black lines. Lateral epidermal cells (“seam” cells) are yellow. Double-headed arrows indicate the extension in anterior-posterior length of a cell as the embryo elongates. (B and C) Stills from DIC time-lapse movies of control and cdc-42(MZ) embryos shown at 30-min intervals. Genotypes were confirmed by single-embryo PCR after imaging. Arrows in C point to extruding cells. See Video 1. (D) Phenotypic classes of arrested embryos from DIC time-lapse imaging experiments (n = 39). (E) Rates of elongation in control (n = 13) and Class III (n = 9) embryos. Fold elongation was measured as schematized. t = 0 represents the comma stage. Values are the mean ± SD. Data for D and E were pooled from eight independent imaging experiments. P-values were calculated using a Mann-Whitney U test. ***, P ≤ 0.001. Bars, 5 µm.
C. elegans AJs contain highly conserved components, including the transmembrane homophilic adhesion protein HMR-1/E-cadherin and the cytoplasmic catenins HMP-1/α-catenin and HMP-2/β-catenin, which interact with the HMR-1/E-cadherin cytoplasmic tail and link it to actin microfilaments (Costa et al., 1998; Korswagen et al., 2000; Kwiatkowski et al., 2010). Null mutations in hmr-1/E-cadherin, hmp-1/α-catenin, or hmp-2/β-catenin cause microfilaments to detach from AJs as epidermal cells elongate, leading to developmental arrest and epidermal rupture (Costa et al., 1998). In addition to α-catenin and β-catenin, the p120 catenin JAC-1 also binds to the cytoplasmic tail of HMR-1/E-cadherin (Pettitt et al., 2003). Although JAC-1 is not essential in C. elegans (Klompstra et al., 2015), its depletion enhances the phenotype of weak mutations in hmp-1 (Pettitt et al., 2003), indicating that JAC-1 is an important regulator of AJ function.
AJs form through a two-step process of polarization and junction maturation. These events occur during the middle of embryogenesis, when epithelial precursor cells undergo a mesenchymal-to-epithelial transition (MET). During the polarization step of MET, clusters of AJ proteins found along the lateral membrane concentrate at the apicolateral region of the cell (Leung et al., 1999; McMahon et al., 2001; Achilleos et al., 2010). Concomitantly, polarity regulators begin to occupy distinct subdomains at the cell surface: the adaptor protein PAR-6 localizes apically, the scaffolding protein PAR-3 concentrates at AJs, the Discs large homologue DLG-1 accumulates at the basal side of AJs, and the Scribble protein LET-413 localizes to basolateral surfaces (Legouis et al., 2000; Bossinger et al., 2001; Firestein and Rongo, 2001; Köppen et al., 2001; McMahon et al., 2001; Aono et al., 2004; Achilleos et al., 2010). Whereas PAR-3 mediates polarization of other epithelial cell types in C. elegans (Köppen et al., 2001; Aono et al., 2004; Achilleos et al., 2010), epidermal cells polarize through an unknown PAR-3–independent mechanism. Junction maturation requires PAR-6, DLG-1, and LET-413; embryos lacking any of these polarity regulators arrest during elongation (Legouis et al., 2000; Bossinger et al., 2001; Firestein and Rongo, 2001; Köppen et al., 2001; McMahon et al., 2001; Totong et al., 2007).
One important regulator of AJs that has not been examined during epidermal cell MET or elongation is the Rho GTPase CDC-42. Active CDC-42 can interface with PAR proteins by binding directly to the PAR-6 CRIB (CDC-42/Rac interactive binding) domain (Gotta et al., 2001; Aceto et al., 2006), and interfering with CDC-42 function in many epithelial cell types disrupts polarity or the turnover of AJ components, leading to epithelial defects (Tepass, 2012; Duquette and Lamarche-Vane, 2014; Mack and Georgiou, 2014). For example, Cdc42 is required for apical membrane differentiation in the blastoderm of Drosophila melanogaster embryos (Hutterer et al., 2004) and in epithelial MDCK cells grown in 3D culture (Martin-Belmonte et al., 2007). Removing Cdc42 in fully polarized Drosophila epithelia causes defects in AJ organization by altering endocytosis or exocytosis, depending on the tissue (Georgiou et al., 2008; Harris and Tepass, 2008; Leibfried et al., 2008). CDC-42 can also regulate junction stability by controlling actin polymerization and turnover. For instance, multiple actin-nucleating factors, such as the Arp2/3 complex and formins, function downstream of CDC-42 to influence F-actin organization at junctions (Otani et al., 2006; Verma et al., 2012; Phng et al., 2015; Rao and Zaidel-Bar, 2016). In turn, junctional F-actin can modulate AJs by affecting endocytosis, or by physically controlling E-cadherin clustering (Collinet and Lecuit, 2013; Truong Quang et al., 2013; Wu et al., 2015).
Because CDC-42 is widely distributed in cells and regulates many different events (Erickson and Cerione, 2001), its activity at specific subcellular locations is often locally controlled. Like other Rho GTPases, CDC-42 is active when bound to GTP and inactive when bound to GDP. RhoGEFs activate CDC-42 and other Rho GTPases by promoting GTP binding, whereas RhoGAPs function as inhibitors by promoting GTP hydrolysis (Bos et al., 2007). Studies using constitutively active and dominant negative versions of CDC-42 have shown that its activity must be regulated to ensure normal junction formation and maintenance (Kroschewski et al., 1999; Rojas et al., 2001; Bruewer et al., 2004; Elbediwy et al., 2012), indicating that RhoGEFs and RhoGAPs are likely to influence CDC-42 activity at junctions. In cultured mammalian epithelial cells, CDC-42 activity is tuned at different stages of junction biogenesis by a small number of GEFs and GAPs, including the RhoGEFs Tuba and Dbl3/ARHGEF21 (Otani et al., 2006; Zihni et al., 2014) and the RhoGAPs Rich1 and SH3BP1 (Wells et al., 2006; Elbediwy et al., 2012). However, it remains unclear which RhoGEFs or RhoGAPs function in vivo to regulate CDC-42 activity as junctions remodel during morphogenesis.
Here, we show that, unexpectedly, CDC-42 is dispensable for polarization and junction maturation during MET in the C. elegans epidermis, but is essential during elongation to control the polarized cell shape changes and junctional actin dynamics of epidermal cells. We identify the conserved RhoGAP PAC-1/ARHGAP21, which localizes specifically to AJs and likely functions with its binding partner PICC-1/CCDC85A-C, as a negative regulator of CDC-42 activity at these sites. pac-1 mutant embryos form ectopic, dynamic, actin extensions at AJs, exhibit increased AJ protein levels, and fail to elongate in a sensitized genetic background. Our findings reveal that RhoGAP-mediated CDC-42 regulation enables tight control over actin organization and AJ protein levels in vivo, allowing epithelial cells to remodel their junctions efficiently as cells change shape.
Results
CDC-42 is required for proper ventral enclosure and embryo elongation
C. elegans has a single cdc-42 homologue, which is expressed ubiquitously in embryos, larvae, and adults (Anderson et al., 2008; Armenti et al., 2014b; Neukomm et al., 2014). Within the embryo, CDC-42 protein arises from both maternal and zygotic sources. Reducing levels of maternal CDC-42 results in a loss of polarity and defects in spindle orientation in early embryos, culminating in embryonic lethality (Gotta et al., 2001; Kay and Hunter, 2001; Anderson et al., 2008). We examined cdc-42 function in later embryos by using a degron-based strategy to deplete maternal CDC-42 protein from the early embryo just after its function in the one-cell embryo is complete, while simultaneously eliminating zygotic cdc-42 expression with a cdc-42 null allele, gk388 (Anderson et al., 2008). Proteins tagged with the PIE-1 zinc finger 1 (ZF1) domain are recognized by the E3 ligase adaptor ZIF-1, which causes the tagged proteins to degrade rapidly from early embryonic somatic cells after the one-cell stage (Reese et al., 2000; DeRenzo et al., 2003; Nance et al., 2003). However, because ZIF-1 appears to be present only in early embryos (Armenti et al., 2014b), ZF1-tagged proteins that are expressed zygotically at later embryonic stages do not degrade. CDC-42 fused at its N-terminus with HA tags and the ZF1 domain (Anderson et al., 2008) largely rescued the lethality of cdc-42 mutants (84% [389/463] of cdc-42[gk388]; ha-zf1-cdc-42 embryos were viable) and degraded rapidly from early embryonic somatic cells before its zygotic expression began during the middle of embryogenesis (Fig. S1, A–B; Anderson et al., 2008). To obtain embryos lacking cdc-42 activity, we allowed cdc-42; ha-zf1-cdc-42/+ hermaphrodites to self-fertilize (Fig. S1 C, Strategy I). One quarter of the resulting progeny should lack the ha-zf1-cdc-42 transgene, and because the maternal HA-ZF1-CDC-42 protein is degraded, these embryos will be left with no maternal or zygotic source of CDC-42 (Fig. S1 C, right). Hereafter, we refer to such embryos as cdc-42(MZ). Of the self-progeny of cdc-42; ha-zf1-cdc-42/+ heterozygotes, 24% (116/479) died before hatching. PCR genotyping showed that 84% (15/18) of randomly selected dead embryos were cdc-42(MZ), whereas the remaining 16% (3/18) contained the ha-zf1-cdc-42 transgene but failed to rescue. Thus, although ha-zf1-cdc-42 transgene largely rescues the cdc-42(gk388) mutant phenotype, it does not do so completely. All healthy genotyped L1 larvae contained the ha-zf1-cdc-42 transgene (19/19), whereas a small number of cdc-42(MZ) embryos died soon after hatching into sickly larvae. We conclude that cdc-42 has an essential function during the second half of embryonic development, after zygotic cdc-42 expression begins.
We used 3D time-lapse differential interference contrast (DIC) imaging to examine the dynamics of ventral enclosure and elongation in live cdc-42(MZ) embryos (Fig. 1, B and C; and Video 1). Of 135 embryos imaged from a cdc-42; ha-zf1-cdc-42/+ mother, 39 (29%) did not hatch. Based on the results of our genotyping, 86% of unhatched embryos are predicted to be cdc-42(MZ) embryos, whereas the remaining small percentage of arrested embryos are predicted to contain the ha-zf1-cdc-42 transgene. Arrested embryos were assigned to three phenotypic classes according to the final stage of morphogenesis they completed (Fig. 1 D): Class I embryos (17/39, 44%) failed to complete ventral enclosure and therefore arrested before elongation; Class II embryos ruptured in the head ventral region early in elongation, typically at the 1.5-fold stage (8/39, 20%; Fig. 1 C); Class III embryos arrested later during elongation, at or after the twofold stage, with extruded cells (14/39, 36%). Embryos that arrested during elongation also elongated at a slower rate than embryos that developed to hatching (Fig. 1 E). Based on these observations and our genotyping data, we conclude that loss of cdc-42 activity causes significant morphogenetic defects that are apparent during ventral enclosure and elongation, and that cause embryonic arrest before elongation is complete.
