Hippocampal CA1 region neurons specifically target latrophilin-2 (Lphn2), an adhesion-type GPCR, to dendritic spines in the stratum lacunosum-moleculare. In this study, Lphn2 controls assembly of excitatory synapses formed by presynaptic entorhinal cortex afferents but not by Schaffer-collateral afferents, suggesting a synaptic recognition function.
Abstract
Synapse assembly likely requires postsynaptic target recognition by incoming presynaptic afferents. Using newly generated conditional knock-in and knockout mice, we show in this study that latrophilin-2 (Lphn2), a cell-adhesion G protein–coupled receptor and presumptive α-latrotoxin receptor, controls the numbers of a specific subset of synapses in CA1-region hippocampal neurons, suggesting that Lphn2 acts as a synaptic target-recognition molecule. In cultured hippocampal neurons, Lphn2 maintained synapse numbers via a postsynaptic instead of a presynaptic mechanism, which was surprising given its presumptive role as an α-latrotoxin receptor. In CA1-region neurons in vivo, Lphn2 was specifically targeted to dendritic spines in the stratum lacunosum-moleculare, which form synapses with presynaptic entorhinal cortex afferents. In this study, postsynaptic deletion of Lphn2 selectively decreased spine numbers and impaired synaptic inputs from entorhinal but not Schaffer-collateral afferents. Behaviorally, loss of Lphn2 from the CA1 region increased spatial memory retention but decreased learning of sequential spatial memory tasks. Thus, Lphn2 appears to control synapse numbers in the entorhinal cortex/CA1 region circuit by acting as a domain-specific postsynaptic target-recognition molecule.
Introduction
Synapse formation and maintenance play key roles in the remarkable specificity of neuronal wiring in the brain and are likely controlled, at least in part, by cell-adhesion molecules that bridge the synaptic cleft between pre- and postsynaptic neurons (Akins and Biederer, 2006; Zipursky and Sanes, 2010; Missler et al., 2012; Yogev and Shen, 2014). Whereas axon guidance is largely mediated by secreted factors (Goodhill, 2016; Seiradake et al., 2016), synaptic target recognition is mediated at least in part by trans-cellular interactions via cell-adhesion molecules (Akins and Biederer, 2006; Zipursky and Sanes, 2010; Missler et al., 2012). Combinatorial use of diverse trans-synaptic cell-adhesion molecules is proposed to both generate the signals for synapse assembly and to provide the basis for the specificity of synaptic connections. Understanding the molecular determinants that guide synaptic specificity is of interest in neuroscience because these determinants ultimately control the assembly of neural circuits.
Latrophilins (Lphns) comprise a family of three cell-adhesion molecules (Lphn1–Lphn3; gene symbols Adgrl1–Adgrl3) that were identified as putative synaptic receptors for α-latrotoxin (Davletov et al., 1996; Krasnoperov et al., 1996; Sugita et al., 1998), a toxin from black widow spider venom that induces massive uncontrolled neurotransmitter release from presynaptic nerve terminals (Südhof, 2001). Lphns belong to the class of adhesion G protein–coupled receptors (GPCRs) that are characterized by large extracellular sequences composed of different N-terminal domains followed by a canonical GPCR autoproteolysis-inducing (GAIN) domain and the typical seven-transmembrane regions of GPCRs (Krasnoperov et al., 1997; Lelianova et al., 1997; Sugita et al., 1998; Langenhan et al., 2013, 2016; Hamann et al., 2015). Lphns form intercellular junctions by interacting in trans via their extracellular domains with at least three other cell-adhesion molecules implicated in synaptic function, namely neurexins, teneurins, and fibronectin leucine-rich transmembrane proteins (FLRTs; Silva et al., 2011; Boucard et al., 2012, 2014; O’Sullivan et al., 2012; Lu et al., 2015). Via these interactions, Lphns participate in multifaceted trans-cellular molecular networks that could contribute to the specificity of synaptic connections. The canonical GAIN domain of Lphn and other adhesion GPCRs mediates their constitutive autocatalytic cleavage at the GPCR proteolysis site just N-terminal to the first transmembrane region (Araç et al., 2012). Although it is unclear whether all adhesion GPCRs engage in G protein–mediated signaling, at least some adhesion GPCRs appear to function as GPCRs (Paavola and Hall, 2012; Langenhan et al., 2013). Thus, Lphns have the potential to act both as cell-adhesion molecules and as signaling receptors at synapses.
The domain structure of Lphns indicates that they function via multiple extra- and intracellular interactions, but their physiological role remains uncharacterized. In Caenorhabditis elegans, the LAT-1 Lphn homologue is essential for organ development (Langenhan et al., 2009, 2013; Müller et al., 2015). In vertebrates, Lphn1 and Lphn3 are primarily expressed postnatally in the brain, suggesting a brain-specific function, whereas Lphn2 is ubiquitously expressed during development, indicating a broader developmental role consistent with that of LAT-1 in C. elegans (Boucard et al., 2014). Experiments with Lphns in cultured neurons suggested a presynaptic function as predicted by their identification as α-latrotoxin receptors (Silva et al., 2011; O’Sullivan et al., 2012, 2014), but initial analyses of constitutive Lphn1 knockout (KO) mice failed to uncover a major synaptic phenotype (Tobaben et al., 2002).
In this study, we focused on Lphn2, the only ubiquitously expressed Lphn isoform in vertebrates. Using newly generated conditional and constitutive knock-in and KO mice, we found that constitutive KO of Lphn2 causes embryonic lethality, suggesting that Lphn2 is required for embryonic development. We show that mice with brain-specific deletion of Lphn2, however, are viable, suggesting that the embryonic function of Lphn2 is caused by Lphn2 expression in nonneural tissues. Moreover, we show that in the brain, Lphn2 plays a defined role during synapse assembly. Specifically, we found in hippocampal CA1-region pyramidal neurons that Lphn2 functions as a postsynaptic, not a presynaptic, cell-adhesion molecule as predicted from its identification as a putative α-latrotoxin receptor. We demonstrate that Lphn2 is selectively targeted to spine synapses in the distal apical dendrites of the stratum lacunosum-moleculare (SLM) of CA1-region pyramidal neurons. These distal dendrites are notable for receiving synaptic inputs from layer III entorhinal cortex neurons (Kitamura et al., 2015). Deletion of Lphn2 from pyramidal CA1-region neurons leads to a selective loss of these synaptic inputs. Behavioral analyses of mice lacking Lphn2 in CA1-region neurons revealed that entorhinal synapses on SLM spines are dispensable for spatial memory but are required for learning temporal sequences of spatial tasks. Our data reveal a central role for Lphn2 as a representative adhesion GPCR in directing synapse formation to a specific dendritic domain of a pyramidal neuron, suggesting that Lphn2 serves as a target-recognition molecule for presynaptic afferents from the entorhinal cortex.
Results
Generation of Lphn2-mVenus conditional knock-in (cKI), Lphn2 conditional KO (cKO), and Lphn2 constitutive KO mice
Using homologous recombination in embryonic stem cells, we generated knock-in mice in which an artificial exon encoding a full-length Lphn2-mVenus fusion protein was inserted into the intron after the first coding exon of the Lphn2 gene (Fig. 1, A and B; and Fig. S1). At the same time, we flanked both the new and the regular first coding exon of the Lphn2 gene with loxP sites to allow conditional deletion, and inserted different pairs of frt sites such that site-directed Flp-mediated recombination randomly produced either Lphn2-mVenus cKI mice or traditional Lphn2 cKO mice. Per design, both of these alleles are conditional and can be converted into Lphn2 KO alleles by Cre recombinase (Figs. 1 A and S1). Thus, we produced cKI mice that express Lphn2-mVenus for tracking endogenously expressed Lphn2 protein, and we also produced cKO mice for conditional and constitutive deletion of Lphn2 using Cre recombinase.
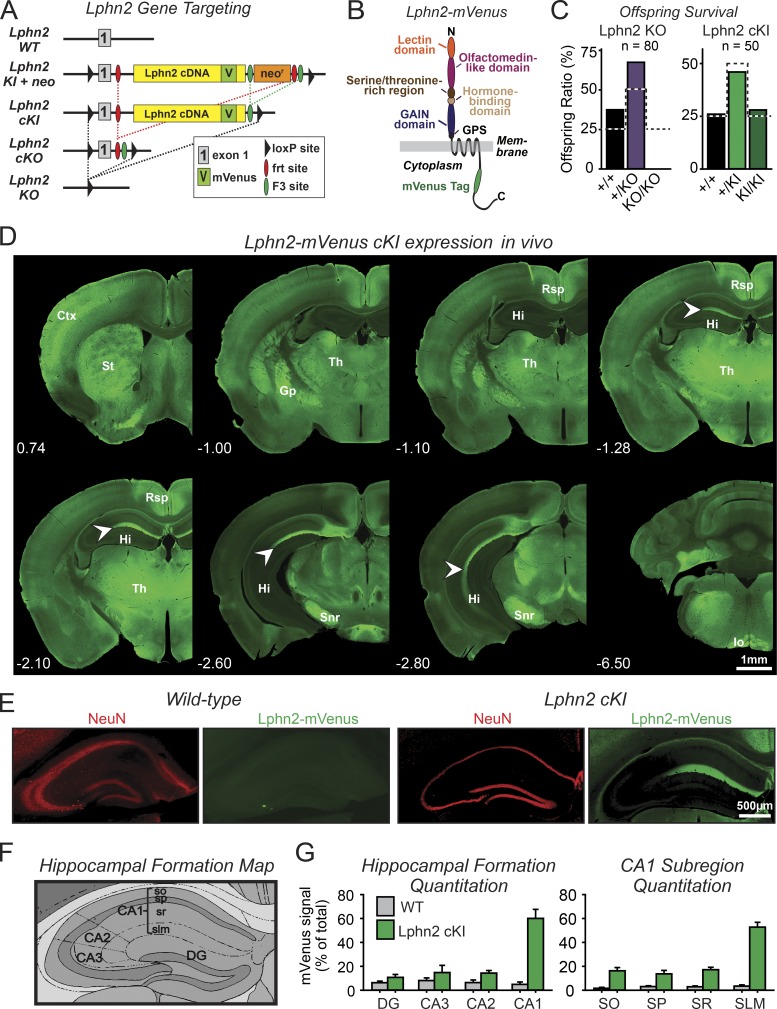
Figure 1.
Development of conditional Lphn2-mVenus knock-in and Lphn2 KO mice. Lphn2-mVenus protein localized to discrete brain nuclei and subcellular domains including the SLM of the hippocampal CA1 region. (A) Gene targeting strategy for Lphn2-mVenus cKI and Lphn2 cKO and constitutive KO mice. (B) Domain architecture of Lphn2-mVenus protein. GPS, GPCR proteolysis site. (C) Constitutive KO of Lphn2 caused embryonic lethality, whereas the Lphn2-mVenus knock-in (KI) did not impair survival (summary graphs of surviving offspring from heterozygous matings; dotted lines indicate survival expected from Mendelian inheritance). (D) Lphn2 mVenus expression in vivo. Coronal sections of the brain from an adult Lphn2-mVenus cKI mouse were labeled using GFP antibodies to detect Lphn2-mVenus protein (green). Indicated coronal section distances are in millimeters relative to Bregma. Arrowheads indicate spine- or shaft-associated Lphn2 puncta. Ctx, cortex; Gp, globus pallidus; Hi, hippocampus; Io, inferior olive; Rsp, retrosplenial cortex; Snr, substantia nigra pars reticulata; St, striatum; Th, thalamus. Note that one of the panels is presented again in Fig. 3 A. (E) Representative immunohistochemistry images of WT and Lphn2-mVenus cKI coronal sections of the hippocampus stained for GFP (to label Lphn2-mVenus) and NeuN (to label neuronal nuclei). Note that a GFP/NeuN merged image of this panel is presented again in Fig. 3 B. (F) Topographical map of the major regions and subregions of the hippocampal formation (CA3, CA2, and CA1, equivalent regions; DG, dentate gyrus; stratum oriens [so], stratum pyramidale [sp], stratum radiatum [sr], and SLM, CA1–3 subregions). Image adapted from the Allen Institute for Brain Science. (G) Quantitation of Lphn2-mVenus signal in indicated hippocampal regions (left) and CA1-region subregions (right). Graphs show means ± SEM (WT, n = 3; cKI, n = 4 mice at ~P30).
To determine whether the constitutive Lphn2 KO or the Lphn2-mVenus cKI impair survival, we monitored offspring from heterozygous matings. No surviving homozygous Lphn2 KO mice were detected (Fig. 1 C). Homozygous Lphn2 cKI mice, however, were generated at the expected Mendelian ratios, suggesting that although the constitutive Lphn2 KO is lethal, the Lphn2-mVenus fusion protein produced by cKI mice is functional and that the mVenus tag attached to Lphn2 in these mice does not impede the normal localization of Lphn2 (Fig. 1 C). Because Lphn2-mVenus cKI mice only express one particular splice variant of Lphn2 and alternative splicing of Lphn2 regulates at least some ligand interactions (Boucard et al., 2014), the viability of mice expressing only this splice variant implies that this splice variant is sufficient for the essential functions of Lphn2.
Lphn2 is differentially expressed in the brain
A brainwide analysis of coronal brain sections from Lphn2-mVenus cKI mice revealed that Lphn2 is not ubiquitously expressed. Rather, we observed that Lphn2-mVenus was restricted to specific brain areas that had no apparent commonality, e.g., the retrosplenial cortex, thalamus, and substantia nigra pars reticulata (Fig. 1 D). Maybe most striking was the localization of Lphn2-mVenus to the SLM in the CA1 region of the hippocampus (Fig. 1, E–G). Thus, Lphn2 is not uniformly present in all brain regions but is differentially expressed in defined subregions and cell types.
