TPR nucleoporins form the nuclear pore complex basket. The fission yeast TPR Alm1 is required for localization of the proteasome to the nuclear envelope, which is in turn required for kinetochore homeostasis and proper chromosome segregation.
Abstract
Kinetochores (KTs) are large multiprotein complexes that constitute the interface between centromeric chromatin and the mitotic spindle during chromosome segregation. In spite of their essential role, little is known about how centromeres and KTs are assembled and how their precise stoichiometry is regulated. In this study, we show that the nuclear pore basket component Alm1 is required to maintain both the proteasome and its anchor, Cut8, at the nuclear envelope, which in turn regulates proteostasis of certain inner KT components. Consistently, alm1-deleted cells show increased levels of KT proteins, including CENP-CCnp3, spindle assembly checkpoint activation, and chromosome segregation defects. Our data demonstrate a novel function of the nucleoporin Alm1 in proteasome localization required for KT homeostasis.
Introduction
Centromeres are specialized regions of chromatin that constitute the site of kinetochore (KT) assembly required for mitotic spindle attachment. Fission yeast centromeres are structurally complex, similar to those of metazoa, and contain repetitive elements (outermost repeats [otr]) flanking a central domain composed of a central core (cnt) surrounded by innermost repeats (imr; Takahashi et al., 1992; Pidoux and Allshire, 2005). Centromeric chromatin is transcriptionally silenced. However, whereas transcriptional silencing at imr/otr depends on the canonical heterochromatin marks (H3K9me2) and HP1/Swi6 binding, transcriptional silencing at cnt depends on the centromeric histone H3 variant CENP-ACnp1 (Pidoux and Allshire, 2005; Castillo et al., 2007). Cnp1, together with the noncanonical nucleosome complex CENP-S–T–W–X (SpMhf1, SpCnp20, SpNew1, and SpMhf2), defines the landmark of centromeric chromatin structure (Stoler et al., 1995; Takahashi et al., 2000, 2005). Cnp1 directly interacts with CENP-CCnp3, which in turn recruits several factors, including CENP-LFta1, CENP-KSim4, or the monopolin complex Pcs1–Mde4, required for KT assembly and function (Cohen et al., 2008; Tanaka et al., 2009; Carroll et al., 2010; Folco et al., 2015).
KTs can be functionally divided into two domains: the inner KT, which constitutes the interface with centromeric chromatin, and the outer KT, which constitutes the microtubule (MT)-binding interface (Santaguida and Musacchio, 2009). During mitosis, each KT must establish correct interactions with spindle MTs. Erroneous KT–MT attachments are detected by the spindle assembly checkpoint (SAC) and corrected by aurora B kinase. If not corrected, erroneous attachments might result in chromosome missegregation and aneuploidy, which contributes to chromosomal instability in cancer cells (Cimini et al., 2003, 2006). Both centromeric chromatin and proper KT structure are required for correct chromosome segregation (Goshima et al., 1999; Bernard et al., 2001; Nonaka et al., 2002; Pidoux et al., 2003; Pidoux and Allshire, 2005; Gregan et al., 2007; Tanaka et al., 2009). Similarly, the appropriate balance of KT proteins must be strictly regulated to ensure proper KT assembly and function (Castillo et al., 2007).
The 26S proteasome is a large, evolutionary conserved protease complex responsible for the ATP-dependent degradation of polyubiquitinated proteins. This proteolytic system plays a pivotal role in cellular quality control and protein homeostasis. Proteasomal degradation takes place in the cytoplasm and also inside the nucleus, where the proteasome is enriched (Wilkinson et al., 1998; Voges et al., 1999). It is known that the levels and distribution of centromeric CENP-ACnp1 are regulated by proteasomal degradation, restricting its localization to centromeric chromatin (Collins et al., 2004; Moreno-Moreno et al., 2006; Ranjitkar et al., 2010; Kitagawa et al., 2014). Interestingly, Cut8, a nuclear envelope (NE) protein required for proteasome function and localization to the NE, is also required for Cnp1 centromeric localization (Tatebe and Yanagida, 2000; Takeda and Yanagida, 2005; Kitagawa et al., 2014), suggesting that proteasome localization at the NE is functionally linked to proper centromere–KT composition. The NE further contributes to the quality control and protein homeostasis of KT components through the ubiquitin–proteasome pathway (Furth et al., 2011; Kriegenburg et al., 2014). However, to date, it is still unknown how the NE influences proteasome localization and regulation to ensure proper KT–centromere stoichiometry and function.
The NE is spanned by the nuclear pore complexes (NPCs), composed of ~30 different types of proteins called nucleoporins (Asakawa et al., 2014; Kabachinski and Schwartz, 2015). The classical view of the NPCs as mediators of nucleocytoplasmic transport has been further extended to many other cellular functions such as regulation of gene expression and genome stability (Palancade et al., 2007; Capelson et al., 2010; Kalverda and Fornerod, 2010; Yang et al., 2015). Associated with the NPCs are the translocated promoter region (TPR) nucleoporins, which are evolutionary conserved large coiled-coil (CC) proteins that localize at the nucleoplasmic side of the NPC, forming a specific structure called the nuclear basket (Cordes et al., 1997; Frosst et al., 2002; Asakawa et al., 2014). TPRs form filaments that connect adjacent NPCs and project into the nucleoplasm, serving as a binding platform for chromatin, transcription factors, and cell cycle regulators (Galy et al., 2000; Niepel et al., 2013; Umlauf et al., 2013; Rajanala et al., 2014). In human cells, TPR contributes to a variety of nuclear processes, such as small ubiquitin–related modifier (SUMO) homeostasis (Schweizer et al., 2013), spatiotemporal regulation of SAC components (Rodriguez-Bravo et al., 2014), and the exclusion of heterochromatin from NPCs (Krull et al., 2010). In addition, in Saccharomyces cerevisiae, the TPR proteins Mlp1 and Mlp2 are involved in anchoring transcriptionally active genes at the NPC (Luthra et al., 2007; Tan-Wong et al., 2009) and mRNA export (Niepel et al., 2013) and are required for maintenance of spindle pole body (SPB) integrity and telomere structure (Hediger et al., 2002; Zhao et al., 2004; Niepel et al., 2005). In Schizosaccharomyces pombe, there are two members of the TPR family of nucleoporins, Nup211 and Alm1 (Bae et al., 2009; DeGrasse et al., 2009). Whereas Nup211 plays an essential role in the regulation of mRNA export, the function of Alm1 has not been characterized to date.
Here, we show that the TPR nucleoporin Alm1 is required to maintain the proper localization of the proteasome anchor and regulator Cut8 and the proteasome subunits Mts2 and Mts4 to the NE. Cells lacking alm1 show an altered stoichiometry of KT proteins, including Cnp3, and DNA segregation defects. Together, these results strongly suggest that Alm1 is required for proper 26S proteasome localization at the NE, and this regulation is critical to maintain correct KT stoichiometry.
Results
Alm1 localizes to the NE
TPR nucleoporins are large CC proteins that localize at the nucleoplasmic side of the NPC, forming the nuclear basket. Alm1 showed extensive regions of predicted CCs (Fig. S1 A). Moreover, Alm1-Tomato localized as bright dots at the nuclear periphery (Fig. S1 B) and partially colocalized with the nucleoporin Nup107-GFP (Fig. S1 C). This partial colocalization suggests that Alm1 also occupies areas that are devoid of NPCs, similar to what has been shown for S. cerevisiae Mlp1 and Mlp2 (Strambio-de-Castillia et al., 1999; Niepel et al., 2013). The localization of Alm1-Tomato mostly overlapped with Nup211-GFP (Fig. 1 A). Furthermore, in the absence of Alm1, a fraction of Nup211 lost its NE localization and accumulated in the nucleoplasm (Fig. 1 B), suggesting that both proteins interact at the NPC. It has previously been reported that Alm1 localizes at the SPBs and the medial ring (Jiménez et al., 2000). Careful analysis of the localization of Alm1-Tomato expressed from its own promoter showed only a transient colocalization of Alm1 with the SPB, and we never observed any Alm1-Tomato signal at the medial ring (Fig. S1 D).
Figure 1.
Alm1 localizes to the NE and NPC, and its absence results in chromosome segregation defects. (A) Alm1-Tomato colocalizes with Nup211-GFP. Maximal projection of three z sections is shown. (Inset) Magnification of the indicated nucleus. Bars, 2.5 µm. Fluorescence intensity levels along the nuclear rim of the selected nucleus from an arbitrary starting point. (B) Images of WT and alm1Δ cells expressing Nup211-GFP. Mean fluorescence intensity of Nup211-GFP at the NE and in the nucleoplasm. n = 30. (C) WT and alm1Δ cells expressing the NE marker Cut11-GFP. Arrow indicates smaller daughter nuclei. Time is indicated in minutes. Ratio of the perimeter of sister nuclei. n = 25. (D) Minichromosome loss assay. Three independent experiments. n > 1,500. (E) Images of WT and alm1Δ mitotic cells expressing Sid2-Tomato and Mis6-GFP. Time between frames is 1 min. Magnifications of the regions indicated by dashed boxes are shown below. Bar, 2.5 µm. Asterisk marks asynchronous KT segregation. Arrow indicates lagging KT. Bars, 5 µm. A.U., arbitrary units. Error bars represent SD. ***, P < 0.001; ****, P < 0.0001.
Thus, as it has been reported for members of the Mlp/TPR family from yeast to human (Zimowska et al., 1997; Strambio-de-Castillia et al., 1999; Xu et al., 2007), the fission yeast TPR nucleoporin Alm1 localizes to the NPC and the nuclear rim.
alm1 deletion results in chromosome missegregation
The localization of Alm1 at the NE prompted us to analyze NE morphology in alm1Δ cells. Whereas we did not find severe alterations in nuclear morphology in alm1Δ cells, we did observe that 40% of mitotic alm1Δ cells presented daughter nuclei of different sizes, which was never observed in WT cells (Fig. 1 C, arrow). We then monitored DNA segregation by in vivo microscopy using histone H3 (hht2) tagged with GFP. We observed that 31.8% of mitotic alm1Δ cells (n = 22) showed lagging masses of DNA during anaphase B, of which approximately half of them resulted in aneuploidy, a phenotype that was never observed in WT cells (n = 25; Fig. S2 A). Chromosome missegregation defects in alm1Δ cells were further confirmed by minichromosome loss assays (Fig. 1 D).
The appearance of lagging chromosomes during anaphase B is indicative of erroneous KT–MT attachments (Gregan et al., 2007; Rumpf et al., 2010). Thus, we monitored KT behavior in cells coexpressing the KT marker Mis6-GFP and the SPB marker Sid2-Tomato. In WT cells, KTs appeared as two to six dots that oscillated between the two SPBs during prophase–metaphase. Just before anaphase onset, the KTs achieved bipolar attachment and simultaneously segregated to the spindle poles (Fig. 1 E; Nabeshima et al., 1998; Tournier et al., 2004; Courtheoux et al., 2007). However, in alm1Δ cells, KTs appeared more extended along the metaphase spindle compared with WT cells, and their pole to pole oscillations seemed to be more dynamic, which suggested that MT–KT interactions might be unstable in alm1Δ (Fig. 1 E, dashed boxes). We also noticed that segregation of sister chromatids at anaphase frequently occurred in an uncoordinated manner. In WT cells, all of the KTs were segregated to the spindle poles simultaneously, whereas in alm1Δ cells, after the segregation of most of the KTs, we still could observe one to two KTs in the spindle that are usually segregated 1–2 min later (Fig. 1 E, asterisk). We further confirmed the presence of lagging KTs in alm1Δ cells during anaphase B (Fig. 1 E, arrow).