CDC-42 is dispensable for polarity establishment and junction maturation
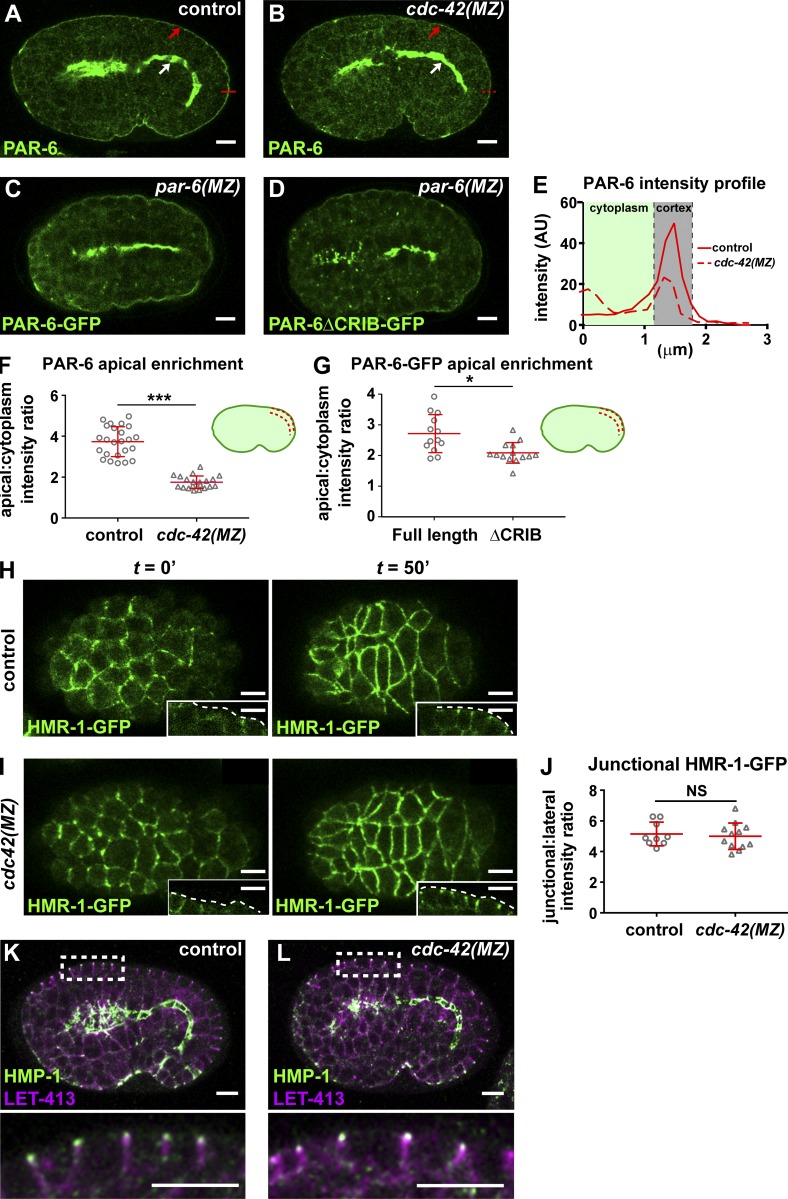
We next tested if there were defects in epidermal cell polarization or junction maturation in cdc-42(MZ) embryos that could explain their arrest phenotype. CDC-42 is thought to promote epithelial cell polarization by recruiting PAR-6 to the apical surface; this interaction occurs through the PAR-6 CRIB domain, which binds directly to active CDC-42 (Joberty et al., 2000; Johansson et al., 2000; Lin et al., 2000; Qiu et al., 2000; Gotta et al., 2001; Hutterer et al., 2004; Aceto et al., 2006). Unexpectedly, PAR-6 still accumulated at the apical membrane of epidermal cells (Fig. 2 B, red arrow) in cdc-42(MZ) embryos, even though the apical to cytoplasmic ratio of PAR-6 immunostaining in the epidermis of cdc-42(MZ) mutants was decreased twofold compared with control embryos (Fig. 2, A, E, and F). To rule out the possibility that a trace amount of HA-ZF1-CDC-42 beyond our limit of detection remained in cdc-42(MZ) embryos and was sufficient to recruit PAR-6, we examined the localization of a previously characterized form of PAR-6 with mutations in the CRIB domain that make it unable to bind CDC-42 (Aceto et al., 2006). PAR-6ΔCRIB-GFP, expressed from endogenous regulatory sequences and examined in a par-6(MZ) mutant background, localized similarly to endogenous PAR-6 in cdc-42(MZ) mutants, with strong apical enrichment in epithelial cells that was less pronounced but still evident in epidermal cells (compared with PAR-6-GFP in wild-type; Fig. 2, C, D, and G). However, par-6(MZ); par-6ΔCRIB-gfp embryos arrested before the 1.7-fold stage of elongation (27/27), in contrast with par-6(MZ); par-6-gfp embryos, which nearly all survived (95%, 232/243). Thus, whereas neither CDC-42 nor the PAR-6 CRIB domain is essential for PAR-6 apical enrichment, both contribute to enriching PAR-6 apically within epidermal cells, and the CRIB domain is needed for PAR-6 function.
Figure 2.
cdc-42 is not required for epidermal cell polarity or junction maturation. (A and B) PAR-6 staining of control embryos (cdc-42 mutants rescued with ha-zf1-cdc-42) and cdc-42(MZ) embryos. Red arrows indicate PAR-6 enrichment at the apical domain in epidermis; white arrows indicate enrichment at the apical domain in intestine. Red lines show t where the intensity profile shown in E was taken. (C and D) PAR-6-GFP and PAR-6ΔCRIB-GFP imaged live in par-6(MZ) embryos. (E) Intensity profile of PAR-6 in control (representative of n = 24) and in cdc-42(MZ) (representative of n = 20) epidermal cells (taken from micrographs shown in A and B). (F) Quantification of apical membrane to cytoplasm intensity ratio of PAR-6 immunostaining in the epidermis of control (n = 24) and cdc-42(MZ) (n = 20) embryos. Individual data points from two independent experiments were pooled. (G) Quantification of apical membrane to cytoplasm intensity ratio in the epidermis of PAR-6-GFP (n = 13) and PAR-6ΔCRIB-GFP (n = 14) in live par-6(MZ) embryos. Individual data points from two independent experiments were pooled. (H–I) Frames from time-lapse movies of control and cdc-42(MZ) embryos expressing HMR-1-GFP, showing polarization and junction maturation of dorsal epidermal cells. Insets show a side view of lateral epidermal cells, in which apical enrichment of HMR-1-GFP to junctions is observed by 50 min. t = 0 is defined as the initiation of polarization, when HMR-1-GFP puncta are first detected in the intestine. See Videos 2 and 3. (J) Quantification of junctional enrichment of HMR-1-GFP in epidermal cells at t = 50 min (control, n = 9 embryos; cdc-42(MZ), n = 12 embryos). (K and L) Immunostaining of junctional HMP-1 and basolateral LET-413 in control and cdc-42(MZ) embryos. Insets below are of boxed region. Line and error bars in graphs (F, G, and J) indicate mean ± SD. P-values were calculated using a Mann-Whitney U test. *, P ≤ 0.05; ***, P ≤ 0.001. NS, P > 0.05. Bars, 5 µm.
Because PAR-6 is required for junction maturation, we asked whether the decreased apical enrichment of PAR-6 in cdc-42(MZ) epidermal cells blocked or slowed junction formation or maturation. We examined junctions when epidermal cells first polarized via MET using fluorescence time-lapse imaging of two junction proteins: the E-cadherin HMR-1-GFP expressed from a knockin allele, and the Discs large protein DLG-1-RFP expressed from an integrated transgene. To genotype live cdc-42(MZ) embryos, we created a functional zf1-yfp-cdc-42 knockin allele, placed this in trans to the cdc-42(gk388) null mutation, and selected YFP-negative self-progeny embryos (cdc-42[gk388] homozygotes; Fig. S1 D, Strategy II). Such embryos had no detectable levels of YFP, indicating that maternal ZF1-YFP-CDC-42 degradation was efficient (Fig. S1, D and E). To visualize HMR-1-GFP during junction formation, we acquired an image stack from the dorsal surface to the center of the embryo. This allowed us to observe MET en face (in epidermal cells) and in cross section (in epidermal and intestinal cells). At initial stages of polarization in control embryos, the intensity of HMR-1-GFP was low and uneven along contacts at the apicolateral edges of epidermal cells (Fig. 2 H and Video 2). As cells polarized, HMR-1-GFP localized evenly at apicolateral AJs. Junction formation in cdc-42(MZ) embryos was indistinguishable from control embryos (Fig. 2 I), and the junctional to lateral ratio of HMR-1-GFP in polarized epidermal cells was not significantly different (Fig. 2 J). As in epidermal cells, intestinal epithelial cells of cdc-42(MZ) embryos showed normal polarization of HMR-1-GFP during MET (Video 3). We obtained similar results for DLG-1-RFP, which, like HMR-1-GFP, initially formed puncta at epithelial cell–cell contacts before puncta coalesced into a continuous apicolateral belt-like structure (Fig. S2, A–B; and Video 4).
We examined additional markers to determine if cdc-42 is dispensable for other aspects of epidermal cell polarization. PAR-3-GFP still accumulated apically in cdc-42(MZ) epidermal cells, albeit with a mild but significant decrease in apical enrichment compared with wild-type (Fig. S2, C–E). Immunostaining for endogenous proteins at AJs (HMP-1), basal junctions (DLG-1), and the basolateral membrane (LET-413) did not reveal any mislocalization in cdc-42(MZ) mutant embryos (Fig. 2, K and L; and Fig. S2, F and G). Altogether, our live-imaging experiments with fluorescently tagged junction proteins and examination of endogenous polarity proteins in fixed embryos demonstrate that, surprisingly, cdc-42 is not required for epidermal cell polarization or junction maturation during the process of MET.
CDC-42 regulation is important for elongation
We next addressed the consequences of increasing cdc-42 activity. In contrast with cdc-42 loss of function, overexpressing cdc-42 using a functional gfp-cdc-42 transgene (Neukomm et al., 2014) did not cause any significant defects or lethality in an otherwise wild-type background (Table 1). However, gfp-cdc-42 overexpression decreased embryonic survival in a genetically sensitized hmp-1(fe4) background, which has been used to identify genes that contribute to AJ function (Pettitt et al., 2003; Cox-Paulson et al., 2012; Lynch et al., 2012). The fe4 mutation compromises the ability of HMP-1/α-catenin to bind F-actin (Maiden and Hardin, 2011) and results in 52% of embryos arresting during elongation, whereas 48% survive through embryogenesis (Pettitt et al., 2003; Table 1). However, only 19% of hmp-1(fe4); gfp-cdc-42 embryos survived, hatching into misshapen larvae similar to hmp-1(fe4) larvae (Table 1). 12% of hmp-1(fe4); gfp-cdc-42 embryos arrested at ventral enclosure (Class I), whereas the remainder developed dorsal epidermal bulges and arrested during elongation, reaching at most the twofold stage (Fig. S3, A and E; and Video 5). This enhancement of lethality was similar to that caused by overexpressing rho-1 (RhoA), which also decreased survival of hmp-1(fe4) mutants to 19%, whereas overexpressing ced-10 (Rac) had no effect on hmp-1(fe4) lethality (Table 1). The effect of overexpressing rho-1 is not surprising because RHO-1 activity must be attenuated by RhoGAP activity in epidermal cells to control tension during elongation (Diogon et al., 2007). These findings suggest that the activity of CDC-42, like RHO-1, might be regulated to ensure proper embryo elongation.
Table 1. Genetic interactions with hmp-1.
| Genotypea | Embryo survival (n) | hmp-1 enhancementb |
|---|---|---|
| % | ||
| Wild type | 98 (555) | N/A |
| gfp-cdc-42 | 99 (387) | N/A |
| gfp-rho-1 | 99 (537) | N/A |
| gfp-ced-10 | 88 (566) | N/A |
| hmp-1(fe4) | 48 (1248) | N/A |
| hmp-1(fe4); gfp-cdc-42 | 19 (273) | Yes |
| hmp-1(fe4); gfp-rho-1 | 19 (160) | Yes |
| hmp-1(fe4); gfp-ced-10 | 46 (434) | No |
| pac-1 | 93 (866) | N/A |
| hmp-1(fe4); pac-1 | 1 (633) | Yes |
| hmp-1(fe4); pac-1; mCherry-pac-1 | 41 (444) | No |
| hmp-1(fe4); pac-1; mCherry-pac-1(R984A) | 0 (479) | Yes |
| picc-1 | 97 (437) | N/A |
| hmp-1(fe4); picc-1 | 14 (498) | Yes |
| pac-1; picc-1 | 92 (573) | N/A |
| jac-1 | 99 (641) | N/A |
| hmp-1(fe4); jac-1 | 0 (726) | Yes |
| jac-1; pac-1 | 70 (918) | N/A |
All mutant alleles are functional nulls except the hypomorphic hmp-1(fe4) allele.
Yes, P < 0.0001, using the Fisher exact test; No, P > 0.01; N/A, not applicable.
RhoGAP PAC-1 localizes to AJs and regulates elongation
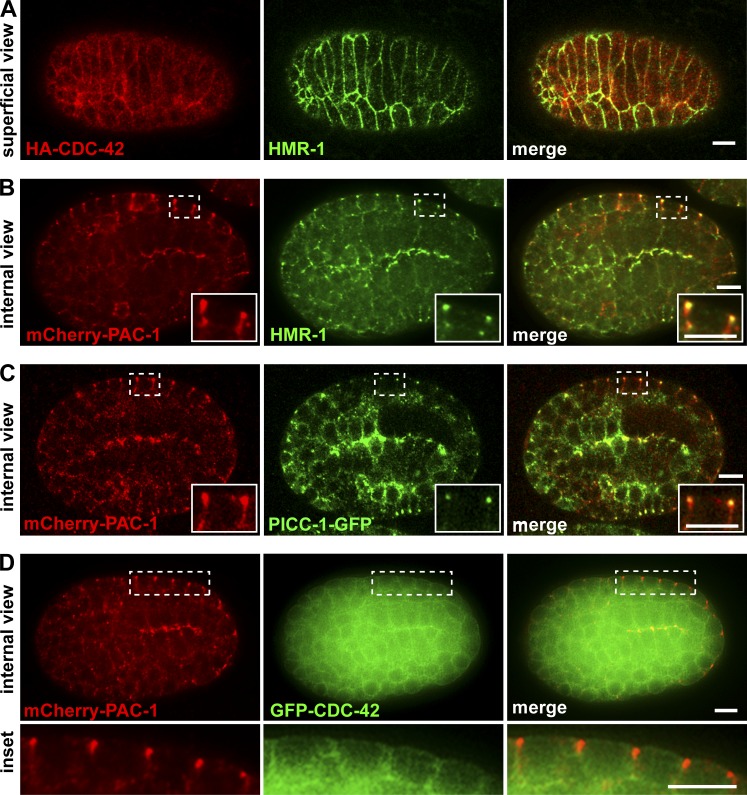
To determine where CDC-42 might function to regulate elongation, we examined the localization of a functional HA-CDC-42 fusion protein (Anderson et al., 2008) within epidermal cells. HA-CDC-42 was enriched at the plasma membrane, including AJs, which we visualized by costaining for HMR-1 (Fig. 3 A). Because epidermal cell AJs must remodel as the cells change shape during elongation, and given the genetic interactions we observed between overexpressed cdc-42 and hmp-1(fe4) mutants, we searched for candidate regulators of CDC-42 activity that localize to AJs. In early embryos, the RhoGAP PAC-1 (human ARHGAP21) interacts physically with proteins that bind to the cytoplasmic tail of HMR-1/E-cadherin, where it functions to inhibit CDC-42 activity and control cell polarity (Anderson et al., 2008; Klompstra et al., 2015). A functional mCherry-PAC-1 fusion protein expressed from a recombineered fosmid was detected in epithelial cells and concentrated at junctions (Fig. 3 B). mCherry-PAC-1 colocalized nearly completely with HMR-1, but was significantly more apical than DLG-1, which is found basal to AJs (Fig. S4, A and B), indicating that PAC-1 is present at AJs. mCherry-PAC-1 also overlapped with the more broadly localized GFP-CDC-42 at AJs (Fig. 3 D), raising the possibility that PAC-1 inhibits a junctional pool of CDC-42 during elongation.