Postsynaptic deletion of Lphn2 impairs synapse numbers in cultured hippocampal neurons
Imaging of hippocampal neurons cultured from Lphn2-mVenus cKI mice demonstrated punctate fluorescence patterns along dendrites, suggesting that Lphn2 is localized to synapses (Fig. 2 A). To explore a potential synaptic function of Lphn2, we sparsely transfected cultured hippocampal neurons at 4 d in vitro (DIV) with plasmids encoding mutant (ΔCre used as a control) or WT Cre recombinase. Transfections targeted <5% of all neurons, allowing us to probe for a cell-autonomous postsynaptic phenotype because most neurons surrounding a transfected cell would be WT (Fig. 2 B). We then analyzed the spine density in ΔCre and Cre-transfected neurons at DIV 14–16 using cotransfected cytosolic GFP as a means to visualize spines. Remarkably, we observed a significant loss of spines (∼40%) in Lphn2-deficient neurons (Fig. 2, C and D).
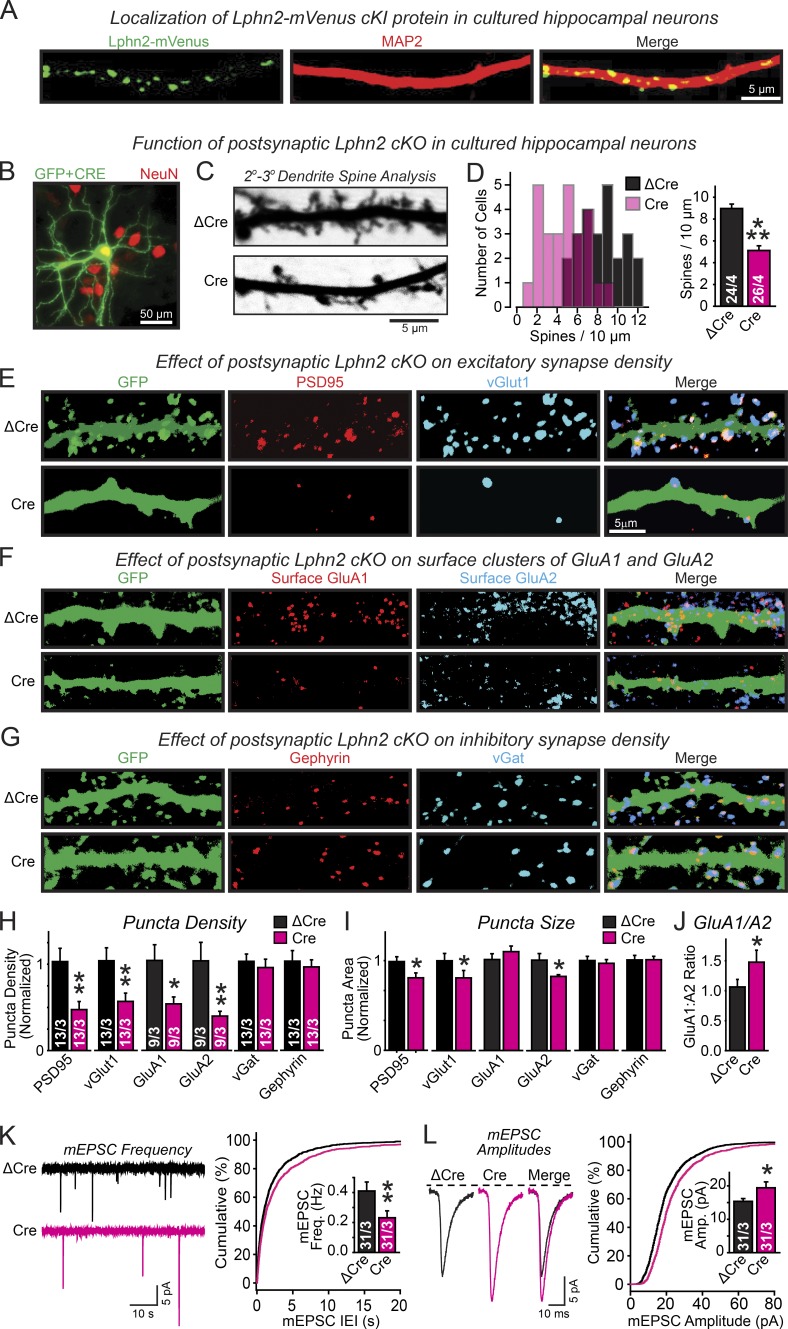
Figure 2.
Lphn2 functions in cultured hippocampal neurons as a postsynaptic protein that is selectively essential for excitatory synapses. (A) Lphn2-mVenus localization probed in hippocampal neurons cultured from Lphn2-mVenus cKI mice by immunocytochemistry for GFP (labeling the mVenus-tagged Lphn2; green) and MAP2 (red). (B) Representative image of hippocampal neurons cultured from Lphn2 cKO mice and sparsely transfected with Cre-EGFP and cytosolic GPF to visualize neuron morphology. Transfections were performed at DIV 4 and analyzed at DIV 14–16 (green, GFP; red, NeuN counterstain to label of neuronal nuclei). (C) Representative images of dendrites from neurons cotransfected with GFP and either inactive (ΔCre) or active Cre recombinase (Cre). (D) Postsynaptic deletion of Lphn2 by sparse transfection of cultured hippocampal Lphn2 cKO neurons with Cre (using ΔCre as a control transfection) decreases the spine density (left, histogram of the number of neurons vs. spine density; right, summary graph of mean spine density). (E–G) Representative images of neurons analyzed by dual immunofluorescence labeling for the excitatory synapse markers PSD95 and vGlut1 (E), for surface AMPARs (GluA1 and GluA2; F), and for the inhibitory synapse markers gephyrin and vGat (G). (H–J) Summary graphs of the density (H) and apparent size of synaptic puncta (I) as well as of the ratio of synaptic puncta size for GluA1 compared with GluA2 (J) analyzed on secondary/tertiary dendrites in Lphn2-deficient (Cre) and control neurons (ΔCre) labeled as described in E–G. (K and L) Lphn2 deletion decreases the frequency of spontaneous mEPSCs (K; left, representative traces; right, cumulative distribution of interevent intervals [inset, mean event frequencies]), but increases the mEPSC amplitude (L; left, mean of mEPSC event traces; right, cumulative distributions of event amplitudes [inset, mean amplitudes]). Data are means ± SEM; numbers of neurons/independent cultures examined are shown in the graphs. Statistical analyses used Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. IEI, interevent interval.
To test whether the loss of spines in Lphn2-deficient hippocampal neurons corresponded with a decrease in synapse density, we stained sparsely Cre- or ΔCre-transfected hippocampal neurons from Lphn2 cKO mice for excitatory and inhibitory synapse markers (Fig. 2, E–J). Consistent with the decrease in spine density, we observed a similarly dramatic decrease on Lphn2-deficient neurons in the density of synaptic puncta labeled by the excitatory markers PSD95 (∼60% decrease) and vGlut1 (∼50% decrease). Moreover, we detected a corresponding decrease of the density of excitatory synaptic puncta imaged by labeling for two surface-exposed AMPA-type glutamate receptors (AMPARs), GluA1 (∼50% decrease) and GluA2 (∼60% decrease; Fig. 2 H). We observed no change in the density of inhibitory synaptic puncta monitored by staining cultured neurons with antibodies to vesicular GABA transporter (vGat) and gephyrin (Fig. 2 H). We also measured the apparent sizes of the synaptic puncta (which reflect a combination of antigen concentration and true synapse size) and observed a small but significant decrease in apparent size of PSD95-, vGlut1-, and GluA2-positive puncta (Fig. 2 I). Owing to a relatively larger decrease in GluA2 than GluA1 signals, Lphn2-deficient neurons exhibited an increase in GluA1/GluA2 staining ratio, suggesting a change in the synaptic AMPAR composition (Fig. 2 J).
The decrease in synapse numbers in Lphn2-deficient neurons was accompanied by a large decrease (∼45%) in the frequency of spontaneous miniature excitatory postsynaptic currents (EPSCs; mEPSCs; Fig. 2 K) and by a smaller increase in mEPSC amplitudes (∼25%; Fig. 2 L). These data viewed together suggest that the Lphn2 KO causes loss of approximately half of the excitatory synapses, with a possibly compensatory strengthening and a shift in the synaptic AMPAR composition of the residual excitatory synapses (Fig. 2 J).
Lphn2 is targeted to dendritic spines in the SLM in CA1-region pyramidal neurons
As α-latrotoxin receptors, Lphns are thought to be presynaptic (Südhof, 2001). Thus, the postsynaptic function of Lphn2 in cultured neurons in synapse formation was surprising. Because cultured neurons can exhibit nonnative properties, we next studied the function of Lphn2 in situ in the CA1 region of the hippocampus. We chose to study this brain area because in brain sections, Lphn2-mVenus appeared to be concentrated in the SLM of the CA1 region (Fig. 3, A and B).
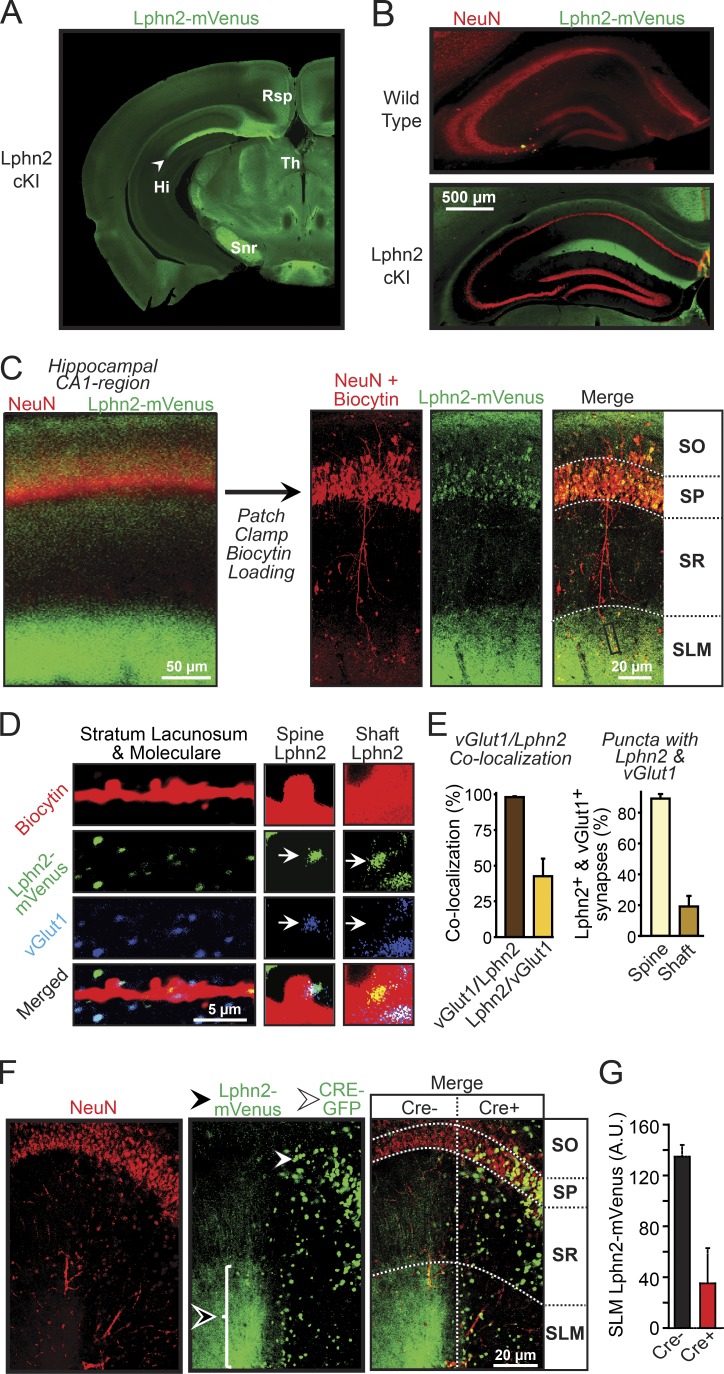
Figure 3.
Postsynaptic Lphn2 is specifically targeted to and essential for dendritic spines in the SLM of CA1-region pyramidal neurons. (A) Representative coronal brain section (−2.6 mm from Bregma) from Lphn2-mVenus cKI mice stained for mVenus analyzed at ~P30. Note that this image is same as one of the panels in Fig. 1 D. Hi, hippocampus; Rsp, retrosplenial cortex; Snr, substantia nigra pars reticulate; Th, thalamus. (B) Representative coronal sections of the hippocampus from WT and Lphn2-mVenus cKI mice stained for mVenus and the neuronal nuclear protein NeuN. Note that this image is an overlay of the same images shown in Fig. 1 E. (C) Biocytin loading of individual pyramidal neurons to analyze dendritic spines (left, image of a CA1-region hippocampal section labeled for NeuN and Lphn2-mVenus; right, similar images of sections that were additionally stained for biocytin after a single pyramidal neuron had been filled with biocytin via a patch pipette). (D) High-magnification image of the distal dendrites in the SLM of a biocytin-filled pyramidal CA1-region neuron from a Lphn2-mVenus cKI mouse (left), and further enlarged images (2.5×) of spines and shafts from these dendrites (right). CA1-region sections were stained for biocytin (red), mVenus (green), and vGluT1 (blue) after the filling of an individual pyramidal neuron with biocytin via the patch pipette. Arrows indicate spine- or shaft-associated puncta. (E) Quantification of the percent vGlut1-positive puncta that were also Lphn2 positive (left bar in left graph) or vice versa (right bar in left graph) and of the distribution of synaptic puncta positive for both Lphn2-mVenus and vGlut1 on dendritic spines or shafts (right graph; n = 2 mice). (F) Representative hippocampal sections from mice after lentiviral Cre recombinase expression in patches of CA1-region neurons using stereotactic injections at P21. Sections were stained for NeuN and GFP (detects both Lphn2-mVenus expression in the SLM [black arrowhead] as well as nuclear Cre-EGFP in infected cells [white arrowhead]). Note that Lphn2-mVenus signal is removed from SLM areas that are connected to pyramidal neurons expressing Cre-EGFP. Horizontal dotted lines indicate boundary lines for the subregions of the CA1 region; the vertical dotted line indicates Cre− and Cre+ regions. (G) Quantification of the Lphn2-mVenus signal in the SLM associated with CA1-region neurons lacking (Cre−) or containing Cre recombinase (Cre+; means ± SEM; n = 4 mice at ~P30). SO, stratum oriens; SP, stratum pyramidale; SR, stratum radiatum.