To analyze the behavior of a single KT pair in vivo, we used a strain that harbors a tandem array of lacO DNA repeats at the lys1 gene located close to the centromere of chromosome I (cen1). This lacO array allows binding of a GFP-LacI fusion protein (Nabeshima et al., 1998). In alm1Δ cells, we observed a prolonged metaphase in which the distance between the two lys1-GFP signals is transiently increased compared with WT cells (Fig. S2 B, arrows and dashed boxes).
Thus, collectively, these results show that alm1∆ cells display abnormal KT behavior during metaphase and are affected in chromosome segregation, suggesting that, in the absence of alm1, either KTs or spindle MTs are altered.
To test whether the mitotic defects in alm1∆ cells are the result of a defective mitotic spindle, we followed spindle formation and behavior by expressing the α-tubulin subunit Atb2 tagged with GFP. In S. pombe, mitotic phases can be distinguished by the spindle dynamics. Phase I initiates with spindle formation (prophase). During phase II, the spindle maintains a constant length of ~2.5–3 µm (metaphase) and culminates with KT segregation to the spindle poles (anaphase A). Finally, during phase III, the spindle elongates, segregating the sister nuclei (anaphase B; Fig. S2, C and D; Nabeshima et al., 1998). We did not observe any significant alteration in spindle formation and function in alm1Δ compared with WT cells. We found, however, that 66.7% of alm1Δ cells (n = 30) presented a delay at the metaphase to anaphase transition (phases I and II; 10.9 ± 0.1 min in WT cells vs. 15.1 ± 0.9 min in alm1Δ; Fig. S2, C–E).
alm1+ deletion leads to SAC activation
The SAC is a mitotic surveillance mechanism that monitors correct attachment between KTs and spindle MTs. This pathway delays the metaphase to anaphase transition until all KTs are properly attached to MTs of the mitotic spindle. The SAC is composed of the kinases Bub1 and Mps1 (Mph1 in S. pombe) and the Mad1, Mad2, Mad3, and Bub3 proteins. The inhibitory signal originates at unattached or tensionless KTs. Here, SAC proteins accumulate and prevent anaphase onset by inhibiting the ubiquitin ligase anaphase-promoting complex (APC)/cyclosome through binding to the APC activator Cdc20 (Musacchio and Salmon, 2007). alm1Δ showed a sustained localization of both Mad2-GFP and Bub1-GFP at KTs during mitosis (Fig. S3, A and B), which is indicative of SAC activation.
A conserved function of the TPR proteins is the spatiotemporal regulation of the SAC components Mad1 and Mad2 (Lee et al., 2008; De Souza et al., 2009; Schweizer et al., 2013). During interphase, both Mad1 and Mad2 localize to the NE. In mitosis, however, Mad1/Mad2 are released from the NE and relocalize to the nucleoplasm and KTs, where they are essential for SAC activation. Mlp/TPR proteins are required for correct localization of Mad2 to the NE during interphase and to KTs during mitosis. In addition, in human cells, TPR is required for full SAC activation in unperturbed mitosis and in response to MT damage (Schweizer et al., 2013; Rajanala et al., 2014). We found that during interphase, Mad2-GFP was not recruited to the NE in alm1Δ, which shows that this function is conserved in S. pombe (Fig. S3 C). The localization of Mad2-GFP at KTs during mitosis, however, was not affected (Fig. S3 A). Furthermore, we found that alm1Δ cells were able to respond to MT damage, as treatment with the MT inhibitor methyl benzimidazol-2-yl carbamate (MBC) induced a delay at the metaphase to anaphase transition and the accumulation of Mad2-GFP at KTs, as seen in WT cells (Fig. S3 E). Moreover, double mutants of alm1Δ and either bub1Δ or mad2Δ showed a negative genetic interaction, indicating that Mad2 and Bub1 become critical for alm1Δ survival (Fig. S3 F).
Aurora B kinase is required for the correction of erroneous attachments and for SAC functionality (Petersen et al., 2001; DeLuca et al., 2006; Knowlton et al., 2006; Zhang et al., 2007; Vanoosthuyse and Hardwick, 2009). To examine whether the appearance of lagging chromosomes in alm1∆ might result from deregulation of aurora kinase (Ark1), we analyzed its localization and activity. We found that both localization to KTs and the kinase activity of Ark1 were unaffected in alm1∆ relative to WT cells (Fig. S3, G and H). Importantly, we found a synergistic phenotype in the double mutant comprising the alm1Δ and ark1-t7 thermosensitive (ts) allele at the permissive temperature (Fig. S3 I), with increased frequency of lagging chromosomes and DNA missegregation events (Fig. S3 J).
Thus, the presence of an apparently normal mitotic spindle and the strong dependency of SAC functionality for survival suggest that alm1Δ cells might be defective in KT–MT capture or stability.
Analysis of genetic interactions of alm1Δ by synthetic genetic array (SGA)
Mutants affecting centromere or KT function usually display DNA segregation defects and are sensitive to MT perturbation (Takahashi et al., 1994; Ekwall et al., 1999; Pidoux et al., 2003). We found that alm1Δ is sensitive to MT perturbation, as growth was inhibited in the presence of 15 µg/ml of the MT-depolymerizing drug thiabendazole (TBZ) compared with the WT strain (Fig. 2 A).
Figure 2.
SGA assay based on TBZ sensitivity identifies genetic factors that contribute to maintain KT structure and functionality. (A) TBZ sensitivity assay of alm1Δ mutant. clr4Δ mutant was used as a positive control (Bernard et al., 2001). (B) Flow-through of the SGA to screen for genetic interactors of alm1Δ mutant in TBZ. WT and alm1Δ query strains were crossed with the Bioneer haploid deletion mutant library (v. 3) and spotted on YES- and TBZ-containing plates. Examples of colony growth on YES and YES + TBZ plates. The growth of the single and double mutants was quantified in both media, and we compared the ratio with the median ratio. Blue indicates small colony size, and yellow indicates large colony size (see Materials and methods). (C) Clustering analysis showing one of the clusters of deletion mutants that have a synthetic effect on TBZ sensitivity with alm1Δ. For each experiment, four replicates were performed. Clustering analysis showing one group of genes with no sensitivity to TBZ, neither in the single nor the double mutant with alm1Δ. Blue indicates synthetic interaction, yellow indicates suppressive interaction, black indicates no interaction, and gray indicates the absence of data. (D) GO enrichment analysis for biological processes of all of the genes identified in the three synthetic clusters. The table contains the percentage between the total number of genes of the indicated GO group and the total number of genes of S. pombe (upper value), and the percentage between the number of gene deletion mutants of the synthetic clusters belonging to the mentioned GO group and the total number of gene deletion mutants of the synthetic clusters (lower value). (E) Tetrad dissection analysis of crosses between alm1Δ and strains with the indicated genotypes. Double mutants between alm1Δ and the indicated mutant backgrounds are encircled.
To gain further insights into the cellular function of Alm1, we screened for genetic interactions by performing an SGA assay (Tong et al., 2001). To enrich for genetic interactions related to KT function, we performed the SGA in conditions of MT perturbation (i.e., in the presence of TBZ; Fig. 2 B and see the SGA assay section of Materials and methods). The analysis of the sensitivity of single and double mutants toward TBZ revealed several mutant clusters that displayed increased sensitivity in combination with alm1Δ (Fig. 2 C and Fig. S4 A). Gene ontology (GO) enrichment analysis of these synthetic clusters showed an enrichment in genes encoding for KT components like spc19, dis2, or ask1; SAC components like bub1, mad1, and mad2; proteins involved in ubiquitin-dependent proteolysis like the ubiquitin-protease upb2; and chromatin regulators (Fig. 2 D). Given the enrichment in KT-encoding genes and the fact that most of the components of this structure are essential, we analyzed genetic interactions of alm1Δ with several KT ts alleles, which were not present in the deletion library (Fig. 2 E). After tetrad dissection, we indeed found viable spores only for the WT and single mutants, whereas alm1Δ showed synthetic lethality with mutants of centromere–KT factors, such as cnp3, mis6, mis12, or mhf1 (Fig. 2 E). Furthermore, we also observed a strong genetic interaction between alm1Δ and the GFP-tagged versions of the outer KT proteins Ndc80 and Nuf2 (Fig. 2 E). Together, these results point to a functional relationship between alm1 and centromere and KT genes.
alm1Δ mutant shows abnormal accumulation of centromere–KT proteins
The synthetic lethality between alm1Δ and mutants in centromere and KT components prompted us to analyze the localization and intensity levels of several of these proteins in alm1Δ by video microscopy (Fig. 3, A–C). Although the levels of Cnp1 were unaffected in alm1Δ cells (Fig. 3, A and B), we found increased levels of the centromere–KT proteins Mhf2, Cnp20, Mde4, Fta1, and especially Cnp3, with a 1.8-fold increase in alm1Δ compared with the WT (Fig. 3, A–C). The increased signal of Cnp3 at KTs in the alm1Δ mutant correlated with an increase in total Cnp3 protein (Fig. 3 D). The mRNA levels, however, were comparable to the WT strain (Fig. 3 E), suggesting that the increased protein levels of Cnp3 in alm1Δ cells were not the result of transcriptional deregulation but instead to altered protein stability or turnover.
Figure 3.
alm1Δ mutant shows altered accumulation of centromere and KT proteins. (A) WT (green lectin stained) and alm1Δ cells expressing Cnp1-mCherry (top) or Cnp3-Tomato (bottom). Bars, 5 µm. (B and C) Mean fluorescence intensity in alm1Δ cells relative to WT cells of the indicated proteins (Cnp1-mCherry, Mhf1-GFP, Mhf2-GFP, Cnp20-GFP, Pcs1-GFP, Mde4-GFP, Fta1-GFP, Sim4-Tomato, and Cnp3-Tomato) at KTs during interphase. n = 50. (D) Western blot analysis of total Cnp3-GFP protein from WT and alm1Δ cells using anti-GFP mAb to detect Cnp3-GFP (top) and TAT1 as a loading control (bottom). Positions of molecular mass markers are indicated in kilodaltons. (E) RT-qPCR analysis of act1, cnp1, and cnp3 mRNA levels in WT and alm1Δ cells. Three biological repeats were performed. (B, C, and E) Error bars represent SD. (F) RT-qPCR analysis of centromere I (dh, dg, imr, and cnt) transcript levels in WT and alm1Δ mutant (normalized to act1 transcript levels). n = 5. Error bars represent SEM. (G) ChIP-qPCR analysis of Cnp3-GFP and H3K9me levels. ChIP data have been normalized to act1 and are shown relative to the maximal enrichment in WT cells at the cen1 region. *, P < 0.05; **, P < 0.001; ***, P < 0.001.
Overexpression of Cnp1 was previously shown to cause its spreading from the centromeric cnt to the adjacent heterochromatin, resulting in altered pericentromeric silencing and chromosome missegregation (Castillo et al., 2007). We therefore analyzed the chromatin state at centromeres by measuring transcript levels of heterochromatin repeats (imr, dg, and dh) and cnt by reverse transcriptase assays combined with quantitative PCR (qPCR; Fig. 3 F). This analysis revealed a reduction in transcripts originating from the imr and otr repeats in alm1Δ cells compared with WT cells. Surprisingly, there was a threefold increase in transcripts derived from the cnt region of centromere I (Fig. 3 F), indicating that this chromatin region is derepressed in alm1Δ. We performed chromatin immunoprecipitation (ChIP) experiments using endogenously expressed Cnp3-GFP and found that Cnp3 distribution at the centromeric region in alm1Δ was indistinguishable from the WT cells (Fig. 3 G), indicating that the excess of Cnp3 protein in alm1Δ cells does not seem to increase the chromatin-bound fraction.
Collectively, these experiments demonstrate that alm1Δ shows abnormal accumulation of several KT proteins, including Cnp3. We further found that the absence of Alm1 alters the expression of centromeric transcripts; however, these changes are not caused by lateral spreading of Cnp3 into pericentromeric chromatin.
