Cell spreading relies on the coordinated and dynamic regulation of integrin-mediated adhesions and actin structures. Böttcher et al. show that in the periphery of adhering cells kindlin-2 binds paxillin to activate Rac1 and the Arp2/3 complex to allow for Rac1-mediated membrane protrusions.
Abstract
Cell spreading requires the coupling of actin-driven membrane protrusion and integrin-mediated adhesion to the extracellular matrix. The integrin-activating adaptor protein kindlin-2 plays a central role for cell adhesion and membrane protrusion by directly binding and recruiting paxillin to nascent adhesions. Here, we report that kindlin-2 has a dual role during initial cell spreading: it binds paxillin via the pleckstrin homology and F0 domains to activate Rac1, and it directly associates with the Arp2/3 complex to induce Rac1-mediated membrane protrusions. Consistently, abrogation of kindlin-2 binding to Arp2/3 impairs lamellipodia formation and cell spreading. Our findings identify kindlin-2 as a key protein that couples cell adhesion by activating integrins and the induction of membrane protrusions by activating Rac1 and supplying Rac1 with the Arp2/3 complex.
Introduction
Cell migration and cell spreading are multistep processes involving protrusion of the plasma membrane, induction of new adhesions to the underlying substratum, and maturation and turnover of adhesion sites (Petrie et al., 2009; Devreotes and Horwitz, 2015). The different processes critically rely on the coordinated and dynamic regulation of integrin-mediated adhesions and actin structures, e.g., the formation of nascent adhesions (NAs) and branched actin networks in lamellipodia, and the assembly of stress fibers that connect focal adhesions (FAs) further toward the middle and rear of spread cells. Lamellipodia are smooth and narrow projections of the plasma membrane that extend along the cell edges and are initiated by the actin nucleation activity of the Arp2/3 complex (Pollard and Borisy, 2003). The canonical Arp2/3 complex consists of seven subunits (Machesky et al., 1994; Welch et al., 1997; Winter et al., 1997; Bugyi and Carlier, 2010), binds to the sides of already existing actin filaments, and triggers the growth of new actin branches. The actin nucleation activity of the Arp2/3 complex is induced by members of the Wiskott–Aldrich syndrome protein family, including WASP and WAVE (Mullins et al., 1998; Rohatgi et al., 1999; Winter et al., 1999; Rouiller et al., 2008), whose activity in turn is controlled by small Rho-like GTPases, including Rac1 and Cdc42 (Takenawa and Suetsugu, 2007).
The physical coupling of the branched actin network to the ECM occurring in lamellipodia and membrane protrusions of isotropically spreading cells is achieved by integrin-mediated adhesions that initially form as small, short-lived NAs at or near the edge of protruding membranes. Once formed, they either disassemble or mature in an actomyosin-dependent manner into large and long-lived FAs (Vicente-Manzanares and Horwitz, 2011). The induction of integrin-mediated adhesions requires an integrin-activation step characterized by the conformational shift of the unbound, low-affinity (inactive) state to the bound, high-affinity (active) state, which is followed by integrin clustering to stabilize integrin–ligand complexes and the assembly of a large multiprotein network that enables signaling. The two cytosolic adaptor proteins talin and kindlin bind to β integrin cytoplasmic domains and induce and/or maintain integrin-mediated cell–extracellular matrix adhesion. The prevalent view is that talin and kindlin cooperate to induce integrin activation (Han et al., 2006; Moser et al., 2008; Theodosiou et al., 2016) and clustering (Cluzel et al., 2005; Ye et al., 2013). An additional function of kindlin is to induce membrane protrusions during early, isotropic cell spreading by directly binding and recruiting paxillin to NAs, which in turn leads to FAK and Rac1 activation (Theodosiou et al., 2016).
Arp2/3–driven membrane protrusion and integrin-mediated adhesion to the ECM in NAs are tightly coupled and depend on each other. It has been shown that Arp2/3 can be recruited to adhesion sites through transient interactions with vinculin (DeMali et al., 2002; Chorev et al., 2014) and FAK (Serrels et al., 2007; Swaminathan et al., 2016).
Talin is unable to induce circumferential membrane protrusions during isotropic spreading in the absence of kindlin-2 (Theodosiou et al., 2016). Because kindlin-2 recruits paxillin and FAK, which in turn was shown to induce Rac1 activation and membrane protrusion, we hypothesized that by circumventing the Rac1 activation defect in kindlin-deficient cells, cell spreading should efficiently be induced. In this study, we tested this hypothesis and further characterized the kindlin-2–paxillin complex using cross-linking proteomics. The findings of our studies are discussed here.
Results
Kindlin-2 directly binds paxillin through the PH and F0 domains
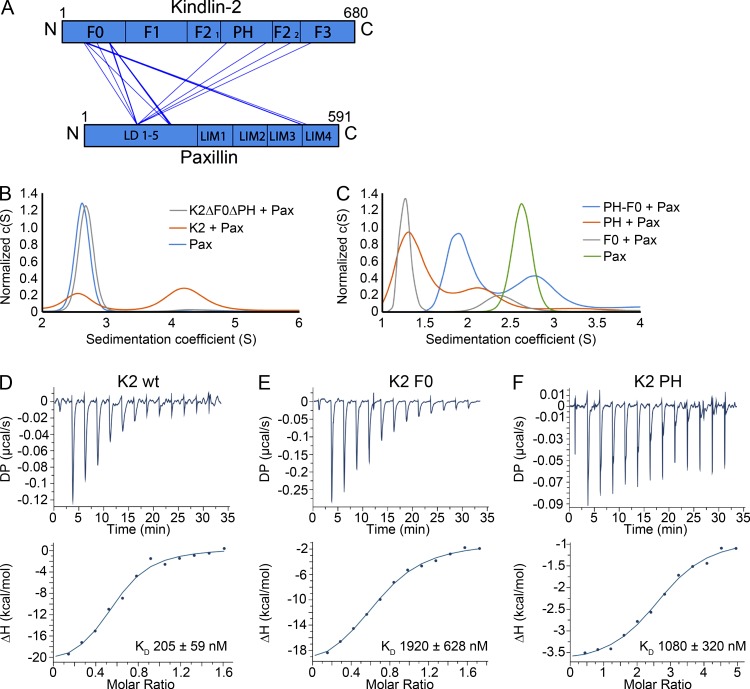
In a previous study, we reported a direct, Zn2+-dependent interaction between the pleckstrin homology (PH) domain of kindlin-2 and the Lin-11, Isl-1, and Mec-3 (LIM3) domain of paxillin by size-exclusion chromatography and pull-down experiments (Theodosiou et al., 2016). Furthermore, we found that the absence of the PH domain in kindlin-2 leads to low levels of paxillin in NAs but to normal levels in mature FAs of fibroblasts (Theodosiou et al., 2016), indicating that paxillin recruitment to FAs occurs either in a kindlin-independent manner or through additional, unrecognized paxillin-binding sites in kindlin. To test the latter possibility, we performed cross-linking mass spectrometry (XL-MS) experiments of recombinant kindlin-2–paxillin complexes by cross-linking the amine groups of lysine side chains with an isotopically coded bissulfosuccinimidyl suberate (Leitner et al., 2010). Cross-linked peptides were identified by tandem mass spectrometry (MS) and used to assemble a map of the inter- and intraprotein cross-links of the kindlin-2–paxillin complex (Figs. 1 A and S1 A and Supplemental dataset). We identified cross-links between the N-terminal LD motifs of paxillin and the PH domain of kindlin-2. In addition, we also observed multiple cross-links between the F0 domain of kindlin-2 and the LIM3/4 domains and the LD motifs of paxillin, suggesting that the PH as well as the F0 domains contribute to kindlin-2 binding to paxillin.
Figure 1.
Cross-linking identifies two distinct kindlin-2–paxillin interaction sites. (A) Cross-link map of the kindlin-2–paxillin interaction. Cross-links are detected between the kindlin-2 PH domain and the paxillin LD motifs and between the kindlin-2 F0 domain and paxillin LD motifs and LIM3-domain region. Intraprotein cross-links are not depicted for clarity. F0, F1, F2, and F3 part of the FERM domain; C, C terminus; LD, LD motifs; LIM, Lin-11, Isl-1 and Mec-3 domain; N, N terminus. (B) Analytical ultracentrifugation profiles of Atto520-labeled paxillin (Pax) in complex with wild-type kindlin-2 and K2ΔF0ΔPH. (C) Complex formation of paxillin with different Atto520-labeled kindlin-2 subdomains determined by analytical ultracentrifugation. (D–F) Isothermal titration calorimetry measurements of paxillin bound to K2 wild type (D), K2 F0 (E), or K2 PH (F). c(S), continuous sedimentation coefficient distribution; DP, differential power.
We verified the XL-MS results by analytical ultracentrifugation, in which fluorescent labeling of one of the two interaction partners was used at a time to detect the corresponding absorption wavelengths. The in vitro reconstituted kindlin-2–paxillin complex sedimented at 4.3 S, whereas recombinant kindlin-2 lacking the F0 and PH domains failed to form a detectable complex with paxillin (Fig. 1 B). Recombinant F0 and PH domains and a hybrid F0–PH domain of kindlin-2 were able to bind paxillin (Fig. 1 C). Interestingly, the sedimentation coefficients of paxillin associated with the F0 or PH domain of kindlin-2 were lower than the sedimentation coefficient of paxillin alone, indicating that the impact of the weight increase on the sedimentation coefficient is compensated by an induced compaction of paxillin upon complex formation with either the F0 or PH domain.
Next, we performed affinity measurements using isothermal titration calorimetry (ITC). The affinity of paxillin for full-length kindlin-2 was 205 ± 59 nM, whereas the affinities of paxillin to the F0 or the PH domain alone were lower and ranged between 1,920 ± 628 and 1,080 ± 320 nM, respectively (Fig. 1, D–F). Addition of EDTA abolished the interactions with paxillin, indicating that not only binding of full-length kindlin-2 but also binding of the isolated F0 or PH domain is Zn2+ ion dependent (Fig. S1, B–D). Altogether, these data point to the existence of a second, previously unnoticed paxillin-binding domain in kindlin-2 and suggest that both domains bind concurrently to increase the affinity of kindlin-2 to paxillin.