The SLM of the hippocampal CA1 region receives inputs from layer III of the entorhinal cortex, which account for a large percentage of excitatory synapses on spines of SLM dendrites (Spruston, 2008; Suh et al., 2011; Takács et al., 2012). To explore whether Lphn2 localizes to these spines, we patched CA1-region pyramidal neurons in acute slices from Lphn2-mVenus cKI mice and filled the neurons with biocytin. We then used confocal microscopy to examine the distribution of Lphn2 in hippocampal sections that were stained for biocytin to enable mapping of the dendrites of individual neurons (Fig. 3 C). In addition, we labeled the sections for NeuN and for vGlut1 as markers for neuronal nuclei and excitatory synapses, respectively (Fig. 3 D).
We found that Lphn2-mVenus was highly concentrated in puncta that are adjacent to dendrites within the SLM (Fig. 3, D and E). A large fraction (∼45%) of Lphn2-positive puncta also contained vGlut1, whereas all vGlut1-positive puncta in the SLM also contained Lphn2. The Lphn2-positive puncta that exhibit no vGlut1 colabeling may represent inhibitory synapses or nonsynaptic puncta, or their vGlut1 signal may be below threshold or out of the plane of the section. More than 80% of Lphn2/vGlut1 double-positive puncta were localized to dendritic spines (Fig. 3, D and E).
We next asked whether Lphn2 is pre- or postsynaptic. To address this question, we took advantage of the conditional nature of the Lphn2-mVenus cKI, which allows deleting Lphn2-mVenus using Cre recombinase (Fig. 1 A). We stereotactically injected lentiviruses expressing EGFP-tagged Cre recombinase into the CA1 region of adolescent mice (Figs. 3 F and S2 A). The limited lentiviral infections caused deletion of Lphn2-mVenus in patches of CA1-region neurons (Fig. 3 F). As a result, we were able to analyze in a single section the localization of Lphn2-mVenus relative to Cre recombinase expression, which can be identified by the nuclear EGFP signal of tagged Cre recombinase. We found that in areas of nuclear Cre recombinase expression, Lphn2 in the SLM was nearly ablated, whereas it was retained in adjacent noninfected areas (Fig. 3, F and G). Thus, Lphn2 is also postsynaptic in vivo and is specifically targeted to the spines of a restricted dendritic domain of CA1-region pyramidal neurons, namely the SLM. These results uncover an unexpected molecular sorting process targeting postsynaptic Lphn2 to specific dendritic domains in CA1-region pyramidal neurons.
Lphn2 controls spine development in SLM dendrites of CA1-region pyramidal neurons
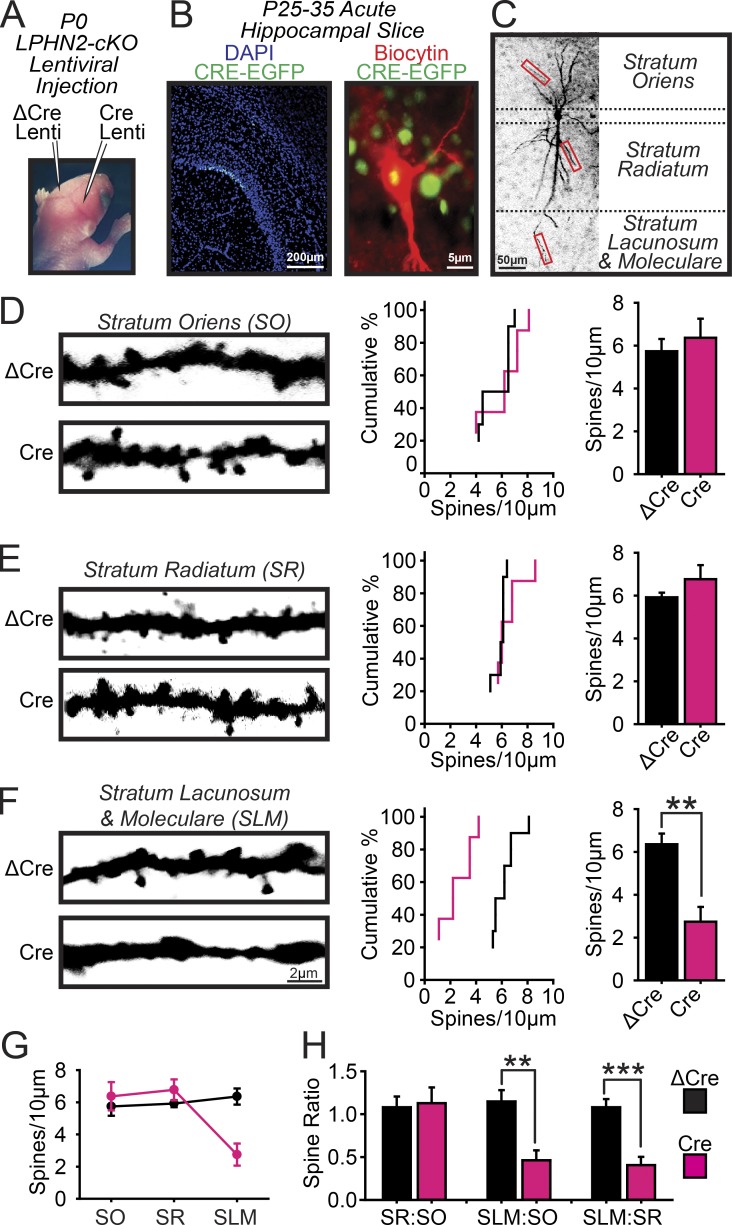
To explore the functional role of postsynaptic Lphn2, we examined the morphological and physiological consequences of Lphn2 deletions in CA1-region neurons. We stereotactically injected low-titer lentiviruses into the CA1 regions of newborn mice to achieve sparse infections, allowing us to analyze postsynaptic and cell-autonomous Lphn2 functions. In these experiments, active (Cre) and mutant inactive Cre recombinase (ΔCre, as a control) were expressed in opposing hemispheres of the same mice (Figs. 4 A and S2 B). At 4–6 wk of age, we prepared acute slices from injected mice, filled individual infected CA1-region pyramidal neurons with biocytin via a patch pipette (Fig. 4 B), and analyzed a neuron’s dendritic spines in all three layers (Fig. 4 C). We found that the spine density was unchanged by the Lphn2 deletion in the stratum oriens and stratum radiatum of CA1-region pyramidal neurons but was decreased dramatically (∼65%) in the SLM (Fig. 4, D–H). Thus, the Lphn2 deletion selectively ablated spines in the SLM.
Figure 4.
Postsynaptic deletion of Lphn2 ablates dendritic spines in the SLM but not the stratum oriens or stratum radiatum of CA1-region pyramidal neurons. (A–C) Experimental strategy. (A) The CA1 regions of opposite hemispheres in newborn Lphn2 cKO mice were stereotactically injected with lentiviruses expressing Cre (test) or ΔCre (control). (B) An individual pyramidal neuron was filled with biocytin via a patch pipette in acute hippocampal slices from injected mice (left, low-magnification image of sparsely CRE-EGFP–infected hippocampus; right, high-magnification view of the soma of a lentivirally infected biocytin-filled neuron surrounded by other lentivirally infected neurons with nuclear EGFP fluorescence). (C) Delineation of dendritic segments that were analyzed by quantitative immunohistochemistry (boxes indicate example areas where spine analysis was performed). (D–F) Lphn2 deletion decreases the spine density on CA1 pyramidal neuron dendrites selectively in the SLM. Spines were analyzed on dendrites in the stratum oriens (D), stratum radiatum (E), and SLM (F; left, representative confocal images of secondary dendrites; middle, cumulative probability plots of the number of spines per 10 µm dendrite; right, summary graphs of spine densities; n = 5 ΔCre and 4 Cre mice at P25–35). (G) Plot of the spine density in dendrites of pyramidal neurons from the CA1 region of the hippocampus shown in absolute densities as a function of the subregions in which the dendrites are present. (H) Ratio analysis of CA1 pyramidal neurons across the CA1 subregions including stratum oriens, stratum pyramidale, stratum radiatum, and SLM. Means ± SEM; statistical analyses were performed using Student’s t test. **, P < 0.01; ***, P < 0.001.
CA1-region KO of Lphn2 selectively impairs synaptic inputs from the entorhinal cortex but not from the CA3 region
We next asked whether CA1-region pyramidal neurons experience a selective loss of synaptic inputs from entorhinal cortex afferents that specifically form synapses on spines in the SLM (Spruston, 2008; Suh et al., 2011; Takács et al., 2012) as would be expected from the decrease in synaptic spines in CA1-region SLM dendrites.
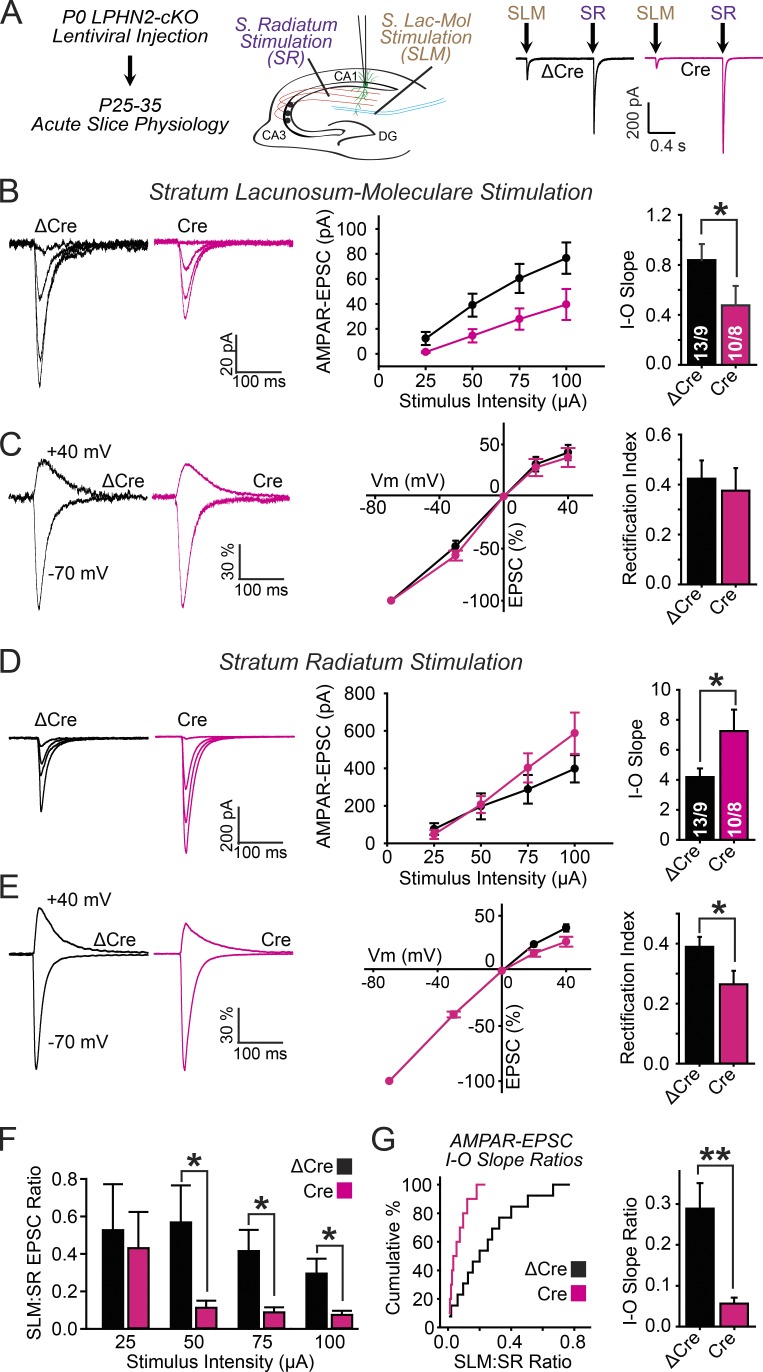
We addressed this question using similar postsynaptic deletions of Lphn2 by stereotaxic injection of lentiviruses expressing inactive (ΔCre) or active Cre recombinase into the CA1 region of newborn mice as described for the morphological studies (Fig. 4 A). We then monitored pharmacologically isolated AMPAR-mediated EPSCs elicited by two differentially positioned stimulating electrodes (Fig. 5 A). The first electrode was placed in the stratum radiatum proximal to the CA3 region to stimulate Schaffer-collateral axons that emanate from CA3-region pyramidal neurons and mediate excitatory synaptic inputs onto CA1-region pyramidal neurons in the stratum oriens and stratum radiatum. The second electrode was placed in the SLM proximal to the cortex to stimulate perforant-path axons that come from entorhinal cortex neurons and form excitatory synapses onto SLM dendritic spines, which contain postsynaptic Lphn2. AMPAR-EPSCs for both pathways were recorded sequentially in the same neurons, separated by 3-s interstimulus intervals to monitor both synaptic inputs onto a single CA1 pyramidal neuron. We measured the input–output relationships of the stimulus intensity versus AMPAR-EPSC amplitude to assess synaptic strength and also examined whether the Lphn2 KO influenced the voltage dependence of AMPAR-EPSCs.