Cnp3 proteostasis is regulated by the proteasome
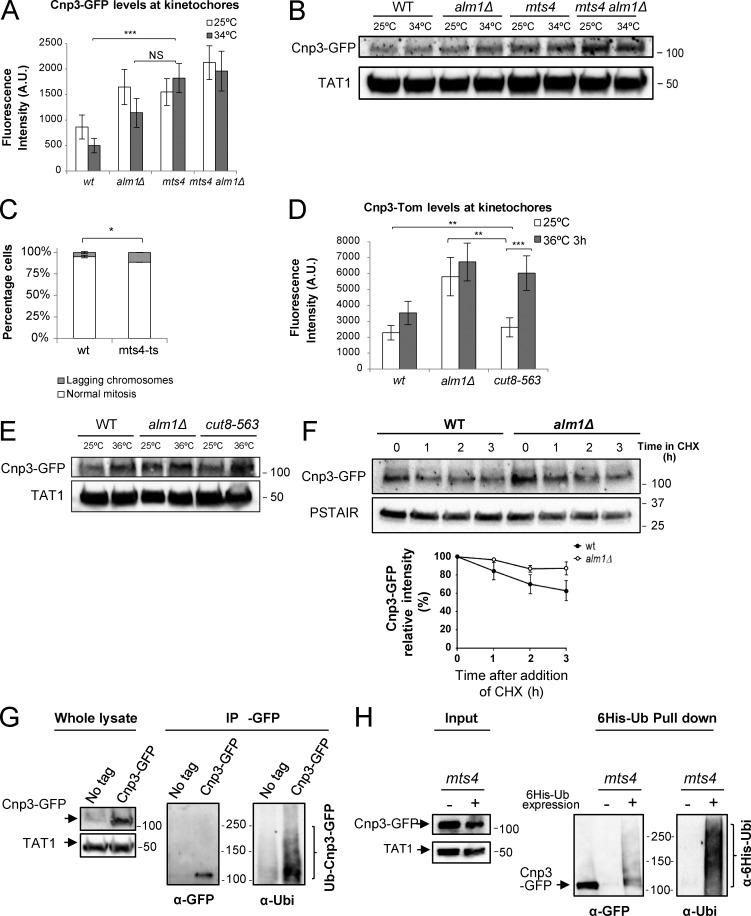
Previous studies have shown that centromeric levels and distribution of CENP-ACnp1 are regulated locally through proteasomal degradation (Collins et al., 2004; Moreno-Moreno et al., 2006; Ranjitkar et al., 2010; Kitagawa et al., 2014). To test whether Cnp3 stability is regulated by proteasomal degradation, we first analyzed Cnp3 protein levels in a ts mutant of the non-ATPase Mts4, a subunit of the 19S regulatory particle of the proteasome (Wilkinson et al., 1997). Microscopic analysis of KT Cnp3-GFP levels in the mts4 mutant revealed that the Cnp3-GFP signal is increased compared with WT cells, even at the permissive temperature, reaching similar levels to those found in the alm1Δ mutant (Fig. 4 A and Fig. S4 B). Consistently, the mts4 mutant showed a twofold increase of lagging chromosomes compared with WT cells at the permissive temperature (Fig. 4 C). The double mutant alm1Δ mts4 showed a further increase in Cnp3 levels at KTs at the permissive temperature (Fig. 4 A and Fig. S4 B). These results are consistent with the total Cnp3-GFP protein levels observed by Western blot analysis (Fig. 4 B).
Figure 4.
Proteasome function and localization are required for stoichiometric accumulation of Cnp3 at KTs. (A) Centromeric Cnp3-GFP levels in the indicated backgrounds and temperatures. n = 50. (B) Western blot analysis of total Cnp3-GFP, using anti-GFP mAb to detect Cnp3-GFP and TAT1 as a loading control. (C) Quantification of mitotic defects (lagging KTs during anaphase) of WT and mts4 cells at 25ºC by in vivo fluorescence microscopy, using Cnp3-GFP as a KT marker. n > 140. (D) Quantification of centromeric Cnp3-Tomato levels in the indicated backgrounds and temperatures. n = 50. (E) Western blot analysis of total Cnp3-GFP protein in the indicated strains, using anti-GFP mAb to detect Cnp3-GFP and antitubulin (TAT1) mAb as a loading control. (F) Cnp3 protein stability in WT and alm1Δ cells in the presence of CHX. Cnp3-GFP was detected by immunoblotting with anti-GFP mAb, and anti-PSTAIR mAb was used as a loading control. Quantification of Cnp3-GFP protein stability. Cnp3-GFP band intensities were quantified using ImageJ and normalized to PSTAIR signals. Relative intensity at time 0 was set up as 100% in each case. Error bars represent SD from three independent experiments. (G) mts4 expressing Cnp3-GFP and untagged mts4 cells were grown to midlog phase at 25ºC and then shifted to 36ºC for 3 h. Samples were collected and subjected to anti-GFP immunoprecipitation (IP). Whole lysate (left) was immunoblotted with anti-GFP mAb to detect Cnp3-GFP and antitubulin (TAT1) mAb as a loading control. Immunoprecipitated proteins were immunoblotted with anti-GFP mAb to detect Cnp3-GFP and antiubiquitin pAb to detect ubiquitinated proteins. (H) mts4 cells overexpressing Cnp3-GFP or Cnp3-GFP and His6-ubiquitin were grown in EMM to midlog phase at 25ºC and then shifted to 36ºC for 3 h. Samples were collected, and polyubiquitinated proteins were purified with Ni2+–nitrilotriacetic acid beads in denaturing conditions. Ubiquitinated proteins were detected by immunoblotting with antiubiquitin pAb (right), and ubiquitinated forms of Cnp3 were detected by immunoblotting using anti-GFP mAb (left). A fraction of the whole cell extract (left) with an equal amount of total protein was immunoblotted with anti-GFP mAb to detect Cnp3-GFP and antitubulin (TAT1) mAb as a loading control. Positions of molecular mass makers are indicated in kilodaltons. Graphs represent mean and SD. A.U., arbitrary units. *, P < 0.05; **, P < 0.001; ***, P < 0.001.
In S. pombe, the 26S proteasome is enriched within the nucleus as seen in animal cells (Amsterdam et al., 1993; Enenkel et al., 1998; Wilkinson et al., 1998; Russell et al., 1999). Cut8 is required to anchor the proteasome to the NE in the fission yeast (Tatebe and Yanagida, 2000; Takeda and Yanagida, 2005; Takeda et al., 2011), and cut8 mutant phenotypes are similar to those of proteasome mutants such as mts4. We therefore analyzed the levels of centromeric Cnp3-GFP in the cut8-563-ts mutant. Although this mutant showed elevated centromeric Cnp3 levels already at permissive temperature, we observed a further twofold increase at restrictive temperature (Fig. 4 D and Fig. S4 C), which is consistent with total Cnp3-GFP protein levels (Fig. 4 E). We were not able to generate the double mutant of alm1Δ and cut8-563, suggesting that the combination of both deficiencies is synthetically lethal.
Next, we analyzed Cnp3 levels in WT and alm1Δ cells in the presence of the protein synthesis inhibitor cycloheximide (CHX). As a control, we analyzed the levels of cyclin B/Cdc13, a known substrate of the proteasome. We found that Cnp3 is unstable in WT cells, although it has a significantly longer half-life than cyclin B, suggesting that only a pool of Cnp3 is degraded (Fig. 4 F and Fig. S4 D). Importantly, the Cnp3 decay was delayed in alm1Δ cells compared with WT cells (Fig. 4 F). We did not observe significant changes in the turnover of cyclin B between WT and alm1Δ cells under these experimental conditions (Fig. S4 D).
To examine whether Cnp3 is ubiquitinated, we immunoprecipitated native Cnp3 from mts4 mutant cells and probed the immunoblot with an antibody against ubiquitin, which revealed the typical smear of polyubiquitinated proteins (Fig. 4 G and see the Ubiquitin pull-down section of Materials and methods). Using a complementary approach, we coexpressed Cnp3-GFP and 6His-tagged ubiquitin in mts4 mutant cells and performed pull-down experiments for His-tagged ubiquitin under denaturing conditions. We analyzed the fraction enriched in ubiquitinated proteins by anti-GFP immunoblotting, in which we detected a smear of slower migrating ubiquitinated forms of Cnp3-GFP (Fig. 4 H). Thus, both experiments are consistent with Cnp3 being ubiquitinated in vivo. Altogether, these data indicate that Cnp3 is regulated by ubiquitin-dependent proteasomal degradation.
Alm1 is required for proper localization of the 26S proteasome to the NE
Both Cut8 and Alm1 localize to the NE (Fig. 5 A; Takeda and Yanagida, 2000, Takeda and Yanagida, 2005; Takeda et al., 2011). To test whether the absence of Alm1 affects Cut8 localization, we analyzed Cut8 distribution in the alm1Δ background by live-cell microscopy. Intriguingly, we found that alm1Δ displays reduced mean levels of Cut8-GFP at the NE (Fig. 5 B). The total levels of Cut8 protein, however, were similar in alm1Δ and WT cells (Fig. 5 C). As it was known that Cut8 is required for proper proteasome nuclear localization, we tested whether the absence of Alm1 also affects the localization of the 26S proteasome. To this end, we analyzed the localization of the proteasomal subunits Mts2 and Mts4 by immunostaining. Notably, we found that both proteasome subunits fail to properly localize to the NE in the alm1Δ background (Fig 5, D–G).
Figure 5.
Alm1 is required for proper accumulation of Cut8 and the proteasome to the NE. (A) WT cells expressing Cut8-GFP and Alm1-Tomato. Images are maximal projections of three z sections. (B) WT (red lectin stained) and alm1Δ cells expressing Cut8-GFP. Cut8-GFP intensity levels at the NE in WT and alm1Δ cells. Graphs represent mean and SD. n = 69. (C) Western blot analysis of total Cut8-GFP protein of WT and alm1Δ cells using anti-GFP mAb to detect Cut8-GFP and anti-PSTAIR mAb as a loading control. (D) Brightfield and immunofluorescence images of WT and alm1Δ cells expressing Mts2-8Myc, using anti-Myc antibodies against Mts2-8Myc (green) and DAPI to stain DNA (blue). (E) Western blot analysis of total Mts2-8Myc protein in WT and alm1Δ cells using anti-Myc antibodies to detect Mts2-8Myc and anti-PSTAIR antibodies as a loading control. (F) Brightfield and immunofluorescence images of WT and alm1Δ cells expressing Mts4-13Myc, using anti-Myc antibodies against Mts4-13Myc (green) and DAPI to stain DNA (blue). (G) Western blot analysis of total Mts4-13Myc protein in WT and alm1Δ cells using anti-Myc antibodies to detect Mts4-13Myc and anti-PSTAIR antibodies as a loading control. Positions of molecular mass markers are indicated in kilodaltons. Bars, 5 µm. A.U., arbitrary units. ***, P < 0.001.
Together, these results show that proteasome localization to the NE is altered in alm1Δ cells, which might affect the degradation of specific substrates.
An excess of Cnp3 impairs chromosome segregation
We reasoned that if the chromosome missegregation defects observed in alm1Δ cells are caused by abnormal accumulation of Cnp3 at KTs, the overexpression of this protein in WT cells would result in similar chromosome segregation defects. To test this hypothesis, we used a strain in which an additional copy of Cnp3-GFP was ectopically expressed under the control of the medium strength nmt41 promoter (pINT-cnp3-GFP) in WT and alm1Δ Cnp3-GFP strains (Fennessy et al., 2014). Western blot analysis of total Cnp3-GFP protein in conditions of moderate overexpression (rich medium) showed increased levels of Cnp3-GFP in both WT and alm1Δ backgrounds (Fig. 6 A). Consistently, using live-cell microscopy, we observed a 2.4- and 1.4-fold increase in centromeric Cnp3-GFP intensity signal in WT and alm1Δ strains, respectively (Fig. 6, B and C). Furthermore, using the same Cnp3 overexpression conditions, we also found an increase in chromosome segregation defects, especially an increase in lagging chromosomes for both WT and alm1Δ cells (Fig. 6, D and E, asterisk and arrows).