The kindlin-2 PH and F0 domains are required for cell adhesion and spreading
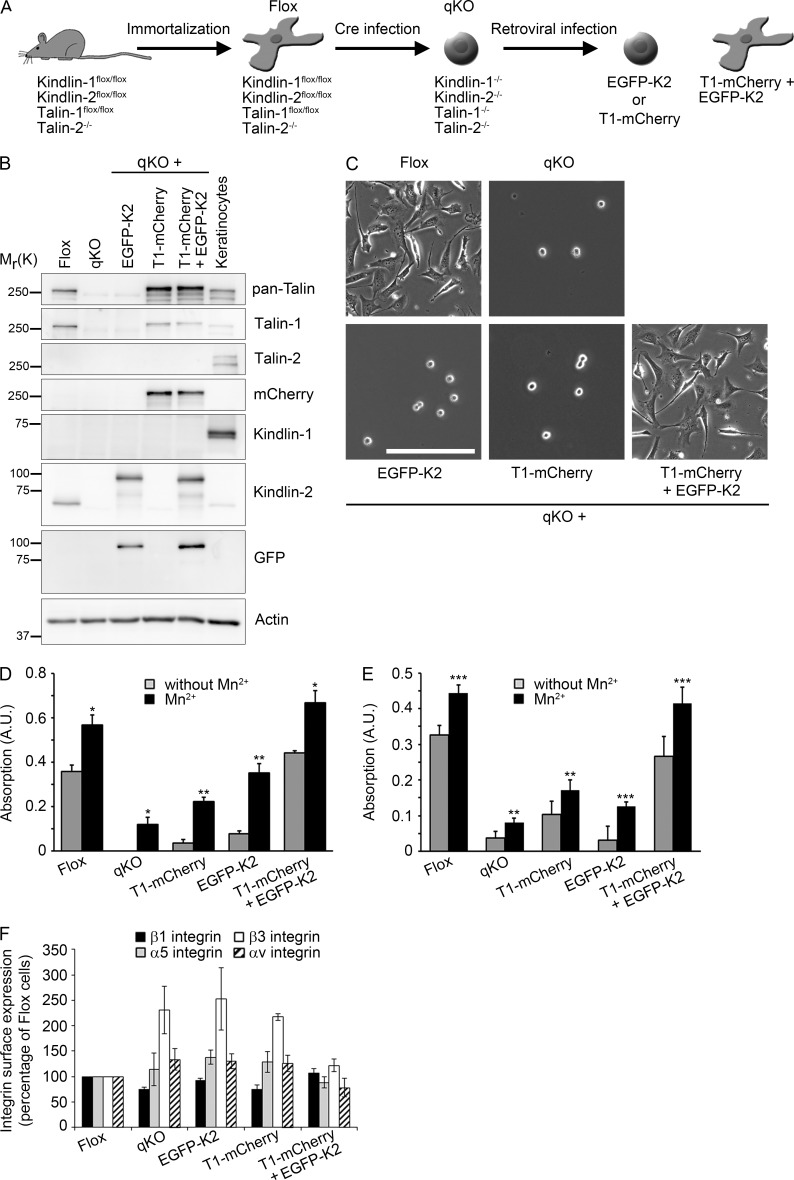
To characterize kindlin-2–paxillin binding in cells, we isolated, immortalized and cloned kidney fibroblasts from mice carrying floxed kindlin-1 (Fermt1flox/flox), kindlin-2 (Fermt2flox/flox), Tln1 alleles, and nullizygous Tln2 alleles (Flox cells; Fig. 2 A). To directly compare kindlin-2 and talin-1 functions in the same cellular background, the floxed alleles were deleted by adenoviral expression of Cre recombinase, resulting in kindlin-1, kindlin-2, talin-1, and talin-2–deficient (quadruple knockout [qKO]) cells, and reconstituted with C-terminally mCherry-tagged talin-1 (T1-mCherry), N-terminally EGFP-tagged kindlin-2 (EGFP-K2), or a combination of both (T1-mCherry + EGFP-K2).
Figure 2.
Kindlin and talin cooperate to establish cell adhesion. (A) Scheme for the generation of qKO cells from mouse kidney lacking talin-1/-2 as well as kindlin-1/-2. qKO fibroblasts were retrovirally transduced with EGFP-kindlin-2 (EGFP-K2), talin-1-mCherry (T1-mCherry), or a combination of both. (B) Western blot of Flox, qKO, and EGFP-K2, T1-mCherry–expressing cells. Keratinocyte lysates served to control kindlin-1 and talin-2 expression. (C) Phase contrast images of the indicated cell lines. Bar, 200 µm. (D) Quantification of cell adhesion on FN 30 min after seeding in the absence or presence of 5 mM Mn2+ (n = 3 independent experiments). Error bars indicate SEM; significance was calculated between untreated cells and the corresponding Mn2+-treated cells (t test). (E) Quantification of cell adhesion on VN 30 min after seeding in the absence or presence of 5 mM Mn2+ (n = 4 independent experiments). Error bars indicate SEM; significance was calculated between untreated cells and the corresponding Mn2+-treated cells (t test). *, P < 0.05; **, P < 0.01; ***, P < 0.001. (F) Quantification of cell surface expression of different integrin subunits on Flox, qKO, and EGFP-K2, T1-mCherry–reexpressing cells measured by flow cytometry. The integrin levels were normalized to the levels on Flox cells (n = 3 independent experiments). Error bars indicate SD.
Cre treatment deleted the floxed Tln1 and floxed Fermt1/2 genes (Fig. 2 B), resulted in cell rounding (Fig. 2 C), and abolished adhesion of the resulting qKO cells to fibronectin (FN) and vitronectin (VN; Fig. 2, D and E). Reexpression of T1-mCherry or EGFP-K2 did not rescue the severe adhesion defects of qKO cells when plated on FN or VN, whereas coexpression of T1-mCherry together with EGFP-K2 rescued cell adhesion and spreading (Fig. 2, B–E). Importantly, loss of endogenous talin and/or kindlin as well as reexpression of T1-mCherry and/or EGFP-K2 did not change surface levels of the FN-binding integrins α5β1 and αvβ3. In fact, α5, αv and β3 levels were increased in qKO and single reconstituted cells (Fig. 2 F).
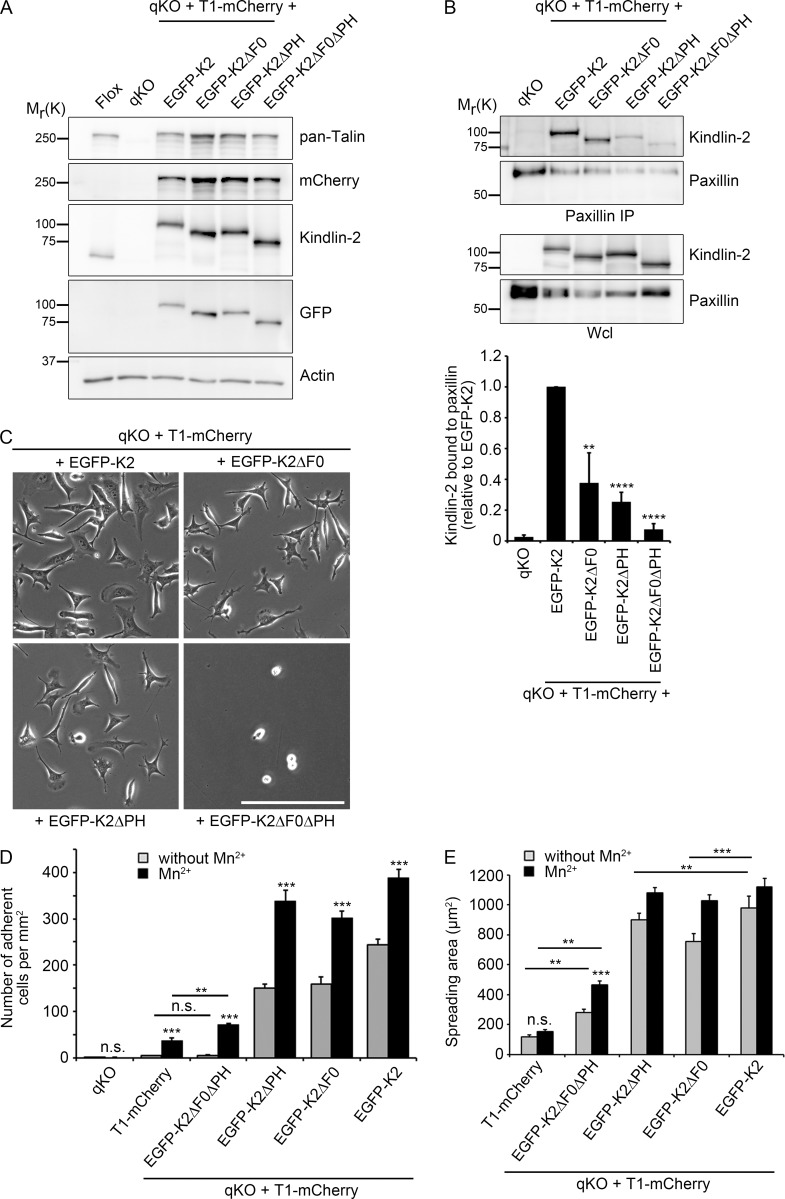
Next, we expressed either EGFP-K2 or EGFP-tagged kindlin-2 truncation mutants that lacked either the F0 (EGFP-K2ΔF0) or the PH domain (EGFP-K2ΔPH) or both domains (EGFP-K2ΔF0ΔPH) in T1-mCherry cells (Fig. 3 A) and performed coimmunoprecipitation (co-IP) assays. The experiments revealed that paxillin coprecipitated EGFP-K2, whereas the co-IP of EGFP-K2ΔF0 or EGFP-K2ΔPH were reduced and of the K2ΔF0ΔPH abolished (Fig. 3 B), confirming that both the F0 and PH domains of kindlin-2 contribute to paxillin binding.
Figure 3.
Kindlin-2 requires paxillin binding to induce cell adhesion and spreading. (A) Western blot of parental Flox cells, qKO cells, and qKO cells expressing T1-mCherry and EGFP-tagged kindlin-2 (K2), K2ΔF0, K2ΔPH, or K2ΔF0ΔPH. (B) Co-IP of endogenous paxillin and densitometric analysis of Western blots to determine paxillin binding to EGFP-tagged kindlin-2 (K2), K2ΔF0, K2ΔPH, or K2ΔF0ΔPH. Graph shows kindlin-2 binding to paxillin relative to EGFP-K2. Wcl, whole cell lysate (n = 3 independent experiments). Error bars indicate SD; significance was calculated between EGFP-K2 and the kindlin-2 deletion variants (t test). (C) Phase contrast images of indicated cell lines. Bar, 200 µm. (D) Quantification of adhesion to FN of qKO cells expressing T1-mCherry alone or in combination with EGFP-tagged kindlin-2 (K2), K2ΔF0, K2ΔPH, or K2ΔF0ΔPH for 30 min in the absence or presence of 5 mM Mn2+ (n = 4 independent experiments). Error bars indicate SEM; significance was calculated between untreated cells and the corresponding Mn2+-treated cells (t test). (E) Quantification of the cell area after spreading on FN for 30 min in the absence and presence of 5 mM Mn2+ (>50 cells counted in two independent experiments). Error bars indicate SEM; significance is indicated (t test). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n.s., not significant.
