The DNA helicase BLM can have both pro- and anti-recombination effects. Patel et al. show that in cells lacking the homologous recombination repair factors BRCA1, XRCC2, or BRCA2, BLM prevents the assembly of stable RAD51 nucleoprotein filaments, leading to genomic instability and reduced cell viability.
Abstract
The BLM gene product, BLM, is a RECQ helicase that is involved in DNA replication and repair of DNA double-strand breaks by the homologous recombination (HR) pathway. During HR, BLM has both pro- and anti-recombinogenic activities, either of which may contribute to maintenance of genomic integrity. We find that in cells expressing a mutant version of BRCA1, an essential HR factor, ablation of BLM rescues genomic integrity and cell survival in the presence of DNA double-strand breaks. Improved genomic integrity in these cells is linked to a substantial increase in the stability of RAD51 at DNA double-strand break sites and in the overall efficiency of HR. Ablation of BLM also rescues RAD51 foci and HR in cells lacking BRCA2 or XRCC2. These results indicate that the anti-recombinase activity of BLM is of general importance for normal retention of RAD51 at DNA break sites and regulation of HR.
Introduction
Individuals with biallelic mutations in the BLM gene are affected by Bloom syndrome (BS), a heritable condition associated with developmental abnormalities and susceptibility to a range of malignancies at an early age (Ellis et al., 1995). The BLM gene product is a helicase of the RECQ family with roles in DNA replication and repair. BLM protein acts at several steps of the homologous recombination (HR) pathway for DNA double-strand break (DSB) repair (Larsen and Hickson, 2013). First, BLM, along with the endonuclease Dna2, contributes to resection of DNA DSBs to generate a single-stranded intermediate that is bound by replication protein A (RPA) and RAD51 (Gravel et al., 2008; Nimonkar et al., 2008, 2011). The RAD51 nucleoprotein filament then pairs with matching sequence in a homologous DNA template, leading to strand invasion and creation of a “D-loop” structure. This process can be inhibited by BLM, representing a potential anti-recombinogenic effect of the protein (van Brabant et al., 2000; Hu et al., 2001; Wu and Hickson, 2003; Bachrati et al., 2006; Bugreev et al., 2007). After resynthesis of DNA across the break site, BLM resolves heteroduplex recombination intermediates by dissolving Holliday junctions, restoring separate DNA duplexes (Wu and Hickson, 2003). The ability of BLM to dissolve Holliday junctions limits the frequency of genetic exchanges between homologous sequences during HR. This is consistent with a marked increase in sister chromatid exchanges (SCEs) in BS cells (Chaganti et al., 1974; Hu et al., 2001).
The ability of BLM to limit crossover resolution of HR intermediates has been suggested to represent its key activity in limiting genomic instability (Luo et al., 2000). According to this model, the absence of BLM leads to an excessive number of loss-of-heterozygosity events owing to increased crossover recombination, which leads to malignancy. BS cells also show an increase in chromosome breaks and rearrangements, potentially indicating that BLM provides one or more additional repair activities (Chu et al., 2010). This activity may be related to the pro-recombinogenic role of BLM during DSB resection or an anti-recombinogenic effect around the time of D-loop formation.
In this study, we use a genetic approach to test whether pro- or anti-recombinogenic activities of BLM are most relevant for maintenance of genomic integrity in mammalian cells. We find that BLM contributes significantly to genomic instability in cells in which key HR factors are missing, suggesting that the anti-recombinogenic role of BLM has the potential to exert a significant influence on the efficiency of HR in cancer cells. BLM appears to exert this effect by displacing RAD51 from resected DNA intermediates in a process that is dependent on BLM helicase activity but does not require association with DNA topoisomerase IIIα.