Figure 6.
Cnp3 overexpression phenocopies alm1Δ segregation defects. (A) Western blot analysis of total Cnp3-GFP using anti-GFP mAb or antitubulin (TAT1) mAb as a loading control. (B) Images of WT and alm1Δ cells expressing Cnp3-GFP from the endogenous locus and WT and alm1Δ cells expressing an additional copy of Cnp3-GFP from the medium-strength promoter nmt41 (pINT-Cnp3-GFP) in repressed conditions (YES media). (C) Centromeric Cnp3-GFP intensity levels in the indicated strains. n = 50. (D) Quantification of mitotic defects (lagging KTs during anaphase B). n = 50. (E) Time-lapse fluorescence images of the indicated strains. Time between frames is 2 min. Red asterisk denotes asynchronous KT segregation, and arrows denote lagging KTs. (F) Fluorescence images of alm1Δ cells expressing Cnp3-GFP from the endogenous promoter (marked with red lectins) and alm1Δ cells expressing Cnp3-GFP from the nmt81 promoter under repressed conditions (EMM + thiamine). (G) Western blot analysis of total Cnp3-GFP protein from cultures of the indicated genotypes grown in EMM + thiamine, using anti-GFP mAb or antitubulin (TAT1) mAb as a loading control. Molecular mass is given in kilodaltons. (H) Quantification of centromeric Cnp3-GFP intensity levels. n = 50 cells. (I) Quantification of mitotic defects (lagging KTs during anaphase B) of the indicated strains. n = 50. Graphs represent mean and SD. Bars, 5 µm. A.U., arbitrary units. *, P < 0.05; ***, P < 0.001.
We next tested whether a decrease in Cnp3 levels would rescue alm1Δ phenotypes. To this end, we inserted the thiamine-repressible nmt81 promoter at the endogenous cnp3 promoter locus in the alm1Δ background. Under repressed conditions (+ thiamine), this promoter rendered reduced Cnp3 levels (Fig. 6, F–H). This, however, only partially suppressed the chromosome segregation defects of alm1Δ (Fig 6 I). Thus, these data suggest that an excess of Cnp3 is one of the reasons for aberrant chromosome segregation seen in the absence of Alm1.
Discussion
The nucleoporins of the TPR family are the main structural element of the nuclear basket (Cordes et al., 1997; Bangs et al., 1998; Kosova et al., 2000; Frosst et al., 2002). As described for orthologues in other organisms, the fission yeast TPR Alm1 shows extensive regions of predicted CC domains, and we demonstrate that Alm1 indeed localizes at the nuclear rim, where it partially colocalizes with the NPC (Fig. S1, A–C). Alm1 colocalizes with Nup211, the other member of the TPR family of nucleoporins, in S. pombe (Fig. 1 A). Furthermore, Nup211 localization at the NE is decreased in the absence of Alm1 (Fig. 1 B), which suggests that they likely form a complex at the nuclear basket, as has been demonstrated for Mlp1 and Mlp2 in S. cerevisiae (Strambio-de-Castillia et al., 1999; Niepel et al., 2005).
In this work, we show that the absence of Alm1 causes a significant delay in the metaphase to anaphase transition because of SAC activation and the appearance of lagging chromosomes during anaphase B, which eventually leads to chromosome missegregation (Fig. 1, C–E; Fig. S2; and Fig. S3, A and B). A conserved function of the TPR proteins is the spatiotemporal regulation of the SAC through recruitment of Mad1 and Mad2 to the NE during interphase (Lee et al., 2008; Lince-Faria et al., 2009; Schweizer et al., 2013; Rodriguez-Bravo et al., 2014). We find that Alm1 is also required for the recruitment of Mad2 at the NE during interphase (Fig. S3 C). This indicates that this particular function of TPR nucleoporins has been conserved during evolution. In animal cells, depletion of TPR results in an accelerated anaphase onset, with decreased localization of Mad2 at KTs, weakened SAC response, and the appearance of lagging chromosomes (Lee et al., 2008; Lince-Faria et al., 2009; Rodriguez-Bravo et al., 2014). However, the alm1Δ mutant shows a delayed anaphase onset as a result of SAC activation, and Mad2 accumulates at KTs in unperturbed mitosis as well as in the presence of the MT-depolymerizing drug MBC (Fig. S3, A and E). This suggests that Alm1 is not required for SAC functionality, but that instead alm1Δ cells are dependent on this pathway for survival (Fig. S3 F). Thus, the chromosome missegregation observed in alm1Δ might result from misfunctional KTs, as the mitotic spindle is apparently normal in alm1Δ cells (Fig. S2, C–E).
Proper centromere and KT structure and function are crucial to avoid erroneous MT–KT attachments (Nonaka et al., 2002; Gregan et al., 2007). For instance, inactivation of Cnp1 affects the loading of other KT proteins, such as Cnp20 or Mis6 (Castillo et al., 2007; Tanaka et al., 2009; Folco et al., 2015), and overexpression of Cnp1 leads to increased levels of centromeric Cnp3, Mal2, and Sim4 (Castillo et al., 2007), resulting in chromosome missegregation. The alm1Δ mutant shows increased levels of several KT proteins, including of Cnp3 (Fig. 3, A–C), which is not the result of deregulated transcription (Fig. 3 E). The increased level of Cnp3 at KTs observed in alm1Δ is not likely the result of its direct binding to centromeric DNA, as we did not observe an increase of centromere-associated Cnp3 by ChIP (Fig. 3 G). The C-terminal domain of CENP-CCnp3 contains two regions that are highly conserved from yeast to humans that are called Mif2p homology domain II (also known as CENP-C motif) and III. The Mif2 homology domain II is responsible for CENP-C binding to centromeric DNA (Trazzi et al., 2002, 2009), whereas the Mif2 homology domain III displays multiple activities, including recruitment of other KT components. Importantly, the Mif2 homology domain III is able to mediate CENP-C self-association in vivo, allowing the formation of higher order structures such as homodimers and homooligomers (Sugimoto et al., 1997; Cohen et al., 2008; Trazzi et al., 2009). Therefore, it seems conceivable that the excess of Cnp3 protein in alm1Δ cells may cause its ectopic accumulation at KTs through binding to other Cnp3 molecules mediated by the Mif2 homology domain III. In S. pombe, Cnp3 binds through the N-terminal domain to Fta1, a component of the Sim4 complex required for proper KT function (Tanaka et al., 2009). Consistently, we observed increased levels of centromeric Fta1-GFP in the alm1Δ background (Fig. 3 C), suggesting that an excess of Cnp3 might drive the ectopic accumulation of other KT components.
Cnp3 is one of the substrates degraded in an Alm1- and proteasome-dependent manner (Fig. 4). However, we found that in WT cells, the Cnp3 turnover is less prominent than that of Cdc13 in the presence of CHX (Fig. 4 F and Fig. S4 D), suggesting that only a fraction of Cnp3 is degraded by the proteasome, similar to what has been shown for Cnp1 regulation by proteasomal degradation at centromeres (Kitagawa et al., 2014). In our experimental conditions, the absence of Alm1 did not significantly affect Cdc13 degradation (Fig. S4 D). Whether the different requirement of Alm1 for Cnp3 and Cdc13 degradation is the result of spatial specificity in proteasomal degradation or a consequence of different kinetics of degradation of both proteins remains an open question.
Overexpression of Cnp3 is sufficient to cause chromosome missegregation in WT cells (Fig. 6, D and E). However, a decrease of Cnp3-GFP levels only partially rescues chromosome segregation defects of alm1Δ cells (Fig. 6 I), suggesting that the imbalance of other centromere or KT factors (Fig. 3, B and C) may also contribute to the impaired KT function.
Among the synthetic genetic interactions obtained from the SGA analysis, we found that mutants with defects in KT components and SAC regulators are enriched (Fig. 2 D). This finding is in agreement with our observation of aberrant KT behavior, SAC activation and dependency, and chromosome missegregation in alm1Δ cells. We also obtained genes with roles in chromatin organization. These include eaf6, which encodes a subunit of the Mst2 histone acetyltransferase complex that regulates histone turnover at centromeres and prevents heterochromatin spreading into euchromatic sites (Wang et al., 2015). We further found fft3, which encodes an ATP-dependent chromatin remodeler required to prevent euchromatin formation at centromeres (Strålfors et al., 2011). Interestingly, Fft3 has also been involved in heterochromatin maintenance and NE attachment of subtelomeric regions (Steglich et al., 2015). Intriguingly, we found that transcription at centromeric chromatin is affected in alm1Δ cells (Fig. 3 F). Whether this phenotype is caused by deficient chromatin regulation pathways or is a consequence of altered KT proteins will be further investigated. Finally, we also found genes involved in ubiquitin-dependent proteolysis, including ubp2 that encodes a deubiquitinating enzyme. ScUbp2 counteracts ubiquitination of the proteasome-associated ubiquitin receptor Rpn10 (Sato et al., 2005). Because ubiquitination of Rpn10 inhibits its capacity to bind to substrates and affects proteasome activity, the lack of Ubp2 might additionally increase cellular Cnp3 levels, providing a likely explanation of the synthetic genetic interaction with the alm1Δ mutant.
In fission yeast, the three centromeres cluster together and are anchored at the NE in interphase cells. During mitosis, they are released from the NE and then attached by the incipient spindle during prophase and metaphase. S. pombe has a closed mitosis in which the NE does not disassemble. Thus, except for a brief period during early mitosis, most of the cell cycle centromeres stay in contact with the NE. Intriguingly, the proteasome is also enriched at the nuclear periphery (Wilkinson et al., 1998), and CENP-ACnp1 has been shown to be regulated by proteasomal degradation (Collins et al., 2004; Moreno-Moreno et al., 2006; Ranjitkar et al., 2010; Kitagawa et al., 2014). We have shown that Cnp3 is also regulated by the proteasome. In particular, Cnp3 levels are increased in mutants affected in proteasomal degradation, such as mts4 or cut8-563 mutants (Fig. 4, A–E), and ubiquitinated forms of Cnp3 can be detected when proteasomal degradation is compromised (Fig 4, G and H). Importantly, we find that Alm1 is required to maintain the proteasome and its anchor Cut8 at the NE (Fig. 5, B, D, and F). Thus, Alm1-dependent proteasome localization at the NE is required for Cnp3 proteostasis.
Cut8 has a membrane-binding domain (Takeda et al., 2011), and its enrichment at the nuclear periphery is regulated by the ubiquitin-conjugating enzyme Rhp6 (Rad6), the ubiquitin ligase Ubr1, and the proteasome itself (Takeda and Yanagida, 2005). Thus, although Alm1 may not be directly involved in the recruitment of Cut8 and the proteasome, it may provide a platform for the assembly or tethering of functional proteasomal complexes at the NE. Interestingly, ScMlp1 and ScMlp2 have been isolated in a complex with the NE-associated protein Esc1 (Niepel et al., 2013). Esc1 is involved in a variety of nuclear functions such as telomere NE anchoring and silencing (Taddei et al., 2004), DNA repair (Therizols et al., 2006), or SUMO homeostasis (Lewis et al., 2007), and, recently, it has been shown to bind to the proteasome (Niepel et al., 2013). Therefore, it has been proposed that Mlp1 and Mlp2 are part of a protein–protein interaction network at the NE that includes the SPB, messenger RNPs, chromatin silencing factors, and the proteasome via Esc1 (Niepel et al., 2013). Further studies will contribute to the understanding of the role of proteasomes at the NE.
Our study reveals a novel role of the TPR nucleoporin Alm1 in the homeostasis of KT proteins through the spatial regulation of the proteasome and highlights the emergent role of the NPC in the maintenance of genome integrity.