In line with our previous finding (Theodosiou et al., 2016), qKO cells expressing either T1-mCherry or EGFP-K2 failed to adhere to FN (Fig. 2 D). Reexpression of EGFP-K2 in T1-mCherry expressing cells fully rescued adhesion to FN, whereas reexpression of EGFP-K2ΔF0 or EGFP-K2ΔPH only partially rescued adhesion and reexpression of EGFP-K2ΔF0ΔPH almost completely abolished cell adhesion and spreading (Fig. 3, C and D, gray bars without Mn2+). Furthermore, EGFP-K2, EGFP-K2ΔF0, and EGFP-K2ΔPH localized to FAs in kindlin-2–deficient T1-mCherry cells, whereas the few adhering EGFP-K2ΔF0ΔPH expressing T1-mCherry cells remained round and failed to form discernable FAs (Fig. S2).
To bypass integrin activation, we treated cells with Mn2+, which binds to the integrin ectodomain and induces the conformational changes of activated integrins (Mould et al., 1995). The experiments revealed that expression of EGFP-K2ΔF0 and EGFP-K2ΔPH almost completely rescued adhesion of T1-mCherry–expressing cells to FN. In contrast, EGFP-K2ΔF0ΔPH expression only slightly increased adhesion in the presence of Mn2+ (Fig. 3 D).
To test whether the EGFP-K2ΔF0ΔPH expressing cells also exhibit defects in integrin outside-in signaling, we analyzed spreading of cells seeded for 30 min on FN in the presence or absence of Mn2+. In the absence of Mn2+, T1-mCherry cells expressing EGFP-K2ΔF0 or EGFP-K2ΔPH spread less compared with T1-mCherry cells expressing EGFP-K2 (Fig. 3 E). Expression of EGFP-K2ΔF0ΔPH in T1-mCherry cells increased the spreading area much less efficiently than expression of EGFP-K2ΔF0 or EGFP-K2ΔPH in T1-mCherry (Fig. 3 E). Altogether, these findings indicate that the absence of either the F0 or PH domain of kindlin-2 impairs cell spreading, whereas the lack of both domains further curbs spreading.
Active Rac1 requires kindlin-2 to induce cell spreading
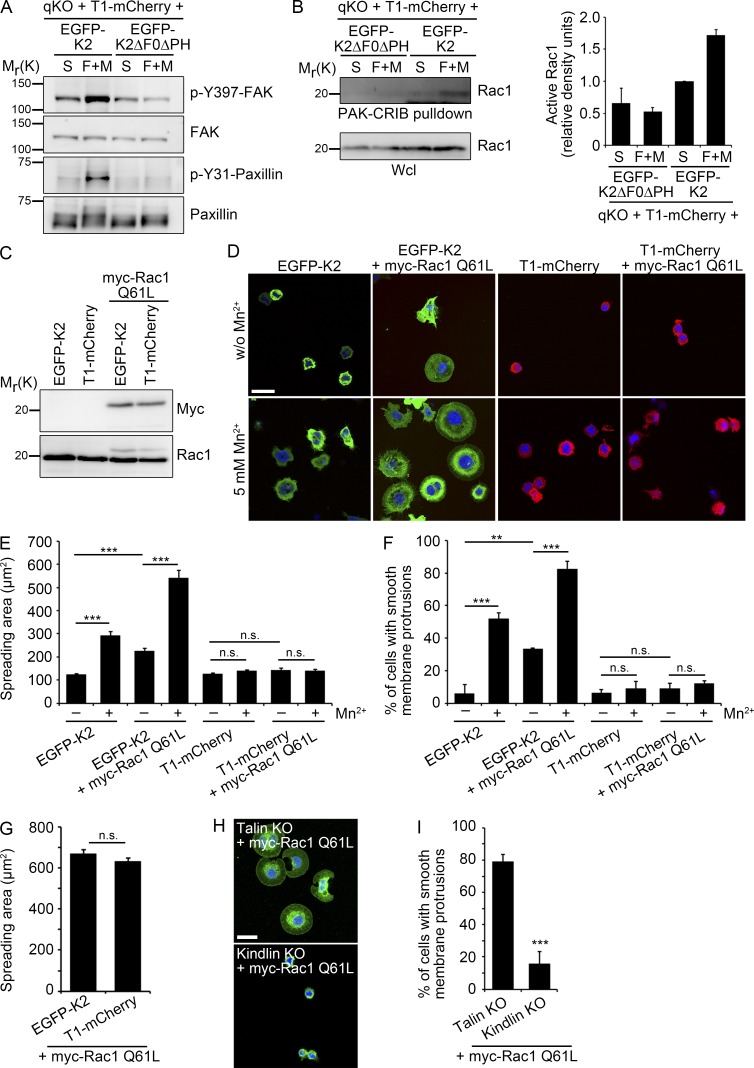
We previously reported that expression of kindlin-2, but not talin-1, enables isotropic cell spreading in the presence of Mn2+ and serum (Theodosiou et al., 2016). Our findings so far revealed that binding of the F0 and PH domains of kindlin-2 to paxillin is required for normal adhesion and spreading of T1-mCherry–expressing cells. Moreover, the absence of F0 and PH domains in kindlin-2 abrogated activation of FAK (Fig. 4 A), which in turn assembles the FAK–Src–p130Cas–Dock180 complex to induce Rac1-mediated cell spreading (Schlaepfer et al., 2004; Brami-Cherrier et al., 2014; Zhang et al., 2014). In line with this finding, the levels of active, GTP-bound Rac1 did not increase in Mn2+-treated T1-mCherry cells adhering to FN and expressing EGFP-K2ΔF0ΔPH (Fig. 4 B).
Figure 4.
The kindlin–paxillin complex induces Rac1 activation and cell spreading. (A) Levels of phosphorylated FAK and paxillin in cell lines kept in suspension (S) or seeded on FN for 30 min in the presence of Mn2+ and serum (F+M). (B) Western blot and densitometric analysis (n = 2) of Rac1 activation in cell lines kept in suspension (S) or seeded on FN for 15 min in the presence of Mn2+ and serum (F+M). The bar chart shows results that were quantified and normalized against total Rac1 levels. (C) Western blot of T1-mCherry and EGFP-K2 cells expressing constitutively active myc-tagged Rac1 (myc-Rac1 Q61L). (D) Fluorescence images of T1-mCherry and EGFP-K2 cells transduced with or without myc-Rac1 Q61L and allowed to spread for 30 min on FN in the absence or presence of 5 mM Mn2+. DAPI was used to stain nuclei. Bar, 25 µm. (E and F) Quantification of spreading area (E) and membrane protrusions (F) of FN-seeded T1-mCherry and EGFP-K2 cells expressing myc-Rac1 Q61L and treated with or without 5 mM Mn2+ 30 min after seeding the cells on FN (n = 3 independent experiments; >90 cells/condition). Error bars indicate SEM; n.s., not significant. (G) Quantification of spreading area 30 min after plating of PLL-seeded T1-mCherry and EGFP-K2 cells expressing myc-Rac1 Q61L (n = 3 independent experiments; >130 cells/condition). Error bars indicate SEM; n.s., not significant. (H) Immunofluorescence images of talin KO and kindlin KO cells transduced with myc-Rac1 Q61L and allowed to spread for 20 min on FN in the presence of 5 mM Mn2+. Cells were stained with an antibody against β1 integrin. DAPI was used to stain nuclei. Bar, 20 µm. (I) Quantification of membrane protrusions of FN-seeded talin KO and kindlin KO cells expressing myc-Rac1 Q61L and treated with 5 mM Mn2+ 20 min after plating (n = 3 independent repeats; >250 cells/condition). Error bars indicate SD. **, P < 0.01; ***, P < 0.001; n.s., not significant.
Next, we tested whether a constitutively active myc-tagged Rac1 Q61L is able to induce spreading of T1-mCherry or K2-EGFP cells. To this end, we retrovirally transduced the cells with Rac1 Q61L (Fig. 4 C) and seeded them on FN without serum or growth factors and in the presence or absence of Mn2+. In the absence of Mn2+, the sporadic and very weakly adherent EGFP-K2 cells remained small and lacked membrane protrusions, whereas Mn2+ treatment improved cell adhesion and induced circumferential membrane protrusions, which further increased upon expression of Rac1 Q61L (Fig. 4, D–F). Interestingly, T1-mCherry–expressing cells failed to form large membrane protrusions on FN and upon expression of Rac1 Q61L, irrespective of whether Mn2+ was absent or present in the culture medium (Fig. 4, D–F), whereas integrin-independent spreading on poly-l-lysine (PLL) was not affected by the absence of kindlin-2 (Fig. 4 G). Importantly, similar results were obtained upon expression of Rac1 Q61L in our previously published talin knockout (KO) cells that express kindlin-2 and kindlin KO cells that express talin-1 (Theodosiou et al., 2016), excluding clonal effects as cause for the absent membrane protrusions in cells coexpressing talin and Rac Q61L (Fig. 4, H and I). Altogether these findings suggest that kindlin-2, aside from operating upstream of Rac1, also provides an additional function to initiate Rac1-induced membrane protrusions, which is lacking in talin-1–expressing T1-mCherry cells.