Figure 3.
CDC-42, PAC-1, and PICC-1 localization at AJs. (A) Superficial view of HA-CDC-42 and HMR-1 coimmunostaining in the dorsal epidermis of a bean-stage embryo. (B) Internal view of mCherry-PAC-1 and HMR-1 coimmunostaining in a bean-stage embryo showing mCherry-PAC-1 colocalization with HMR-1. (C) Internal view of mCherry-PAC-1 and PICC-1-GFP costaining showing colocalization at junctions. (D) Internal view of mCherry-PAC-1 and GFP-CDC-42 in a live bean-stage embryo. The boxed region of epidermal cells is shown in the inset below. Bars, 5 µm.
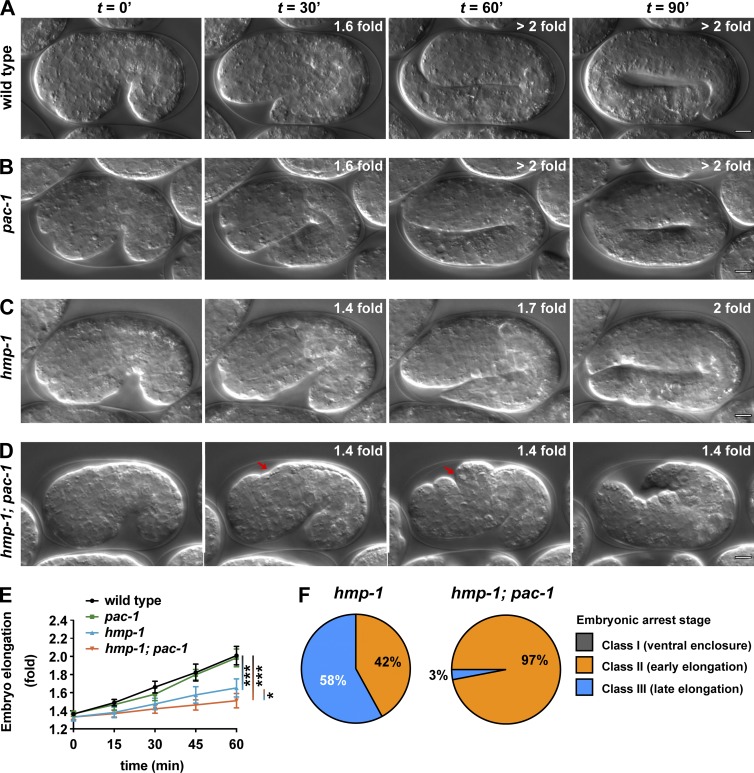
If PAC-1 inhibits CDC-42, then pac-1 mutants, similar to overexpressed cdc-42, should enhance the embryonic lethality of hmp-1(fe4) mutants. pac-1(xn6) contains a premature stop codon predicted to truncate the protein before the RhoGAP domain (Anderson et al., 2008). Although pac-1(xn6) mutants have defects in cell positioning during gastrulation, most embryos recover and hatch with normal morphology (Fig. 4 B, Table 1, and 2; Anderson et al., 2008). In contrast, pac-1(xn6) strongly enhanced the lethality of hmp-1(fe4) mutants (Fig. 4 D and Tables 1 and S1). The majority of hmp-1(fe4); pac-1(xn6) embryos elongated more slowly than hmp-1(fe4) or pac-1(xn6) single mutants (Fig. 4 E). For example, more than half of arrested hmp-1(fe4) mutant embryos reached the twofold stage or greater (Class III, 58%; Fig. 4 F and Video 6), whereas nearly all hmp-1(fe4); pac-1(xn6) embryos arrested before the twofold stage of elongation (Class II, 97%; Fig. 4 F). Although hmp-1(fe4); pac-1(xn6) embryos arrested early in elongation, all embryos completed ventral enclosure and arrested after the appearance of dorsal epidermal bulges (Fig. 4 D, arrow). The elongation phenotype of hmp-1(fe4); pac-1(xn6) embryos was nearly as severe as that of hmp-1(fe4) embryos treated with hmp-1 RNAi, which also formed dorsal bulges and arrested during, or just after, ventral enclosure (Fig. S3 D). The enhanced lethality of hmp-1(fe4); pac-1(xn6) embryos was reversed to the level seen in hmp-1(fe4) alone by expressing wild-type mCherry-PAC-1 from a transgene, but was not affected by expressing comparable levels of mCherry-PAC-1(R984A), which contains a mutation that inactivates the RhoGAP domain (Anderson et al., 2008) without disrupting expression level or localization to AJs (Table 1 and Fig. S5, A and B). Altogether, these findings indicate that the RhoGAP activity of PAC-1 is important for elongation and raise the possibility that PAC-1 regulates AJs by functioning as a RhoGAP that locally inhibits CDC-42.
Figure 4.
pac-1enhances hmp-1(fe4) during elongation. (A–D) Stills from DIC time-lapse movies of wild-type, pac-1, and hmp-1 mutant embryos and hmp-1; pac-1 double mutant embryos. Red arrows indicate dorsal epidermal bulges. See Video 6. (E) Quantification of elongation rates for wild-type (n = 12), pac-1 (n = 10), hmp-1 (n = 7), and hmp-1; pac-1 (n = 12) embryos. t = 0 min represents the comma stage, and values are the mean ± SD. (F) Phenotypic classes of hmp-1 (n = 31) and hmp-1; pac-1 (n = 32) mutant embryos. Data were pooled from three independent imaging experiments for each genotype. P-values were calculated using a Mann-Whitney U test. *, P ≤ 0.05; ***, P ≤ 0.001. Bars, 5 µm.
PAC-1 regulates elongation by inhibiting CDC-42
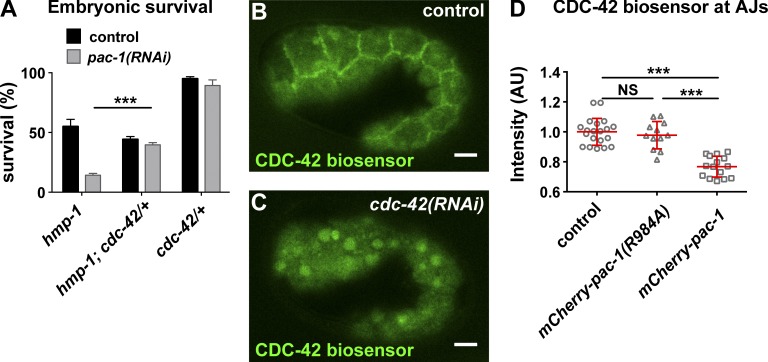
If PAC-1 inhibits CDC-42 through its RhoGAP activity, then CDC-42 should be overactive in embryos lacking PAC-1 activity. As an initial test of this hypothesis, we asked whether reducing cdc-42 gene dosage rescued the enhanced lethality of hmp-1(fe4) caused by pac-1 inactivation. Indeed, hmp-1(fe4); cdc-42/+ embryos were significantly less affected by pac-1 RNAi than hmp-1(fe4) mutants (Fig. 5 A). This finding suggests that CDC-42 is overactive in pac-1(RNAi) embryos, and that overactive CDC-42 contributes to the lethality of hmp-1(fe4) pac-1(RNAi) embryos.
Figure 5.
pac-1 inhibits junctional CDC-42 activity. (A) Comparison of survival of embryos of the indicated genotypes, with the addition of control (empty vector) or pac-1 feeding RNAi. Values are the mean ± SEM from three independent experiments; p-value was calculated using a two-tailed t test with Welch’s correction. (B and C) CDC-42 biosensor localization in embryos treated with control (empty vector) RNAi or partial cdc-42 RNAi. (D) Quantification of CDC-42 biosensor intensity at epidermal AJs in control embryos (n = 20), embryos overexpressing mCherry-pac-1 (n = 15), or embryos overexpressing GAP-dead mCherry-pac-1(R984A) (n = 12). Individual data points from two independent experiments were pooled. Red bar and error bars are mean ± SD. P-values were calculated using a Mann-Whitney U test. ***, P ≤ 0.001; NS, P > 0.05. Bars, 5 µm.
To determine whether CDC-42 is active at AJs, we altered a maternally expressed biosensor consisting of the G protein–binding domain (GBD) of WSP-1/Wasp fused to GFP, which was shown to specifically report on CDC-42 activity (Kumfer et al., 2010) by expressing it from cdc-42 regulatory sequences. In epidermal cells, the CDC-42 biosensor concentrated at AJs (Fig. 5 B). Partial depletion of cdc-42 by RNAi caused the biosensor to relocate from AJs to the cytoplasm and nucleus (Fig. 5 C), demonstrating that it reports on CDC-42 activity. In embryos overexpressing mCherry-PAC-1, biosensor levels at AJs were significantly decreased in comparison with embryos overexpressing GAP-dead mCherry-PAC-1(R984A) (Fig. 5 D). Altogether, these findings indicate that PAC-1 regulates the activity of CDC-42 at AJs to modulate elongation of the embryo.
PAC-1 likely functions with interacting linker protein PICC-1
At sites of contact between early embryonic cells, the coiled-coil protein PICC-1 (CCDC85A-C in humans) couples PAC-1 to HMR-1 by binding to both PAC-1 and the p120 catenin JAC-1 (Klompstra et al., 2015). Mammalian CCDC85B (also known as DIPA) can interact with p120 and localizes to AJs, although its function is unknown (Markham et al., 2014). To determine if PICC-1 might function with PAC-1 at epidermal cell AJs, we first examined the localization of a PICC-1-GFP fusion protein expressed from a recombineered genomic clone. PICC-1-GFP was present at AJs, where it colocalized with mCherry-PAC-1 (Fig. 3 C and Fig. S4 B).
Like pac-1(xn6) mutants, picc-1(xn14) null mutants are viable (Klompstra et al., 2015; Table 1). Given that PAC-1 and PICC-1 have been shown to function together, we asked whether picc-1 also interacts genetically with hmp-1(fe4). Nearly all hmp-1(fe4); picc-1(xn14) and hmp-1(fe4) picc-1(RNAi) embryos arrested, and did so at a similar stage as pac-1(xn6); hmp-1(fe4) embryos (Tables 1 and S1; Fig. S3, B and E; and Video 7). pac-1(xn6); picc-1(xn14) double mutants were also viable, like each single mutant, consistent with the genes functioning together in the same pathway (Table 1). Because PICC-1 links PAC-1 to JAC-1, we also examined genetic interactions between jac-1 and pac-1. jac-1(xn15) null mutants are viable (Klompstra et al., 2015) and enhanced the lethality of hmp-1(fe4) mutants to the same extent as pac-1(xn6) (Tables 1 and S1), with embryos arresting before the twofold stage of elongation (Fig. S3, C and E). However, 30% of jac-1(xn15); pac-1(xn6) double null mutants died. Thus, whereas PAC-1 and JAC-1 may function together at AJs, one or both proteins likely have independent functions that are important for elongation.
AJ protein levels are increased in pac-1 mutant embryos
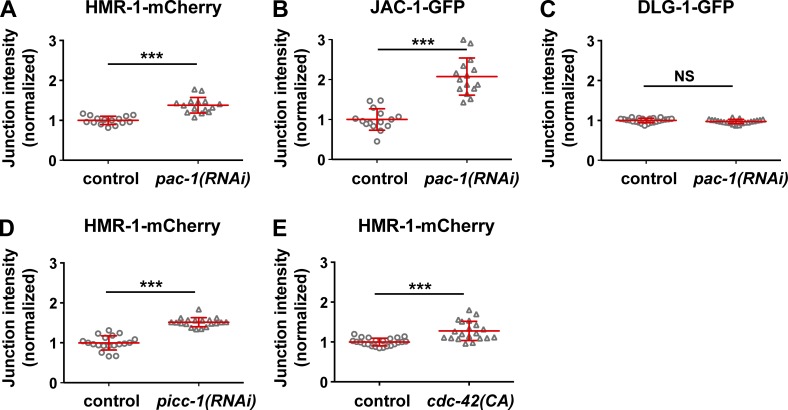
Because PAC-1 localized to AJs and regulated elongation, we asked whether the localization or level of junction proteins was abnormal in embryos lacking pac-1 function. To quantify protein levels, we used functional HMR-1-mCherry, JAC-1-GFP, and HMP-1-GFP fluorescent reporters expressed from transgenes in living embryos. Each protein concentrated at epidermal AJs in both wild-type and pac-1(RNAi) embryos. However, their levels at AJs were significantly increased in pac-1(RNAi) embryos relative to wild-type (Fig. 6, A and B; and Fig. S5 C). To confirm that the observed AJ protein increases were not limited to overexpressed transgenic proteins, we examined HMR-1-GFP expressed from a knockin allele (Marston et al., 2016); HMR-1-GFP showed a similar increase in pac-1(xn6) mutants relative to wild-type (Fig. S5, D). In contrast with AJ proteins, the amount of DLG-1-GFP present at junctions was not increased in pac-1(RNAi) embryos (Fig. 6 C), consistent with the localization of DLG-1 to a separate junctional zone (Fig. S4).