Materials and methods
Yeast strains, genetic procedures, cell culture, and media
The S. pombe strains used in this study are listed in Table S1. Strains expressing GFP-, mCherry-, and tdTomato-tagged (Snaith et al., 2005) proteins and deletion strains were constructed using PCR-based methods and homologous recombination as described previously (Bähler et al., 1998). ura4+, kanMX6 (Kan), hphMX6 (hph), and natMX6 (nat) genes were used as selection markers (Bähler et al., 1998; Sato et al., 2005; Snaith et al., 2005). All tagged strains used in this study are expressed from their endogenous loci unless otherwise stated. Yeast transformation was done by the lithium acetate method (Moreno et al., 1991).
The alm1+ gene was deleted with the ura4+ gene by using the primers Alm1-Ura4.s (5′-TTTCTTCTAACGTGGTATTATTTCTGTACATCGAGGTTGATCACTTACTTCTCGTATACGTACACTGGTGCTTCATCGCCAGGGTTTTCCCAGTCACGAC-3′) and Alm1-Ura4+.a (5′-AAAGTCGTTTTTTTTATTGGAAACCAGAAATAATAATTATGCAGCTAACCTATTTTATTATCTGTTTACAAACTCTAGCGGATAACAATTTCACACAGGA-3′). The primers used to construct the strain alm1-tomato-Nat were Alm1-Tag.s (5′-ACACTAATTCTCCTCCTAAACGGTCCAGTTCAGACGCTGGTATGGATGTTTCCAATGATGTTAAGAAAGCCAAACCCGGACGGATCCCCGGGTTAATTAA-3′) and Alm1-Tag.a (5′-AATTAAAGTCGTTTTTTTTATTGGAAACCAGAAATAATAATTATGCAGCTAACCTATTTTATTATTGTTTACAAACTCTGAATTCGAGCTCGTTTAAAC-3′). Primers to construct the strain nup211-GFP-kan were Nup211_Ctag_FW (5′-CTGCTAAATCCGGCTCCCTTAAAAGACAACGTGACGATGCGAACAAAGGAGGATCCAGTTCGAACCAAAAGAAAGCAAAACGGATCCCCGGGTTAATTAA-3′) and Nup211_Ctag_RV (5′-TTTACTCATGTCATTATTATAAATCATGTTAACTAAATATGAATAGTCCTAAGAGTGATTTATGAACCATATGAAAACATGAATTCGAGCTCGTTTAAAC-3′).
The primers used to construct the strain cnp3-GFP-kan were cnp3.C-tagFor (5′-CTGTTCGACTTTATTATACCCATGCGACCGACACCTTGGAAAATAAAAGAAGGGGAATAGGGGATTTTCCAAACGAACGACGGATCCCCGGGTTAATTAA-3′) and cnp3.C-tagRev (5′-AAATTGATAACAGATAATTAAATTACATAACGACTAAATGACTTTTGTACATCGAAATCCATAACACACTATTAGTATTGGAATTCGAGCTCGTTTAAAC-3′).
The primers used to construct the strain mts4-13Myc-kan were mts4-Forward (5′-CGGAAGCATATACACCTTTGACTTCGTTGGAAGGTATTGTTATTTTAAAAAAAAATACGGAGGACATTGAAATGACCGCTCGGATCCCCGGGTTAATTAA-3′) and mts4-Reverse (5′-GGGCGCTGAATTTCAACAATCCAGTTTGTTTTATTAATAGCAAAGAACTGGAAATCAAGCATTAGTACATAAAAATTGTAGAATTCGAGCTCGTTTAAAC-3′).
To construct the strain pINT-cnp3-GFP-Nat, cnp3 was amplified from genomic DNA using the primers BamH1-Cnp3.F (5′-GGAGGATCCATGACGATGAATGAAACGTCT-3′) and BamH1-Cnp3.R (5′-GTAGGATCCTCGTTCGTTTGGAAAATCC-3′) and cloned into pINTH-41EGFPC plasmid (Fennessy et al., 2014). The strain nmt81-Cnp3-GFP-Nat alm1::ura4+ was constructed by integrating the kanMX6-p81nmt cassette at the cnp3 promoter locus in the alm1::ura4+ Cnp3-GFP-Nat strain. The primers used to amplify the kanMX6-p81nmt cassette were nmt81-Cnp3.F. (5′-AGACAACACTTACGTCCCCATATATTGTTTTGTTTGGCTATTTAGGCTATATTTTTACTTAATGTTTATTTAATAGAATAGAATTCGAGCTCGTTTAAAC-3′) and nmt81-Cnp3.R. (5′-ACGTACCTGCCTACGACACCAATTTCAAAAAACTGATTCTCTCGTTGTCTTGCCGGAATAGCAGACGTTTCATTCATCGTCATGATTTAACAAAGCGACTATA-3′).
Strains harboring Sid2-Tomato, Mis6-GFP, Cut12-GFP, lys1(::LacOp)his7+::LacI-GFP-NLS, pcs1-GFP, or mde4-GFP were derived from original strains provided by F. Chang (University of California, San Francisco, San Francisco, CA), T. Toda (The Francis Crick Institute, London, England, UK), V. Alvarez (Centro Andaluz de Biología del Desarrollo/Universidad Pablo de Olavide [CABD/UPO], Seville, Spain), P. Nurse (The Francis Crick Institute), and J. Gregan (Research Institute of Molecular Pathology, Vienna, Austria), respectively. Additional strains listed in Table S1 were provided by P. Nurse (The Francis Crick Institute), S. Moreno (Instituto de Biología Funcional y Genómica, Salamanca, Spain), J. Petersen (South Australian Health and Medical Research Institute, Adelaide, Australia), Y. Watanabe (Institute of Molecular and Cellular Biosciences, University of Tokyo, Tokyo, Japan), I. Hagan (Cancer Research UK, University of Manchester, Manchester, England, UK), Y. Hiraoka (Advanced ICT Research Institute, Kobe, Japan; National Institute of Information and Communications Technology, Kobe, Japan/Graduate School of Frontier Biosciences, Osaka University, Suita, Japan), O. Niwa (Kazusa DNA Research Institute, Chiba, Japan), and the National Bio-Resource Project (Osaka, Japan).
Strain crosses and sporulation were performed in sporulation agar at 25°C, and genetic dissection was performed using a dissection microscope (MSM 400; Singer Instruments). Standard cell culture procedures and media were used (Moreno et al., 1991). Unless otherwise stated, experiments were performed in rich media (YES) at 25ºC, and cells were grown until exponential midlog phase. For mts4-ts mutant, cells were grown at the permissive temperature of 25ºC and shifted to 34ºC for 1 h. For cut8-563-ts mutant, cells were grown at the permissive temperature of 25ºC and shifted to 36ºC for 3 h.
For sensitivity drop assays, strains were grown in YES to exponential midlog phase at 25ºC. Serial fivefold dilutions were made, starting from an OD of 0.2, and spotted onto YES plates. For TBZ sensitivity assay, cells were spotted onto YES containing DMSO or YES plates containing 15 µg/ml TBZ (T8904; Sigma-Aldrich) and grown at 25ºC and 30ºC. Pictures were taken after incubation for 3–6 d.
For Cnp3-GFP down-regulation, cells expressing Cnp3-GFP under the control of the weak nmt81 promoter were grown in Edinburgh minimal media (EMM) at 25ºC, and then 15 µM thiamine (T4625; Sigma-Aldrich) was added to the culture medium. Cells were further grown for three generations in the presence of thiamine.
Microscopy and image analysis
Live-cell imaging was performed with a spinning-disk confocal microscope (IX-81; Olympus; Evolve camera, Plan Apochromat 100×, 1.4 NA objective; Roper Scientific). Images were acquired with Metamorph software (Molecular Devices) and analyzed with ImageJ (National Institutes of Health). Unless otherwise stated, time-lapse experiments were performed at 25°C in YES, and images are maximal projections of 18 z sections with a step size of 0.3 µm. In brief, cells were collected by centrifugation and immobilized in soybean lectin (L1395; Sigma-Aldrich)–coated 35-mm glass-bottom culture dishes (P35-1.5-10-C; MatTek) or µ-Slide 8-well dishes (80827; Ibid). Conditioned media were used to maintain cells during filming. When two strains were filmed in the same microscope field, one of them was stained with either FITC- (F2401; EY Laboratories) or TRITC-conjugated lectins (T2401; EY laboratories) by incubating 100 µl of cell culture with 1 µl of lectins for 5 min at room temperature. Cells were washed twice with fresh media and immediately mixed and prepared for filming.
For MBC treatment, cells were grown to exponential midlog phase in YES at 25°C and treated with 10 µg/ml MBC (37867-4; Sigma-Aldrich) or DMSO as control 10 min before filming.
Unless otherwise stated, quantification was performed on maximal projections of 18 z sections with a step size of 0.3 µm. Quantification of Nup211-GFP in Fig.1 B was performed on maximal projections of three z sections of 0.3 µm. Quantification of Cut8-GFP in Fig. 5 B was performed in single-central z sections. For fluorescence signal quantification, background was subtracted after measuring intensity in every region of interest.
Minichromosome loss assay
500–1,000 cells from Ade + colonies of WT and alm1Δ backgrounds were plated on YES + 0.15 mg/ml adenine plates and incubated for 3–5 d. The number of colonies with a red sector was counted. The number of chromosome loss events per division is the number of these half-sectored colonies divided by the total number of colonies.
Western blot and kinase assay
Protein extracts for Western blotting were prepared by trichloroacetic acid precipitation. Unless otherwise stated, Western blots were performed from asynchronous cultures. In brief, 45 ml of exponentially growing cultures was quenched by the addition of 5 ml of ice-cold 100% trichloroacetic acid (Sigma-Aldrich). After 30-min incubation on ice, cells were pelleted by centrifugation and washed with 10 ml of ice-cold 100% acetone (Sigma-Aldrich). After removing and drying acetone, cell pellets were resuspended in 500 µl of beating buffer (8 M urea, 50 mM ammonium bicarbonate, and 5 mM EDTA) with protease inhibitors (cOmplete, Mini, EDTA free; 11836170001; Roche) and lysed in a sample preparation system (FastPrep; MP Biomedicals) with acid-washed glass beads (Sigma-Aldrich). Cell extracts were cleared by centrifugation, the supernatant was recovered, and the protein concentration was measured using an RC DC protein assay kit (5000121; Bio-Rad). Protein samples were loaded on 10% TGX-Fast Cast acrylamide gels (161-0183; Bio-Rad) and blotted on nitrocellulose membranes (Bio-Rad). Blots were probed with primary mouse anti-GFP mAb (11814460001; Roche) and either mouse anti–α-tubulin mAb (TAT1; K. Gull, University of Oxford, Oxford, England, UK) or anti-PSTAIR mAb (P7962; Sigma-Aldrich) at a 1:1,000 dilution, followed by the secondary antibody anti–mouse IgG at a 1:2,000 dilution (A3562; Sigma-Aldrich). ECL (Bio-Rad) or Supersignal West Femto (Thermo Fisher) and Chemidoc XRS+ (Bio-Rad) were used for detection.