Results
Ablation of Blm rescues genomic instability and cell survival in Brca1Δ11/Δ11 cells
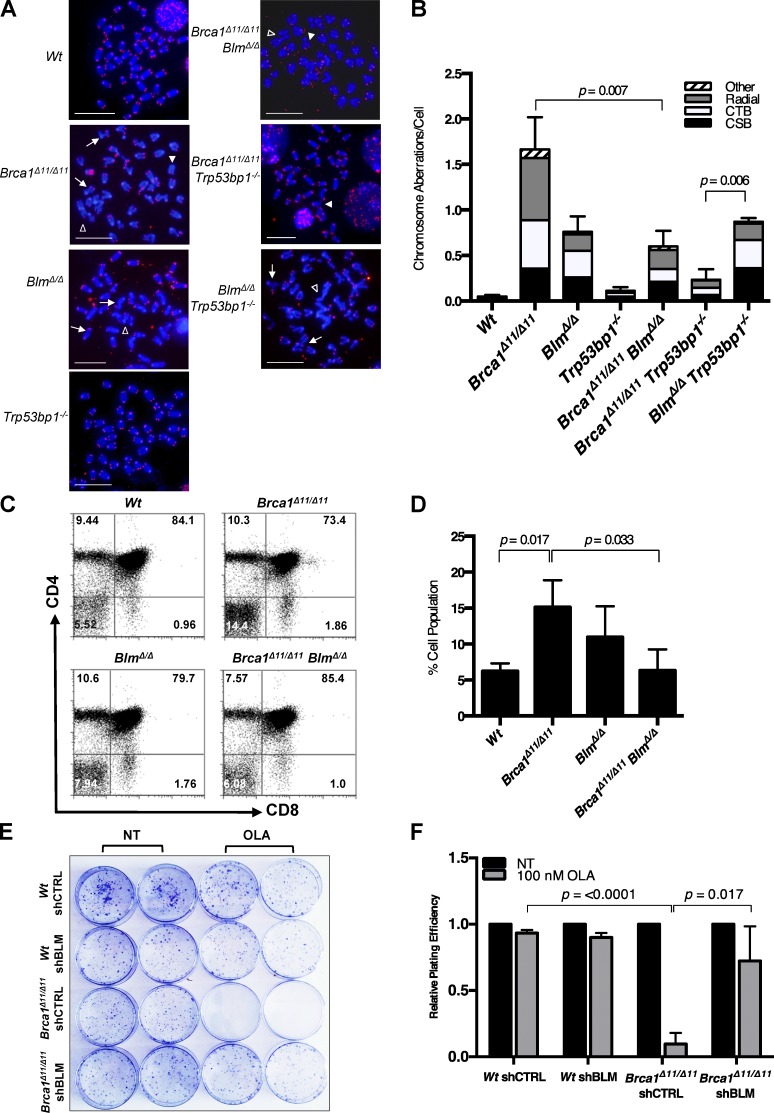
BLM has been shown to act with the endonuclease Dna2 to promote formation of 3′ single-stranded overhangs at DSB sites (Gravel et al., 2008; Nimonkar et al., 2008, 2011). This process contributes to the efficiency of HR; however, the exonuclease Exo1 can perform a similar function in mammalian cells. To evaluate the importance of BLM in generating 3′ single-stranded regions at DSBs, we used mice with conditional deletion of Blm in the B lymphocyte lineage, crossed to Trp53bp1−/− mice (Fig. 1, A and B; and Fig. S1 A; Rickert et al., 1997; Ward et al., 2004; Chester et al., 2006). BlmΔ/Δ cells showed an increased frequency of chromosome aberrations after exposure to either olaparib or mitomycin C, which induce DSBs in dividing cells. Trp53bp1−/− mice lack 53BP1, a negative regulator of DSB resection (Bunting et al., 2010; Chapman et al., 2012; Hakim et al., 2012). We reasoned that increased formation of 3′ single-stranded overhangs at DSBs in Trp53bp1−/− mice might rescue genomic instability arising from loss of the DSB resection activity of BLM. BlmΔ/Δ;Trp53bp1−/− cells showed chromosome instability equivalent to BlmΔ/Δ;Trp53bp1+/+ cells, indicating that removing a barrier to resection is not sufficient to substitute for BLM in the early stages of HR. On the other hand, Brca1Δ11/Δ11;BlmΔ/Δ cells showed a frequency of chromosome aberrations that was significantly lower than that seen in Brca1Δ11/Δ11 cells (Fig. 1, A and B; and Fig. S1, A and B). Brca1Δ11/Δ11 cells normally have a high level of genomic instability that arises at least in part because of a failure to perform HR (Moynahan et al., 1999; Xu et al., 1999; Bunting et al., 2010). This genomic instability is dependent on the presence of BLM, indicating that BLM contributes to the defect in HR in Brca1Δ11/Δ11 cells.
Figure 1.