Figure 6.
pac-1, picc-1, and cdc-42 regulate AJ protein level. (A–E) Quantitative intensity of fluorescently tagged AJ proteins in the indicated genetic background. Control embryos for RNAi experiments were treated with empty vector RNAi. “cdc-42(CA)” is epidermal overexpression of constitutively active HA-CDC-42. Intensity values were normalized such that the mean control value was 1. Individual data points from two independent experiments were pooled. (A) HMR-1-mCherry: control (n = 15) and pac-1(RNAi) (n = 15); (B) JAC-1-GFP: control (n = 15) and pac-1(RNAi) (n = 15); (C) DLG-1-GFP: control (n = 24) and pac-1(RNAi) (n = 21); (D) HMR-1-mCherry: control (n = 19) and picc-1(RNAi) (n = 19); (E) HMR-1-mCherry: control (n = 23) and cdc-42(CA) (n = 20). Red bars and error bars are mean ± SD. P-values were calculated using a Mann-Whitney U test. ***, P < 0.0001; NS, P > 0.05.
Depletion of picc-1 also caused an increase in junctional HMR-1-mCherry levels (Fig. 6 D), further suggesting that PAC-1 and PICC-1 function together. To determine whether the increase in AJ protein levels after loss of PAC-1 or PICC-1 could be caused by an increase in CDC-42 activity, we examined levels of HMR-1-mCherry in embryos expressing constitutively active CDC-42 specifically in the epidermis (“cdc-42[CA]” embryos; Fig. S5 E). HMR-1-mCherry remained concentrated at AJs in cdc-42(CA) embryos, but its levels were increased relative to wild-type (Fig. 6 E; compare with Fig. 6 A). We conclude that an increase in CDC-42 activity in epidermal cells is sufficient to augment levels of AJ proteins at junctions, suggesting that this is the basis for the increased AJ protein levels observed in pac-1(RNAi) and picc-1(RNAi) embryos.
CDC-42 activity levels affect junctional actin dynamics during elongation
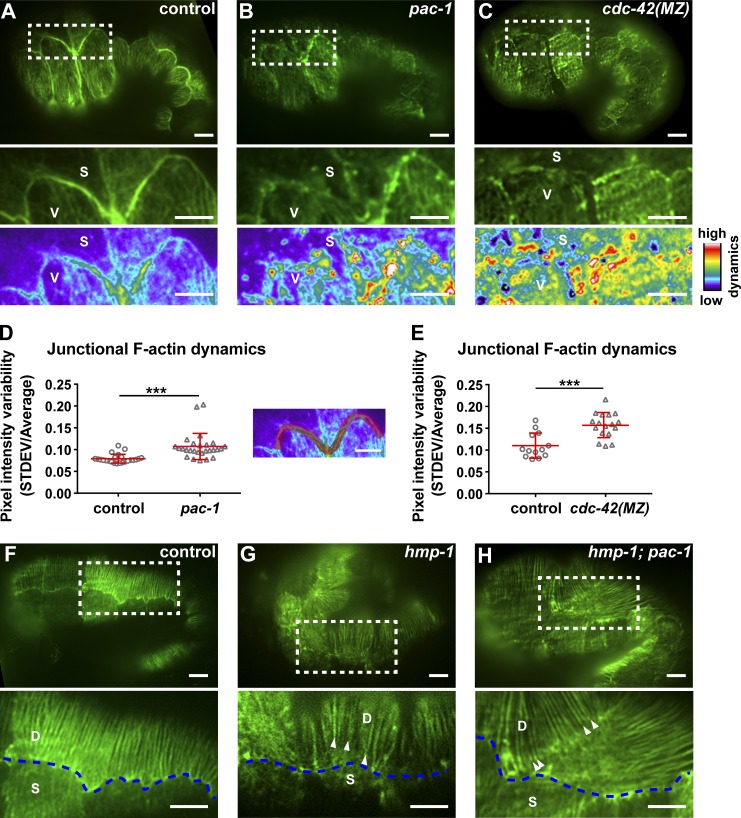
Because CDC-42 can also regulate actin polymerization, we analyzed junctional F-actin in pac-1(xn6) embryos. To determine whether junctional F-actin is aberrant, we expressed LifeAct-GFP within epidermal cells and captured high frame rate near–total internal reflection fluorescence (TIRF) movies during elongation (Fig. 7). In control embryos, actin filaments at AJs formed thick bundles that did not change their morphology (Fig. 7 A and Video 8). However, in pac-1(xn6) embryos, we observed multiple dynamic junction-associated actin extensions protruding in various directions from the junctional actin belt over the same imaging interval (Fig. 7 B and Video 8). To quantify actin dynamics, we calculated the SD of LifeAct-GFP intensity at each pixel throughout the imaging period, which was normalized to the mean pixel intensity to adjust for variation in transgene expression level. This value, displayed as a heat map in images (Fig. 7, A–C), reports on the dynamics of F-actin extensions at a given location. Compared with control embryos, F-actin in pac-1(xn6) mutant embryos was significantly more dynamic at epidermal AJs (Fig. 7, B and D). F-Actin also appeared less concentrated at junctions and significantly more dynamic in cdc-42(MZ) embryos (Fig. 7, C and E), suggesting that cycling of cdc-42 activity is needed for proper F-actin organization and dynamics at AJs.
Figure 7.
pac-1and cdc-42 regulate junctional actin morphology and dynamics. (A–C, top) Near-TIRF high-resolution imaging of LifeAct-GFP in control, pac-1 (see Video 8) and cdc-42(MZ) embryos at 1.3-fold elongation. Insets (middle) show the junctions between the seam cells (S) and ventral cells (V). Insets (bottom) are pseudo-colored images depicting actin dynamics (STDEV/AVERAGE values at each pixel) over the course of the movie (16 s: 512 time points imaging at 32 frames/s). (D and E) Junctional actin dynamics (STDEV/AVERAGE of pixel intensity over 512 time points, imaging at 32 fps) taken from a subset of junctions between seam and ventral cells, as depicted to the right from the graph (D). Embryos in (D) were 1.3-fold: control (n = 23), pac-1 (n = 20); embryos in (E) were 1.2-fold: control (n = 13), cdc-42(MZ) (n = 17). Each data point shows the average value for one embryo. Individual data points from two independent experiments were pooled for each data set. Red bars and error bars are mean ± SD. P-values were calculated using a Mann-Whitney U test. ***, P < 0.001. (F–H, top) CFBs in control, hmp-1(fe4), and hmp-1(fe4); pac-1 embryos. Insets (bottom) show the CFBs in dorsal cells (labeled D) and disconnection of CFBs (arrowheads) from the junction with seam cells (S). Blue dashed line depicts the junction between dorsal (D) and seam (S) cells. Bars, 5 µm.
At a later stage of elongation (beginning at the ∼1.7-fold stage), actin fibers in dorsal and ventral cells become reorganized into circumferential bundles (CFBs; Fig. 7 F; Costa et al., 1998). At this stage, AJs in pac-1(xn6) embryos no longer exhibited ectopic actin extensions (data not depicted). Thus, pac-1 inactivation causes a transient increase in dynamic junctional F-actin extensions during the early stages of elongation—when pac-1(xn6) mutant embryos in the hmp-1(fe4) sensitized background arrest. In 18 of 18 hmp-1(fe4); pac-1(xn6) embryos imaged during elongation, circumferential actin bundles were visibly detached from junctions (Fig. 7 H), and sites of detachment corresponded to regions where bulges of epidermis formed on the dorsal surface. Separation of circumferential actin bundles from AJs occurs in hmp-1 null mutant embryos (Costa et al., 1998), and to a much lesser extent in hmp-1(fe4) hypomorphic mutant embryos (Pettitt et al., 2003; Fig. 7 G), and likely explains why hmp-1(fe4); pac-1(xn6) embryos fail to elongate.
Discussion
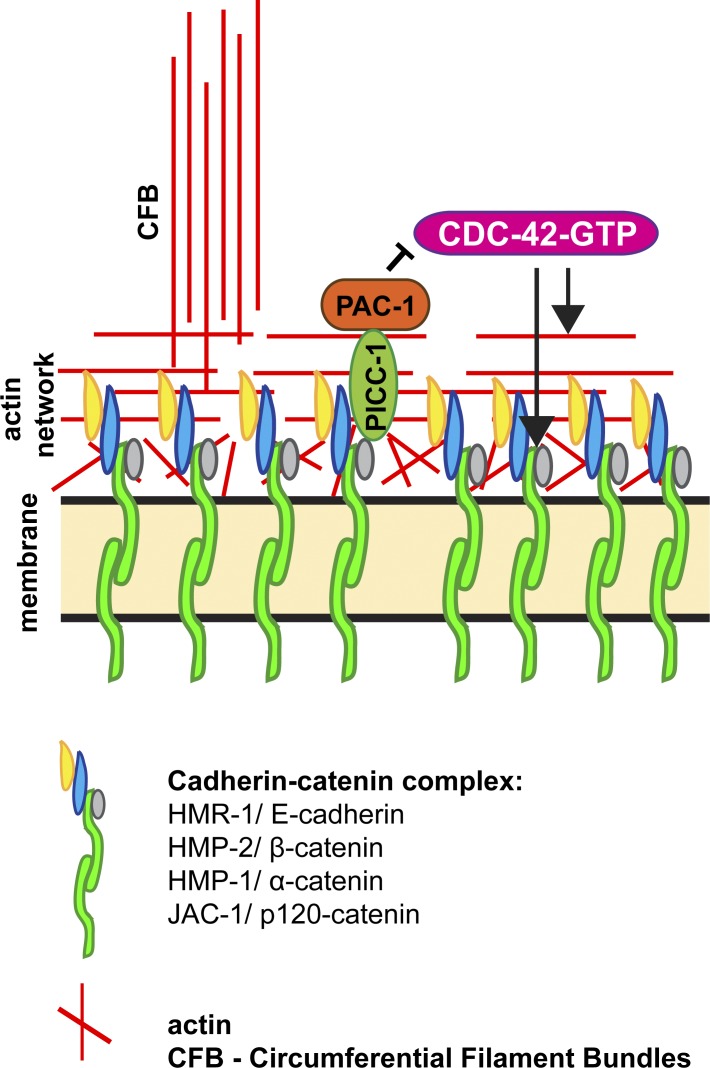
As a direct regulator of PAR polarity proteins, actin polymerization factors, and vesicle trafficking proteins (Mack and Georgiou, 2014), CDC-42 is positioned to control diverse cell biological events that are critical to AJ formation and remodeling. Here, we have taken advantage of two strengths of C. elegans embryos—the ability to remove CDC-42 function acutely before epithelial cells form, and the capability to visualize the dynamics of junction proteins and associated actin microfilaments in living embryos—to define how CDC-42 contributes to junction formation and remodeling during epithelial morphogenesis. Our findings provide two new insights into CDC-42 function and regulation. First, we show that cdc-42 mutant epidermal cells are able to polarize and undergo junction maturation normally during MET, contrasting with interpretations from studies in several other systems. Second, we identify a conserved pathway containing the RhoGAP PAC-1/ARHGAP21 and its binding partner PICC-1/CCDC85A-C, which inhibits CDC-42 activity at AJs (Fig. 8). We show that PAC-1, PICC-1, and appropriate CDC-42 activity are important for proper junctional actin organization and levels of AJ proteins, and contribute to the fidelity of epidermal cell shape changes.
Figure 8.
Model for PAC-1 inhibition of CDC-42 at AJs. See Discussion text for details. Different populations of actin are shown, including junctional actin and CFBs, which align apically to connect one side of the cell to another.
Cdc42 and epithelial polarity
Numerous studies that examined cultured epithelial cell lines or fixed tissues in genetic model organisms have concluded that cdc-42 is required for epithelial cell polarization. For example, in the early Drosophila embryo, expressing dominant-negative Cdc42 blocks apicobasal polarization of the blastoderm epithelium (Hutterer et al., 2004). Similarly, conditional Cdc42 knockout in several mammalian cell types, including 3D-cultured MDCK cells, mouse mammary epithelial cells, the developing kidney, and pancreatic epithelial cells, results in defective apicobasal polarity of tubular epithelia (Martin-Belmonte et al., 2007; Kesavan et al., 2009; Elias et al., 2015; Druso et al., 2016). However, these and other studies did not examine the initial stages of epithelial polarization as it unfolded using time-lapse microscopy, leaving it unclear whether the observed polarity phenotypes result from a failure to initially establish polarity, or rather a rapid loss of polarity caused by defects in its maintenance. Here, we addressed the role of CDC-42 in polarizing C. elegans epithelial cells by removing CDC-42 protein acutely from embryos before precursor cells differentiate into epithelia, and observing polarization and junction maturation live using fluorescent time-lapse microscopy of polarity and junction proteins (HMR-1, DLG-1, PAR-3). These results demonstrate that CDC-42 is dispensable for the epithelia we examined (epidermis and intestine) to establish apicobasal polarity or to form mature junctions. The only defects we noted were a reduced level of apical PAR-6 and PAR-3 enrichment in epidermal cells, although both proteins were still apically localized. To confirm these results and to rule out the possibility that trace amounts of CDC-42 might remain in cdc-42(MZ) mutants to facilitate polarization, we examined the localization of a CRIB mutant form of PAR-6 unable to bind to CDC-42 (Aceto et al., 2006). CRIB mutant PAR-6 still accumulated apically in polarizing epithelia, albeit with reduced apical enrichment in epidermal cells, similar to endogenous PAR-6 in cdc-42(MZ) mutants. A previously published examination of intestinal PAR-6 in larvae partially depleted of cdc-42 using RNAi, as well as of embryos expressing maternal CRIB mutant PAR-6 (in par-6(+) embryos), made similar observations (partial loss of apical PAR-6) and concluded that CDC-42 was required for PAR-6 apical accumulation (Shafaq-Zadah et al., 2012). However, these studies are consistent with our observations using embryos entirely lacking CDC-42 protein, or expressing CRIB mutant PAR-6 in the absence of endogenous PAR-6, and point to a partial requirement for CDC-42 in apical PAR-6 accumulation as well as a redundant, CDC-42–independent mechanism for PAR-6 localization that does not use the CRIB domain. Our findings therefore indicate that CDC-42 is not required for polarity establishment or junction formation in C. elegans epidermal and intestinal epithelial cells.