For Ark1-kinase assay, cdc25-22 and alm1Δ cdc25-22 cells expressing Ark1-PK were grown to midlog phase in YES at 25ºC and then synchronized in G2 by shifting to 36ºC for 4 h. Cells were released to 25ºC for 45 min, and samples were taken every 15 min. Ark1-kinase assay was performed as previously described (Petersen et al., 2001). In brief, 50 ml of cells was harvested by centrifugation, and the pellets were washed with ice-cold STOP buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, and 1 mM NaN3, pH 8.0) and snap-frozen in liquid nitrogen. The cell pellets were resuspended in 200 µl HEN buffer (50 mM Hepes, pH 8.0, 150 mM NaCl, 5 mM EGTA, 5 mM EDTA, 1% NP-40, and 50 mM β-glycerophosphate) with protease inhibitors (cOmplete Mini, EDTA free; 11836170001), and extracts were prepared by glass bead lysis at 4ºC. Cell extracts were cleared by centrifugation. 30 µl Sepharose beads (17-0618-01; GE Healthcare) was coupled with 10 µl of mouse anti-PK mAb (MCA 1360; Serotec) for 1 h at 4ºC in an agitation platform. 95 µl of cleared extract was added to 10 µl of packed beads and incubated at 4ºC for 30 min. The beads with the associated immunoprecipitate were washed three times with 0.5 ml HEN buffer and three times with KAB buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM DTT, and 10 mM MgCl2) and resuspended in 20 µl KAB buffer. Beads were incubated at 32ºC for 5 min, and then 5 µl of substrate mix (20 µM ATP and 5 µg of purified Histone H3; 11034758001; Roche) was added and incubated further for 20 min at 32ºC. The reaction was stopped by the addition of 15 µl 2× SDS-PAGE loading buffer (0.25 M Tris-HCl, pH 6.8, 8% [wt/vol] SDS, 0.004% [wt/vol] bromophenol blue, and 20% [vol/vol] 2-mercaptoethanol) and incubated at 98ºC for 5 min. 25 µl of samples was immunoblotted with mouse anti-PK mAb (provided by I. Hagan) to detect Ark1-Pk and rabbit polyclonal antiphosphoserine 10-histone H3-antibody (provided by I. Hagan). In all experiments, at least three biological repeats were performed.
Cell fixation and immunostaining
For DAPI staining, cells were fixed with 70% cold ethanol, washed with PBS, and resuspended in PBS + 0.2 µg/ml DAPI (Sigma-Aldrich).
Immunofluorescence microscopy was performed as described previously (Hagan and Hyams, 1988). In brief, 10 ml of exponentially growing culture was fixed with freshly prepared 30% paraformaldehyde (294474L; VWR) in PEM buffer (0.1 M Pipes, 2 mM EGTA, and 1 mM MgSO4, pH 6.9) plus glutaraldehyde (G6257; Sigma-Aldrich) at a final concentration of 0.2%, followed by 1-h incubation at room temperature. After washing three times with PEM, the cell wall was digested during 1 h at 37ºC with 2.5 mg/ml zymolyase 20T (120491-1; AMSBIO) in PEMS (PEM + 1.2 M sorbitol); cells were permeabilized with 1% Triton X-100 and quenched with 1 mg/ml sodium borohydride. After washing twice with PEM, cell pellet was resuspended in PEMBAL buffer (PEM buffer, 0.1% sodium azide, and 1% BSA) before antibody incubation. Primary antibody was mouse anti-Myc (9E10; Santa Cruz), used at a dilution of 1:100 for mts2-8Myc and 1:200 for mts4-13Myc. The secondary antibody was goat Alexa Fluor 488–tagged anti–mouse (A11029l; Invitrogen, Molecular Probes), used at a dilution of 1:1,000.
SGA assay
SGA assay based on TBZ sensitivity was performed as described previously (Verrier et al., 2015), using as query strains WT and alm1Δ mutant. Using the Singer RoToR HDA, WT and alm1Δ cells were crossed with 3,400 gene deletion mutants (Bioneer haploid deletion mutant library, v. 3.0) and selected by incubation at 42ºC for 4 d (temperature method; Dixon et al., 2008). After haploid selection, two additional steps of double mutant selection were performed, spotting onto EMM-Ura (PMD0410; ForMedium) and YES (Difco) supplemented with 100 µg/ml G418 (1013027; Invitrogen) for selection of deletion mutants, 100 µg/ml hygromycin (10687-010; Invitrogen) for selection of marker next to centromere, and 100 µg/ml ClonNat (Werner Bioreagents) for selection of alm1Δ mutant. Finally, cells were spotted on YES plates containing TBZ at a final concentration of 10 µg/ml and YES with DMSO as a control. The growth of the single and double mutants was quantified on both media, and the ratio with the median ratio was compared (Barrales et al., 2016). Genetic interactions of double mutants were analyzed based on colony size (area) as a readout of cellular fitness. GO enrichment analysis was performed using the term enrichment service of AmiGO 2 (GO Consortium), searching for biological process without using the Bonferroni correction.
mRNA levels
Cells were grown in liquid YES medium at 25ºC to exponential midlog phase, and total RNA was obtained by using a spin column (RNeasy Mini kit; Qiagen) according to the manufacturer’s instructions. Transcriptional levels of act1, cnp1, and cnp3 were measured by RT-qPCR using iTaq Universal SYBR green one-step kit as indicated (Bio-Rad) in 10 µl of final volume with 5 µl iTaq Universal SYBR green reaction mix, 0.125 µl iScript reverse transcriptase, a mixture of forward and reverse primers (final concentration of 300 nM each), and 500 ng RNA. The primers used were cnp1.F (5´-GTTTGCGCTGGCAATCTACG-3´), cnp1.R (5´-CCTGGCTAATTGCATGTCTCG-3´), cnp3.F (5´-CGTTGAAATGCCAGCAGGAG-3´), cnp3.R (5´-ACTGTGACCTCGATCTTTCCC-3´), act1.F (5´-AAGTACCCCATTGAGCACGG-3´), and act1.R (5´-CAGTCAACAAGCAAGGGTGC-3´).
RT-qPCR
Total RNA was isolated from cells grown to exponential midlog phase in YES at 25ºC using a TRIzol reagent (Life Technologies). After removing DNA contamination from the total RNA with TURBO DNA-free (Applied Biosystems), samples were subjected to RT-PCR analyses using the SuperScript III First-Strand Synthesis SuperMix (Life Technologies) for cDNA synthesis and treated with RNase. RT-qPCR reactions were carried out in 15 µl of volume, with 7.5 µl Light Cycler 480 SYBR green master mix (Roche), 2.5 µl of a mixture of forward and reverse primers (1.5 µM), and 5 µl cDNA, previously diluted. The primers used were act1-4.F (5´-GATTCTCATGGAGCGTGGTT-3´), act1-4.R (5´-CGCTCGTTTCCGATAGTGAT-3´), cen-dh.F (5´-TGAATCGTGTCACTCAACCC-3´), cen-dh.R (5´-TGAATCGTGTCATTCAACCC-3´), cen-dg-1.F (5´-TGCTCTGACTTGGCTTGTCTT-3´), cen-dg-.1.R (5´-CCCTAACTTGGAAAGGCACA-3´), SG1953 (5´-TCGCCGGTAACAAAAGGATCA-3´), SG1954 (5´-GCATTAGACAACTCGTTCGATC-3´), imr. MR35.F (5´-GAGCATGGTGGTGGTTATGGA-3´), and imr. MR36.R (5´-CGACTAAACCGAAAGCCTCGA-3´).
ChIP analyses
ChIP was performed as described in Braun et al. (2011) with minor modifications. In brief, Cnp3-GFP cell cultures were incubated overnight in rich medium at 30°C, diluted in fresh rich medium, and incubated until they reached an OD595 of ∼0.6. Cells were cross-linked with 1% formaldehyde for 10 min at room temperature, and the reaction was quenched by adding glycine to a final concentration of 125 mM. Chromatin was extracted and sheared using a sonicator (QSonica Q800R1; 30 min total, with 30-s on/off cycles). Chromatin from lysates corresponding to 15–30 OD600 of cells was immunoprecipitated with 2 µg antibody (anti-GFP, provided by A. Ladurner, Munich University, Munich, Germany) using Dynabeads Protein G (Life Technologies). Immunoprecipitated DNA was quantified by qPCR using Fast SYBR green master mix (Life Technologies) and a 7500 Fast Real-Time PCR system (Applied Biosystems) using a tilling array of primers covering the centromere (Braun et al., 2011).
Measurement of protein stability
For the analysis of the half-life of Cnp3 and Cdc13, WT and alm1Δ cells were grown in YES at 25ºC to midlog phase, and then 100 µg/ml CHX (01810; Sigma-Aldrich) was added to the cultures. Cells were harvested at the indicated time points, and whole cell extracts were prepared for immunoblotting using mouse anti-GFP mAb (11814460001; Roche) to detect Cnp3-GFP, rabbit anti-Cdc13 pAb (SP4, provided by S. Moreno), and mouse anti-PSTAIR mAb (P7962; Sigma-Aldrich) as a loading control. ImageJ software (version 1.5; National Institutes of Health) was used to quantify Cnp3-GFP and Cdc13 protein levels.
Immunoprecipitation of Cnp3 protein
mts4-ts cells expressing Cnp3-GFP were grown in YES to midlog phase at 25ºC and then shifted to 36ºC for 3 h. 45 min before harvesting, 5 mM N-ethylmaleimide (E3876; Sigma-Aldrich) was added to the culture media to inhibit deubiquitinating enzymes. Cells were harvested by centrifugation, and the pellets were washed with ice-cold STOP buffer and snap-frozen in liquid nitrogen. Native extracts were prepared in PBS with protease inhibitor cocktail (8340; Sigma-Aldrich) and 10 mM PMSF. 6 mg of total protein extracts was incubated with anti–mouse IgG–coated magnetic beads (Dynabeads; 11201D; Thermo Fisher) for 1 h at 4ºC. Beads were washed three times with PBS with protease inhibitors, resuspended in 50 µl SDS-PAGE sample loading buffer (0.25 M Tris-HCl, pH 6.8, 8% [wt/vol] SDS, 0.004% [wt/vol] bromophenol blue, and 20% [vol/vol] 2-mercaptoethanol), and boiled for 5 min. Samples were clarified by centrifugation at 13,000 rpm for 10 min, and 20 µl was immunoblotted with mouse anti-GFP mAb (11814460001; Roche) to detect Cnp3-GFP and rabbit antiubiquitin pAb (sc-9133; Santa Cruz; provided by J.A. Sanchez Alcazar, CABD/UPO) to detect ubiquitinated proteins.
Ubiquitin pull-down
Polyubiquitination analysis was performed as previously described (Shiozaki and Russell, 1997). Cells expressing the endogenous cnp3 tagged with GFP and overexpressing Cnp3-GFP from the medium-strength nmt41 promoter in a single copy (pINT-Cnp3-GFP) were transformed with the pREP1-His6-Ubi plasmid (originally provided by H. Seino, National Institute of Genetics, Mishima, Shizuoka, Japan). Cells were grown in EMM at 25ºC in the absence of thiamine for 22 h and then shifted to 36ºC for 3 h. 45 min before harvesting, 5 mM N-ethylmaleimide (E3876; Sigma-Aldrich) was added to the culture media to inhibit deubiquitinating enzymes. Cells were harvested by centrifugation and washed once with ice-cold STOP buffer, and whole cell extract was prepared in buffer G (6 M guanidine HCl, 0.1 M sodium phosphate, and 50 mM Tris-HCl, pH 8.0). 6 mg of total protein extract (input) was incubated for 1 h with agarose–nitrilotriacetic acid-Ni2+ (P6611; Sigma-Aldrich) at room temperature. The beads were then washed three times with buffer U (8 M urea, 0.1 M sodium phosphate, and 50 mM Tris-HCl, pH 8.0) and once with buffer U with 10 mM imidazole. 50 µl SDS-PAGE sample loading buffer was added to the beads and incubated for 5 min at 100ºC. 20 µl was immunoblotted with mouse anti-GFP mAb (11814460001; Roche), and 20 µl rabbit antiubiquitin pAb (sc-9133; Santa Cruz) inputs with the same amount of total protein was immunoblotted with anti-GFP mAb (Roche) to detect Cnp3-GFP and mouse antitubulin mAb (Tat1; provided by K. Gull) as a loading control.