Ablation of Blm rescues genomic instability, T cell development, and poly (ADP-ribose) polymerase inhibitor sensitivity in Brca1Δ11/Δ11 cells. (A) Metaphase spreads from primary mouse B lymphocyte cells stained with DAPI and Cy3-labeled telomeric probe. The arrows point to chromatid breaks, closed arrowheads point to chromosome breaks, and open arrowheads point to radial chromosomes. Bars, 10 µm. (B) Quantification of genomic instability in metaphase spreads after 2 µM overnight treatment with the poly (ADP-ribose) polymerase inhibitor olaparib. CSB, chromosome breaks; CTB, chromatid breaks. (C) Flow cytometry data from primary T lymphocyte cells from mice of indicated genotypes stained with CD4 and CD8 antibodies. (D) Quantification of CD4− CD8− double-negative T cells. (E) Clonogenic survival assay after BLM knockdown in WT and BRCA1Δ11/Δ11 cells with no treatment (NT) and chronic treatment with 100 nM Olaparib (OLA), a poly (ADP-ribose) polymerase inhibitor. (F) Quantification of clonogenic survival assay after shBLM in WT and BRCA1Δ11/Δ11 MEFs. Graphs represent mean ± SD of three independent experiments.
We used two assays to test whether the different levels of genomic instability in Brca1Δ11/Δ11, BlmΔ/Δ, and Brca1Δ11/Δ11;BlmΔ/Δ cells affected their ability to divide and proliferate. First, we targeted Brca1 in the thymus using a conditional knockout approach to produce Brca1Δ11/Δ11 thymocytes (Hennet et al., 1995). These Brca1Δ11/Δ11 thymocytes show a partial developmental block at the transition from the CD4− CD8− “double negative” stage to the CD4+ CD8+ “double positive” stage, corresponding to apoptosis induced by genomic instability (Fig. 1, C and D; Mak et al., 2000). In contrast, Brca1Δ11/Δ11;BlmΔ/Δ cells showed a significantly reduced block at the double-negative to double-positive transition. The improved DNA repair seen in Brca1Δ11/Δ11;BlmΔ/Δ cells relative to Brca1Δ11/Δ11 cells is therefore relevant in a physiological setting. Second, we used a colony forming assay to measure the ability of WT and Brca1Δ11/Δ11 mouse embryonic fibroblast (MEF) cells to proliferate in vitro after exposure to olaparib (Fig. 1 E). Whereas Brca1Δ11/Δ11 cells stably expressing a control shRNA showed substantial hypersensitivity to olaparib, knockdown of BLM afforded a significant rescue of cell survival (Fig. 1 F). BLM therefore contributes to cell death in Brca1-deficient cells, in a manner that correlates with improved DSB repair.