What might explain the differences we observe in C. elegans embryonic epithelia and epithelia in other model systems that depend on Cdc42 for polarity? One possibility is that some cells use alternative or redundant polarization pathways. This may very well be the case, as there is increasing evidence that “core” epithelial polarity proteins, for example PAR-3, are required for polarization of some epithelial cell types but not others, even within the same organism (Achilleos et al., 2010). Moreover, deletion of Cdc42 from some cell types, such as keratinocytes or embryonic stem cell–derived endothelial cells, does not inhibit AJ formation (Erasmus et al., 2010; Qi et al., 2011). A formal possibility is that one or more C. elegans Racs function redundantly with CDC-42 in polarization, because in mammals, Rac also binds the PAR-6 CRIB domain (Joberty et al., 2000; Johansson et al., 2000; Lin et al., 2000; Qiu et al., 2000). However, it is unknown whether this interaction occurs in C. elegans, and because CRIB mutant PAR-6 still enriches apically, this interaction would have to use a different mechanism than the CDC-42–PAR-6 interaction. An alternative and nonexclusive possibility is that CDC-42 is required only for the maintenance of epithelial junctions (a conserved role—see next section), which in different cell types and organisms have varying degrees of importance in maintaining polarity. For example, early mouse embryos lacking E-cadherin show strong polarity defects, whereas E-cadherin is not needed for polarity in C. elegans epithelia, and is needed for polarity maintenance but not establishment in the Drosophila blastoderm (Harris and Peifer, 2004; Stephenson et al., 2010; Von Stetina and Mango, 2015). Studies in other model systems, in particular cells lacking cdc-42 before they undergo MET and examined by fluorescence time-lapse imaging, will be needed to help resolve these possibilities.
CDC-42 and junction regulation
CDC-42 has a conserved role in trafficking AJ proteins and regulating actin polymerization at junctions, helping to ensure an appropriate balance between cell adhesion and junction remodeling during morphogenetic events. Here, we showed that loss of CDC-42 affects junction integrity, and alters actin organization and dynamics at AJs. One class of cdc-42(MZ) mutant embryos arrested before, or just after, completion of ventral enclosure. This finding suggests that CDC-42 may contribute to the migration of epidermal cell sheets, or to the genesis of new epidermal AJs that form at the ventral surface as ventral enclosure completes. Both processes might be affected by the changes in actin morphology and dynamics that we observed in cdc-42 mutants. Our observations are consistent with a study showing that loss of the RhoGAP rga-7, which can regulate CDC-42 activity, causes defects in ventral enclosure (Ouellette et al., 2016). However, in contrast with pac-1, rga-7 does not appear to inhibit CDC-42 activity, but rather contributes to the localization of active CDC-42 clusters along junctions of migrating epidermal cells (Ouellette et al., 2016). Another study identified defects in epidermal cell intercalations in embryos with compromised CDC-42 activity, pointing to a potential role in junction regulation or cell movement (Walck-Shannon et al., 2016).
A second class of cdc-42(MZ) mutants completed ventral enclosure and arrested at variable stages of elongation. We consider it unlikely that these elongation defects are an indirect consequence of ventral enclosure defects, because failure to properly form junctions at the ventral surface upon the completion of ventral enclosure typically leads to rupture at the beginning of elongation (Costa et al., 1998). In addition, our analysis of overexpressed gfp-cdc-42, pac-1 mutants, and picc-1 mutants in the hmp-1(fe4) background, revealed roles primarily in elongation but not ventral enclosure. We propose that CDC-42 contributes to both events, and that distinct regulators may be used during ventral enclosure and elongation to control CDC-42 activity sequentially.
Increasing the activity of junctional CDC-42 augmented levels of HMR-1 and its associated catenins at AJs, and also resulted in aberrant junctional F-actin organization and dynamics. Because CDC-42 effectors include proteins that regulate trafficking as well as those that control actin polymerization (Harris and Tepass, 2010), it is possible that these phenotypes are uncoupled and arise independently. Alternatively, given that F-actin is involved in vesicle trafficking and that AJs directly link to junctional microfilaments, it is feasible that the two phenotypes are coupled. It is also unclear whether it is the increase in junction protein levels, and/or the disorganization of F-actin at junctions, that causes elongation defects in hmp-1(fe4); pac-1 mutants. For example, overexpression of Drosophila E-cadherin inhibits tracheal development (Shindo et al., 2008) and wound repair (Hunter et al., 2015), and inhibition of E-cadherin endocytosis results in cell intercalation defects in the early Drosophila embryo (Levayer et al., 2011). In addition, depleting actin-bundling protein EEF1A in human keratinocytes, which augments the actin turnover rate and increases junctional E-cadherin levels, leaves junctions more sensitive to mechanical stress (Erasmus et al., 2016). These and other studies indicate that increased levels of E-cadherin at junctions do not necessarily indicate that junctions are “stronger.” Given the altered organization and dynamics of F-actin at junctions that we observed in pac-1 mutants, it is also likely that F-actin cannot connect as efficiently to AJs as in wild-type embryos.
PAC-1 and PICC-1 homologues localize to AJs in mammals (Sousa et al., 2005; Markham et al., 2014; Van Itallie et al., 2014), and the PAC-1 homologue ARHGAP21 has been shown to inhibit CDC-42 in cultured cells (Barcellos et al., 2013), suggesting that these proteins have deeply conserved roles in regulating junctional CDC-42. Although the in vivo functions of mammalian PAC-1 (ARHGAP21 and ARHGAP23) and PICC-1 (CCDC85A, CCDC85B, and CCDC85C) homologues have not yet been determined, a mutation in mouse Ccdc85C is associated with hemorrhagic hydrocephalus. CCDC85C protein localizes to apical junctions in radial glia lining brain ventricles (Mori et al., 2012), raising the possibility that this birth defect results from improper regulation of AJ proteins or junctional actin during brain development.
Materials and methods
Genetics
Worm strains used in this study are listed in Table S2.
Transgene construction
mCherry-pac-1(R984A)
mCherry-pac-1(R984A) was created by recombineering the R984A mutation into mCherry-pac-1 in the fosmid clone WRM063aE11 (Klompstra et al., 2015).
cdc42P::gfp-zf1-rho-1
cdc42P::gfp-zf1-rho-1 was created using Gateway cloning. Destination vector pJN406, which includes the cdc-42 promoter, gfp-zf1, and unc-119(+) (Anderson et al., 2008), was used with an entry clone containing a full-length rho-1 cDNA, corresponding to WormBase transcript Y51H4A.3a.
par6P::par-6-ΔCRIB-gfp
par6P::par-6-ΔCRIB-gfp was created by cloning an NsiI StuI restriction fragment from the pie-1P::par-6-CM2-GFP plasmid (Aceto et al., 2006) into the same sites of a plasmid expressing par-6-gfp from endogenous regulatory sequences (Nance et al., 2003). The unc-119(+) gene from plasmid pJN254 (Nance et al., 2003) was inserted into the vector NotI site.
All other transgenes were assembled using Gibson cloning (Gibson et al., 2009):
cdc42P::gst-gfp-wsp-1gbd
cdc42P::gst-gfp-wsp-1gbd was assembled from vector pJN405, which includes unc-119(+) and cdc-42P; gst, amplified from pGEX-4T-1; and gfp-wsp-1gbd, amplified from worm strain WH0363 (a gift from K. Kumfer, University of Wisconsin, Madison, WI; Kumfer et al., 2010).
cdc42P::mCherry-ha-ha-pac-1
cdc42P::mCherry-ha-ha-pac-1 was assembled using vector pJN566 (Armenti et al., 2014a); cdc-42P promoter amplified from pJN405; and pac-1 cDNA amplified from pDA7 (pie-1P::gfp-pac-1; Anderson et al., 2008).
cdc42P::mCherry-ha-ha-pac-1(R984A)
cdc42P::mCherry-ha-ha-pac-1(R984A) was assembled in the same manner as cdc42P::mCherry-ha-ha-pac-1, except the pac-1(R984A) sequence was amplified from pDA8 [pie-1P::gfp-pac-1(R984A)] (Anderson et al., 2008).
lin-26P::ha-ha-cdc-42(Q61L)
lin-26P::ha-ha-cdc-42(Q61L) was assembled using vector pJN566 (Armenti et al., 2014a); a 4.1-kb lin-26 promoter (Landmann et al., 2004); and ha-ha-cdc-42(Q61L) amplified from hsp-16P::ha-cdc-42(Q61L) (Anderson et al., 2008).
lin-26P::gfp-nls
lin-26P::gfp-nls was assembled using vector pJN566 (Armenti et al., 2014a) and the 4.1-kb lin-26 promoter (Landmann et al., 2004). The NLS was added to gfp within a primer used for Gibson assembly: 5′-TGGCATGGATGAACTATACAAAatgaccgctccaaagaagaaacgcaaagtaccggtagaaaaaTAGAACCCAGCTTTCTTGTACAAAG-3′ (NLS is in italics).
lin-26P::LifeAct-GFP
lin-26P::LifeAct-GFP was assembled using vector pJN566 (Armenti et al., 2014a) and the 4.1-kb lin-26 promoter (Landmann et al., 2004). LifeAct coding sequences (Riedl et al., 2008) were inserted between the promoter and gfp within a primer used for Gibson assembly: 5′-ATGGGAGTTGCTGATCTTATTAAAAAATTCGAATCTATTTCTAAAGAAGAAATGTATCAATCGTCTTCTTCATCTCCAT-3′ (LifeAct sequence is in italics).
CRISPR knockins
Plasmids for CRISPR/Cas9 genome editing to make par-6(xn60: par-6-zf1-yfp + unc-119) and cdc-42(xn65: zf1-yfp-cdc-42 + unc-119) were constructed as described previously (Dickinson et al., 2013), with the following modifications. For par-6, the guide RNA sequence from plasmid pDD122 was replaced with the sequences (5′-GCACCGCAGCCGCTACAGG-3′) and (5′-GTCCACCTGTAGCGGCTGCGG-3′) to create two single guide RNAs (sgRNAs) that cleave near the par-6 C-terminus (plasmids pJA031 and pJA032). For cdc-42, the sequence (5′-GCCGTCACAGTAATGATCGG-3′) was used in pDD122 to direct cleavage near the cdc-42 N-terminus (plasmid pJA037). Homologous repair plasmids for par-6 (pJA034) and cdc-42 (pJA036) were constructed using Gibson assembly. For par-6, the following DNA segments were assembled in order: 1517 bp upstream of par-6 stop codon (including six silent point mutations adjacent to the predicted sgRNA cut sites) as the left homology arm; zf1-yfp with unc-119; and the 3′-terminal 1508 bp of par-6 genomic sequence as the right homology arm. For cdc-42, the repair plasmid contained the following DNA segments: the 5′-terminal 1511 bp of cdc-42 genomic sequence as the left homology arm; zf1-yfp with unc-119; and 1502 bp downstream of the cdc-42 start codon as the right homology arm (including five silent point mutations adjacent to the predicted sgRNA cut sites), and was amplified from WS5018 strain genomic DNA to include 5′ linker sequence (Neukomm et al., 2014). zf1-yfp with unc-119 flanked by LoxP sites was amplified from plasmid pJN601, which contains LoxP-flanked unc-119 inserted in reverse orientation into a synthetic intron within yfp (Armenti et al., 2014b). The vector backbone for each construct was PCR-amplified from pJN601 using Gibson assembly primers that overlapped with homology arms for par-6 or cdc-42.
par-6(xn60: par-6-zf1-yfp + unc-119) was generated by microinjecting the sgRNA plasmids pJA031 and pJA032 (which also contains Cas9), the homologous repair template pJA034, and plasmid coinjection markers pGH8 (rab-3P::mCherry::unc-54utr; plasmid 19359; Addgene), pCFJ104 (myo-3P::mCherry::unc-54utr; plasmid 19328; Addgene), pCFJ90 (myo-2P::mCherry::unc-54utr; plasmid 19327; Addgene), and pMA122 (peel-1 negative selection; plasmid 34873; Addgene) into unc-119(ed3) mutant worms (Frøkjær-Jensen et al., 2012; Dickinson et al., 2013). Plates containing non-Unc F2 transformants were heat-shocked at 34°C for 4 h to activate PEEL-1 toxin in array-bearing animals, and successfully edited F2 non-Unc animals were confirmed by the absence of mCherry expression in the F2 generation and YFP expression in their progeny. cdc-42(xn65: zf1-yfp-cdc-42 + unc-119) was generated using the same procedure, but with cdc-42 sgRNA and repair construct. Both knockin alleles were functional, but caused a low level of lethality (par-6 [xn60], 88% [387/439] viable; cdc-42[xn65], 85% [258/305] viable).