Graphs and statistical analyses
Graphs and statistical analyses were performed with Prism 5.0 (GraphPad Software) and Excel (Microsoft). Unless otherwise stated, graphs represent the mean, and error bars represent SD. n is the total number of cells scored from at least three independent experiments. Statistical comparison between two groups was performed by unpaired Student’s t test, considering two-tailed p-values exceeding 0.05 to be NS. Asterisks denote P < 0.05 (*), P < 0.001 (**), P < 0.001 (***), and P < 0.0001 (****). Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows the predicted CC structure of Alm1 (A), its localization and colocalization with Nup107 at the nuclear pore (B and C), and the transient colocalization of Alm1 and Cut12 (D). Fig. S2 shows mitotic chromosome segregation of WT and alm1Δ with the histone marker Hht2-GFP (A), Sid2-tomato and lacI-GFP bound to tandem array repeats of lacO at the lys1 locus at chromosome I (B), and spindle dynamics of WT and alm1Δ during mitosis (C–E). Fig. S3 shows localization of Mad2-GFP (A) and Bub1GFP (B) during mitosis in WT and alm1Δ, Mad2-GFP localization during interphase (C), total protein levels by Western blot of Mad2 (D), Mad2-GFP KT localization in MBC metaphase blocked cells (E), and genetic interaction of alm1Δ with mad2Δ and bub1Δ (F). The figure also depicts localization (G) and kinase activity (H) of Ark1 and genetic interaction of alm1Δ and ark1-t7 ts mutant (I and J). Fig. S4 shows four clusters of gene-deletion mutants that have a synthetic effect on TBZ sensitivity with alm1Δ (A), images of WT, alm1Δ, mts4, and alm1Δ mts4 double mutant expressing Cnp3-GFP (B), images of WT, alm1Δ, and cut8-563 ts mutant expressing Cnp3-Tomato (C), and analysis of Cdc13 degradation in the presence of CHX (D). Table S1 contains the list of strains used in this study.
Supplementary Material
Acknowledgments
We would like to thank Paul Nurse’s lab (The Rockefeller University, New York, NY), our colleagues at the CABD, and members of R.R. Daga’s lab and the Cell Cycle group, especially Victor A. Tallada, for helpful discussions , Victor M. Carranco for excellent technical help, and the CABD proteomic and microscopy facility technicians Katherina Garcia and Laura Tomas for their helpful advice.
This work was supported by the Ministerio de Economía y Competitividad of the Spanish government (grant BFU2015-70604-P to R.R. Daga). P. Gallardo was supported by a grant from Universidad Pablo de Olavide (grant Beca Puente PP1-1402).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- CC
- coiled-coil
- CHX
- cycloheximide
- cnt
- central core
- EMM
- Edinburgh minimal media
- GO
- gene ontology
- imr
- innermost repeats
- KT
- kinetochore
- MBC
- methyl benzimidazol-2-yl carbamate
- MT
- microtubule
- NE
- nuclear envelope
- NPC
- nuclear pore complex
- otr
- outermost repeats
- qPCR
- quantitative PCR
- SAC
- spindle assembly checkpoint
- SGA
- synthetic genetic array
- SPB
- spindle pole body
- TBZ
- thiabendazole
- TPR
- translocated promoter region
- ts
- thermosensitive
References
- Amsterdam A., Pitzer F., and Baumeister W.. 1993. Changes in intracellular localization of proteasomes in immortalized ovarian granulosa cells during mitosis associated with a role in cell cycle control. Proc. Natl. Acad. Sci. USA. 90:99–103. 10.1073/pnas.90.1.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa H., Yang H.J., Yamamoto T.G., Ohtsuki C., Chikashige Y., Sakata-Sogawa K., Tokunaga M., Iwamoto M., Hiraoka Y., and Haraguchi T.. 2014. Characterization of nuclear pore complex components in fission yeast Schizosaccharomyces pombe. Nucleus. 5:149–162. 10.4161/nucl.28487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae J.A., Moon D., and Yoon J.H.. 2009. Nup211, the fission yeast homolog of Mlp1/Tpr, is involved in mRNA export. J. Microbiol. 47:337–343. 10.1007/s12275-009-0125-7 [DOI] [PubMed] [Google Scholar]
- Bähler J., Wu J.Q., Longtine M.S., Shah N.G., McKenzie A. III, Steever A.B., Wach A., Philippsen P., and Pringle J.R.. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951. [DOI] [PubMed] [Google Scholar]
- Bangs P., Burke B., Powers C., Craig R., Purohit A., and Doxsey S.. 1998. Functional analysis of Tpr: identification of nuclear pore complex association and nuclear localization domains and a role in mRNA export. J. Cell Biol. 143:1801–1812. 10.1083/jcb.143.7.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrales R.R., Forn M., Georgescu P.R., Sarkadi Z., and Braun S.. 2016. Control of heterochromatin localization and silencing by the nuclear membrane protein Lem2. Genes Dev. 30:133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P., Maure J.F., Partridge J.F., Genier S., Javerzat J.P., and Allshire R.C.. 2001. Requirement of heterochromatin for cohesion at centromeres. Science. 294:2539–2542. 10.1126/science.1064027 [DOI] [PubMed] [Google Scholar]
- Braun S., Garcia J.F., Rowley M., Rougemaille M., Shankar S., and Madhani H.D.. 2011. The Cul4-Ddb1(Cdt)2 ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell. 144:41–54. 10.1016/j.cell.2010.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M., Doucet C., and Hetzer M.W.. 2010. Nuclear pore complexes: guardians of the nuclear genome. Cold Spring Harb. Symp. Quant. Biol. 75:585–597. 10.1101/sqb.2010.75.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C.W., Milks K.J., and Straight A.F.. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189:1143–1155. 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A.G., Mellone B.G., Partridge J.F., Richardson W., Hamilton G.L., Allshire R.C., and Pidoux A.L.. 2007. Plasticity of fission yeast CENP-A chromatin driven by relative levels of histone H3 and H4. PLoS Genet. 3:e121 10.1371/journal.pgen.0030121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D., Moree B., Canman J.C., and Salmon E.D.. 2003. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J. Cell Sci. 116:4213–4225. 10.1242/jcs.00716 [DOI] [PubMed] [Google Scholar]
- Cimini D., Wan X., Hirel C.B., and Salmon E.D.. 2006. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 16:1711–1718. 10.1016/j.cub.2006.07.022 [DOI] [PubMed] [Google Scholar]
- Cohen R.L., Espelin C.W., De Wulf P., Sorger P.K., Harrison S.C., and Simons K.T.. 2008. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell. 19:4480–4491. 10.1091/mbc.E08-03-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K.A., Furuyama S., and Biggins S.. 2004. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr. Biol. 14:1968–1972. 10.1016/j.cub.2004.10.024 [DOI] [PubMed] [Google Scholar]
- Cordes V.C., Reidenbach S., Rackwitz H.R., and Franke W.W.. 1997. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J. Cell Biol. 136:515–529. 10.1083/jcb.136.3.515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtheoux T., Gay G., Reyes C., Goldstone S., Gachet Y., and Tournier S.. 2007. Dynein participates in chromosome segregation in fission yeast. Biol. Cell. 99:627–637. 10.1042/BC20070047 [DOI] [PubMed] [Google Scholar]
- DeGrasse J.A., DuBois K.N., Devos D., Siegel T.N., Sali A., Field M.C., Rout M.P., and Chait B.T.. 2009. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics. 8:2119–2130. 10.1074/mcp.M900038-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., and Salmon E.D.. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- De Souza C.P., Hashmi S.B., Nayak T., Oakley B., and Osmani S.A.. 2009. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol. Biol. Cell. 20:2146–2159. 10.1091/mbc.E08-08-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S.J., Fedyshyn Y., Koh J.L., Prasad T.S., Chahwan C., Chua G., Toufighi K., Baryshnikova A., Hayles J., Hoe K.L., et al. . 2008. Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proc. Natl. Acad. Sci. USA. 105:16653–16658. 10.1073/pnas.0806261105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K., Cranston G., and Allshire R.C.. 1999. Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics. 153:1153–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C., Lehmann A., and Kloetzel P.M.. 1998. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 17:6144–6154. 10.1093/emboj/17.21.6144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennessy D., Grallert A., Krapp A., Cokoja A., Bridge A.J., Petersen J., Patel A., Tallada V.A., Boke E., Hodgson B., et al. . 2014. Extending the Schizosaccharomyces pombe molecular genetic toolbox. PLoS One. 9:e97683 10.1371/journal.pone.0097683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco H.D., Campbell C.S., May K.M., Espinoza C.A., Oegema K., Hardwick K.G., Grewal S.I., and Desai A.. 2015. The CENP-A N-tail confers epigenetic stability to centromeres via the CENP-T branch of the CCAN in fission yeast. Curr. Biol. 25:348–356. 10.1016/j.cub.2014.11.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosst P., Guan T., Subauste C., Hahn K., and Gerace L.. 2002. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J. Cell Biol. 156:617–630. 10.1083/jcb.200106046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth N., Gertman O., Shiber A., Alfassy O.S., Cohen I., Rosenberg M.M., Doron N.K., Friedler A., and Ravid T.. 2011. Exposure of bipartite hydrophobic signal triggers nuclear quality control of Ndc10 at the endoplasmic reticulum/nuclear envelope. Mol. Biol. Cell. 22:4726–4739. 10.1091/mbc.E11-05-0463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V., Olivo-Marin J.C., Scherthan H., Doye V., Rascalou N., and Nehrbass U.. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 403:108–112. 10.1038/47528 [DOI] [PubMed] [Google Scholar]
- Goshima G., Saitoh S., and Yanagida M.. 1999. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 13:1664–1677. 10.1101/gad.13.13.1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J., Riedel C.G., Pidoux A.L., Katou Y., Rumpf C., Schleiffer A., Kearsey S.E., Shirahige K., Allshire R.C., and Nasmyth K.. 2007. The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr. Biol. 17:1190–1200. 10.1016/j.cub.2007.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I.M., and Hyams J.S.. 1988. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 89:343–357. [DOI] [PubMed] [Google Scholar]
- Hediger F., Dubrana K., and Gasser S.M.. 2002. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J. Struct. Biol. 140:79–91. 10.1016/S1047-8477(02)00533-6 [DOI] [PubMed] [Google Scholar]
- Jiménez M., Petit T., Gancedo C., and Goday C.. 2000. The alm1+ gene from Schizosaccharomyces pombe encodes a coiled-coil protein that associates with the medial region during mitosis. Mol. Gen. Genet. 262:921–930. 10.1007/PL00008660 [DOI] [PubMed] [Google Scholar]
- Kabachinski G., and Schwartz T.U.. 2015. The nuclear pore complex--structure and function at a glance. J. Cell Sci. 128:423–429. 10.1242/jcs.083246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B., and Fornerod M.. 2010. Characterization of genome-nucleoporin interactions in Drosophila links chromatin insulators to the nuclear pore complex. Cell Cycle. 9:4812–4817. 10.4161/cc.9.24.14328 [DOI] [PubMed] [Google Scholar]
- Kitagawa T., Ishii K., Takeda K., and Matsumoto T.. 2014. The 19S proteasome subunit Rpt3 regulates distribution of CENP-A by associating with centromeric chromatin. Nat. Commun. 5:3597 10.1038/ncomms4597 [DOI] [PubMed] [Google Scholar]
- Knowlton A.L., Lan W., and Stukenberg P.T.. 2006. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 16:1705–1710. 10.1016/j.cub.2006.07.057 [DOI] [PubMed] [Google Scholar]
- Kosova B., Panté N., Rollenhagen C., Podtelejnikov A., Mann M., Aebi U., and Hurt E.. 2000. Mlp2p, a component of nuclear pore attached intranuclear filaments, associates with nic96p. J. Biol. Chem. 275:343–350. 10.1074/jbc.275.1.343 [DOI] [PubMed] [Google Scholar]