Enhanced RAD51 stability at DNA break sites and HR in Brca1Δ11/Δ11 cells after deletion of Blm
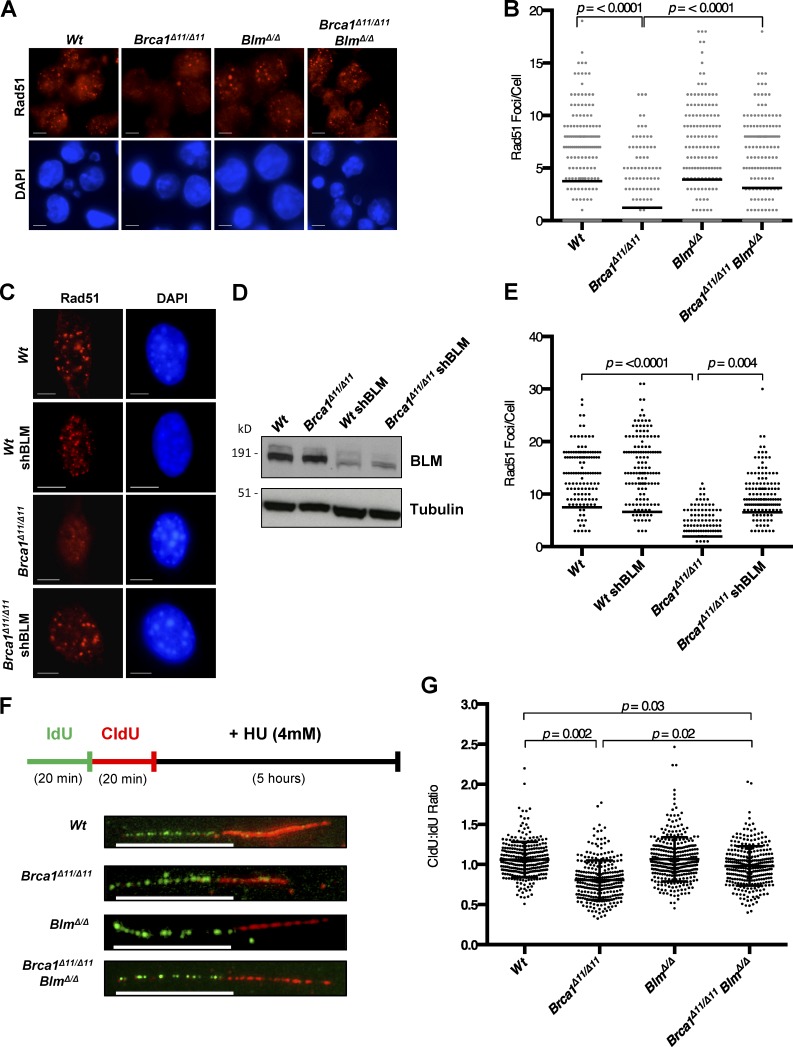
To understand how BLM impacts DNA repair in Brca1Δ11/Δ11 cells, we measured nuclear ionizing radiation–induced foci (IRIF) of RAD51, which mark sites of HR-mediated DNA repair. Brca1Δ11/Δ11 cells are defective for IRIF of RAD51 (Bhattacharyya et al., 2000; Huber et al., 2001), but we found that the proportion of cells exhibiting RAD51 foci was equivalent to WT in Brca1Δ11/Δ11;BlmΔ/Δ B cells (Fig. 2, A and B). RAD51 foci were also observed in Brca1Δ11/Δ11 MEFs after shRNA-mediated knockdown of BLM, whereas cells expressing control shRNA showed a defect in RAD51 foci formation (Fig. 2, C–E). BLM therefore appears to limit RAD51 assembly at DSBs in Brca1Δ11/Δ11 cells. Several factors that regulate the assembly and recruitment of RAD51 at break sites have also been shown to regulate RAD51-mediated protection of DNA replication forks (Schlacher et al., 2012; Higgs et al., 2015; Leuzzi et al., 2016; Sato et al., 2016). Using a DNA fiber assay, we found that BLM affected fork protection (Fig. 2, F and G). As previously reported, Brca1Δ11/Δ11 cells show a defect in fork protection relative to WT cells after hydroxyurea treatment (Ray Chaudhuri et al., 2016). Deletion of Blm in Brca1Δ11/Δ11 cells afforded a partial rescue of fork degradation, which may be related to the ability of BLM to regulate the stability of RAD51 at nascent replication tracts after replication stress.
Figure 2.
Enhanced Rad51 foci after ablation of Blm in Brca1Δ11/Δ11 cells. (A) Immunofluorescence analysis of Rad51 IRIF in primary B cells from mice of indicated genotypes after IR. (B) Quantification of Rad51 IRIF in primary B cells. (C) Immunofluorescence analysis of Rad51 IRIF in MEF cells with indicated genotypes and shRNA knockdown after IR. (D) Western blot analysis of knockdown efficiency. (E) Quantification of Rad51 IRIF in MEF cells. (F) Experimental scheme and representative images of replication fork degradation analysis after 20 min iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) labeling, respectively, and exposure to 4 mM hydroxyurea (HU) for 5 h. (G) DNA fiber analysis denoting the mean ratios of CldU/IdU label lengths. Graphs represent mean ± SD of three independent experiments. Bars, 10 µm.