Worm transformation
cdc42P::gfp-zf1-rho-1, mCherry-pac-1(R984A), and cdc42P::gst-gfp-gbd(wsp-1) were integrated into unc-119(ed3) worms using biolistic transformation (Praitis et al., 2001).
The following transgenes were injected into wild-type worms to produce extrachromosomal arrays (Mello et al., 1991): cdc42P::mCherry-ha-ha-pac-1 (10 ng/µl + 80 ng/µl pRF4); cdc42P::mCherry-ha-ha-pac-1(R984A) (10 ng/µl + 80 ng/µl pRF4); lin-26P::ha-ha-cdc-42(Q61L) (1 ng/µl + 90 ng/µl pRF4); and lin-26P::nls-gfp (1 ng/µl + 90 ng/µl pRF4). pRF4 contains the dominant rol-6(su1006) allele (Mello et al., 1991).
cdc-42(MZ) embryos
Strategy I (Fig. S1 C)
cdc-42(gk388)/cdc-42(gk388); xnIs83 (ha-ha-zf1-ha-cdc-42)/+ hermaphrodites were allowed to self-fertilize; 25% of F2 progeny are expected to be cdc-42(MZ): cdc-42(gk388) mutant embryos that express maternal HA-HA-ZF1-HA-CDC-42 only in very early embryos before it is degraded. cdc-42(MZ) were distinguished from the control embryos by staining with anti-HA antibody or by single-embryo PCR genotyping using primers 5′-GCAAGTTGTTTGGTACGGAAAACCG-3′ and 5′-TGCATAGTCTGGCACGTCGTATG-3′.
Strategy II (Fig. S1 D)
cdc-42(gk388)/cdc-42(xn65: zf1-yfp-cdc-42) hermaphrodites were allowed to self-fertilize; 25% of F2 progeny are expected to be cdc-42(MZ): cdc-42(gk388) mutants that express maternal YFP-ZF1-CDC-42 only in very early embryos before it is degraded. cdc-42(MZ) were distinguished from rescued control embryos by the absence of YFP-ZF1-CDC-42. Genotyping of cdc-42(MZ) embryos also carrying a gfp transgene was performed on the SP5 confocal, capturing narrow bandpass emission after 488-nm (GFP) and 514-nm (YFP) excitation to separate the fluorescent proteins.
par-6(MZ) embryos
par-6(MZ); par-6Δcrib-gfp embryos were obtained by allowing par-6(tm1425)/par-6(xn60: par-6-zf1-yfp); par-6Δcrib-gfp/+ hermaphrodites to self-fertilize; 25% of F2 embryos are expected to be par-6(MZ): par-6(tm1425)/par-6(tm1425) that express maternal PAR-6-ZF1-YFP only during very early embryonic stages; 75% of these mutants should have the par-6Δcrib-gfp transgene. par-6(MZ) embryos were distinguished from control sibling embryos by the absence of PAR-6-ZF1-YFP.
Western blots
100 synchronized young adult worms were picked into M9 buffer, placed on a rotator, and washed once an hour for five hours. The worm pellet was frozen in liquid nitrogen and thawed on ice, and 50 µl 2 × LDS sample buffer (Invitrogen) containing 200 mM dithiothreitol was added. Samples were vortexed until worm corpses were completely dissolved. The lysate was heated at 90°C for 10 min and centrifuged. Lysates were loaded in 10% Bis-Tris SDS-PAGE gels (Invitrogen) for immunoblot analysis. Primary antibodies used for analysis were mouse anti–α-tubulin 1:30,000 (clone B-5-1-2; Sigma-Aldrich) and rat anti-RFP 1:1,000 (clone 5F8; http://antibodies-online.com). Primary antibodies were detected with HRP-conjugated secondary antibodies and the ECL Prime kit (Amersham).
RNAi
For jac-1 RNAi, base pairs 1546 to 2640 corresponding to WormBase transcript Y105C5B.21b were cloned into vector pPD129.36 (Timmons and Fire, 1998). For hmp-1 RNAi, base pairs 2163 to 2856 of WormBase transcript R13H4.4a were cloned into vector pPD129.36. For pac-1 RNAi, base pairs 3706 to 4585 of Wormbase transcript C04D8.1a were used, corresponding to the C04D8.1 feeding library clone (Kamath and Ahringer, 2003). For picc-1 RNAi, 1.6 kb corresponding to the entire WormBase transcript F29G9.2a was cloned into pPD129.36. For cdc-42 RNAi, the entire cdc-42 coding sequence (Wormbase R07G3.1 transcript) in vector pPD129.36 was used (Aceto et al., 2006).
RNAi constructs were transformed into Escherichia coli strain HT115, and experiments were performed using the feeding method (Timmons et al., 2001). L4 larvae were placed on RNAi plates seeded with bacterial cultures grown for 8–9 h at 37°C in selective media. Seeded plates were left at room temperature for 12–20 h for induction. Empty vector pPD129.36 was used as a negative control in all experiments. Worms were fed for 36–40 h at 25°C, and were either transferred to new RNAi plates to lay eggs (to assay embryonic lethality) or chopped for fixation and staining of embryos. For cdc-42 RNAi, embryos were examined after 30 h of feeding at 20°C.
Each RNAi experiment included a control with GFP- or mCherry-fused target protein to check for knockdown efficiency.
Determining embryonic lethality
Seven to ten adult worms were placed on seeded plates and allowed to lay eggs for six hours at 23°C. Worms were subsequently removed, and eggs were counted. After 24 h, the number of unhatched eggs was counted. Experiments were performed in triplicate.
Immunostaining
Embryos were placed in 7 µl water on poly-l-lysine–coated slides, freeze-cracked on dry ice, fixed in methanol and paraformaldehyde, and stained as described (Anderson et al., 2008). The following primary antibodies and dilutions were used: rabbit anti-HMR-1 1:10,000 (Klompstra et al., 2015), mouse anti–HMP-1 1:10 (Costa et al., 1998), mouse anti–PSD-95 (recognizes DLG-1) 1:200 (Affinity BioReagents), rabbit anti-GFP 1:1,000 (Ab6556.25; AbCam), rat anti-mCherry 1:100 (clone 5F8; http://antibodies-online.com), rabbit anti–PAR-6 1:20 (Schonegg and Hyman, 2006), mouse anti-HA 1:1,000 (clone 16B12; Covance), and rabbit anti–LET-413 1:5,000 (Aono et al., 2004). The following secondary antibodies were used: Alexa Fluor 488 anti-rabbit IgG (H+L) 1:1,000 (Molecular Probes), Cy3 anti-rat 1:100 (Jackson ImmunoResearch), Cy3 anti-mouse IgG (subclasses 1+2a+2b+3) 1:250 (Jackson ImmunoResearch), Alexa Fluor 647 anti-mouse IgG (H+L) 1:100 (Jackson ImmunoResearch), and Alexa Fluor 647 anti-rabbit 1:200 (Molecular Probes).
Embryo mounting
For live imaging, embryos were mounted on 4% agar pads. For scoring elongation phenotypes and rates, multiple two-cell embryos were mounted together, allowed to develop for 3 h at room temperature, and imaged simultaneously in the same field of view. cdc-42(MZ) embryos were imaged together with control embryos in the same field of view until control embryos hatched. For LifeAct-GFP near-TIRF imaging, embryos were mounted in ∼4 µl H2O containing 100–150 polystyrene beads (14.92 µm diameter; NT29N; Bangs Labs) to flatten the embryo surface.
Imaging
DIC movies were acquired using an AxioImager (Zeiss), 63× 1.4 NA or 40× 1.3 NA objective, DIC optics, an Axiocam MRM camera, and AxioVision software. Timelapse images were acquired every 3 min for 7–9 h at 23°C, with six z-planes spaced at 2-µm intervals.
Fluorescence images for Fig. 3 (B and D), Fig. S1 (A–C), Fig. S2 (A–D), Fig. S4 A, and Fig. S5 E were acquired on an AxioImager with 63× 1.4 NA or 40× 1.3 NA objective and a camera (model C10600-10B-H, S. 160522; Hamamatsu). Images were deconvolved using the constrained iterative method in AxioVision software. The sum of 7–10 z-sections taken at 0.3-µm intervals is shown. All other fluorescent images (except LifeAct-GFP) were acquired using an SP5 confocal microscope (Leica), 63× 1.4 NA oil-immersion objective for fixed samples, 63× 1.2 NA water-immersion lens for live imaging, 488-, 561-, 594-, or 633-nm laser, 4× zoom, and 0.7-µm z-interval. For intensity measurements, 1.5-fold live embryos were imaged using HyD detectors and the photon-counting mode.
LifeAct-GFP images were acquired on an inverted Eclipse-Ti microscope (Nikon) with TIRF illuminator and Perfect Focus, 488 nm laser, 100× 1.49 NA oil-immersion objective, iXon3 897 EMCCD camera (Andor) with a ET525/50M filter (Chroma Technology). Laser illumination angle and intensity were controlled by NIS-Elements Software (Nikon). For near-TIRF imaging (Robin et al., 2014), the laser illumination angle was the same for all experiments and was chosen to achieve the highest signal intensity with nearly even illumination. Images were magnified by 1.5× and taken at 32 frames/s.
Image analysis
All measurements were performed using ImageJ (National Institutes of Health). For measuring elongation rates, the length of the eggshell was determined and used as a reference for onefold elongation. The zero time point for all measurements was at the bean stage.
For the intensity profile in Fig. 2 E, a line one pixel in width and 3 µm in length length was drawn perpendicular to the apical epidermal membrane as shown in Fig. 2 (A and B). The intensity of each pixel along the line was plotted using the Plot Profile ImageJ plug-in.
The apical membrane to cytoplasm intensity ratio in epidermal cells was measured in bean-stage embryos. A single focal plane image was selected from a z-stack. A line one pixel in width and 20 µm in length was drawn along the apical membrane, and a second line was drawn in the cytoplasm ∼2 µm below the apical membrane, as shown in Fig. 2 F. Mean pixel intensity values along each line were calculated using the ImageJ measuring tool. Each data point on the graph represents the ratio between the mean intensity value at the apical membrane divided by the mean intensity value in the cytoplasm measured for one embryo.
The junctional to lateral intensity ratio for HMR-1-GFP (Fig. 2 J) was measured by drawing a line along the lateral membrane and then dividing the intensity measured in the junctional area by the mean intensity along the lateral domain.
Pearson correlation coefficient colocalization tests were performed on deconvolved images using the JACoP plug-in in ImageJ.
The efficiency of degradation of YFP-ZF1-CDC-42 was quantified in the epidermis of 1.5-fold embryos. Photon counting mode on the Leica SP5 confocal was used for imaging. Embryos were selected using the freehand selection tool on an image produced by z-projection of two confocal z-sections. Background intensity was subtracted from the mean intensity value.
The intensity of AJ components at junctions was measured at the 1.5-fold stage. A z-projection of the sum of the three top z-sections was measured for each embryo. A duplicate of this image was used to create a mask, which was filtered with an FFT Bandpass filter (to filter out structures smaller than two pixels) and thresholded. The mask was then used to select junctions on original nonprocessed image for measurement. The “Analyze Particles” plug-in was used to measure intensities.
The intensity of the CDC-42 biosensor at junctions was measured at the 1.5-fold stage by drawing a line across the junction and measuring the maximal intensity. For each embryo, four measurements were taken.
LifeAct-GFP images in Fig. 7 were obtained by averaging 200 frames and applying the Unsharp Mask filter (radius 1 pixel and mask weight 0.7). In Fig. 7 D, embryos at the same developmental stage (1.3-fold elongation) and in the same orientation (lateral and ventral rows of cells up) were analyzed. In Fig. 7 E, embryos at 1.2- and 1.3-fold elongation were analyzed. An image stack of 512 time points was processed by running a mean of 16 frames to reduce noise, using linear contrast enhancement with no saturated pixels, and subtracting background. SD was calculated for each pixel through time and then divided by a mean intensity of this pixel to normalize. The output of this calculation was an image that was pseudo-colored as a heat map using LUT. To measure actin remodeling at the junctions, a segmented line 1 µm in width was drawn along the junction between two ventral cells (18 and 20, Worm Atlas) and their neighboring seam cells.
For plotting image quantification and statistical analysis, mean values for each embryo were copied to Prism 6.
Statistics
Statistical analysis was performed in GraphPad Prism 6. Statistical tests, number of embryos, and number of experiments are indicated in the figure legends. No statistical method was used to predetermine the sample size. The investigators were not blinded during experiments and outcome assessment.