Figure 5.
Postsynaptic Lphn2 deletion in CA1-region pyramidal neurons impairs excitatory synaptic transmission of entorhinal cortex inputs but enhances excitatory synaptic transmission of CA3-region Schaffer collateral inputs. (A) Experimental design. Lentiviruses expressing Cre (test) and ΔCre (control) were stereotactically injected into the CA1 region of P0 Lphn2 cKO mice (see Fig. 4 A). Dual-input patch-clamp recordings on the same lentivirally infected CA1-region pyramidal neurons in acute hippocampal slices at P25–35–aged mice were performed using independent stimulating electrodes that excited synaptic inputs from the entorhinal cortex (placed on the SLM proximal to the entorhinal cortex) and from CA3-region Schaffer collaterals (placed on the stratum radiatum proximal to the CA3 region). (B) Measurements of synaptic strength via input/output (I-O) curves (left, representative EPSC traces; middle, summary plot of the EPSC amplitudes as a function of the stimulus current; right, summary graph of fitted linear input/output slopes). (C) Rectification index analysis of AMPAR-EPSCs (left, representative EPSC traces monitored in neurons held at −70 or +40 mV; middle, summary plots of EPSC amplitudes as a function of the holding potential normalized to the EPSC amplitude recorded at a −70-mV holding potential; right, summary graphs of the rectification index calculated as the ratio of EPSC amplitudes recorded at +40 vs. −70 mV holding potentials). VM, membrane voltage. (D and E) Same as B and C, except that AMPAR-EPSCs were recorded in the same neurons as in B and C as a function of stratum radiatum stimulation. (F) Summary graphs of the ratio of AMPAR-EPSC amplitudes recorded for SLM and stratum radiatum inputs at the indicated electrode stimulation strengths. (G, left) SLM to stratum radiatum input/output slope ratio plotted as cumulative distributions; (right) SLM to stratum radiatum input/output slope ratio summary graph. Plots and graphs shown are means ± SEM; the numbers of neurons/animals examined are shown in the graphs. Statistical analyses were performed using Student’s t test. *, P < 0.05; **, P < 0.01.
We found that the CA1 region KO of Lphn2 massively impaired synaptic responses induced by stimulation of entorhinal cortex afferents, as revealed by an almost 50% decrease in the slope of the input/output curves (Fig. 5 B). We observed no changes in the voltage dependence of the remaining SLM EPSCs (Fig. 5 C). Conversely, we found that the KO of Lphn2 increased the strength of synaptic connections formed by Schaffer collaterals emanating from CA3-region neurons as revealed by a nearly 100% increase in the input/output curves (Fig. 5 D). At these synapses, the voltage dependence of AMPAR-EPSCs exhibited increased rectification (Fig. 5 E). These changes in Schaffer-collateral synapses in Lphn2-deficient neurons may reflect a homeostatic response to the loss of entorhinal cortex inputs. Plots of the ratio of EPSC amplitudes (Fig. 5 F) of the slopes of the input/output relations (Fig. 5 G) between entorhinal cortex- and Schaffer collateral–derived synapses illustrate the shift in synaptic weight from entorhinal cortex inputs to Schaffer collateral inputs. These data are consistent with our results in cultured hippocampal neurons in which the Lphn2 KO not only decreased synapse numbers but also increased the proportion of GluA2-lacking AMPARs, which exhibit higher channel conductance (Fig. 2, I and K; Isaac et al., 2007). Collectively, our data indicate that Lphn2 localizes to a restricted dendritic domain in CA1-region neurons, where it functions in a compartmentalized fashion to selectively control assembly of synapses from entorhinal cortex afferents.
Deletion of hippocampal Lphn2 impairs spatial–temporal learning
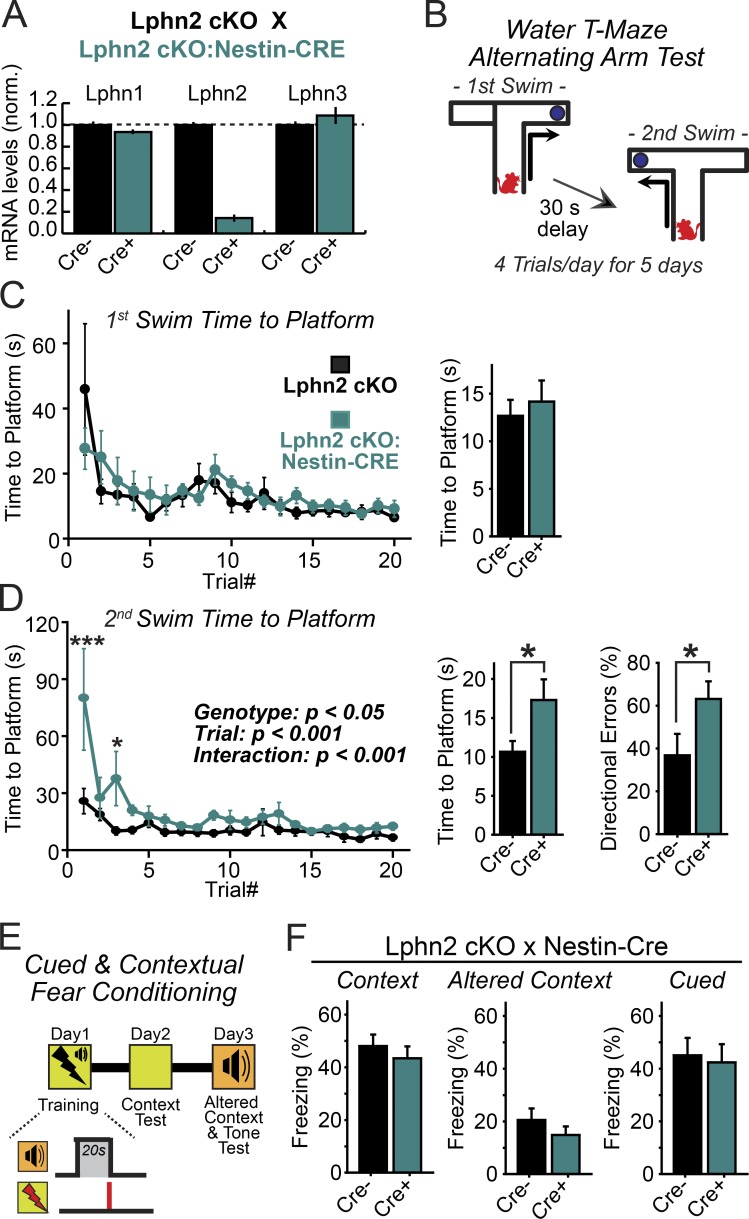
Lastly, we explored the behavioral consequence of the Lphn2 deletion in mice. Because constitutive deletion of Lphn2 was embryonically lethal (Fig. 1 C), we were unable to test constitutive KO mice. However, we found that deletion of Lphn2 from the entire brain by crossing Lphn2 cKO mice with a Cre recombinase driver line under control of the Nestin promoter (Nestin-Cre) did not cause lethality, although it ablated Lphn2 expression from brain as expected and induced a modest decrease in body weight (Fig. S3). This result indicates that the constitutive Lphn2 KO causes lethality owing to a non-neuronal function of Lphn2, consistent with the broad expression of Lphn2 in non-neuronal tissues such as the heart, liver, and kidneys (Boucard et al., 2014) and the role of LAT-1 in planar polarity in C. elegans (Langenhan et al., 2009; Müller et al., 2015).
We systematically examined the behavior of brain-specific Lphn2 KO mice. We detected no major impairments in a series of broad tests that monitor open field behavior, anxiety, motor coordination, or prepulse inhibition (Fig. S3). Because direct inputs from medial entorhinal cortex layer III neurons onto CA1 pyramidal cells are implicated in spatial learning and other forms of episodic memory (Remondes and Schuman, 2004; Suh et al., 2011), we hypothesized that ablation of Lphn2-dependent synapses in the CA1 region may impair spatial learning. However, when we measured contextual fear conditioning, we again observed no difference between littermate control and brain-specific Lphn2 KO mice (Fig. 6, E and F).
Figure 6.
Lphn2 is dispensable in the brain for spatial and contextual memory formation but is essential for establishing temporal memory sequences. (A) Lphn2 cKO female mice were crossed to male Lphn2 cKO (Nestin-Cre) mice to generate offspring that were homozygous for Lphn2 cKO and either negative (CRE−) or positive (CRE+) for the Cre transgene. Age-matched adult mice brains (>P30) were analyzed for Lphn1–3 mRNA expression by quantitative RT-PCR analysis (n = 4 mice). (B) Experimental design for the alternating water T-maze memory test. Mice were trained for 5 d four times a day to first swim into the right arm of a water T-maze and subsequently into the left arm after a 30-s delay. (C) Summary plots (left) and graphs (right; averaged across all trials) of the time to platform during the first T-maze swim. (D) Summary plots (left) and graphs (center; averaged across all trials) of the time to platform and directional error rate (right; averaged across all trials) during the second swim. Animals analyzed were littermate control mice or mice with Nestin-Cre recombinase–induced deletion of Lphn2 from brain (n = 8 littermate mice; P60–90). Summary plots: statistical analyses were performed using repeated-measure ANOVA with post hoc comparisons. Summary graphs: statistical analyses were performed using Student’s t test. *, P < 0.05; ***, P < 0.001. (E) Experimental design for fear memory analyses using mice with transgenic Nestin-Cre recombinase–mediated Lphn2 deletion from the entire brain. (F) Normal cued and contextual fear memory in both an altered and training context in littermate control mice or mice lacking Lphn2 in brain (n = 11 littermate mice; P60–90). Plots and graphs shown are means ± SEM.
These experiments suggest that the brain-specific Lphn2 deletion leaves most overall functions of the brain intact. However, recent work suggests that synapses formed by entorhinal cortex afferents on CA1-region pyramidal neurons in the SLM are specifically required for episodic memory tasks involving a temporal delay and/or a behavioral switch (Suh et al., 2011). To test whether the brain-specific Lphn2 deletion may produce a similar deficit, we examined a new cohort of brain-specific Lphn2 KO mice with a hippocampal-dependent spatial–temporal memory task that involves a 30-s gap in a sequential water T-maze assay (Fig. 6 B). The T-maze was set up with the left arm blocked and an escape platform positioned on the end of the right arm, forcing mice to learn a relatively simple spatial paradigm. Brain-specific Lphn2 KO mice exhibited no impairment in performance during the first swim in this one-armed maze (Fig. 6 C), a measure of their swimming ability and of the spatial memory of the escape platform. These mice failed, however, in learning a simplified sequential spatial task. After a 30-s delay upon completion of the first swim, the left arm of the T-maze was opened up, and the escape platform was moved to the left arm. With both arms now open, mice were asked to learn that the location of the platform had switched to the opposite arm. Brain-specific Lphn2 KO mice learned this second swimming task more slowly than controls, as evidenced both by the time to platform and the error rate (Fig. 6 D).
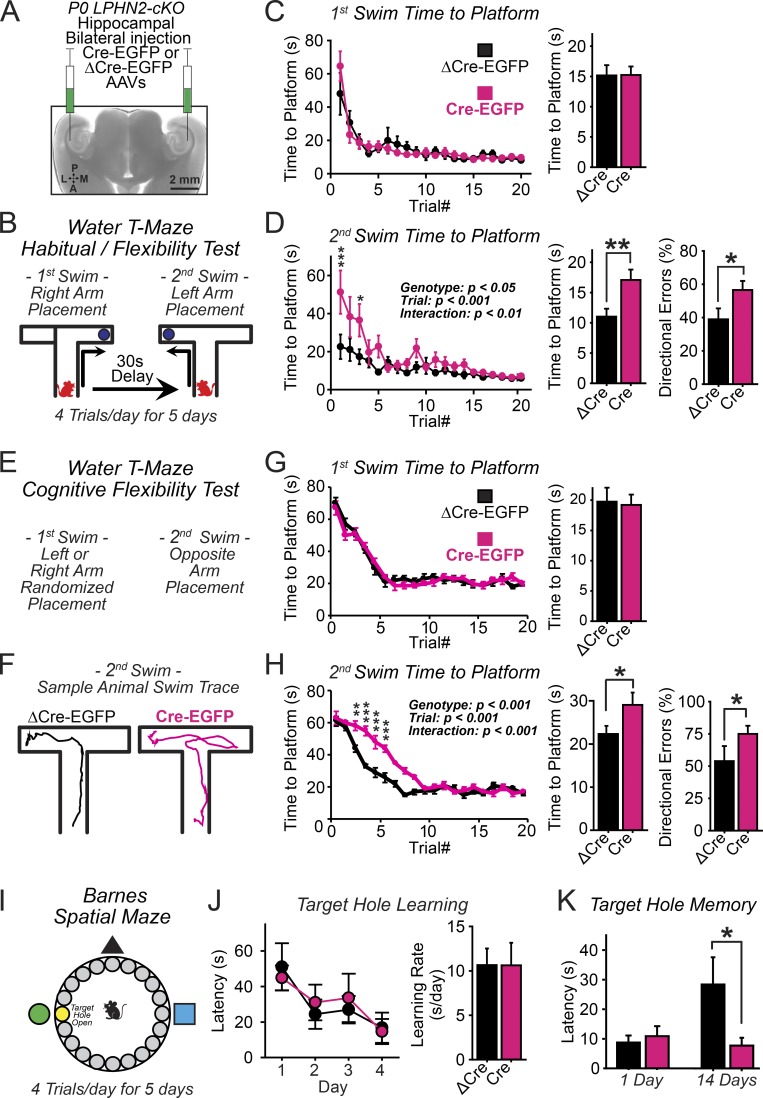
Is the impairment in spatial–temporal sequence learning in Lphn2-deficient mice a result of the role of Lphn2 at SLM synapses in the hippocampal CA1 region? To specifically delete Lphn2 in the CA1 region of the hippocampus, we stereotactically injected adeno-associated viruses (AAVs) encoding inactive (ΔCre) or active Cre recombinase (Cre) bilaterally into the CA1 regions of newborn mice and examined these mice behaviorally at 2–3 mo of age (Fig. 7 A). We used the same spatial–temporal water T-maze assay described above and found a performance deficit in the second swim navigation task similar to that of brain-specific Lphn2 KO mice (Fig. 7, B–D).