Kindlin-2 associates with the Arp2/3 complex
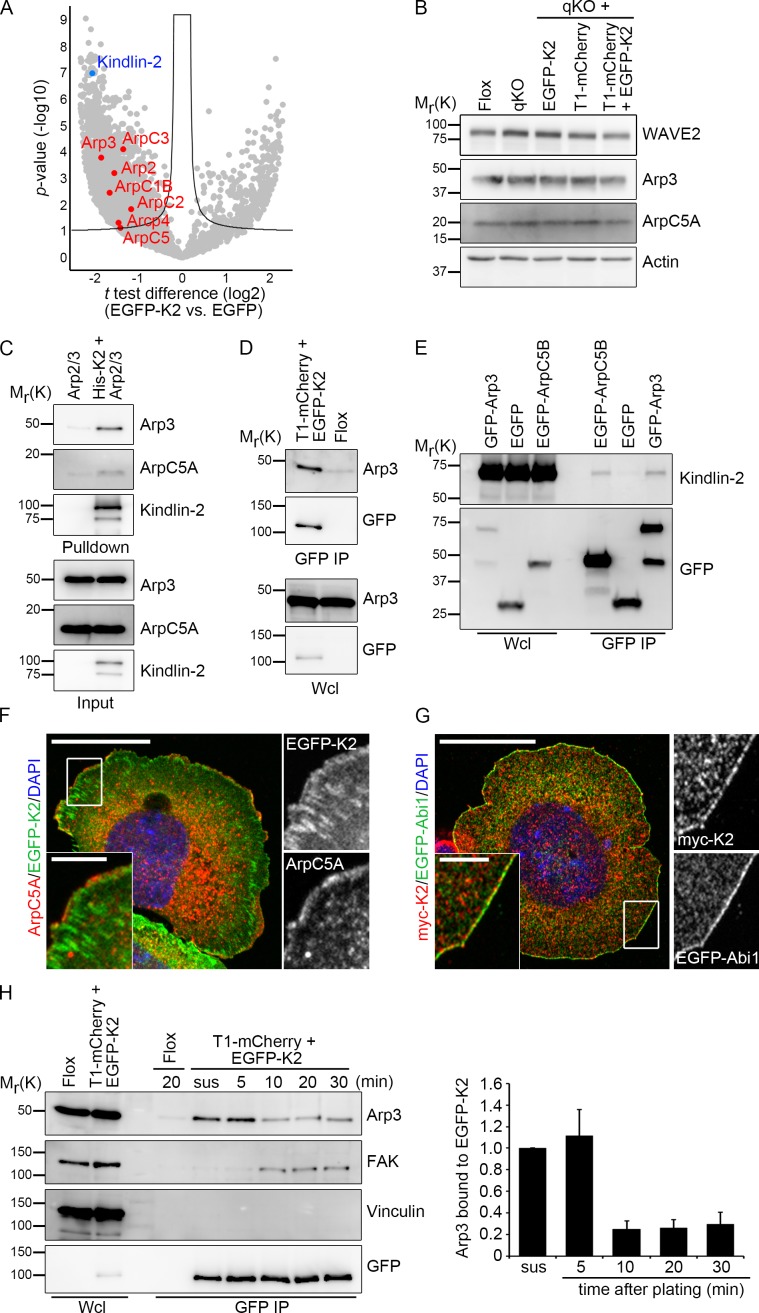
Because kindlin-2, but not talin-1, was able to induce stable membrane protrusions upon expression of Rac1 Q61L, we hypothesized that kindlin-2 harbors an additional functional feature that operates in parallel to the activation of FAK/Rac1. To identify new functional properties of kindlin-2, we screened for novel interaction partners by immunoprecipitation of GFP-tagged kindlin-2 followed by MS. Among proteins that were precipitated with kindlin-2 were subunits of the Arp2/3 complex (Fig. 5 A), which triggers the circumferential membrane protrusion with the characteristic smooth rim during early cell spreading (Suraneni et al., 2012, 2015). Although Western blots revealed similar levels of Arp2/3 and Arp2/3–activating WAVE2 protein in EGFP-K2 and T1-mCherry cells (Fig. 5 B), chemical inhibition of Arp2/3 (Nolen et al., 2009) strongly reduced the formation of lamellipodial protrusions in Mn2+-treated EGFP-K2 cells (Fig. S3 A), highlighting the crucial role of Arp2/3 for kindlin-2–mediated formation of membrane protrusions. Hence, we decided to further analyze the relationship of the two proteins.
Figure 5.
Kindlin-2 binds and recruits the Arp2/3 complex to NAs. (A) Volcano plot showing the t test difference of label-free MS quantification of protein LFQ intensity (log2) of EGFP-K2 versus GFP (control) immunoprecipitates plotted against the p-value (−log10) resulting from a one-sided t test (n = 3 independent experiments for both groups). Kindlin-2 is highlighted in blue and components of the Arp2/3 complex in red. The black curve indicates the significance cutoff (false discovery rate, 0.05; S0:1). (B) Protein levels of WAVE2 and Arp2/3 complex subunits in the indicated cell lines. (C) Ni-NTA pull down of His-tagged K2 incubated with purified Arp2/3 complex followed by Western blot for kindlin-2 and the Arp2/3 complex components Arp3 and ArpC5A. (D) Immunoprecipitation of GFP-kindlin-2 to determine binding to Arp3. (E) Coimmunoprecipitation of Arp2/3 complex subunits and endogenous kindlin-2 from Flox cells. (F) Immunostaining of ArpC5A (red) in T1-mCherry + EGFP-K2–expressing cells seeded on FN-coated coverslips for 20 min before immunostaining. DAPI was used to stain nuclei. Bars: (main) 20 µm; (insets) 4 μm. (G) Colocalization of myc-tagged kindlin-2 and EGFP-Abi1 in cells seeded on FN-coated coverslips for 12 min before fixation and immunostaining with antibodies against myc (red). DAPI was used to stain the nuclei. Bars: (main) 20 µm; (insets) 4 µm. (H) Western blot and densitometric analysis of GFP-immunoprecipitations from EGFP-K2 cells kept in suspension (sus) or seeded into FN-coated dishes for indicated time points (in minutes). Graph shows Arp3 binding to EGFP-K2 relative to suspension cells (n = 3 independent experiments). Error bars indicate SD.
Consistent with the proteomic data, in vitro pull-down assays revealed that recombinant His-tagged kindlin-2 (His-K2) bound the purified Arp2/3 protein complex (Fig. 5 C). Furthermore, GFP-K2 coimmunoprecipitated the Arp2/3 complex (Fig. 5 D), and conversely, Arp2/3 complex members were able to coimmunoprecipitate endogenous kindlin-2 (Fig. 5 E). Finally, kindlin-2 colocalized with the Arp2/3 complex and the WAVE complex component Abi1 in membrane protrusions of spreading cells, but not in FAs (Fig. 5, F and G).
Next, we determined the kinetics of the interaction between kindlin-2 and the Arp2/3 complex before and after plating T1-mCherry + EGFP-K2–expressing cells on FN. The co-IP experiments revealed that the interaction of kindlin-2 with Arp3 was highest in suspended, nonadherent cells and in the first 5 min after seeding and sharply decreased ∼10 min after seeding (Fig. 5 H). Interestingly, neither vinculin nor FAK coimmunoprecipitated with kindlin in suspended and early spreading cells (Fig. 5 H). Furthermore, the association of kindlin-2 and Arp2/3 remained unaffected in vinculin-deficient and FAK-depleted cells (Fig. S3, B and C), which altogether suggests that kindlin-2–Arp2/3 complexes exist independently of vinculin–Arp2/3 (DeMali et al., 2002; Chorev et al., 2014) and FAK–Arp2/3 complexes (Serrels et al., 2007; Swaminathan et al., 2016) to induce membrane protrusions from newly assembled adhesion sites at the periphery of the plasma membrane.
The integrity of the Arp2/3–kindlin-2 complex is required for cell spreading
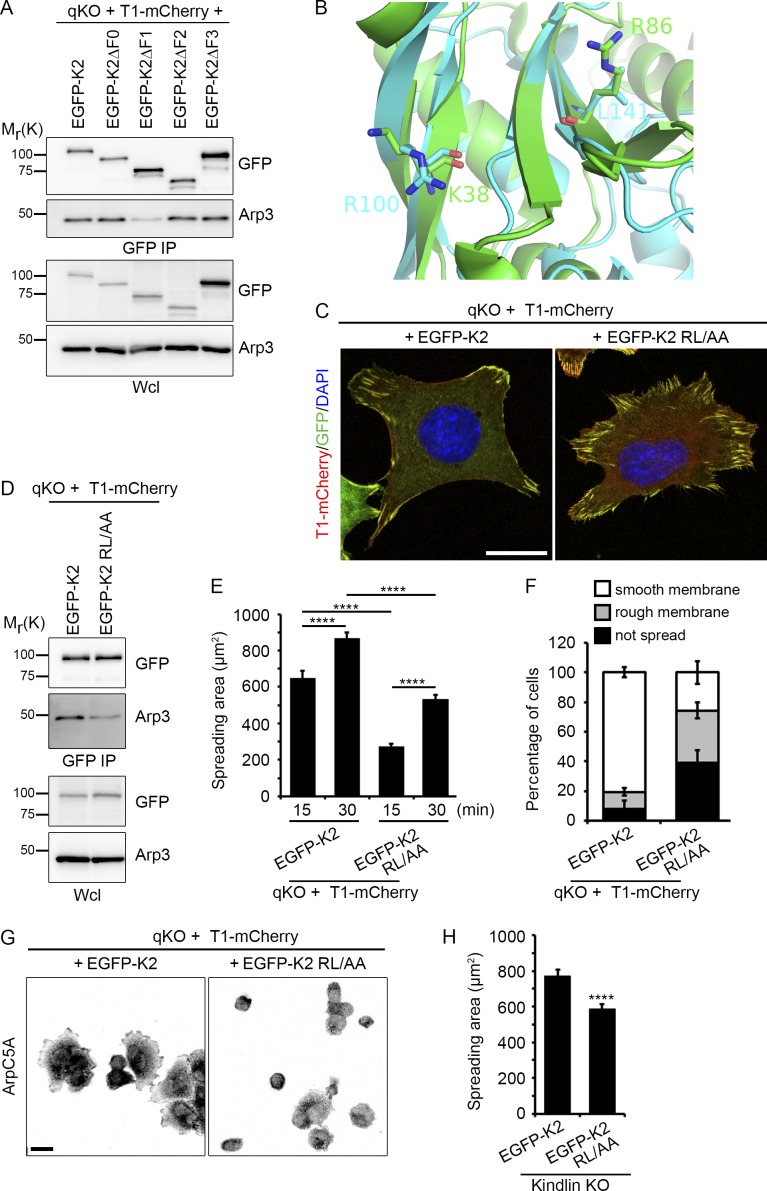
The interaction between FAK and Arp2/3 is mediated by lysine-38 and arginine-86 located in the F1 subdomain of the four-point-one, ezrin, radixin, moesin (FERM) domain of FAK and the Arp2/3 complex (Serrels et al., 2007). We also observed that the F1 subdomain of kindlin-2 was required to efficiently coimmunoprecipitate Arp3 (Fig. 6 A). A superposition of the FAK FERM F1 domain and a model of the mouse kindlin-2 F1 domain suggested that arginine-100 and leucine-141 of kindlin-2 occupy the positions of lysine-38 and arginine-86 in FAK (Fig. 6 B). To test this hypothesis, we generated qKO cells stably expressing T1-mCherry and an EGFP-tagged kindlin-2, in which arginine-100 and leucine-141 were substituted with alanine residues (EGFP-K2 RL/AA). Although EGFP-K2 RL/AA localized to talin-1–containing adhesion structures and allowed cell adhesion (Fig. 6 C), EGFP-K2 RL/AA coimmunoprecipitated less Arp3 than EGFP-K2 (Fig. 6 D).