Online supplemental material
Fig. S1 shows strategies for obtaining cdc-42 mutants. Fig. S2 shows markers of polarization and junction maturation in cdc-42(MZ) embryos. Fig. S3 shows elongation and arrest phenotypes of embryos in genetic interaction experiments. Fig. S4 shows quantification of PAC-1 colocalization with junction proteins. Fig. S5 shows expression of GAP-dead PAC-1 and constitutively active CDC-42, and additional quantification of AJ protein levels in pac-1(xn6) embryos. Table S1 shows genetic interactions between hmp-1(fe4) embryos and embryos treated with indicated RNAi. Table S2 shows the list of strains used in this study. Video 1 shows elongation in control and cdc-42 embryos. Video 2 shows junction formation and maturation of HMR-1-GFP in control and cdc-42 embryos. Video 3 shows polarization of HMR-1-GFP in the intestine of control and cdc-42 embryos. Video 4 shows junction maturation of DLG-1-RFP in control and cdc-42 embryos. Video 5 shows elongation in a hmp-1(fe4); gfp-cdc-42 embryo. Video 6 shows elongation in control, pac-1(xn6), hmp-1(fe4), and hmp-1(fe4); pac-1(xn6) embryos. Video 7 shows elongation in a hmp-1(fe4); picc-1 embryo. Video 8 shows junctional actin dynamics in control and pac-1(xn6) embryos.
Supplementary Material
Acknowledgments
We thank Michael Hengartner for providing the gfp-cdc-42 strain, Bob Goldstein for the hmr-1-gfp knockin strain, Kenneth Kemphues for the par-3-gfp strain, Michel Labouesse for dlg-1-rfp strains, and Kraig Kumfer for the plasmid used to make the CDC-42 biosensor. Some strains were provided by the Caenorhabditis Genetis Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). Thanks to members of the Nance laboratory for critical reading of the manuscript; and Ed Munro, François Robin, and Michael Cammer for assistance with near-TIRF imaging.
Funds for this work were provided by National Institutes of Health grants R01GM098492, R01GM078341, and R35GM118081 (to J. Nance), and by American Hospital Association postdoctoral fellowship 14POST18740059 (to Y. Zilberman).
The authors declare no competing financial interests.
Author contributions: Y. Zilberman, D.C. Anderson, and J. Nance conceived of the experiments. J. Abrams contributed the xn60 and xn65 strains, and Y. Zilberman and D.C. Anderson performed all other experiments. Y. Zilberman, D.C. Anderson, and J. Nance analyzed and interpreted data, and Y. Zilberman and J. Nance wrote the manuscript.
Footnotes
Abbreviations used:
- AJ
- adherens junction
- CFB
- circumferential bundle
- CRIB
- CDC-42/Rac interactive binding
- DIC
- differential interference contrast
- MET
- mesenchymal-to-epithelial transition
- sgRNA
- single guide RNA
- TIRF
- total internal reflection fluorescence
- ZF1
- zinc finger 1
References
- Aceto D., Beers M., and Kemphues K.J.. 2006. Interaction of PAR-6 with CDC-42 is required for maintenance but not establishment of PAR asymmetry in C. elegans. Dev. Biol. 299:386–397. 10.1016/j.ydbio.2006.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilleos A., Wehman A.M., and Nance J.. 2010. PAR-3 mediates the initial clustering and apical localization of junction and polarity proteins during C. elegans intestinal epithelial cell polarization. Development. 137:1833–1842. 10.1242/dev.047647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D.C., Gill J.S., Cinalli R.M., and Nance J.. 2008. Polarization of the C. elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science. 320:1771–1774. 10.1126/science.1156063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono S., Legouis R., Hoose W.A., and Kemphues K.J.. 2004. PAR-3 is required for epithelial cell polarity in the distal spermatheca of C. elegans. Development. 131:2865–2874. 10.1242/dev.01146 [DOI] [PubMed] [Google Scholar]
- Armenti S.T., and Nance J.. 2012. Adherens junctions in C. elegans embryonic morphogenesis. Subcell. Biochem. 60:279–299. 10.1007/978-94-007-4186-7_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenti S.T., Chan E., and Nance J.. 2014a Polarized exocyst-mediated vesicle fusion directs intracellular lumenogenesis within the C. elegans excretory cell. Dev. Biol. 394:110–121. 10.1016/j.ydbio.2014.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenti S.T., Lohmer L.L., Sherwood D.R., and Nance J.. 2014b Repurposing an endogenous degradation system for rapid and targeted depletion of C. elegans proteins. Development. 141:4640–4647. 10.1242/dev.115048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos K.S., Bigarella C.L., Wagner M.V., Vieira K.P., Lazarini M., Langford P.R., Machado-Neto J.A., Call S.G., Staley D.M., Chung J.Y., et al. . 2013. ARHGAP21 protein, a new partner of α-tubulin involved in cell-cell adhesion formation and essential for epithelial-mesenchymal transition. J. Biol. Chem. 288:2179–2189. 10.1074/jbc.M112.432716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.L., Rehmann H., and Wittinghofer A.. 2007. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 129:865–877. 10.1016/j.cell.2007.05.018 [DOI] [PubMed] [Google Scholar]
- Bossinger O., Klebes A., Segbert C., Theres C., and Knust E.. 2001. Zonula adherens formation in Caenorhabditis elegans requires dlg-1, the homologue of the Drosophila gene discs large. Dev. Biol. 230:29–42. 10.1006/dbio.2000.0113 [DOI] [PubMed] [Google Scholar]
- Bruewer M., Hopkins A.M., Hobert M.E., Nusrat A., and Madara J.L.. 2004. RhoA, Rac1, and Cdc42 exert distinct effects on epithelial barrier via selective structural and biochemical modulation of junctional proteins and F-actin. Am. J. Physiol. Cell Physiol. 287:C327–C335. 10.1152/ajpcell.00087.2004 [DOI] [PubMed] [Google Scholar]
- Chisholm A.D., and Hardin J.. 2005. Epidermal morphogenesis. WormBook. 1–22. 10.1895/wormbook.1.35.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinet C., and Lecuit T.. 2013. Stability and dynamics of cell-cell junctions. Prog. Mol. Biol. Transl. Sci. 116:25–47. 10.1016/B978-0-12-394311-8.00002-9 [DOI] [PubMed] [Google Scholar]
- Costa M., Raich W., Agbunag C., Leung B., Hardin J., and Priess J.R.. 1998. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J. Cell Biol. 141:297–308. 10.1083/jcb.141.1.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Paulson E.A., Walck-Shannon E., Lynch A.M., Yamashiro S., Zaidel-Bar R., Eno C.C., Ono S., and Hardin J.. 2012. Tropomodulin protects α-catenin-dependent junctional-actin networks under stress during epithelial morphogenesis. Curr. Biol. 22:1500–1505. 10.1016/j.cub.2012.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cram E.J. 2014. Mechanotransduction in C. elegans morphogenesis and tissue function. Prog. Mol. Biol. Transl. Sci. 126:281–316. 10.1016/B978-0-12-394624-9.00012-9 [DOI] [PubMed] [Google Scholar]
- DeRenzo C., Reese K.J., and Seydoux G.. 2003. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature. 424:685–689. 10.1038/nature01887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D.J., Ward J.D., Reiner D.J., and Goldstein B.. 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 10:1028–1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diogon M., Wissler F., Quintin S., Nagamatsu Y., Sookhareea S., Landmann F., Hutter H., Vitale N., and Labouesse M.. 2007. The RhoGAP RGA-2 and LET-502/ROCK achieve a balance of actomyosin-dependent forces in C. elegans epidermis to control morphogenesis. Development. 134:2469–2479. 10.1242/dev.005074 [DOI] [PubMed] [Google Scholar]
- Druso J.E., Endo M., Lin M.C., Peng X., Antonyak M.A., Meller S., and Cerione R.A.. 2016. An essential role for Cdc42 in the functioning of the adult mammary gland. J. Biol. Chem. 291:8886–8895. 10.1074/jbc.M115.694349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette P.M., and Lamarche-Vane N.. 2014. Rho GTPases in embryonic development. Small GTPases. 5:e972857 10.4161/sgtp.29716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbediwy A., Zihni C., Terry S.J., Clark P., Matter K., and Balda M.S.. 2012. Epithelial junction formation requires confinement of Cdc42 activity by a novel SH3BP1 complex. J. Cell Biol. 198:677–693. 10.1083/jcb.201202094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias B.C., Das A., Parekh D.V., Mernaugh G., Adams R., Yang Z., Brakebusch C., Pozzi A., Marciano D.K., Carroll T.J., and Zent R.. 2015. Cdc42 regulates epithelial cell polarity and cytoskeletal function during kidney tubule development. J. Cell Sci. 128:4293–4305. 10.1242/jcs.164509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erasmus J., Aresta S., Nola S., Caron E., and Braga V.M.M.. 2010. Newly formed E-cadherin contacts do not activate Cdc42 or induce filopodia protrusion in human keratinocytes. Biol. Cell. 102:13–24. 10.1042/BC20090048 [DOI] [PubMed] [Google Scholar]
- Erasmus J.C., Bruche S., Pizarro L., Maimari N., Pogglioli T., Tomlinson C., Lees J., Zalivina I., Wheeler A., Alberts A., et al. . 2016. Defining functional interactions during biogenesis of epithelial junctions. Nat. Commun. 7:13542 10.1038/ncomms13542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J.W., and Cerione R.A.. 2001. Multiple roles for Cdc42 in cell regulation. Curr. Opin. Cell Biol. 13:153–157. 10.1016/S0955-0674(00)00192-7 [DOI] [PubMed] [Google Scholar]
- Firestein B.L., and Rongo C.. 2001. DLG-1 is a MAGUK similar to SAP97 and is required for adherens junction formation. Mol. Biol. Cell. 12:3465–3475. 10.1091/mbc.12.11.3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M.W., Ailion M., and Jorgensen E.M.. 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods. 9:117–118. 10.1038/nmeth.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou M., Marinari E., Burden J., and Baum B.. 2008. Cdc42, Par6, and aPKC regulate Arp2/3-mediated endocytosis to control local adherens junction stability. Curr. Biol. 18:1631–1638. 10.1016/j.cub.2008.09.029 [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. III, and Smith H.O.. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Gotta M., Abraham M.C., and Ahringer J.. 2001. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr. Biol. 11:482–488. 10.1016/S0960-9822(01)00142-7 [DOI] [PubMed] [Google Scholar]
- Harris K.P., and Tepass U.. 2008. Cdc42 and Par proteins stabilize dynamic adherens junctions in the Drosophila neuroectoderm through regulation of apical endocytosis. J. Cell Biol. 183:1129–1143. 10.1083/jcb.200807020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K.P., and Tepass U.. 2010. Cdc42 and vesicle trafficking in polarized cells. Traffic. 11:1272–1279. 10.1111/j.1600-0854.2010.01102.x [DOI] [PubMed] [Google Scholar]
- Harris T.J.C., and Peifer M.. 2004. Adherens junction-dependent and -independent steps in the establishment of epithelial cell polarity in Drosophila. J. Cell Biol. 167:135–147. 10.1083/jcb.200406024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter M.V., Lee D.M., Harris T.J., and Fernandez-Gonzalez R.. 2015. Polarized E-cadherin endocytosis directs actomyosin remodeling during embryonic wound repair. J. Cell Biol. 210:801–816. 10.1083/jcb.201501076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer A., Betschinger J., Petronczki M., and Knoblich J.A.. 2004. Sequential roles of Cdc42, Par-6, aPKC, and Lgl in the establishment of epithelial polarity during Drosophila embryogenesis. Dev. Cell. 6:845–854. 10.1016/j.devcel.2004.05.003 [DOI] [PubMed] [Google Scholar]
- Joberty G., Petersen C., Gao L., and Macara I.G.. 2000. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2:531–539. 10.1038/35019573 [DOI] [PubMed] [Google Scholar]
- Johansson A., Driessens M., and Aspenström P.. 2000. The mammalian homologue of the Caenorhabditis elegans polarity protein PAR-6 is a binding partner for the Rho GTPases Cdc42 and Rac1. J. Cell Sci. 113:3267–3275. [DOI] [PubMed] [Google Scholar]
- Kamath R.S., and Ahringer J.. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 30:313–321. 10.1016/S1046-2023(03)00050-1 [DOI] [PubMed] [Google Scholar]
- Kay A.J., and Hunter C.P.. 2001. CDC-42 regulates PAR protein localization and function to control cellular and embryonic polarity in C. elegans. Curr. Biol. 11:474–481. 10.1016/S0960-9822(01)00141-5 [DOI] [PubMed] [Google Scholar]
- Kesavan G., Sand F.W., Greiner T.U., Johansson J.K., Kobberup S., Wu X., Brakebusch C., and Semb H.. 2009. Cdc42-mediated tubulogenesis controls cell specification. Cell. 139:791–801. 10.1016/j.cell.2009.08.049 [DOI] [PubMed] [Google Scholar]
- Klompstra D., Anderson D.C., Yeh J.Y., Zilberman Y., and Nance J.. 2015. An instructive role for C. elegans E-cadherin in translating cell contact cues into cortical polarity. Nat. Cell Biol. 17:726–735. 10.1038/ncb3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köppen M., Simske J.S., Sims P.A., Firestein B.L., Hall D.H., Radice A.D., Rongo C., and Hardin J.D.. 2001. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat. Cell Biol. 3:983–991. 10.1038/ncb1101-983 [DOI] [PubMed] [Google Scholar]