Figure 7.
Hippocampal specific elimination of Lphn2 expression impairs learning of temporal memory sequences but enhances long-term spatial memory. (A) Experimental strategy. The CA1 region of the hippocampus of newborn Lphn2 cKO mice was bilaterally injected with inactive (ΔCre) or active (Cre) Cre recombinase–expressing AAVs, and mice were analyzed behaviorally at 2–3 mo of age. (B) Design of the alternating water T-maze habit forming cognitive flexibility test. Mice were trained to first swim into the right arm of a water T-maze and then after a 30-s delay into the left arm (5 d of training with four trials daily). (C) Summary plots (left) and graphs (right; averaged across all trials) of the time to platform during the first T-maze swim. (D) Summary plots (left) and graphs (center; averaged across all trials) of the time to platform and directional error rate (right; averaged across all trials) during the second swim (n = 9 littermate mice at P60–90). Summary plots: statistical analyses were performed using repeated-measure ANOVA with post hoc comparisons. Summary graphs: statistical analyses were performed using Student’s t test. (E) Design of the water T-maze cognitive flexibility test. Same as B, but directions were randomly altered between trials. (F) Representative mouse paths during the second swim recorded using video tracking software. (G) Summary plots (left) and graphs (right; averaged across all trials) of the time to platform during the first T-maze swim. (H) Summary plots (left) and graphs (center; averaged across all trials) of the time to platform and directional error rate (right; averaged across all trials) during the second swim. (n = 10 ΔCre and 10 Cre mice; P60–90). Summary plots: statistical analyses were performed using repeated-measure ANOVA with post hoc comparisons. Summary graphs: statistical analyses were performed using Student’s t test. (I) Experimental design of the Barnes maze test. (J) Summary plot (left) and graph of the target-hole learning rate, calculated as the slope of a fitted linear regression. (K) Summary graph of the target-hole memory at 1 and 14 d after training, analyzed in mice with AAV injections into the hippocampal CA1 region (n = 9 littermate mice; P60–90). Statistical analyses were performed using Student’s t test. Plots and graphs shown are means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The protocol we used for the water T-maze assay did not necessarily measure cognitive flexibility, but it also measured habit formation given the predictable left versus right arm spatial navigation rules. As a further test, we performed a variation of this assay in which the direction of the assay was randomly switched between the right and left arms (Fig. 7, E–H). In this increasingly difficult assay, animals lacking hippocampal CA1-region Lphn2 again exhibited a significant delay in learning the second swim navigation task (Fig. 7 H). We further measured these animals for any alteration in spatial learning and memory processing but found no effect in exploratory behavior measured in an open field or on novel object recognition (Fig. S4). Thus, loss of Lphn2-dependent synapses in the SLM has no effect on spatial learning itself but impairs learning memory tasks involving temporal switching, suggesting the possibility that memories are more strongly formed initially.
Finally, to further test this conclusion in another behavioral paradigm, we examined CA1-region specific Lphn2 KO mice in the Barnes maze (Fig. 7 I; Barnes, 1979). We observed normal spatial learning during an initial 4-d training phase in which the mouse learned the location of a target escape hole, confirming the finding that Lphn2 is not required for spatial reference memory (Fig. 7 J). When we tested the same mice 2 wk later, however, control mice had largely forgotten the location of the target hole, whereas mice with a CA1-region Lphn2 deletion retained the memory of the location of the target hole (Fig. 7 K). This finding indicates that memories are indeed more long lasting in Lphn2-deficient brain, which may account for the cognitive flexibility impairment that is observed in the water T-maze.
Discussion
Synaptic cell-adhesion molecules have been studied intensely, leading to the recognition that synapses may be shaped by the interplay of a large number of different trans-synaptic interactions. Lphns are candidate synaptic cell-adhesion molecules that may contribute to synapse formation and/or specification, but their functions have remained elusive. Lphns are particularly interesting because they are GPCRs that double up as cell-adhesion molecules and because they have been shown to bind to α-latrotoxin, suggesting that they may be receptors for this presynaptic toxin (Südhof, 2001). To probe the function of Lphns, we focus in this study on Lphn2 as the only Lphn isoform that is developmentally required (Fig. 1 C). We generated new genetic models that allow visualization of Lphn2 in vivo and in culture by tagging endogenous Lphn2 with mVenus in vivo using a knock-in, enabling us to demonstrate that surprisingly, Lphn2 is a postsynaptic cell-adhesion molecule. Our data show that both in cultured hippocampal neurons and in hippocampal pyramidal CA1-region neurons in vivo, Lphn2 is essential for a subset of synapses, which in vivo are concentrated in the dendritic domain of the SLM in CA1 neurons. Given this spatially segregated localization and function of Lphn2, we propose that Lphn2 acts as a postsynaptic target-recognition molecule that is selectively essential for assembly and/or maintenance of entorhinal cortex synaptic inputs but is dispensable for assembly of CA3-derived synapses onto these CA1 pyramidal neurons. Lphn2 could potentially perform its essential role in hippocampal synapses by mediating synapse recognition and assembly and/or by contributing a synapse maintenance signal. Regardless of Lphn2’s mechanism of action, however, our data establish that Lphn2 acts as a synaptic molecule that is essential for a specific subset of synapses in a defined circuit.
The conclusion that Lphn2 is a postsynaptic adhesion GPCR that is specifically targeted to spine synapses in one particular dendritic domain in vivo is based on two independent lines of evidence: the localization of the Lphn2-mVenus knock-in protein and the electrophysiological analysis of neurons after conditional postsynaptic KO of Lphn2. Although it is conceivable that the Lphn2-mVenus knock-in altered the function of Lphn2, this seems unlikely given that the knock-in expresses Lphn2-mVenus from the endogenous gene and fully rescues the lethality of the constitutive Lphn2 deletion. The results obtained by imaging Lphn2-mVenus were validated with the electrophysiological experiments, which confirm that Lphn2 functions postsynaptically and that the Lphn2 KO selectively impairs synaptic inputs to the dendritic domain to which Lphn2-mVenus localizes (SLM), but not to dendritic domains (stratum oriens and stratum radiatum) that exhibit little Lphn2-mVenus presence.
We hypothesize that postsynaptic Lphn2 acts as a synaptic target-recognition molecule in the hippocampus by interacting with presynaptic cell-adhesion molecules such as teneurins, FLRTs, and/or neurexins. These presynaptic cell-adhesion molecules are known to form trans-synaptic complexes with Lphns (Silva et al., 2011; Boucard et al., 2012, 2014; O’Sullivan et al., 2012; Lu et al., 2015) and have been implicated in neural circuit development and synaptic organization processes (Südhof, 2008; Hong et al., 2012; Mosca et al., 2012; O’Sullivan et al., 2012; Reissner et al., 2013; Mosca, 2015; de Wit and Ghosh, 2016). It is possible that the Lphn2 localization specificity we describe in this study is established by its activation upon binding one or a combination of these ligands.
The role of a GPCR in mediating synapse assembly is unexpected, suggesting the possibility that trans-cellular cell-adhesion may be transduced into a synapse assembly signal mediated by local G protein signaling. Moreover, the postsynaptic function of Lphn2 is surprising given that Lphns were thought to be presynaptic α-latrotoxin receptors (Krasnoperov et al., 1997; Lelianova et al., 1997), suggesting that Lphns may either act as α-latrotoxin receptors in a complex with presynaptic interactors such as neurexins (which are themselves α-latrotoxin receptors; Sugita et al., 1999) or that Lphns can be variably pre- or postsynaptic. Consistent with a postsynaptic location, Lphns are known to avidly interact with postsynaptic Shank scaffolding proteins (Tobaben et al., 2000; Kreienkamp et al., 2002), which may contribute to their targeting or signal transduction (Sheng and Kim, 2000).
An essential feature of the mammalian brain is its ability to perceive and learn patterns of events as they occur in time. The hippocampus has a central role in this process by deconstructing spatial and temporal information via uniquely organized synaptic connections that are both intra- and interhippocampal and that integrate incoming information from other brain regions such as the entorhinal cortex (Eichenbaum, 2004; Pastalkova et al., 2008; MacDonald et al., 2011; Suh et al., 2011; Igarashi et al., 2014; Kitamura et al., 2015). What is extraordinary about the hippocampus and its neural circuitry is the spatially organized topography of its synaptic connections (Spruston, 2008). Axonal projections and synaptic connections from defined cell types are organized into discrete layers, making the hippocampus an ideal model system for understanding the formation and organization of neural circuits. However, molecular players that control the assembly of discrete hippocampal circuits have yet to be identified. Using Lphn2 cKO mice, we could selectively alter the balance of synaptic inputs on CA1 neurons in vivo, allowing us to test the role of synapses formed by entorhinal cortex afferents on CA1-region pyramidal neurons in behavior. We demonstrate that these synapses are not essential for spatial learning as such but are required for a sequential spatial learning task.
The function of the entorhinal cortex–hippocampal CA1-region synapses in enabling sequential temporal switching, i.e., flexibility, in spatial learning generally agrees with the proposed role of entorhinal inputs in temporal associative learning (Eichenbaum, 2004; Pastalkova et al., 2008; MacDonald et al., 2011; Suh et al., 2011; Kitamura et al., 2015). In addition to identifying a molecular mechanism involved in a specific synapse formation event, our results thus confirm with a new postsynaptic approach the notion that the circuit formed by entorhinal cortex layer III neurons with CA1-region pyramidal neurons is not required for the formation of spatial memories, but is essential for remembering memory tasks involving sequential temporal stages.
Materials and methods
Generation of Lphn2 mutant mice
For the generation of Lphn2 mutant mice, we used a dual strategy (“knock-in–first” strategy) that allowed us to (a) generate mice in which a cDNA encoding mouse Lphn2 with a C-terminal mVenus tag was inserted (cKI mice), and to (b) generate Lphn2 floxed mice that are subject to conditional deletion using the Cre/loxP system (cKO mice). The generation of the knock-in allele made it only possible to express a single splice version of Lphn2 as a coding DNA sequence. We therefore decided on a cDNA encoding a 1,487-aa large product (compare NCBI reference sequence NP_001074767) that includes the previously described mini-exon between the lectin and olfactomedin domains (Boucard et al., 2014). The monomeric Venus tag was placed between leucine (position 1,311) and threonine (position 1,312), increasing the size of the Lphn2-mVenus protein to 1,724 aa. The targeting vector was designed so that a single loxP site was introduced ∼500 bp upstream of the ATG-bearing exon comprising the translational start of Lphn2 and a “knock-in–first cassette” ∼230 bp downstream of the respective exon in intronic sequences, with low conservation across different species. The aforementioned knock-in cassette consisted of two chained sets of frt site isoforms (frt and F3 sites; Branda and Dymecki, 2004), which flanked parts of the Engrailed 2 splice acceptor (a strategy basing on vectors available from the Canadian Centre for Modeling Human Disease) fused to the Lphn2-mVenus cDNA, a poly adenylation signal, an artificial spacer sequence, the PGK neoresistance gene, and a 3′ loxP site. The chained set of heterologous frt sites led to the generation of either a constitutive Lphn2cKI allele without the selection cassette or the Lphn2cKO allele. Because of the placement of the loxP sites, also the Lphn2cKI allele was subject to Cre-mediated deletion. The targeting vector was flanked by 4.3-kb (5′) and 4-kb (3′) homology regions, and a diphtheria toxin minigene was inserted for negative selection. Embryonic stem cell culture and selection for homologous recombination was performed using a 129S6xC57BL/6J hybrid mouse embryonic stem cell line. Gene targeting was performed by C. Guo in the Transgenic Facility at Janelia Farms Howard Hughes Medical Institute campus.
Positive homologous recombination and recombination after breeding to Flp-deleter (Rodríguez et al., 2000) and cytomegalovirus-promoter Cre-deleter mice (Schwenk et al., 1995) were confirmed by Southern blotting and PCR. Primers used for genotyping to distinguish WT, Lphn2KI + neo, Lphn2cKI, Lphn2cKO, and Lphn2KO alleles included MX11603 (5′-CCCATTTTCCTAAGAGGAACGCCACGCTAG-3′), MX11604 (5′-GTGTGATGATCAGAGTAACAGCAGTGTATC-3′), MX11618 (5′-TGCAGAGTGTGGCAGATGTTGCTGCAC-3′), and MX11622 (5′-GATCCAATCCCTGGCATGACTGTCTTC-3′). The following oligonucleotide combinations were used to test for the presence of different alleles (amplicon size is shown in parentheses): MX11603/604 Lphn2WT (266 bp), Lphn2cKO and Lphn2cKI (inclusion of 5′ loxP site, 333 bp), MX11603/618 Lphn2KO (286 bp), and MX11622/618 (386 bp) as a 3′ alternative to test for Lphn2cKI.
Primers that have been used to generate probes for Southern blotting analysis included MX10452 (5′-GAGTCTGGGATTGGACCCTCCTAGCAG-3′), MX10453 (5′-TTGAAACAGGCTCAAGTAGGTCAGAC-3′), MX10571 (5′-GTTCGATTTCACAGCCTAGCTAGCGTGC-3′), and MX10572 (5′-TTACATATCTTGAGCTAGTAACGAAATG-3′).