Figure 6.
Lamellipodia formation and cell spreading requires kindlin-2 binding to Arp2/3. (A) Western blot of GFP immunoprecipitates from the indicated EGFP-K2 mutant–expressing cell lines kept in suspension. (B) Superposition of in silico–modeled murine kindlin-2 F1 domain (cyan) and the FERM domain of avian FAK (PDB: 4CYE, green). The residues K38 and R86 responsible for Arp2/3 interaction in the F1 domain of FAK correspond to residues R100 and L141 in the kindlin-2 F1 domain, as indicated. (C) Localization of T1-mCherry and EGFP-tagged kindlin-2 variants in T1-mCherry–expressing cells seeded for 2 h on FN. DAPI was used to stain nuclei. Bar, 20 µm. (D) Western blot of GFP immunoprecipitates from EGFP-K2 and EGFP-K2 RL/AA cell lines kept in suspension. (E) Quantification of spreading area of T1-mCherry cells expressing EGFP-K2 or EGFP-K2 RL/AA seeded for 15 min or 30 min on FN (n = 3 independent experiments; >90 cells/condition). Error bars indicate SEM. (F) Classification into nonspreading or spreading cells with smooth or rough membrane protrusions of T1-mCherry cells expressing either EGFP-K2 or EGFP-K2 RL/AA and seeded for 30 min on FN. (G) T1-mCherry cells expressing either EGFP-K2 or EGFP-K2 RL/AA were plated for 30 min on FN and then fixed and stained with antibodies against Arpc5A. Bar, 20 µm. (H) Quantification of spreading area of kindlin KO cells expressing either EGFP-K2 or EGFP-K2 RL/AA and seeded for 15 min onto FN (n = 2 independent experiments; 89 cells per condition). Error bars indicate SEM. ****, P < 0.0001.
A time course analysis of cell spreading showed that 30 min after cell seeding, only approximately half of EGFP-K2 RL/AA–expressing cells were spread (Fig. 6, E and F; and Fig. S4, A and B), and only 22% formed ArpC5A-positive, protrusive membranes (22% ± 5%, n = 162) compared with 80% spread EGFP-K2 expressing cells, of which 70% displayed ArpC5A-positive membrane protrusions (70% ± 4%, n = 93; Fig. 6, F and G). A similar spreading defect was observed upon expression of EGFP-K2 RL/AA in an independent kindlin KO cell line (Theodosiou et al., 2016), although the differences were less pronounced (Fig. 6 H). Altogether, these findings indicate that the Arp2/3–kindlin-2 complex promotes the production of protrusive membranes during early cell spreading and lamellipodia formation.
Discussion
In the present study, we report two major findings. First, we identified a previously unrecognized interaction between the kindlin-2 F0 domain and the LIM3/4 domains as well as the LD motifs of paxillin using cross-linking proteomics. A deletion mutant of kindlin-2 lacking the F0 and PH domain failed to bind paxillin and localize to adhesion sites, suggesting that both sites contribute to paxillin binding and integrin-ligand binding, possibly through the recruitment of proteins to the kindlin-2–paxillin complex.
Second, we observed that constitutively active Rac1 (Rac1 Q61L) induced isotropic spreading with circumferential membrane protrusions in Mn2+-treated kindlin/talin–null cells solely reexpressing fluorescently tagged kindlin-2 (EGFP-K2), but not talin-1 (T1-mCherry), cells. This finding strongly suggests that kindlin-2 provides an essential function in addition to Rac1 activation that is not provided by talin-1. MS-based interactome screening revealed that kindlin-2 associates with the Arp2/3 complex, and coimmunostaining revealed colocalization of the two proteins in the smooth rim of the protruding membrane. The accumulation of a functional Arp2/3 complex at the periphery of spreading cells requires a direct association of kindlin-2 with the Arp2/3 complex. Consistently, disruption of this interaction by introducing point mutations into the F1 subdomain of kindlin-2 caused impaired membrane protrusion. The kindlin-2 structure (Kammerer et al., 2017; Li et al., 2017) supports the model in which arginine-100 and leucine-141 of kindlin-2 occupy the positions of lysine-38 and arginine-86 in FAK with respect to Arp2/3 binding. Of note, however, the experimental structure indicates that leucine-141 in kindlin-2 is partly masked by a flexible loop within the kindlin-2 F1 module that locates into a groove between the F1 and F3 modules. It is therefore possible that although the K2 RL/AA substitutions do not diminish binding to β1 integrin tails and ILK, they may destabilize the structure of the Arp2/3 interaction site and hence contribute to the decreased Arp2/3 binding observed in our experiments.
The kinetic interaction studies suggest that kindlin-2–Arp2/3 complexes are preassembled in the cytoplasm and recruited to nascent integrin adhesion sites where the kindlin-2–paxillin complex induces Rac1-WAVE–mediated activation of the Arp2/3 complex and the formation of smooth membrane protrusions. Although Arp2/3 levels were unchanged in T1-mCherry cells and Arp2/3 was previously shown to associate with vinculin and FAK (DeMali et al., 2002; Serrels et al., 2007; Chorev et al., 2014; Swaminathan et al., 2016), which both bind talin, the expression of Rac1 Q61L in T1-mCherry–expressing cells was not sufficient to induce cell spreading and Arp2/3–mediated formation of lamellipodia. It is therefore possible that talin-1 requires a signal (e.g., from kindlin-2 or kindlin-2–bound paxillin) to become functionalized and contribute to cell spreading with Arp2/3 bound to vinculin or FAK. Clearly, the cross talk between kindlin-2 and talin-1 during adhesion and initial spreading requires further cell biological and biochemical investigations.
It is conceivable that kindlin-2 contributes to cell spreading with additional signals or properties. Such a function of kindlin-2 could be clustering of active Rac1 GTPases at the plasma membrane. Phagocytosis was shown to require localized actin polymerization and membrane remodeling, which critically depends on Rac1 activation and Rac1 recruitment to and clustering at the plasma membrane (Castellano et al., 2000). Kindlin-3 has recently been shown to induce clustering of ligand-occupied integrins (Ye et al., 2013). If this function is conserved among all kindlins, then kindlin-2 may aggregate Rac1, helping Rac1 to exceed a threshold required for inducing lamellipodia-driven cell spreading.
In summary, our findings suggest that the preassembled kindlin-2–Arp2/3 complex is recruited to early adhesion sites in the cell periphery to induce initial cell spreading. As soon as the task is accomplished, other FA proteins, including FAK and vinculin, control Arp2/3 activity. It is possible that each FA protein assembles a specific Arp2/3 complex equipped with specific properties (Chorev et al., 2014; Abella et al., 2016) that are determined by subunit isoform compositions and/or additional proteins present in the different complexes (Serrels et al., 2007; Chorev et al., 2014; Abella et al., 2016; Swaminathan et al., 2016). Future studies are required to define whether kindlin-2 associates with the canonical Arp2/3 complex or whether kindlin-2 assembles a specific Arp2/3 complex that initiates but does not complete cell spreading.
Materials and methods
Mouse strains and cell lines
The floxed kindlin-1 (Fermt1flox/flox), kindlin-2 (Fermt2flox/flox), and talin-1 (Tln1flox/flox) mouse strains and the constitutive talin-2–null (Tln2−/−) mouse strain (Nieswandt et al., 2007; Conti et al., 2009; Rognoni et al., 2014; Theodosiou et al., 2016) were intercrossed to isolate kidney fibroblasts from 21-d-old mice. Cells were immortalized by retroviral transduction of the SV40 large T antigen and then cloned (Flox cells). To obtain cells lacking talin-1 and talin-2 as well as kindlin-1 and kindlin-2 (qKO cells) the parental Flox cells with similar integrin surface profiles were adenovirally transduced with Cre. To generate talin and kindlin rescue cell lines, the qKO cells were retrovirally transduced with cDNAs coding for mCherry-tagged talin-1 (T1-mCherry) and EGFP-tagged full-length kindlin-2 (EGFP-K2) or EGFP-tagged kindlin-2 truncation mutants that lacked the F0 domain (EGFP-K2ΔF0), PH domain (EGFP-K2ΔPH), or both domains (EGFP-K2ΔF0ΔPH). Fibroblasts lacking either talins (talin KO) or kindlins (kindlin KO) and myc-kindlin-2 (myc-K2)–reexpressing cells have been described previously (Theodosiou et al., 2016). Vinculin flox (Vincfl/fl) and vinculin-deficient (Vinc−/−) fibroblasts were a gift from C. Grashoff (Max Planck Institute of Biochemistry, Martinsried, Germany; Austen et al., 2015). For stably depleting FAK expression, qKO cells were retrovirally transduced with shRNA target sequences against mouse FAK (5′-GTCCAACTATGAAGTATTA-3′ and 5′-GGTCCAATGACAAGGTATA-3′). All cell lines were cultured in DMEM supplemented with 10% FCS and penicillin/streptomycin.
Transient and stable transfection/transduction
To generate stable cell lines, vesicular stomatitis virus G pseudotyped retroviral vectors were produced by transient transfection of 293T (human embryonic kidney) cells. 48 and 72 h after transfection of the viral packaging plasmids, viral particles were harvested by collecting the cell culture medium. After filtering the collected medium through 0.45-µm filters, viral particles were pelleted by ultracentrifugation at 20,300 rpm for 2 h with a SW 32 Ti rotor (Beckman Coulter) and resuspended in 45 µl cold Hank’s balanced salt solution (14175046; Thermo Fisher Scientific) per 15-cm dish. Cells were transiently transfected with Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Flow cytometry
Flow cytometry was performed with a FACSCanto TMII cytometer (BD Biosciences) equipped with FACS DiVa software (BD Biosciences). Fibroblasts were incubated with primary antibodies diluted in PBS + 1% BSA for 1 h on ice and washed with cold PBS + 1% BSA before incubation with the secondary antibody for 45 min on ice. Data analysis was performed with the FlowJo program (version 9.4.10).