- Kriegenburg F., Jakopec V., Poulsen E.G., Nielsen S.V., Roguev A., Krogan N., Gordon C., Fleig U., and Hartmann-Petersen R.. 2014. A chaperone-assisted degradation pathway targets kinetochore proteins to ensure genome stability. PLoS Genet. 10:e1004140 10.1371/journal.pgen.1004140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S., Dörries J., Boysen B., Reidenbach S., Magnius L., Norder H., Thyberg J., and Cordes V.C.. 2010. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 29:1659–1673. 10.1038/emboj.2010.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Sterling H., Burlingame A., and McCormick F.. 2008. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 22:2926–2931. 10.1101/gad.1677208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A., Felberbaum R., and Hochstrasser M.. 2007. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J. Cell Biol. 178:813–827. 10.1083/jcb.200702154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lince-Faria M., Maffini S., Orr B., Ding Y., Cláudia Florindo C.E., Sunkel C.E., Tavares A., Johansen J., Johansen K.M., and Maiato H.. 2009. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J. Cell Biol. 184:647–657. 10.1083/jcb.200811012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra R., Kerr S.C., Harreman M.T., Apponi L.H., Fasken M.B., Ramineni S., Chaurasia S., Valentini S.R., and Corbett A.H.. 2007. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J. Biol. Chem. 282:3042–3049. 10.1074/jbc.M608741200 [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., and Nurse P.. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823. 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Moreno-Moreno O., Torras-Llort M., and Azorín F.. 2006. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 34:6247–6255. 10.1093/nar/gkl902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A., and Salmon E.D.. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393. 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa T., Straight A.F., Murray A., Chikashige Y., Yamashita Y.M., Hiraoka Y., and Yanagida M.. 1998. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell. 9:3211–3225. 10.1091/mbc.9.11.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M., Strambio-de-Castillia C., Fasolo J., Chait B.T., and Rout M.P.. 2005. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J. Cell Biol. 170:225–235. 10.1083/jcb.200504140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M., Molloy K.R., Williams R., Farr J.C., Meinema A.C., Vecchietti N., Cristea I.M., Chait B.T., Rout M.P., and Strambio-De-Castillia C.. 2013. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol. Biol. Cell. 24:3920–3938. 10.1091/mbc.E13-07-0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N., Kitajima T., Yokobayashi S., Xiao G., Yamamoto M., Grewal S.I., and Watanabe Y.. 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4:89–93. 10.1038/ncb739 [DOI] [PubMed] [Google Scholar]
- Palancade B., Liu X., Garcia-Rubio M., Aguilera A., Zhao X., and Doye V.. 2007. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Mol. Biol. Cell. 18:2912–2923. 10.1091/mbc.E07-02-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J., Paris J., Willer M., Philippe M., and Hagan I.M.. 2001. The S. pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J. Cell Sci. 114:4371–4384. [DOI] [PubMed] [Google Scholar]
- Pidoux A.L., and Allshire R.C.. 2005. The role of heterochromatin in centromere function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:569–579. 10.1098/rstb.2004.1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A.L., Richardson W., and Allshire R.C.. 2003. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 161:295–307. 10.1083/jcb.200212110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanala K., Sarkar A., Jhingan G.D., Priyadarshini R., Jalan M., Sengupta S., and Nandicoori V.K.. 2014. Phosphorylation of nucleoporin Tpr governs its differential localization and is required for its mitotic function. J. Cell Sci. 127:3505–3520. 10.1242/jcs.149112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitkar P., Press M.O., Yi X., Baker R., MacCoss M.J., and Biggins S.. 2010. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol. Cell. 40:455–464. 10.1016/j.molcel.2010.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Bravo V., Maciejowski J., Corona J., Buch H.K., Collin P., Kanemaki M.T., Shah J.V., and Jallepalli P.V.. 2014. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell. 156:1017–1031. 10.1016/j.cell.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpf C., Cipak L., Schleiffer A., Pidoux A., Mechtler K., Tolić-Nørrelykke I.M., and Gregan J.. 2010. Laser microsurgery provides evidence for merotelic kinetochore attachments in fission yeast cells lacking Pcs1 or Clr4. Cell Cycle. 9:3997–4004. 10.4161/cc.9.19.13233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S.J., Steger K.A., and Johnston S.A.. 1999. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J. Biol. Chem. 274:21943–21952. 10.1074/jbc.274.31.21943 [DOI] [PubMed] [Google Scholar]
- Santaguida S., and Musacchio A.. 2009. The life and miracles of kinetochores. EMBO J. 28:2511–2531. 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Dhut S., and Toda T.. 2005. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 22:583–591. 10.1002/yea.1233 [DOI] [PubMed] [Google Scholar]
- Schweizer N., Ferrás C., Kern D.M., Logarinho E., Cheeseman I.M., and Maiato H.. 2013. Spindle assembly checkpoint robustness requires Tpr-mediated regulation of Mad1/Mad2 proteostasis. J. Cell Biol. 203:883–893. 10.1083/jcb.201309076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K., and Russell P.. 1997. Stress-activated protein kinase pathway in cell cycle control of fission yeast. Methods Enzymol. 283:506–520. 10.1016/S0076-6879(97)83040-6 [DOI] [PubMed] [Google Scholar]
- Snaith H.A., Samejima I., and Sawin K.E.. 2005. Multistep and multimode cortical anchoring of tea1p at cell tips in fission yeast. EMBO J. 24:3690–3699. 10.1038/sj.emboj.7600838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich B., Strålfors A., Khorosjutina O., Persson J., Smialowska A., Javerzat J.P., and Ekwall K.. 2015. The Fun30 chromatin remodeler Fft3 controls nuclear organization and chromatin structure of insulators and subtelomeres in fission yeast. PLoS Genet. 11:e1005101 10.1371/journal.pgen.1005101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S., Keith K.C., Curnick K.E., and Fitzgerald-Hayes M.. 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9:573–586. 10.1101/gad.9.5.573 [DOI] [PubMed] [Google Scholar]
- Strålfors A., Walfridsson J., Bhuiyan H., and Ekwall K.. 2011. The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLoS Genet. 7:e1001334 10.1371/journal.pgen.1001334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel G., and Rout M.P.. 1999. Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol. 144:839–855. 10.1083/jcb.144.5.839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Kuriyama K., Shibata A., and Himeno M.. 1997. Characterization of internal DNA-binding and C-terminal dimerization domains of human centromere/kinetochore autoantigen CENP-C in vitro: role of DNA-binding and self-associating activities in kinetochore organization. Chromosome Res. 5:132–141. 10.1023/A:1018422325569 [DOI] [PubMed] [Google Scholar]
- Taddei A., Hediger F., Neumann F.R., Bauer C., and Gasser S.M.. 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 23:1301–1312. 10.1038/sj.emboj.7600144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Murakami S., Chikashige Y., Funabiki H., Niwa O., and Yanagida M.. 1992. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell. 3:819–835. 10.1091/mbc.3.7.819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamada H., and Yanagida M.. 1994. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell. 5:1145–1158. 10.1091/mbc.5.10.1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Chen E.S., and Yanagida M.. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 288:2215–2219. 10.1126/science.288.5474.2215 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Takayama Y., Masuda F., Kobayashi Y., and Saitoh S.. 2005. Two distinct pathways responsible for the loading of CENP-A to centromeres in the fission yeast cell cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:595–607. 10.1098/rstb.2004.1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., and Yanagida M.. 2005. Regulation of nuclear proteasome by Rhp6/Ubc2 through ubiquitination and destruction of the sensor and anchor Cut8. Cell. 122:393–405. 10.1016/j.cell.2005.05.023 [DOI] [PubMed] [Google Scholar]
- Takeda K., Tonthat N.K., Glover T., Xu W., Koonin E.V., Yanagida M., and Schumacher M.A.. 2011. Implications for proteasome nuclear localization revealed by the structure of the nuclear proteasome tether protein Cut8. Proc. Natl. Acad. Sci. USA. 108:16950–16955. 10.1073/pnas.1103617108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Chang H.L., Kagami A., and Watanabe Y.. 2009. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell. 17:334–343. 10.1016/j.devcel.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Tan-Wong S.M., Wijayatilake H.D., and Proudfoot N.J.. 2009. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 23:2610–2624. 10.1101/gad.1823209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebe H., and Yanagida M.. 2000. Cut8, essential for anaphase, controls localization of 26S proteasome, facilitating destruction of cyclin and Cut2. Curr. Biol. 10:1329–1338. 10.1016/S0960-9822(00)00773-9 [DOI] [PubMed] [Google Scholar]
- Therizols P., Fairhead C., Cabal G.G., Genovesio A., Olivo-Marin J.C., Dujon B., and Fabre E.. 2006. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J. Cell Biol. 172:189–199. (published erratum appears in J. Cell Biol. 2006. 172:951). 10.1083/jcb.200505159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Pagé N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. . 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 294:2364–2368. 10.1126/science.1065810 [DOI] [PubMed] [Google Scholar]
- Tournier S., Gachet Y., Buck V., Hyams J.S., and Millar J.B.. 2004. Disruption of astral microtubule contact with the cell cortex activates a Bub1, Bub3, and Mad3-dependent checkpoint in fission yeast. Mol. Biol. Cell. 15:3345–3356. 10.1091/mbc.E04-03-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trazzi S., Bernardoni R., Diolaiti D., Politi V., Earnshaw W.C., Perini G., and Della Valle G.. 2002. In vivo functional dissection of human inner kinetochore protein CENP-C. J. Struct. Biol. 140:39–48. 10.1016/S1047-8477(02)00506-3 [DOI] [PubMed] [Google Scholar]
- Trazzi S., Perini G., Bernardoni R., Zoli M., Reese J.C., Musacchio A., and Della Valle G.. 2009. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS One. 4:e5832 10.1371/journal.pone.0005832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D., Bonnet J., Waharte F., Fournier M., Stierle M., Fischer B., Brino L., Devys D., and Tora L.. 2013. The human TREX-2 complex is stably associated with the nuclear pore basket. J. Cell Sci. 126:2656–2667. 10.1242/jcs.118000 [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V., and Hardwick K.G.. 2009. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 19:1176–1181. 10.1016/j.cub.2009.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier L., Taglini F., Barrales R.R., Webb S., Urano T., Braun S., and Bayne E.H.. 2015. Global regulation of heterochromatin spreading by Leo1. Open Biol. 5:150045 10.1098/rsob.150045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D., Zwickl P., and Baumeister W.. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015–1068. 10.1146/annurev.biochem.68.1.1015 [DOI] [PubMed] [Google Scholar]
- Wang J., Reddy B.D., and Jia S.. 2015. Rapid epigenetic adaptation to uncontrolled heterochromatin spreading. eLife. 4:e06179 10.7554/eLife.06179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson C.R., Wallace M., Seeger M., Dubiel W., and Gordon C.. 1997. Mts4, a non-ATPase subunit of the 26 S protease in fission yeast is essential for mitosis and interacts directly with the ATPase subunit Mts2. J. Biol. Chem. 272:25768–25777. 10.1074/jbc.272.41.25768 [DOI] [PubMed] [Google Scholar]
- Wilkinson C.R., Wallace M., Morphew M., Perry P., Allshire R., Javerzat J.P., McIntosh J.R., and Gordon C.. 1998. Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 17:6465–6476. 10.1093/emboj/17.22.6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.M., Rose A., Muthuswamy S., Jeong S.Y., Venkatakrishnan S., Zhao Q., and Meier I.. 2007. NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell. 19:1537–1548. 10.1105/tpc.106.049239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.J., Asakawa H., Haraguchi T., and Hiraoka Y.. 2015. Nup132 modulates meiotic spindle attachment in fission yeast by regulating kinetochore assembly. J. Cell Biol. 211:295–308. 10.1083/jcb.201501035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lan W., Ems-McClung S.C., Stukenberg P.T., and Walczak C.E.. 2007. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol. Biol. Cell. 18:3264–3276. 10.1091/mbc.E07-01-0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wu C.Y., and Blobel G.. 2004. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167:605–611. 10.1083/jcb.200405168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimowska G., Aris J.P., and Paddy M.R.. 1997. A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J. Cell Sci. 110:927–944. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.