Southern blot probes were generated by PCR using the following oligonucleotide combinations: MX10452/453 (5′) and MX10571/572 (3′). All oligonucleotides were purchased from Integrated DNA Technologies. The original mouse line (Lphn2KI+neo allele) has been submitted to The Jackson Laboratory Mouse Repository for distribution (B6; 129S6-Adgrl2tm/sud/J; Jackson Laboratory stock number 023401).
We attempted to breed all alleles to homozygosity. Lphn2KO/KO mice, however, were embryonic lethal. Lphn2cKI/cKI and Lphn2cKO/cKO mice were viable. Mice were weaned at 21 d of age and housed in groups of two to five on a 12-h light/dark cycle with food and water ad libidum. Stanford Animal Housing Facility: all procedures conformed to National Institutes of Health Guidelines for the Care and Use of Laboratory Mice and were approved by the Stanford University Administrative Panel on Laboratory Animal Care.
Virus production
Lentiviruses
Nuclear-localized EGFP-Cre and EGFP-ΔCre fusion proteins deliverable by lentiviruses were from previously described vectors (Kaeser et al., 2011). For production of lentiviruses, the lentiviral expression vector and three helper plasmids (pRSV-REV, pMDLg/pRRE, and vesicular stomatitis virus G protein) were cotransfected into HEK293T cells (ATCC) at 6, 2, 2, and 2 µg of DNA per 25 cm2 culture area, respectively. Transfections were performed using the calcium-phosphate method. Media with viruses were collected at 48 h after transfection, centrifuged at 500 g for 5 min to pellet cellular debris, filtered (0.45 µm pore size), and ultracentrifuged at 55,000 g for 1.5 h. Pellets were resuspended in MEM at 1/500 of the initial volume, aliquoted, and stored at −80°C.
AAVs
For hippocampal targeting in subsequent behavioral experiments, we used an AAV-DJ strain that had a demonstrated high-infectivity efficiency in vivo (Xu et al., 2012). AAV vectors were constructed from an empty cloning vector where the expression cassette was as follows: left inverted terminal repeat of AAV2, cytomegalovirus promoter, and β-globin intron, and multiple cloning site, hGH poly A sequence, and right inverted terminal repeat. EGFP, inactive (ΔCre), and active (Cre) Cre recombinase were inserted into the multiple cloning sites. AAV plasmids were cotransfected with pHelper and pRC-DJ into HEK293T cells. 72 h after transfection, cells were harvested, lysed, and run on an iodixanol gradient by ultracentrifugation at 400,000 g for 2 h. The 40% iodixanol fraction containing AAV was collected, concentrated, and washed in a 100K molecular weight cutoff ultracon filter. The infectious titer of virus was measured by infecting HEK293T cells with serial dilutions and then was used for stereotaxic infections at 107 infectious U/µl.
Stereotaxic injections
P0 Lphn2 cKO mutant mice were anesthetized for 2 min in ice, and concentrated AAV or lentiviruses were injected with a glass pipette using an infusion pump (Harvard Apparatus). The hippocampus was bilaterally targeted using the following coordinates from Lambda: anterior–posterior, +0.55 mm; medial–lateral, ±1.95 mm; and dorsal–ventral, −2.0 mm. Flow rate was 0.15 µl/min, and injected volume was 0.8–1 µl. Efficiency and localization of viral expression was confirmed by nuclear EGFP expression of inactive and active EGFP–Cre recombinase fusion protein expression by histology.
Electrophysiology
For whole-cell patch clamp physiology experiments, the patch pipettes were pulled from borosilicate glass capillary tubes (TW150-4; World Precision Instruments) using a PC-10 pipette puller (Narishige). The resistance of pipettes filled with intracellular solution varied between 3–5 MΩ. Synaptic currents were monitored with a MultiClamp 700B amplifier (Molecular Devices). The frequency, duration, and magnitude of the extracellular stimulus were controlled with a Model 2100 Isolated Pulse Stimulator (A-M Systems, Inc.) synchronized with Clampex 9 data acquisition software (Molecular Devices). For excitatory voltage-clamp recordings (AMPAR-EPSC and mEPSC), a whole-cell pipette solution was used containing 135 mM Cs-methanesulfonate, 15 mM CsCl, 8 mM NaCl, 10 mM tetraethylammonium-Cl, 10 mM Hepes, 0.2 mM EGTA, 0.3 mM Na-GTP, 4 mM Na-ATP, 0.1 mM spermine, and 10 mM QX-314, pH 7.4 (adjusted with CsOH). The bath solution of artificial cerebral spinal fluid (ACSF) contained 140 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1.3 mM MgCl2, 10 mM Hepes, and 10 mM glucose, pH 7.4 (adjusted with NaOH). AMPAR-EPSCs were pharmacologically isolated by adding the NMDA-receptor blocker AP-5 (50 µM) and the GABAA-receptor blocker picrotoxin (50 µM) to the extracellular bath solution. AMPAR-EPSCs and mEPSC recordings were performed while holding the cell at −70 mV. Spontaneous mEPSCs were monitored in the presence of tetrodotoxin (500 nM) to block action potentials at −70 mV holding potential. Synaptic currents were sampled at 10 kHz and analyzed offline using Clampfit 9 (Molecular Devices) software. Miniature events were analyzed using the template matching search and a minimal threshold of 5 pA, and each event was visually inspected for inclusion or rejection by an experimenter blind to the recording condition.
For acute slice electrophysiology, lentiviruses were injected into P0 mice, and infected CA1 pyramidal neurons were analyzed at P25–35. Horizontal hippocampal slices (300 µm) were prepared by cutting in ice-cold solution containing 85 mM NaCl, 75 mM sucrose, 2.5 mM KCl, 1.3 mM NaH2PO4, 24 mM NaHCO3, 0.5 mM CaCl2, 4 mM MgCl2, and 25 mM d-glucose saturated with 95% O2/5% CO2. Slices were transferred to a holding chamber containing ACSF: 126 mM NaCl, 2.5 mM KCl, 1 mM NaH2PO4, 26.2 mM NaHCO3, 2.5 mM CaCl2, 1.3 mM MgSO4-7H2O, 11 mM d-glucose, and ∼290 mM mOsm. Slices were allowed to recover at 31.5°C for 30 min followed by holding at room temperature for >1 h. Acute slices were transferred to a recording chamber continuously superfused with oxygenated ACSF (1.5 ml/min) maintained at 30.5°C. Neurons were clamped at −70 mV, and two pathways of extracellular-evoked AMPAR-EPSCs in hippocampal slices were monitored. AMPAR-EPSCs were evoked by electrical stimulation by nichrome electrodes positioned at the stratum radiatum proximal to CA3 and the SLM proximal to the entorhinal cortex. The current–voltage relationship of AMPA-R EPSCs was measured at holding potentials from −70 mV to +40 mV.
Antibodies
For immunohistochemistry and immunocytochemistry experiments, the following primary antibodies were used: GluA1 (1:4; rabbit polyclonal; PC246; EMD Millipore), GluA2 (1:30; mouse monoclonal; MAB397; EMD Millipore), PSD95 (1:200; mouse monoclonal; 124-011; Synaptic Systems), gephyrin (1:200; mouse monoclonal; 147-111; Synaptic Systems), vGat (1:500; guinea pig polyclonal; 131-004; Synaptic Systems), vGlut1 (1:500; guinea pig polyclonal; AB5905; EMD Millipore), GFP (1:1,000 immunofluorescence; 1:2,000 immunoblot; rabbit polyclonal; A-11122; Invitrogen), and NeuN (1:500; mouse monoclonal; MAB377; EMD Millipore). The following secondary polyclonal antibodies with fluorophores (Invitrogen) were used: anti–rabbit, anti–mouse, and anti–guinea pig Alexa Fluor 488/546/633 (1:500).
Culture and transfection of hippocampal neurons
Hippocampal neurons were cultured from Lphn2-cKO mice essentially as described previously (Maximov et al., 2007). In brief, primary cortical neurons were isolated from P0–1 mice, dissociated by papain digestion, and plated on Matrigel (BD)-coated glass coverslips in 24-well plates. The neurons were cultured for 14–18 DIV in MEM (Gibco) supplemented with B27 (Gibco), glucose, transferrin, FBS, and Ara-C.
Neuronal transfection was performed 3–4 d after plating using a Ca2+-phosphate method to achieve sparse delivery of plasmids to isolated neurons that could be identified by their GFP expression. A DNA/Ca2+-phosphate precipitate was prepared by mixing the following (per well; 24-well plate): 2 µg of DNA, 2 µl of 2 M CaCl2, and distilled H2O to a volume of 15 µl. DNA mixture was added dropwise under constant vortex to an equal volume of 2× Hepes-buffered saline (274 mM NaCl, 10 mM KCl, 1.4 mM Na2HPO4, 15 mM d-glucose, and 42 mM Hepes, pH 7.05). The precipitate was allowed to form for 10–15 min at room temperature before addition to the cultures. Cultured neurons were placed in serum-free MEM (0.5 ml per well) supplemented with NMDA receptor blocker AP-5 (50 µM) and 10 mM MgCl2. The original conditioned media was saved. 30 µl of the DNA/calcium phosphate precipitate were added dropwise to each well. Dishes were returned to 5% CO2 incubator at 37°C for 30 min. The incubation was stopped by washing the cells twice with 1 ml per well of MEM. The saved conditioned medium was added back to each well, and the cells were returned to the CO2 incubator. The transfection efficiency typically ranged from 1 to 5%.
Immunocytochemistry
Culture neurons
Neurons were fixed with 4% PFA in PBS for 10 min at room temperature, permeabilized by brief exposure to cold methanol (−20°C), followed by 5 min in 0.1% Triton X-100/PBS. Cells were placed in blocking buffer (10% donkey serum/PBS) for 1 h, incubated with diluted primary antibodies in 2% donkey serum/PBS for 1 h, washed 3× for 5 min in PBS, incubated with diluted secondary antibodies in 2% donkey serum/PBS for 1 h, washed 3× in PBS, and mounted on uncharged UltraClear microslides (Denville Scientific) using DAPI Fluoromount-G (Southern Biotech).
GluA1 and GluA2 surface labeling
GluA1 and GluA2 surface labeling was performed essentially as described previously (Aoto et al., 2013). In brief, culture neurons were washed with PBS containing 0.5 mM CaCl2 and 1 mM MgCl2 (PBSMC) with 4% sucrose. Neurons were preincubated at 37°C for 5 min with primary antibodies against GluA1 to allow labeling of surface AMPA receptors, washed with ice-cold PBSMC, fixed with 4% PFA + 4% sucrose for 15 min, and then blocked in a detergent-free blocking solution (PBS with 2% normal goat serum [Sigma-Aldrich] and 0.02% sodium azide) for 1 h followed by secondary antibody incubation at room temperature for 1 h, and then cells were mounted and imaged. GluA1/GluA2 ratio was calculated as (GluA1 puncta density * area * intensity)/(GluA2 puncta density * area * intensity) for the same stretch of analyzed dendrite.
Imaging of cultured hippocampal neurons
For neuron cell culture experiments, dendritic morphology was visualized by sparse calcium phosphate transfection of a GFP-expressing construct (L316). Neurons were selected for imaging and analysis based on pyramidal neuron morphology exhibited by prominent apical dendrites as well as the presence of spine structures protruding off dendrites. Images were acquired using a 60× objective with 4× digital zoom on an A1 Eclipse Ti confocal microscope with constant image settings operated by NIS-Elements Advanced Research v4.5 acquisition software (Nikon). Z stack images were converted to maximal projection images and analyzed using MetaMorph Software (Molecular Devices) with synaptic puncta quantified for puncta density per 10 µm of dendrite, size, and intensity.
Morphological analyses of hippocampal CA1-region neurons
For CA1-region pyramidal neuron morphological analyses, neurons from acute hippocampal slices (described in the Electrophysiology section) were filled for 10–15 min with 0.2% biocytin (Sigma-Aldrich) in whole-cell pipette solution. After filling neurons, the patch pipette was slowly withdrawn, carefully monitoring for resealing of the membrane. Slices were then fixed with 4% PFA in PBS for 1–2 h at room temperature and washed 3× for 5 min in PBS. For immunohistochemistry experiments without biocytin loading, 50 µm coronal serial sections were directly placed in fixative buffer. Slices were then permeabilized and blocked with 10% normal goat serum and 0.5% Triton X-100 in PBS for 1 h. If immunohistochemistry was to be performed, slices were then incubated with primary antibody diluted in 2% normal goat serum and 0.01% Triton X-100 in PBS for >2 h. Slices were washed 3× for 5 min in PBS and then incubated with secondary antibody diluted in 2% normal goat serum and 0.01% Triton X-100 in PBS for >2 h. Biocytin-filled neurons were also visualized at this step with the addition of 2 µg/ml streptavidin Alexa Fluor 555 conjugate (S21381; Molecular Probes). Slices were washed 3× for 5 min in PBS and then mounted on uncharged UltraClear microslides (Denville Scientific) using DAPI Vectashield (Vector Laboratories). Dendrites from three neuronal regions were selected for imaging and analysis, including the stratum oriens, stratum radiatum, and SLM. Z stacks of images were taken using a 40× objective with 4× digital zoom on an A1 Eclipse Ti confocal microscope. Spine analysis was performed on selected secondary/tertiary dendrites, with spines manually identified and quantified for density in NIS-Elements Advanced Research v4.5 analysis software. Each neuron was analyzed across multiple dendritic branches, pooling data to calculate mean values.