Antibodies and inhibitors
The following antibodies or molecular probes were used at indicated concentrations for Western blot, immunofluorescence (IF), or flow cytometry (FACS): rabbit anti–kindlin-1 (homemade; Ussar et al., 2008) Western blot: 1:5,000; mouse anti–kindlin-2 (MAB2617; Millipore) Western blot: 1:1,000; mouse anti-talin (8D4; Sigma) Western blot: 1:1,000; mouse anti–talin-1 (ab57758; Abcam) Western blot: 1:2,000; mouse anti–talin-2 (ab105458; Abcam) Western blot: 1:2,000; mouse anti-paxillin (610051; BD Transduction Laboratories) Western blot: 1:1,000; rabbit anti-actin (A-2066; Sigma) Western blot: 1:1,000; mouse anti-Arp3 (A5979; Sigma) Western blot: 1:1,000; mouse anti-ArpC5 (Olazabal et al., 2002) Western blot: 1:10, IF 1:2; hamster anti–integrin β1-488 (102211; BioLegend) FACS: 1:200; rat anti–integrin β1 (MAB1997; Chemicon) FACS: 1:400; rabbit anti–integrin β1 (homemade; Azimifar et al., 2012) IF: 1:400, hamster anti–integrin β3-biotin (553345; PharMingen) FACS: 1:200; rat anti–integrin α5-biotin (557446; PharMingen) FACS: 1:200; rat anti–integrin αv-biotin (551380; PharMingen) FACS: 1:200; rat IgG2a isotype control (13-4321; eBioscience) FACS: 1:200; rabbit anti-GFP (A11122; Invitrogen) Western blot: 1:2,000; rabbit anti-Cherry (PM005; MBL International) Western blot: 1:1,000; mouse anti-myc (4A6; Millipore) Western blot: 1:1,000, IF 1:300; mouse anti–Rac1 (R56220; BD Biosciences) Western blot: 1:1,000; and rabbit anti-WAVE2 (3659; Cell Signaling) Western blot: 1:1,000.
The following secondary antibodies were used: goat anti–rabbit Alexa Fluor 488 (A11008), goat anti–mouse Alexa Fluor 488 (A11029), goat anti–rat Alexa Fluor 488 (A11006), goat anti–mouse Alexa Fluor 546 (A11003), donkey anti–mouse Alexa Fluor 647 (A31571), goat anti–rabbit Alexa Fluor 647 (A21244; all from Thermo Fisher Scientific) FACS: 1:500, IF: 1:500; streptavidin-Cy5 (016170084) FACS: 1:400; goat anti–rat HRP (712035150; both from Dianova) Western blot: 1:10,000, donkey anti–rabbit Cy3 (711-165-152; Jackson ImmunoResearch) IF: 1:500, goat anti–mouse HRP (172-1011), and goat anti–rabbit HRP (172-1019; both from Bio-Rad) Western blot: 1:10,000.
Phalloidin–Alexa Fluor 647 (A22287; Thermo Fisher Scientific) and DAPI (Sigma) were used to stain F-actin and nuclei, respectively. The Arp2/3 complex inhibitors CK-666 (SML0006; Sigma) and CK-869 (C9125; Sigma) were dissolved in dimethyl sulfoxide at 50 mg/ml.
Plasmids, constructs, and expression and purification of recombinant proteins
Mouse kindlin-2 complementary DNAs (full length: amino acids 1–680, K2ΔF0: deletion of amino acids 1–91, K2ΔF1: deletion of amino acids 96–272, K2ΔF2: deletion of amino acids 271–559, K2ΔF3: deletion of amino acids 571–680, K2ΔPH: deletion of amino acids 381–476, K2ΔF0ΔPH: deletions of amino acids 1–91 and 381–476, and K2 RL/AA, in which R100 and L141 were substituted with alanine) were cloned into pEGFP-C1 vector (Clontech) using XhoI and BamHI sites. For retrovirus-mediated expression, cDNAs of EGFP-K2, EGFP-K2ΔF0, EGFP-K2ΔF1, EGFP-K2ΔF2, EGFP-K2ΔF3, EGFP-K2ΔPH, EGFP-K2ΔF0ΔPH, and EGFP-K2 RL/AA were inserted between NheI and BamHI sites of the pRetroQ-AcGFP-C1 (Clontech) vector. Stable expression of myc-Kindlin2 cDNA was achieved with the sleeping beauty transposase system using ITR-Puro (+) plasmid containing the mouse kindlin-2 cDNA. Myc-tagged Rac1 Q61L was cloned into the retroviral vector pCLMFG to generate stable cell lines. EGFP-tagged variants of Abi1 and the Arp2/3 complex subunit ArpC5B were as described previously (Lai et al., 2008). Constructs of murine paxillin (full length: amino acids 1–557) and kindlin-2 (full length: amino acids 1–680, K2 F0: amino acids 1–97, K2 PH: amino acids 386–496, K2ΔPH: deletion of amino acids 372–500, K2ΔF0ΔPH: deletions of amino acids 1–97 and 372–500 and K2 F0-PH: amino acids 1–97 and 386–496 fused by a flexible linker [SGGGGTSGGGG]) were cloned into the AgeI–XhoI site of pCoofy17 (Max Planck Institute of Biochemistry core facility), yielding proteins N-terminally tagged with a 10× histidine tag followed by a SUMO3 tag. Production of recombinant proteins in Escherichia coli Rosetta cells (Millipore) was induced by addition of IPTG to a final concentration of 0.2 mM at 18°C for 24 h. After cell lysis and clarification of the supernatant, the proteins were purified by Ni-NTA affinity chromatography (Qiagen). Eluate fractions containing the protein of interest were pooled, cleaved with SenP2 protease, and subsequently purified by size exclusion chromatography (Superdex 200 16/60; GE Healthcare).
The Arp2/3 complex was affinity purified from pig brain as described previously (Block et al., 2012). In brief, the brain was homogenized with a blender in the presence of a fivefold excess (vol/vol) of ice-cold extraction buffer containing 20 mM Tris-HCl, pH 7.5, 25 mM KCl, 2 mM DTT, 1 mM MgCl2, 0.5 mM EDTA, and 0.1 mM ATP and the homogenate centrifuged at 20,000 g for 45 min. The cleared supernatant was loaded on a GST-WAVE1-VCA (amino acids 492–559) glutathione Sepharose column, and after extensive washing of the column with extraction buffer, the remaining non–Arp2/3 proteins were eluted by 0.2 M KCl in extraction buffer. The Arp2/3 complex was subsequently eluted with 0.2 M MgCl2 in extraction buffer and further purified by size exclusion chromatography using an Äkta Purifier System equipped with a HiLoad 26/600 Superdex 200 column (GE Healthcare) equilibrated with elution buffer. Fractions containing the Arp2/3 complex were pooled, dialyzed against a storage buffer (150 mM KCl, 1 mM DTT, 55% glycerol, and 20 mM Hepes, pH 7.4), and stored at −20°C.
Chemical cross-linking and MS of the kindlin-2–paxillin complex
The kindlin-2–paxillin complex was purified on a size exclusion column (SEC650; Bio-Rad) equilibrated with XL buffer (25 mM Hepes, pH 8, 300 mM NaCl, 5% glycerol, 0.05% Tween 20, 1 mM tris[2-carboxyethyl]phosphine [TCEP], and 10 µM ZnCl2). The used fraction showed a 1:1 stoichiometry of kindlin-2 and paxillin and a concentration of 60 µg/ml. This solution was supplemented with 10 µM of isotopically light (d0)– and heavy (d12)–labeled bissulfosuccinimidyl suberate cross-linker (Creative Molecules Inc.) and allowed to react on ice for 18 h before stopping the cross-linking reaction with 5 mM ammonium bicarbonate. Cross-linked proteins were enzymatically digested, and cross-linked peptides were identified by tandem MS as reported previously (Herzog et al., 2012). In brief, cross-linked proteins were denatured by adding two sample volumes of 8 M urea and reduced by incubating with 5 mM TCEP (Thermo Fisher Scientific) at 35°C for 15 min. Proteins were alkylated with 10 mM iodoacetamide (Sigma-Aldrich) for 35 min at RT in the dark. Proteins were proteolytically digested by adding lysyl endopeptidase (1/50 [wt/wt]; Wako) for 2 h at 35°C followed by the addition of trypsin (1/50 [wt/wt]; Promega) overnight at a final concentration of 1 M urea. Proteolysis was stopped by the addition of 1% (vol/vol) trifluoroacetic acid. Acidified peptides were purified by reversed-phase chromatography on C18 columns (Sep-Pak, Waters). Eluates were dried and reconstituted in 20 µl of mobile phase (water/acetonitrile/trifluoroacetic acid, 75:25:0.1) and cross-linked peptides were enriched on a Superdex Peptide PC 3.2/30 column (GE Healthcare) at a flow rate of 25 µl/min. Fractions of the cross-linked peptides were dried and reconstituted in 20 µl 2% acetonitrile and 0.2% formic acid and analyzed by liquid chromatography coupled to tandem MS using a LTQ Orbitrap Elite (Thermo Fisher Scientific) instrument. The cross-link fragment ion spectra were searched and the peptides identified by the open-source software xQuest (Walzthoeni et al., 2012). False discovery rates calculated by xProphet were ≤0.05, and results were filtered according to the following parameters: delta score ≤0.85, MS1 tolerance window of −4 to 4 ppm, and score >22.
Analytical ultracentrifugation
To qualitatively determine the binding of different kindlin-2 constructs to paxillin, the proteins of interest were labeled using Atto520-NHS-ester (ATTOTec) according to the manufacturer’s instructions. The measurements were conducted using an analytical Optima XL-I centrifuge (Beckman-Coulter) at 14°C and 50,000 rpm/201,600 g, and sedimentation data were recorded at the wavelength of maximum absorption of the dye. The data were analyzed using SedFit (Schuck, 2000) in continuous sedimentation coefficient distribution mode.
ITC
Quantitative ITC measurements were conducted using a PeaqITC instrument (Malvern) at a constant jacket temperature of 14°C. Proteins were rebuffered in 20 mM Tris, pH 7.5, 200 mM NaCl, 1 mM TCEP, and 10 µM ZnCl2. 12 × 3 µl of the respective kindlin-2 construct was injected into the measurement cell containing paxillin. To show the Zn2+ dependence of the interaction, 5 mM EDTA was added to the sample cell. All data were analyzed using MicroCal PeaqITC Analysis software.