- Korswagen H.C., Herman M.A., and Clevers H.C.. 2000. Distinct beta-catenins mediate adhesion and signalling functions in C. elegans. Nature. 406:527–532. 10.1038/35020099 [DOI] [PubMed] [Google Scholar]
- Kroschewski R., Hall A., and Mellman I.. 1999. Cdc42 controls secretory and endocytic transport to the basolateral plasma membrane of MDCK cells. Nat. Cell Biol. 1:8–13. [DOI] [PubMed] [Google Scholar]
- Kumfer K.T., Cook S.J., Squirrell J.M., Eliceiri K.W., Peel N., O’Connell K.F., and White J.G.. 2010. CGEF-1 and CHIN-1 regulate CDC-42 activity during asymmetric division in the Caenorhabditis elegans embryo. Mol. Biol. Cell. 21:266–277. 10.1091/mbc.E09-01-0060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski A.V., Maiden S.L., Pokutta S., Choi H.J., Benjamin J.M., Lynch A.M., Nelson W.J., Weis W.I., and Hardin J.. 2010. In vitro and in vivo reconstitution of the cadherin-catenin-actin complex from Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 107:14591–14596. (published erratum appears in Proc. Natl. Acad. Sci. USA. 108: 4264) 10.1073/pnas.1007349107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmann F., Quintin S., and Labouesse M.. 2004. Multiple regulatory elements with spatially and temporally distinct activities control the expression of the epithelial differentiation gene lin-26 in C. elegans. Dev. Biol. 265:478–490. 10.1016/j.ydbio.2003.09.009 [DOI] [PubMed] [Google Scholar]
- Legouis R., Gansmuller A., Sookhareea S., Bosher J.M., Baillie D.L., and Labouesse M.. 2000. LET-413 is a basolateral protein required for the assembly of adherens junctions in Caenorhabditis elegans. Nat. Cell Biol. 2:415–422. 10.1038/35017046 [DOI] [PubMed] [Google Scholar]
- Leibfried A., Fricke R., Morgan M.J., Bogdan S., and Bellaiche Y.. 2008. Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-cadherin endocytosis. Curr. Biol. 18:1639–1648. 10.1016/j.cub.2008.09.063 [DOI] [PubMed] [Google Scholar]
- Leung B., Hermann G.J., and Priess J.R.. 1999. Organogenesis of the Caenorhabditis elegans intestine. Dev. Biol. 216:114–134. 10.1006/dbio.1999.9471 [DOI] [PubMed] [Google Scholar]
- Levayer R., Pelissier-Monier A., and Lecuit T.. 2011. Spatial regulation of Dia and Myosin-II by RhoGEF2 controls initiation of E-cadherin endocytosis during epithelial morphogenesis. Nat. Cell Biol. 13:529–540. 10.1038/ncb2224 [DOI] [PubMed] [Google Scholar]
- Lin D., Edwards A.S., Fawcett J.P., Mbamalu G., Scott J.D., and Pawson T.. 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2:540–547. 10.1038/35019592 [DOI] [PubMed] [Google Scholar]
- Lynch A.M., Grana T., Cox-Paulson E., Couthier A., Cameron M., Chin-Sang I., Pettitt J., and Hardin J.. 2012. A genome-wide functional screen shows MAGI-1 is an L1CAM-dependent stabilizer of apical junctions in C. elegans. Curr. Biol. 22:1891–1899. 10.1016/j.cub.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack N.A., and Georgiou M.. 2014. The interdependence of the Rho GTPases and apicobasal cell polarity. Small GTPases. 5:10 10.4161/21541248.2014.973768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden S.L., and Hardin J.. 2011. The secret life of α-catenin: moonlighting in morphogenesis. J. Cell Biol. 195:543–552. 10.1083/jcb.201103106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham N.O., Doll C.A., Dohn M.R., Miller R.K., Yu H., Coffey R.J., McCrea P.D., Gamse J.T., and Reynolds A.B.. 2014. DIPA-family coiled-coils bind conserved isoform-specific head domain of p120-catenin family: potential roles in hydrocephalus and heterotopia. Mol. Biol. Cell. 25:2592–2603. 10.1091/mbc.E13-08-0492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston D.J., Higgins C.D., Peters K.A., Cupp T.D., Dickinson D.J., Pani A.M., Moore R.P., Cox A.H., Kiehart D.P., and Goldstein B.. 2016. MRCK-1 Drives Apical Constriction in C. elegans by Linking Developmental Patterning to Force Generation. Curr. Biol. 26:2079–2089. 10.1016/j.cub.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Belmonte F., Gassama A., Datta A., Yu W., Rescher U., Gerke V., and Mostov K.. 2007. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell. 128:383–397. 10.1016/j.cell.2006.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon L., Legouis R., Vonesch J.L., and Labouesse M.. 2001. Assembly of C. elegans apical junctions involves positioning and compaction by LET-413 and protein aggregation by the MAGUK protein DLG-1. J. Cell Sci. 114:2265–2277. [DOI] [PubMed] [Google Scholar]
- Mello C.C., Kramer J.M., Stinchcomb D., and Ambros V.. 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10:3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori N., Kuwamura M., Tanaka N., Hirano R., Nabe M., Ibuki M., and Yamate J.. 2012. Ccdc85c encoding a protein at apical junctions of radial glia is disrupted in hemorrhagic hydrocephalus (hhy) mice. Am. J. Pathol. 180:314–327. 10.1016/j.ajpath.2011.09.014 [DOI] [PubMed] [Google Scholar]
- Nance J., Munro E.M., and Priess J.R.. 2003. C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development. 130:5339–5350. 10.1242/dev.00735 [DOI] [PubMed] [Google Scholar]
- Neukomm L.J., Zeng S., Frei A.P., Huegli P.A., and Hengartner M.O.. 2014. Small GTPase CDC-42 promotes apoptotic cell corpse clearance in response to PAT-2 and CED-1 in C. elegans. Cell Death Differ. 21:845–853. 10.1038/cdd.2014.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani T., Ichii T., Aono S., and Takeichi M.. 2006. Cdc42 GEF Tuba regulates the junctional configuration of simple epithelial cells. J. Cell Biol. 175:135–146. 10.1083/jcb.200605012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette M.H., Martin E., Lacoste-Caron G., Hamiche K., and Jenna S.. 2016. Spatial control of active CDC-42 during collective migration of hypodermal cells in Caenorhabditis elegans. J. Mol. Cell Biol. 8:313–327. 10.1093/jmcb/mjv062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt J., Cox E.A., Broadbent I.D., Flett A., and Hardin J.. 2003. The Caenorhabditis elegans p120 catenin homologue, JAC-1, modulates cadherin-catenin function during epidermal morphogenesis. J. Cell Biol. 162:15–22. 10.1083/jcb.200212136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phng L.K., Gebala V., Bentley K., Philippides A., Wacker A., Mathivet T., Sauteur L., Stanchi F., Belting H.G., Affolter M., and Gerhardt H.. 2015. Formin-mediated actin polymerization at endothelial junctions is required for vessel lumen formation and stabilization. Dev. Cell. 32:123–132. 10.1016/j.devcel.2014.11.017 [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., and Austin J.. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 157:1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Liu J., Wu X., Brakebusch C., Leitges M., Han Y., Corbett S.A., Lowry S.F., Graham A.M., and Li S.. 2011. Cdc42 controls vascular network assembly through protein kinase Cι during embryonic vasculogenesis. Arterioscler. Thromb. Vasc. Biol. 31:1861–1870. 10.1161/ATVBAHA.111.230144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu R.G., Abo A., and Steven Martin G.. 2000. A human homolog of the C. elegans polarity determinant Par-6 links Rac and Cdc42 to PKCzeta signaling and cell transformation. Curr. Biol. 10:697–707. 10.1016/S0960-9822(00)00535-2 [DOI] [PubMed] [Google Scholar]
- Rao M.V., and Zaidel-Bar R.. 2016. Formin-mediated actin polymerization at cell-cell junctions stabilizes E-cadherin and maintains monolayer integrity during wound repair. Mol. Biol. Cell. 27:2844–2856. 10.1091/mbc.E16-06-0429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese K.J., Dunn M.A., Waddle J.A., and Seydoux G.. 2000. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol. Cell. 6:445–455. 10.1016/S1097-2765(00)00043-5 [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. . 2008. LifeAct: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin F.B., McFadden W.M., Yao B., and Munro E.M.. 2014. Single-molecule analysis of cell surface dynamics in Caenorhabditis elegans embryos. Nat. Methods. 11:677–682. 10.1038/nmeth.2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas R., Ruiz W.G., Leung S.M., Jou T.S., and Apodaca G.. 2001. Cdc42-dependent modulation of tight junctions and membrane protein traffic in polarized Madin-Darby canine kidney cells. Mol. Biol. Cell. 12:2257–2274. 10.1091/mbc.12.8.2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röper K. 2015. Integration of cell-cell adhesion and contractile actomyosin activity during morphogenesis. Curr. Top. Dev. Biol. 112:103–127. 10.1016/bs.ctdb.2014.11.017 [DOI] [PubMed] [Google Scholar]
- Schonegg S., and Hyman A.A.. 2006. CDC-42 and RHO-1 coordinate acto-myosin contractility and PAR protein localization during polarity establishment in C. elegans embryos. Development. 133:3507–3516. 10.1242/dev.02527 [DOI] [PubMed] [Google Scholar]
- Shafaq-Zadah M., Brocard L., Solari F., and Michaux G.. 2012. AP-1 is required for the maintenance of apico-basal polarity in the C. elegans intestine. Development. 139:2061–2070. 10.1242/dev.076711 [DOI] [PubMed] [Google Scholar]
- Shindo M., Wada H., Kaido M., Tateno M., Aigaki T., Tsuda L., and Hayashi S.. 2008. Dual function of Src in the maintenance of adherens junctions during tracheal epithelial morphogenesis. Development. 135:1355–1364. 10.1242/dev.015982 [DOI] [PubMed] [Google Scholar]
- Sousa S., Cabanes D., Archambaud C., Colland F., Lemichez E., Popoff M., Boisson-Dupuis S., Gouin E., Lecuit M., Legrain P., and Cossart P.. 2005. ARHGAP10 is necessary for alpha-catenin recruitment at adherens junctions and for Listeria invasion. Nat. Cell Biol. 7:954–960. 10.1038/ncb1308 [DOI] [PubMed] [Google Scholar]
- Stephenson R.O., Yamanaka Y., and Rossant J.. 2010. Disorganized epithelial polarity and excess trophectoderm cell fate in preimplantation embryos lacking E-cadherin. Development. 137:3383–3391. 10.1242/dev.050195 [DOI] [PubMed] [Google Scholar]
- Tepass U. 2012. The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu. Rev. Cell Dev. Biol. 28:655–685. 10.1146/annurev-cellbio-092910-154033 [DOI] [PubMed] [Google Scholar]
- Timmons L., and Fire A.. 1998. Specific interference by ingested dsRNA. Nature. 395:854 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Timmons L., Court D.L., and Fire A.. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 263:103–112. 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Totong R., Achilleos A., and Nance J.. 2007. PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells. Development. 134:1259–1268. 10.1242/dev.02833 [DOI] [PubMed] [Google Scholar]
- Truong Quang B.A., Mani M., Markova O., Lecuit T., and Lenne P.F.. 2013. Principles of E-cadherin supramolecular organization in vivo. Curr. Biol. 23:2197–2207. 10.1016/j.cub.2013.09.015 [DOI] [PubMed] [Google Scholar]
- Van Itallie C.M., Tietgens A.J., Aponte A., Fredriksson K., Fanning A.S., Gucek M., and Anderson J.M.. 2014. Biotin ligase tagging identifies proteins proximal to E-cadherin, including lipoma preferred partner, a regulator of epithelial cell-cell and cell-substrate adhesion. J. Cell Sci. 127:885–895. 10.1242/jcs.140475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Han S.P., Michael M., Gomez G.A., Yang Z., Teasdale R.D., Ratheesh A., Kovacs E.M., Ali R.G., and Yap A.S.. 2012. A WAVE2-Arp2/3 actin nucleator apparatus supports junctional tension at the epithelial zonula adherens. Mol. Biol. Cell. 23:4601–4610. 10.1091/mbc.E12-08-0574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina S.E., and Mango S.E.. 2015. PAR-6, but not E-cadherin and β-integrin, is necessary for epithelial polarization in C. elegans. Dev. Biol. 403:5–14. 10.1016/j.ydbio.2015.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong-Brender T.T., Yang X., and Labouesse M.. 2016. C. elegans embryonic morphogenesis. Curr. Top. Dev. Biol. 116:597–616. 10.1016/bs.ctdb.2015.11.012 [DOI] [PubMed] [Google Scholar]
- Walck-Shannon E., Lucas B., Chin-Sang I., Reiner D., Kumfer K., Cochran H., Bothfeld W., and Hardin J.. 2016. CDC-42 orients cell migration during epithelial intercalation in the Caenorhabditis elegans epidermis. PLoS Genet. 12:e1006415 10.1371/journal.pgen.1006415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells C.D., Fawcett J.P., Traweger A., Yamanaka Y., Goudreault M., Elder K., Kulkarni S., Gish G., Virag C., Lim C., et al. . 2006. A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell. 125:535–548. 10.1016/j.cell.2006.02.045 [DOI] [PubMed] [Google Scholar]
- Wu Y., Kanchanawong P., and Zaidel-Bar R.. 2015. Actin-delimited adhesion-independent clustering of E-cadherin forms the nanoscale building blocks of adherens junctions. Dev. Cell. 32:139–154. 10.1016/j.devcel.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Zihni C., Munro P.M., Elbediwy A., Keep N.H., Terry S.J., Harris J., Balda M.S., and Matter K.. 2014. Dbl3 drives Cdc42 signaling at the apical margin to regulate junction position and apical differentiation. J. Cell Biol. 204:111–127. 10.1083/jcb.201304064 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.