Immunolabeling and imaging of whole-brain sections
Mice were anesthetized and perfused transcardially with 5 ml PBS and 50 ml freshly prepared 4% PFA. Brains were dissected and postfixed in 4% PFA overnight at 4°C. Brains were briefly rinsed in PBS, and 50-µm coronal serial sections were collected using a Vibratome VT100S (Leica Biosystems). For immunofluorescence staining, sections were washed 3× in PBS for 10 min under gentle agitation and incubated for 1 h in blocking solution containing 10% horse serum (Thermo Fisher Scientific), 0.2% BSA (Sigma-Aldrich), and 0.5% Triton X-100 in PBS. Subsequently, sections were transferred into carrier solution containing 1% horse serum, 0.2% BSA, and 0.5% Triton X-100 in PBS, to which the primary antibody of choice was added. The sections were incubated overnight at 4°C on an orbital shaker. On the following day, the sections were washed three times with PBS for 10 min each, and secondary antibody staining was performed overnight as in the previous step. On the last day, sections were washed three times in PBS for 10 min each and finally mounted on uncharged UltraClear microslides (Denville Scientific) using DAPI Fluoromount-G (Southern Biotech). Images were collected using a 4× objective on a VS120 slide-scanning microscope (Olympus).
Quantitative RT-PCR analysis
For quantitative RT-PCR brains of adults, age-matched mice (n = 4 per group) were dissected and immediately collected in TRIzol reagent (Invitrogen) for isolation according to the manufacturer’s protocol. Conditions for quantitative RT-PCR assays, including primer/probe sequences, temperature cycling, and data collections, were performed as described previously (Boucard et al., 2014). The Lphn2 assay in this study was designed to determine the presence or absence of exon 1 (Integrated DNA Technologies): probe, 5′-CCCGAATGGTAAGGCTGCTCGC-3′; forward, 5′-GTTTCTCACCGAGTACAGAAGG-3′; and reverse, 5′-CACAGGATAGCTCTCGTCTAAC-3′.
Behavioral analyses
For assessing the behavior of mice with Lphn2 deletions throughout the brain, Lphn2cKO mice were crossed with a Nestin-Cre driver line (Tronche et al., 1999). Progeny were backcrossed to Lphn2 cKO mice to yield Lphn2 cKO/Nestin-Cre+/−−mice, which were crossed with each other to produce Lphn2 cKO littermates with or without the Nestin-Cre transgene, which was genotyped with the following primers: 5′-GCCTGCATTACCGGTCGATGC-3′ and 5′-GTGGCAGATGGGGCGGCAACA-3′. Stereotactic injections of AAVs into the hippocampus for behavioral analyses were performed on litters of Lphn2 cKO mice, such that littermates received either AAVs encoding Cre recombinase or control ΔCre recombinase. Matching sex sets of Cre− and Cre+ littermate mice were housed together until behavior experiments were performed at 2–3 mo of age. Experiments were performed during the 12-h light cycle period. Behavioral testing was performed in the following order: open field, grip test, accelerating rotarod, olfaction assay, Y-maze, tail suspension test, prepulse inhibition, and fear conditioning. Mice were allowed to rest for at least 7 d between tests. Animal experiments were conducted following protocols approved by Administrative Panel on Laboratory Animal Care at Stanford University. The following specific tests were performed:
Water T-maze
The maze was constructed with the following dimensions: start zone, 30 cm length × 10 cm width; and upper perpendicular zone, 70 cm length × 10 cm width. Maze was filled with 10 cm water. The first swim was performed using a one-armed T-maze. With the left arm of the T-maze blocked off, mice were placed at the base of the start zone, requiring a swim to a rescue platform positioned at the end of the right arm of the maze. Upon completion of the first swim, mice were placed in a dry holding chamber for 30 s while the maze was opened to a two-armed maze, and the platform was switched to the opposite arm. Mice were then placed at the base of the start zone to start the second forced swim. Upon reaching the rescue platform, this concluded one trial. Time to swim to the platform was monitored in addition to whether a directional error occurred upon reaching the T junction. A maximum swim time of 180 s was allowed for any given trial. Mice were trained for four consecutive days at five trials per day.
To eliminate a habitual component to the above described water T-maze protocol and to focus on the cognitive flexibility component, a randomized first swim protocol was instead used. The maze used was similar as described above. The first swim was performed similarly as described above using a one-armed T-maze except that the platform was assigned to the left or right arm of the maze in a randomized manner. With a randomized arm of the T-maze blocked off, mice were placed at the base of the start zone, requiring a swim to a rescue platform positioned at the end of the other arm of the maze. Upon completion of the first swim, mice were placed in a dry holding chamber for 30 s while the maze was opened to a two-armed maze, and the platform was switched to the opposite arm. Mice were then placed at the base of the start zone to start the second swim. Upon reaching the rescue platform, this concluded one trial. Time to swim to the platform was monitored in addition to whether a directional error occurred upon reaching the T junction. Mice were trained for four consecutive days at five trials per day.
Barnes Maze
2–3-mo-old mice were put through a modified version of a Barnes maze protocol described previously (Sunyer, 2007). The maze consists of a brightly lit circular open platform (92 cm diameter) with 20 equally spaced holes (hole diameter, 5 cm) along the perimeter. Underneath the designated target hole, an escape box (7 cm deep, 7 cm width, and 10 cm length) was placed. Underneath the remaining holes, false escape boxes lacking depth were placed, made of same color/texture material as the escape box. Extra-maze cues were placed on the surrounding walls to serve as reference cues to learn the position of the target escape hole. To begin the maze, mice were placed in the center of the maze in a holding chamber (15 cm × 15 cm) for 30 s. The chamber was then lifted, and the mice were free to explore the maze with 19 of the 20 holes closed and were assayed for their ability to spatially navigate the maze to find the target escape hole. The target escape box was positioned underneath the maze as a small dark recessed chamber, which the mice naturally sought out, taking advantage of their desire to escape brightly lit and exposed environments. During the initial four-consecutive-day training period, the mice learned the spatial location of the target hole, with four trials conducted per day (∼2-h intertrial interval). Parameters measured included the time to target hole (latency) as well as time spent within the target hole (affinity). Memory of the target hole was measured as a single trial, conducted at 1 d and 14 d after the initial training period. Data acquisition and analysis was performed using BIOSERVE Viewer v6.1 video tracking software (VMware).
Fear conditioning
2–3-mo-old mice were handled daily for 5 d before training. On training day, mice were placed in a fear-conditioning chamber (H10-11M-TC; Coulbourn Instruments) located in the center of a sound-attenuating cubicle (Coulbourn Instruments). The conditioning chamber was cleaned with 10% ethanol to provide a background odor. A ventilation fan provided a background noise at ∼55 dB. After a 2-min exploration period, three tone–footshock pairings separated by 1-min intervals were delivered. The 85-dB 2-kHz tone lasted for 30 s, and the footshocks were 0.75 mA and lasted for 2 s. The footshocks coterminated with the tone. The mice remained in the training chamber for another 30 s before being returned to home cages. In a context test, mice were placed back into the original conditioning chamber for 5 min. The altered-context and tone tests were conducted in a new room. The same conditioning chamber was moved to this room and was modified by changing its metal grid floor to a plastic sheet, white metal side walls to plastic walls decorated with red stripes, and background odor of ethanol to vanilla. The ventilation fan was turned off to reduce background noise. Mice were placed in the altered chamber for 5 min to measure the freeze level in the altered context, and after this 5 min period, a tone (85 dB and 2 kHz) was delivered for 1 min to measure the freeze to tone. The behavior of the mice was recorded with the Freezeframe software and analyzed with Freezeview software (Coulbourn Instruments). Motionless bouts lasting >1 s were considered as freeze.
Open-field force plate analysis
Mice were analyzed on a force-plate actometer as described previously (Fowler et al., 2001). Analyses were performed before other behavioral assays were performed and were initiated by placing mice individually into the center of a force-plate actometer (28 cm × 28 cm) for a period of 30 min, analyzed in 5-min bins.
Grip test
Animals were placed on top of a wire mesh grid. The grid was then shaken lightly three times to cause the mouse to grip the wires and was then turned upside down. The mesh was held ∼20 cm above the home cage bedding, high enough to prevent the mouse from climbing down but not to cause harm in the event of a fall. Three trials per mouse were performed with a 1-min intertrial interval. The time taken by the animal to fall off the grid was recorded using a stopwatch. The mean of the three trials was assigned as the latency of each animal to fall.
Accelerating rotarod
An accelerating rotarod (Med Associates Inc.) was used to assess motor coordination and motor learning. Mice were placed on the rotarod, which slowly accelerated from 6 to 60 rounds per minute over 5 min. Mice were given three trials per day for 3 d with 30–40-min intertrial intervals. Time to fall from the rotarod (latency) was recorded for each trial. Mice that remained on the rotarod for the whole 5-min trial were assigned a 300-s latency.
Olfactory assay
Mice were food deprived 18–14 h before the test. The next day, each mouse was placed in a clean cage (the test cage) and allowed to acclimate for 5 min. The animal was then returned to its home cage, and a food pellet was buried in one of the corners ∼1 cm beneath the surface of the test cage. Then, the mouse was transferred back to the test cage and placed at the opposite corner from where the food pellet was buried. The latency for each mouse to retrieve the food pellet was recorded. Mice that failed to locate the food pellet after 5 min were assigned a 300-s latency.
Tail suspension test
Mice were suspended by their tail for 6 min and videotaped. The time spent immobile was recorded.
Acoustic startle response and prepulse inhibition
Mice were placed in sound-attenuated test chambers (Kinder Scientific) and allowed a 5-min adaptation period before the start of the testing session. Background white noise (65 dB) was present during this adaptation period and throughout the testing session. Testing consisted of 50 acoustic startle trials using five different stimulus intensities (75, 85, 95, 105, and 115 dB) presented 10 times in a pseudo-randomized order so that an equal number of presentations of each stimulus intensity is included in each half of the session and no single intensity is presented more than two times in succession. The stimuli were 50-ms sound bursts separated by pseudorandomized intertrial intervals within a range of 5–30 s (15 s mean), so that the animals did not anticipate the stimulus. The mean magnitude of the startle response for every stimulus intensity was recorded for each animal. The next day, the animals were tested for prepulse inhibition and habituation of the acoustic startle response. After 5 min of adaptation inside the test chambers, mice were presented with four trial types in three blocks. The first and third block consisted of ten 50-ms sound bursts of 115 dB. The second block included four different types of acoustic stimuli in a randomized order: pulse alone (115 dB sound burst for 50 ms) and three separate prepulse + pulse combinations, with prepulse set at three sound levels of 68, 71, and 77 dB for 20 ms, followed by a 50-ms pulse at 115 dB. There was a 100-ms gap between the prepulse and the pulse. A total number of 10 trials under each acoustic stimulus condition were presented, with mean 15-s variable intervals ranging from 5 s to 30 s. As with standard startle testing, trial types and intertrial interval were presented in a quasi-random balanced manner with equal representations of trial types and intervals in each half of the session. The mean magnitude of the startle response from blocks 1 and 3 was used to estimate habituation of the acoustic startle. Percent prepulse inhibition was calculated from the data of the second block as follows: (1 − [mean startle response to prepulse + pulse] ÷ [mean startle response to pulse alone]) * 100.
Novel object recognition
Mice were habituated three times in an empty white box for 15 min, placing the mouse back into the home cage for 5 min between habituations. Two identical objects were placed 10 cm apart in the white box, and mice explored the objects for 5 min. After returning the mice to their home cage for 5 min, they explored the objects for an additional 15 min. Mice were then placed into their home cage for 1 h. One of the objects was exchanged for a novel object, and the mice explored both old and novel objects for 5 min. The amount of time directly interacting with and investigating the old and novel object was recorded.
Data availability
All relevant data presented in this study are available from the authors.
Statistics
Experiments were performed with the experimenter blinded to the conditions being tested. Data are shown as means ± SEM. Student’s t tests were performed as one-tailed t tests, assuming normal distributions. Statistically significant differences are indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Water T-maze behavioral data were analyzed using factorial ANOVA, with genotype or virus as a between-subject factor and trial as a repeated-measure. Genotype, trial, and genotype × trial interactions were analyzed by two-way ANOVA followed by Sidak’s multiple comparisons post hoc tests. Type I error rate was set at α = 0.05 (two-tailed) for all statistical comparisons. When ANOVA results are presented in figures, significant effects are highlighted by bold and italic font. Individual trial significant effects are marked by asterisks in the summary plots, and comparisons that are not significant are not identified.
Online supplemental material
Fig. S1 shows strategy for the generation and validation of Lphn2 mutant mice. Fig. S2 shows experimental strategies used for stereotactic injections of viruses into the hippocampal CA1 region. Fig. S3 shows further analysis of the brain-specific deletion of Lphn2 mediated by crossing Nestin-Cre mice with Lphn2 cKO mice. Fig. S4 shows how deletion of Lphn2 by stereotactic delivery of AAV-Cre viruses into the CA1 region of the hippocampus does not alter open field or novel object recognition behaviors
Supplementary Material
Acknowledgments
This paper was supported by a grant from National Institute of Mental Health (MH052804) and by a fellowship from Autism Speaks (7953) to G.R. Anderson.
The authors declare no competing financial interests.
Author contributions: G.R. Anderson designed, performed, and analyzed most experiments; S. Maxeiner generated and initially characterized Lphn2-KI and Lphn2-cKO mice; R. Sando and T. Tsetsenis performed behavior analyses; G.R. Anderson and T.C. Südhof designed experiments and analyzed data; G.R. Anderson, R.C. Malenka, and T.C. Südhof wrote the paper with input from all of the authors.