Modeling of the kindlin-2 F1 structure
A kindlin-2 structural model was generated by I-tasser (Roy et al., 2010) using the talin-head FERM domain (Protein Data Bank [PDB]: 3IVF; Elliott et al., 2010) as search model. A version of the kindlin-2 F1 domain structure obtained by this approach was aligned to a structure of avian FAK (PDB: 4CYE) using PyMol (Molecular Graphics System, Version 1.6; Schrodinger, LLC). The obtained structural root mean square deviations were 2.259 Å for one molecule in the asymmetric unit and 2.106 Å for the second molecule in the asymmetric unit of 4CYE, respectively.
Immunostaining
For immunostaining, cells were cultured on plastic ibidi µ-Slides (80826) or glass coverslips coated with 10 µg/ml FN (Calbiochem). Cells were routinely fixed with 4% PFA (wt/vol) in PBS (180 mM NaCl, 3.5 mM KCl, 10 mM Na2HPO4, and 1.8 mM K2H2PO4) for 10 min at RT and permeabilized for 10 min with 0.1% Triton X-100/PBS on ice. Background signals were blocked by incubating cells for 1 h at RT in 3% BSA/PBS. Subsequently, they were incubated in the dark with primary and secondary antibodies diluted in 3% BSA/PBS, stained with DAPI, and mounted with Elvanol. Fluorescent images were acquired with a LSM 780 confocal microscope (Zeiss) equipped with 100×/NA 1.46 and 40×/NA 1.4 oil objectives and operated by Zen software (version 2.1; Zeiss). Image acquisition was performed at ambient temperature. Images were processed and analyzed with ImageJ (National Institutes of Health) or Photoshop (Adobe).
Immunoprecipitations and pull-down assays
For immunoprecipitation of paxillin, cells were lysed in β1 lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.05% sodium deoxycholate, and 10 mM EDTA) and incubated with paxillin antibodies for 2 h at 4°C while rotating. Subsequently, lysates were incubated with 50 µl protein A/G Plus Agarose (Santa Cruz Biotechnology) for 2 h at 4°C. After repeated washes with lysis buffer, proteins were eluted from the beads using Laemmli buffer, separated on a 10% SDS-PAGE, and analyzed by Western blotting.
For immunoprecipitation of GFP-tagged proteins, cells were lysed in M-PER (78501; Thermo Fisher Scientific) or β1 lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.05% sodium deoxycholate, and 10 mM EDTA) and proteins were immunoprecipitated using the μMACS GFP Isolation kit (130-091-288; Miltenyi Biotec) for 20 min on ice following the manufacturer’s protocol.
For the Ni pull down, 10 µg recombinant His–kindlin-2 was bound to 30 µl Ni Sepharose (GE Healthcare) in 0.5 ml M-PER buffer (78501; Thermo Fisher Scientific) for 1–2 h at 4°C under rotation. After several washes, the beads were incubated with 12.5 µg purified Arp2/3 complex in 0.5 ml M-PER buffer containing 80 mM imidazole for 2 h at 4°C under rotation. After three washes with M-PER buffer containing 80 mM imidazole, proteins were eluted by boiling the samples for 7 min at 95°C in Laemmli sample buffer and analyzed by Western blot using antibodies against kindlin-2, ArpC5A, and Arp3.
Kindlin-2 interactome analysis by MS
qKO cells expressing T1-mCherry and EGFP-K2 or EGFP were kept in suspension for 20 min at 37°C. Cells were washed in PBS and lysed in M-PER before incubation with 50 µl magnetic beads coupled to monoclonal mouse anti-GFP antibody (130-091-288; Miltenyi Biotec) for 20 min on ice. Cell lysates were added to magnetic columns and washed four times with M-PER buffer and once with wash buffer II containing 20 mM Tris, pH 7.5 (Miltenyi Biotec). Purified proteins were predigested on-column by adding 25 µl 2 M urea in 50 mM Tris, pH 7.5, 1 mM dithiothreitol, and 150 ng Trypsin for 30 min at RT and eluted by adding two times 50 µl 2 M urea in 50 mM Tris, pH 7.5, and 5 mM chloroacetamide. After overnight incubation at RT, the digestion was stopped by adding 1 µl trifluoroacetic acid, and the peptides were purified using C18 stage tips (Rappsilber et al., 2007). The samples were loaded on a 15-cm column packed with 1.9-μm C18 beads (Dr Maisch GmbH) via the nanoLC-1200 autosampler (Thermo Fisher Scientific) and sprayed directly into Q Exactive HF mass spectrometer (Thermo Fisher Scientific). The mass spectrometer was operated in data-dependent mode with the Top N acquisition method. The raw data were processed using the MaxQuant computational platform (Cox and Mann, 2008). The peak lists generated were searched with initial precursor and fragment mass tolerance of 7 and 20 ppm, respectively. Carbamidomethylation of cysteine was used as static modification, and oxidation of methionine and protein N-terminal acetylation was used as variable modification. The peak lists were searched against the UniProt Mouse database, and the proteins and peptides were filtered at a 1% false discovery rate. Proteins were quantified using the MaxLFQ algorithm in MaxQuant (Cox et al., 2014).
Spreading and adhesion assays
Cells were grown to 70% confluence followed by overnight incubation in DMEM containing 0.2% FCS. After detaching with trypsin/EDTA, cells were serum starved for 1 h in adhesion assay medium (10 mM Hepes, pH 7.4, 137 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 2.7 mM KCl, 4.5 g/l glucose, and 3% BSA [wt/vol]) in bacterial dishes. Flat-bottom 96-well plates were coated with 10 µg/ml FN (Calbiochem), 1 µg/ml VN (07180; StemCell), or 5% BSA diluted in PBS. Cells were then plated (40,000 per well) in adhesion buffer supplemented with 8% FCS and 5 mM Mn2+ where indicated and incubated for 30 min at 37°C. After vigorous washing with PBS, cell attachment was measured by fixing and staining with crystal violet staining (0.1% in 20% methanol) in an absorbance plate reader at a wavelength of 570 nm.
For cell spreading, 40,000 cells were seeded on 10 µg/ml FN or PLL (0.01% wt/vol)–coated coverslips, cultured for the indicated time points at 37°C, fixed with 4% PFA (wt/vol) in PBS, and stained with phalloidin–Alexa Fluor 647 and DAPI. At least 10 images were taken using a Zeiss AxioImager Z1 microscope equipped with a 63×/NA 1.4 oil objective, and cell spreading area was measured using AxioVision software (release 4.8.2; Zeiss).
For phase contrast analysis of spreading cells, 50,000 cells were seeded on 10 µg/ml FN-coated six-well plates and cultured at 37°C. Living cells were imaged with an EVOS FL Auto Cell Imaging System (Thermo Fisher Scientific) equipped with an EVOS Onstage Incubator using a 20×/NA 0.4 objective at 37°C. Images were analyzed with ImageJ (National Institutes of Health).
Rac1 GTPase activity
Cells were grown in DMEM containing 0.2% FCS overnight, detached with trypsin–EDTA, and starved for 1 h in adhesion assay medium in bacterial dishes. Cells were then plated on 10 µg/ml FN–coated dishes in serum-free medium containing 5 mM Mn2+. Cell lysis and active Rac1-GTPase pull-down was performed using the active Rac1 Pull-Down and Detection kit (16118; Thermo Fisher Scientific) according to manufacturer’s instructions. The active Rac1 signal was normalized to the total protein level of the GTPase.
Statistical analysis
All experiments were repeated at least three times (as indicated in figure legends). Statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n.s., not significant) was determined by two-tailed unpaired t test and performed with Prism (GraphPad).
Online supplemental material
Fig. S1 shows the intra- and interprotein cross-links between kindlin-2 and paxillin as well as the Zn2+ dependence of kindlin–paxillin binding. Fig. S2 shows the FA recruitment of different kindlin-2 deletions. Fig. S3 displays the kindlin-2–Arp2/3 interaction in the absence of either vinculin or FAK. Fig. S4 shows the spreading behavior of kindlin-2 RL/AA–expressing cells. Supplemental data 1 is a detailed overview of the high-confidence lysine–lysine cross-links of the kindlin-2–paxillin complex.
Supplementary Material
Acknowledgments
We thank Hildegard Reiter for expert technical help and Carsten Grashoff for providing the vinculin KO cells.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 914, project A05), the European Research Council (grant 322652), and the Max Planck Society (R. Fässler).
The authors declare no competing financial interests.
Author contributions: R.T. Böttcher and M. Veldeers carried out the experiments and analyzed the data. P. Rombaut and F. Herzog performed the cross-linking proteomics. R. Zent and M. Theodosiou generated the flox and qKO fibroblasts. J. Faix, T.E. Stradal, and K. Rottner provided tools and analyzed the data. R.T. Böttcher and R. Fässler initiated, supervised, and designed the present study and wrote the manuscript. The manuscript was read and approved by all authors.
Footnotes
Abbreviations used:
- co-IP
- coimmunoprecipitation
- FA
- focal adhesion
- FN
- fibronectin
- IF
- immunofluorescence
- ITC
- isothermal titration calorimetry
- KO
- knockout
- MS
- mass spectrometry
- NA
- nascent adhesion
- PH
- pleckstrin homology
- PLL
- poly-l-lysine
- qKO
- quadruple knockout
- TCEP
- tris(2-carboxyethyl)phosphine
- VN
- vitronectin
- XL-MS
- cross-linking mass spectrometry
References
- Abella J.V.G., Galloni C., Pernier J., Barry D.J., Kjær S., Carlier M.-F., and Way M.. 2016. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat. Cell Biol. 18:76–86. 10.1038/ncb3286 [DOI] [PubMed] [Google Scholar]
- Austen K., Ringer P., Mehlich A., Chrostek-Grashoff A., Kluger C., Klingner C., Sabass B., Zent R., Rief M., and Grashoff C.. 2015. Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat. Cell Biol. 17:1597–1606. 10.1038/ncb3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimifar S.B., Böttcher R.T., Zanivan S., Grashoff C., Krüger M., Legate K.R., Mann M., and Fässler R.. 2012. Induction of membrane circular dorsal ruffles requires co-signalling of integrin-ILK-complex and EGF receptor. J. Cell Sci. 125:435–448. 10.1242/jcs.091652 [DOI] [PubMed] [Google Scholar]
- Block J., Breitsprecher D., Kühn S., Winterhoff M., Kage F., Geffers R., Duwe P., Rohn J.L., Baum B., Brakebusch C., et al. 2012. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr. Biol. 22:1005–1012. 10.1016/j.cub.2012.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brami-Cherrier K., Gervasi N., Arsenieva D., Walkiewicz K., Boutterin M.-C., Ortega A., Leonard P.G., Seantier B., Gasmi L., Bouceba T., et al. 2014. FAK dimerization controls its kinase-dependent functions at focal adhesions. EMBO J. 33:356–370. 10.1002/embj.201386399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugyi B., and Carlier M.-F.. 2010. Control of actin filament treadmilling in cell motility. Annu. Rev. Biophys. 39:449–470. 10.1146/annurev-biophys-051309-103849 [DOI] [PubMed] [Google Scholar]
- Castellano F., Montcourrier P., and Chavrier P.. 2000. Membrane recruitment of Rac1 triggers phagocytosis. J. Cell Sci. 113:2955–2961. [DOI] [PubMed] [Google Scholar]
- Chorev D.S., Moscovitz O., Geiger B., and Sharon M.. 2014. Regulation of focal adhesion formation by a vinculin-Arp2/3 hybrid complex. Nat. Commun. 5:3758 10.1038/ncomms4758 [DOI] [PubMed] [Google Scholar]
- Cluzel C., Saltel F., Lussi J., Paulhe F., Imhof B.A., and Wehrle-Haller B.. 2005. The mechanisms and dynamics of (α)v(β)3 integrin clustering in living cells. J. Cell Biol. 171:383–392. 10.1083/jcb.200503017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti F.J., Monkley S.J., Wood M.R., Critchley D.R., and Müller U.. 2009. Talin 1 and 2 are required for myoblast fusion, sarcomere assembly and the maintenance of myotendinous junctions. Development. 136:3597–3606. 10.1242/dev.035857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., and Mann M.. 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 13:2513–2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMali K.A., Barlow C.A., and Burridge K.. 2002. Recruitment of the Arp2/3 complex to vinculin: Coupling membrane protrusion to matrix adhesion. J. Cell Biol. 159:881–891. 10.1083/jcb.200206043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P., and Horwitz A.R.. 2015. Signaling networks that regulate cell migration. Cold Spring Harb. Perspect. Biol. 7:a005959 10.1101/cshperspect.a005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott P.R., Goult B.T., Kopp P.M., Bate N., Grossmann J.G., Roberts G.C.K., Critchley D.R., and Barsukov I.L.. 2010. The Structure of the talin head reveals a novel extended conformation of the FERM domain. Structure. 18:1289–1299. 10.1016/j.str.2010.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lim C.J., Watanabe N., Soriani A., Ratnikov B., Calderwood D.A., Puzon-McLaughlin W., Lafuente E.M., Boussiotis V.A., Shattil S.J., and Ginsberg M.H.. 2006. Reconstructing and deconstructing agonist-induced activation of integrin αIIbβ3. Curr. Biol. 16:1796–1806. 10.1016/j.cub.2006.08.035 [DOI] [PubMed] [Google Scholar]
- Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F.-U., Ban N., et al. 2012. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 337:1348–1352. 10.1126/science.1221483 [DOI] [PubMed] [Google Scholar]
- Kammerer P., Aretz J., and Fässler R.. 2017. Lucky kindlin: A cloverleaf at the integrin tail. Proc. Natl. Acad. Sci. USA. 114:9234–9236. 10.1073/pnas.1712471114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F.P.L., Szczodrak M., Block J., Faix J., Breitsprecher D., Mannherz H.G., Stradal T.E.B., Dunn G.A., Small J.V., and Rottner K.. 2008. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 27:982–992. 10.1038/emboj.2008.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M., and Aebersold R.. 2010. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics. 9:1634–1649. 10.1074/mcp.R000001-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Deng Y., Sun K., Yang H., Liu J., Wang M., Zhang Z., Lin J., Wu C., Wei Z., and Yu C.. 2017. Structural basis of kindlin-mediated integrin recognition and activation. Proc. Natl. Acad. Sci. USA. 114:9349–9354. 10.1073/pnas.1703064114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L.M., Atkinson S.J., Ampe C., Vandekerckhove J., and Pollard T.D.. 1994. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J. Cell Biol. 127:107–115. 10.1083/jcb.127.1.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M., Nieswandt B., Ussar S., Pozgajova M., and Fässler R.. 2008. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat. Med. 14:325–330. 10.1038/nm1722 [DOI] [PubMed] [Google Scholar]
- Mould A.P., Akiyama S.K., and Humphries M.J.. 1995. Regulation of integrin α 5 β 1-fibronectin interactions by divalent cations. Evidence for distinct classes of binding sites for Mn2+, Mg2+, and Ca2+. J. Biol. Chem. 270:26270–26277. 10.1074/jbc.270.44.26270 [DOI] [PubMed] [Google Scholar]
- Mullins R.D., Heuser J.A., and Pollard T.D.. 1998. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA. 95:6181–6186. 10.1073/pnas.95.11.6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieswandt B., Moser M., Pleines I., Varga-Szabo D., Monkley S., Critchley D., and Fässler R.. 2007. Loss of talin1 in platelets abrogates integrin activation, platelet aggregation, and thrombus formation in vitro and in vivo. J. Exp. Med. 204:3113–3118. 10.1084/jem.20071827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., and Pollard T.D.. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034. 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olazabal I.M., Caron E., May R.C., Schilling K., Knecht D.A., and Machesky L.M.. 2002. Rho-kinase and myosin-II control phagocytic cup formation during CR, but not FcγR, phagocytosis. Curr. Biol. 12:1413–1418. 10.1016/S0960-9822(02)01069-2 [DOI] [PubMed] [Google Scholar]
- Petrie R.J., Doyle A.D., and Yamada K.M.. 2009. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 10:538–549. 10.1038/nrm2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., and Borisy G.G.. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., and Ishihama Y.. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Rognoni E., Widmaier M., Jakobson M., Ruppert R., Ussar S., Katsougkri D., Böttcher R.T., Lai-Cheong J.E., Rifkin D.B., McGrath J.A., and Fässler R.. 2014. Kindlin-1 controls Wnt and TGF-β availability to regulate cutaneous stem cell proliferation. Nat. Med. 20:350–359. 10.1038/nm.3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., and Kirschner M.W.. 1999. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 97:221–231. 10.1016/S0092-8674(00)80732-1 [DOI] [PubMed] [Google Scholar]
- Rouiller I., Xu X.-P., Amann K.J., Egile C., Nickell S., Nicastro D., Li R., Pollard T.D., Volkmann N., and Hanein D.. 2008. The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 180:887–895. 10.1083/jcb.200709092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Kucukural A., and Zhang Y.. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer D.D., Mitra S.K., and Ilic D.. 2004. Control of motile and invasive cell phenotypes by focal adhesion kinase. Biochim. Biophys. Acta. 1692:77–102. 10.1016/j.bbamcr.2004.04.008 [DOI] [PubMed] [Google Scholar]
- Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78:1606–1619. 10.1016/S0006-3495(00)76713-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrels B., Serrels A., Brunton V.G., Holt M., McLean G.W., Gray C.H., Jones G.E., and Frame M.C.. 2007. Focal adhesion kinase controls actin assembly via a FERM-mediated interaction with the Arp2/3 complex. Nat. Cell Biol. 9:1046–1056. 10.1038/ncb1626 [DOI] [PubMed] [Google Scholar]
- Suraneni P., Rubinstein B., Unruh J.R., Durnin M., Hanein D., and Li R.. 2012. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 197:239–251. 10.1083/jcb.201112113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P., Fogelson B., Rubinstein B., Noguera P., Volkmann N., Hanein D., Mogilner A., and Li R.. 2015. A mechanism of leading-edge protrusion in the absence of Arp2/3 complex. Mol. Biol. Cell. 26:901–912. 10.1091/mbc.E14-07-1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan V., Fischer R.S., and Waterman C.M.. 2016. The FAK-Arp2/3 interaction promotes leading edge advance and haptosensing by coupling nascent adhesions to lamellipodia actin. Mol. Biol. Cell. 27:1085–1100. 10.1091/mbc.E15-08-0590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T., and Suetsugu S.. 2007. The WASP-WAVE protein network: Connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8:37–48. 10.1038/nrm2069 [DOI] [PubMed] [Google Scholar]
- Theodosiou M., Widmaier M., Böttcher R.T., Rognoni E., Veelders M., Bharadwaj M., Lambacher A., Austen K., Müller D.J., Zent R., and Fässler R.. 2016. Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. eLife. 5:e10130 10.7554/eLife.10130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussar S., Moser M., Widmaier M., Rognoni E., Harrer C., Genzel-Boroviczeny O., and Fässler R.. 2008. Loss of Kindlin-1 causes skin atrophy and lethal neonatal intestinal epithelial dysfunction. PLoS Genet. 4:e1000289 10.1371/journal.pgen.1000289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., and Horwitz A.R.. 2011. Adhesion dynamics at a glance. J. Cell Sci. 124:3923–3927. 10.1242/jcs.095653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzthoeni T., Claassen M., Leitner A., Herzog F., Bohn S., Förster F., Beck M., and Aebersold R.. 2012. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat. Methods. 9:901–903. 10.1038/nmeth.2103 [DOI] [PubMed] [Google Scholar]
- Welch M.D., Iwamatsu A., and Mitchison T.J.. 1997. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 385:265–269. 10.1038/385265a0 [DOI] [PubMed] [Google Scholar]
- Winter D., Podtelejnikov A.V., Mann M., and Li R.. 1997. The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr. Biol. 7:519–529. 10.1016/S0960-9822(06)00223-5 [DOI] [PubMed] [Google Scholar]
- Winter D., Lechler T., and Li R.. 1999. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr. Biol. 9:501–504. 10.1016/S0960-9822(99)80218-8 [DOI] [PubMed] [Google Scholar]
- Ye F., Petrich B.G., Anekal P., Lefort C.T., Kasirer-Friede A., Shattil S.J., Ruppert R., Moser M., Fässler R., and Ginsberg M.H.. 2013. The mechanism of kindlin-mediated activation of integrin αIIbβ3. Curr. Biol. 23:2288–2295. 10.1016/j.cub.2013.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Moore S.W., Iskratsch T., and Sheetz M.P.. 2014. N-WASP-directed actin polymerization activates Cas phosphorylation and lamellipodium spreading. J. Cell Sci. 127:1394–1405. 10.1242/jcs.134692 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.