Footnotes
Abbreviations used:
- AAV
- adeno-associated virus
- ACSF
- artificial cerebral spinal fluid
- AMPAR
- AMPA-type glutamate receptor
- cKI
- conditional knock-in
- cKO
- conditional KO
- DIV
- d in vitro
- EPSC
- excitatory postsynaptic current
- GPCR
- G protein–coupled receptor
- KO
- knockout
- Lphn
- latrophilin
- mEPSC
- miniature EPSC
- SLM
- stratum lacunosum-moleculare
- vGat
- vesicular GABA transporter
References
- Akins M.R., and Biederer T.. 2006. Cell-cell interactions in synaptogenesis. Curr. Opin. Neurobiol. 16:83–89. 10.1016/j.conb.2006.01.009 [DOI] [PubMed] [Google Scholar]
- Aoto J., Martinelli D.C., Malenka R.C., Tabuchi K., and Südhof T.C.. 2013. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 154:75–88. 10.1016/j.cell.2013.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araç D., Boucard A.A., Bolliger M.F., Nguyen J., Soltis S.M., Südhof T.C., and Brunger A.T.. 2012. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 31:1364–1378. 10.1038/emboj.2012.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.A. 1979. Memory deficits associated with senescence: a neurophysiological and behavioral study in the rat. J. Comp. Physiol. Psychol. 93:74–104. 10.1037/h0077579 [DOI] [PubMed] [Google Scholar]
- Boucard A.A., Ko J., and Südhof T.C.. 2012. High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J. Biol. Chem. 287:9399–9413. 10.1074/jbc.M111.318659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard A.A., Maxeiner S., and Südhof T.C.. 2014. Latrophilins function as heterophilic cell-adhesion molecules by binding to teneurins: regulation by alternative splicing. J. Biol. Chem. 289:387–402. 10.1074/jbc.M113.504779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda C.S., and Dymecki S.M.. 2004. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell. 6:7–28. 10.1016/S1534-5807(03)00399-X [DOI] [PubMed] [Google Scholar]
- Davletov B.A., Shamotienko O.G., Lelianova V.G., Grishin E.V., and Ushkaryov Y.A.. 1996. Isolation and biochemical characterization of a Ca2+-independent alpha-latrotoxin-binding protein. J. Biol. Chem. 271:23239–23245. 10.1074/jbc.271.38.23239 [DOI] [PubMed] [Google Scholar]
- de Wit J., and Ghosh A.. 2016. Specification of synaptic connectivity by cell surface interactions. Nat. Rev. Neurosci. 17:22–35. [DOI] [PubMed] [Google Scholar]
- Eichenbaum H. 2004. Hippocampus: cognitive processes and neural representations that underlie declarative memory. Neuron. 44:109–120. 10.1016/j.neuron.2004.08.028 [DOI] [PubMed] [Google Scholar]
- Fowler S.C., Birkestrand B.R., Chen R., Moss S.J., Vorontsova E., Wang G., and Zarcone T.J.. 2001. A force-plate actometer for quantitating rodent behaviors: illustrative data on locomotion, rotation, spatial patterning, stereotypies, and tremor. J. Neurosci. Methods. 107:107–124. 10.1016/S0165-0270(01)00359-4 [DOI] [PubMed] [Google Scholar]
- Goodhill G.J. 2016. Can Molecular Gradients Wire the Brain? Trends Neurosci. 39:202–211. 10.1016/j.tins.2016.01.009 [DOI] [PubMed] [Google Scholar]
- Hamann J., Aust G., Araç D., Engel F.B., Formstone C., Fredriksson R., Hall R.A., Harty B.L., Kirchhoff C., Knapp B., et al. 2015. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev. 67:338–367. 10.1124/pr.114.009647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W., Mosca T.J., and Luo L.. 2012. Teneurins instruct synaptic partner matching in an olfactory map. Nature. 484:201–207. 10.1038/nature10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K.M., Ito H.T., Moser E.I., and Moser M.-B.B.. 2014. Functional diversity along the transverse axis of hippocampal area CA1. FEBS Lett. 588:2470–2476. 10.1016/j.febslet.2014.06.004 [DOI] [PubMed] [Google Scholar]
- Isaac J.T., Ashby M.C., and McBain C.J.. 2007. The role of the GluR2 subunit in AMPA receptor function and synaptic plasticity. Neuron. 54:859–871. 10.1016/j.neuron.2007.06.001 [DOI] [PubMed] [Google Scholar]
- Kaeser P.S., Deng L., Wang Y., Dulubova I., Liu X., Rizo J., and Südhof T.C.. 2011. RIM proteins tether Ca2+ channels to presynaptic active zones via a direct PDZ-domain interaction. Cell. 144:282–295. 10.1016/j.cell.2010.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T., Macdonald C.J., and Tonegawa S.. 2015. Entorhinal-hippocampal neuronal circuits bridge temporally discontiguous events. Learn. Mem. 22:438–443. 10.1101/lm.038687.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnoperov V.G., Beavis R., Chepurny O.G., Little A.R., Plotnikov A.N., and Petrenko A.G.. 1996. The calcium-independent receptor of alpha-latrotoxin is not a neurexin. Biochem. Biophys. Res. Commun. 227:868–875. 10.1006/bbrc.1996.1598 [DOI] [PubMed] [Google Scholar]
- Krasnoperov V.G., Bittner M.A., Beavis R., Kuang Y., Salnikow K.V., Chepurny O.G., Little A.R., Plotnikov A.N., Wu D., Holz R.W., and Petrenko A.G.. 1997. α-Latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron. 18:925–937. 10.1016/S0896-6273(00)80332-3 [DOI] [PubMed] [Google Scholar]
- Kreienkamp H.-J.J., Soltau M., Richter D., and Böckers T.. 2002. Interaction of G-protein-coupled receptors with synaptic scaffolding proteins. Biochem. Soc. Trans. 30:464–468. [DOI] [PubMed] [Google Scholar]
- Langenhan T., Prömel S., Mestek L., Esmaeili B., Waller-Evans H., Hennig C., Kohara Y., Avery L., Vakonakis I., Schnabel R., and Russ A.P.. 2009. Latrophilin signaling links anterior-posterior tissue polarity and oriented cell divisions in the C. elegans embryo. Dev. Cell. 17:494–504. 10.1016/j.devcel.2009.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenhan T., Aust G., and Hamann J.. 2013. Sticky signaling--adhesion class G protein-coupled receptors take the stage. Sci. Signal. 6:re3 10.1126/scisignal.2003825 [DOI] [PubMed] [Google Scholar]
- Langenhan T., Piao X., and Monk K.R.. 2016. Adhesion G protein-coupled receptors in nervous system development and disease. Nat. Rev. Neurosci. 17:550–561. 10.1038/nrn.2016.86 [DOI] [PubMed] [Google Scholar]
- Lelianova V.G., Davletov B.A., Sterling A., Rahman M.A., Grishin E.V., Totty N.F., and Ushkaryov Y.A.. 1997. α-latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. J. Biol. Chem. 272:21504–21508. 10.1074/jbc.272.34.21504 [DOI] [PubMed] [Google Scholar]
- Lu Y.C., Nazarko O.V., Sando R. III, Salzman G.S., Südhof T.C., and Araç D.. 2015. Structural Basis of Latrophilin-FLRT-UNC5 Interaction in Cell Adhesion. Structure. 23:1678–1691. (published erratum appears in Structure 2016. 24:348) 10.1016/j.str.2015.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C.J., Lepage K.Q., Eden U.T., and Eichenbaum H.. 2011. Hippocampal “time cells” bridge the gap in memory for discontiguous events. Neuron. 71:737–749. 10.1016/j.neuron.2011.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximov A., Pang Z.P., Tervo D.G., and Südhof T.C.. 2007. Monitoring synaptic transmission in primary neuronal cultures using local extracellular stimulation. J. Neurosci. Methods. 161:75–87. 10.1016/j.jneumeth.2006.10.009 [DOI] [PubMed] [Google Scholar]
- Missler M., Südhof T.C., and Biederer T.. 2012. Synaptic cell adhesion. Cold Spring Harb. Perspect. Biol. 4:a005694 10.1101/cshperspect.a005694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca T.J. 2015. On the Teneurin track: a new synaptic organization molecule emerges. Front. Cell. Neurosci. 9:204 10.3389/fncel.2015.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca T.J., Hong W., Dani V.S., Favaloro V., and Luo L.. 2012. Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature. 484:237–241. 10.1038/nature10923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A., Winkler J., Fiedler F., Sastradihardja T., Binder C., Schnabel R., Kungel J., Rothemund S., Hennig C., Schöneberg T., and Prömel S.. 2015. Oriented Cell Division in the C. elegans Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1. PLoS Genet. 11:e1005624 10.1371/journal.pgen.1005624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan M.L., de Wit J., Savas J.N., Comoletti D., Otto-Hitt S., Yates J.R. III, and Ghosh A.. 2012. FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron. 73:903–910. 10.1016/j.neuron.2012.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan M.L., Martini F., von Daake S., Comoletti D., and Ghosh A.. 2014. LPHN3, a presynaptic adhesion-GPCR implicated in ADHD, regulates the strength of neocortical layer 2/3 synaptic input to layer 5. Neural Dev. 9:7 10.1186/1749-8104-9-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paavola K.J., and Hall R.A.. 2012. Adhesion G protein-coupled receptors: signaling, pharmacology, and mechanisms of activation. Mol. Pharmacol. 82:777–783. 10.1124/mol.112.080309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastalkova E., Itskov V., Amarasingham A., and Buzsáki G.. 2008. Internally generated cell assembly sequences in the rat hippocampus. Science. 321:1322–1327. 10.1126/science.1159775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissner C., Runkel F., and Missler M.. 2013. Neurexins. Genome Biol. 14:213 10.1186/gb-2013-14-9-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remondes M., and Schuman E.M.. 2004. Role for a cortical input to hippocampal area CA1 in the consolidation of a long-term memory. Nature. 431:699–703. 10.1038/nature02965 [DOI] [PubMed] [Google Scholar]
- Rodríguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., and Dymecki S.M.. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139–140. 10.1038/75973 [DOI] [PubMed] [Google Scholar]
- Schwenk F., Baron U., and Rajewsky K.. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23:5080–5081. 10.1093/nar/23.24.5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiradake E., Jones E.Y., and Klein R.. 2016. Structural Perspectives on Axon Guidance. Annu. Rev. Cell Dev. Biol. 32:577–608. 10.1146/annurev-cellbio-111315-125008 [DOI] [PubMed] [Google Scholar]
- Sheng M., and Kim E.. 2000. The Shank family of scaffold proteins. J. Cell Sci. 113:1851–1856. [DOI] [PubMed] [Google Scholar]
- Silva J.-P.P., Lelianova V.G., Ermolyuk Y.S., Vysokov N., Hitchen P.G., Berninghausen O., Rahman M.A., Zangrandi A., Fidalgo S., Tonevitsky A.G., et al. 2011. Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc. Natl. Acad. Sci. USA. 108:12113–12118. 10.1073/pnas.1019434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruston N. 2008. Pyramidal neurons: dendritic structure and synaptic integration. Nat. Rev. Neurosci. 9:206–221. 10.1038/nrn2286 [DOI] [PubMed] [Google Scholar]
- Südhof T.C. 2001. alpha-Latrotoxin and its receptors: neurexins and CIRL/latrophilins. Annu. Rev. Neurosci. 24:933–962. 10.1146/annurev.neuro.24.1.933 [DOI] [PubMed] [Google Scholar]
- Südhof T.C. 2008. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 455:903–911. 10.1038/nature07456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S., Ichtchenko K., Khvotchev M., and Südhof T.C.. 1998. alpha-Latrotoxin receptor CIRL/latrophilin 1 (CL1) defines an unusual family of ubiquitous G-protein-linked receptors. G-protein coupling not required for triggering exocytosis. J. Biol. Chem. 273:32715–32724. 10.1074/jbc.273.49.32715 [DOI] [PubMed] [Google Scholar]
- Sugita S., Khvochtev M., and Südhof T.C.. 1999. Neurexins are functional alpha-latrotoxin receptors. Neuron. 22:489–496. [DOI] [PubMed] [Google Scholar]
- Suh J., Rivest A.J., Nakashiba T., Tominaga T., and Tonegawa S.. 2011. Entorhinal cortex layer III input to the hippocampus is crucial for temporal association memory. Science. 334:1415–1420. 10.1126/science.1210125 [DOI] [PubMed] [Google Scholar]
- Sunyer B.S. Patil, H. Höger, and G. Lubec. 2007. Barnes maze, a useful task to assess spatial reference memory in the mice. Protocol Exchange 10.1038/nprot.2007.390 [DOI]
- Takács V.T.T., Klausberger T., Somogyi P., Freund T.F.F., and Gulyás A.I.. 2012. Extrinsic and local glutamatergic inputs of the rat hippocampal CA1 area differentially innervate pyramidal cells and interneurons. Hippocampus. 22:1379–1391. 10.1002/hipo.20974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobaben S., Südhof T.C., and Stahl B.. 2000. The G protein-coupled receptor CL1 interacts directly with proteins of the Shank family. J. Biol. Chem. 275:36204–36210. 10.1074/jbc.M006448200 [DOI] [PubMed] [Google Scholar]
- Tobaben S., Südhof T.C., and Stahl B.. 2002. Genetic analysis of alpha-latrotoxin receptors reveals functional interdependence of CIRL/latrophilin 1 and neurexin 1 alpha. J. Biol. Chem. 277:6359–6365. 10.1074/jbc.M111231200 [DOI] [PubMed] [Google Scholar]
- Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P.C., Bock R., Klein R., and Schütz G.. 1999. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23:99–103. 10.1038/12703 [DOI] [PubMed] [Google Scholar]
- Xu W., Morishita W., Buckmaster P.S., Pang Z.P., Malenka R.C., and Südhof T.C.. 2012. Distinct neuronal coding schemes in memory revealed by selective erasure of fast synchronous synaptic transmission. Neuron. 73:990–1001. 10.1016/j.neuron.2011.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev S., and Shen K.. 2014. Cellular and molecular mechanisms of synaptic specificity. Annu. Rev. Cell Dev. Biol. 30:417–437. 10.1146/annurev-cellbio-100913-012953 [DOI] [PubMed] [Google Scholar]
- Zipursky S.L., and Sanes J.R.. 2010. Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell. 143:343–353. 10.1016/j.cell.2010.10.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data presented in this study are available from the authors.