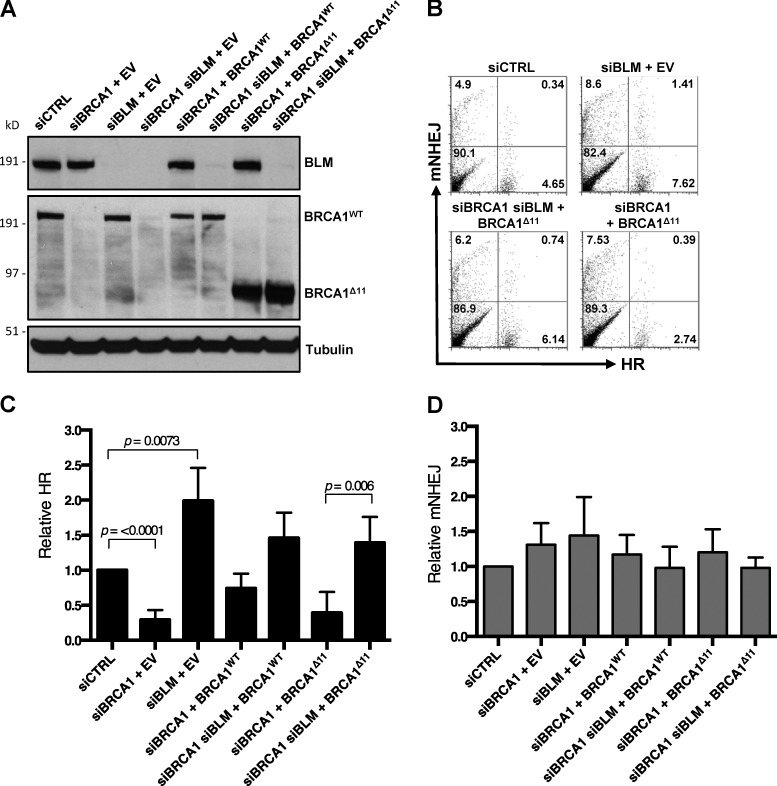
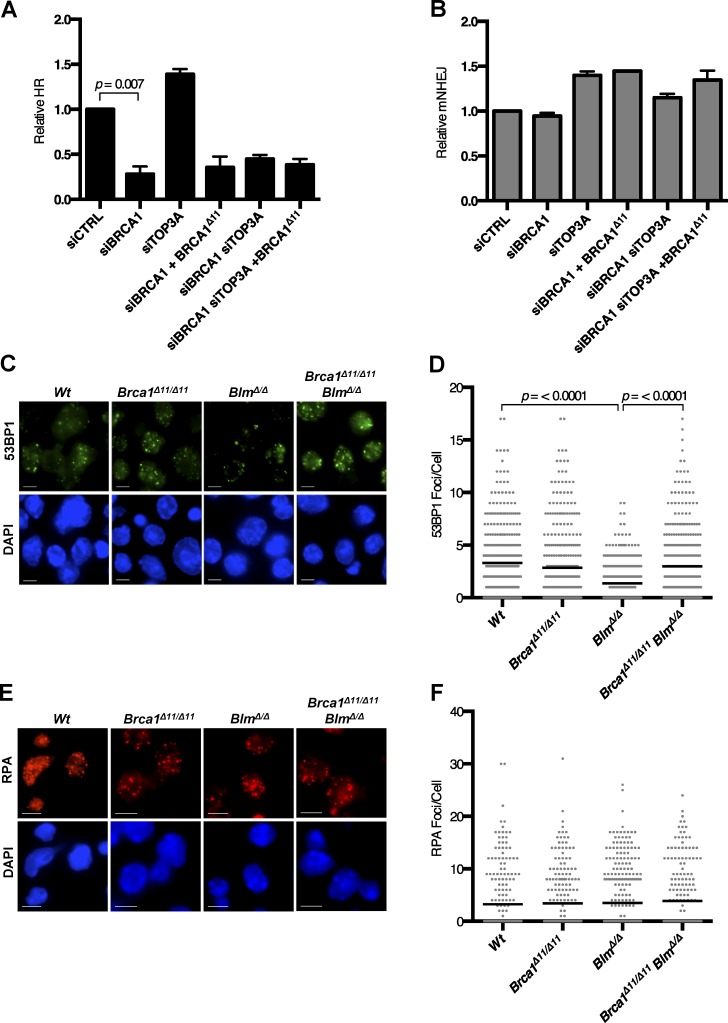
Next, we used U2OS EJ-DR reporter cells to directly measure the importance of BLM for HR efficiency after siRNA-mediated silencing of BLM and BRCA1 (Fig. 3, A and B; Bindra et al., 2013). Consistent with previous studies and our RAD51 foci results (Fig. 2, A–E), we observed that the rate of HR was substantially reduced in cells expressing BRCA1Δ11. The rate of HR showed a highly significant increase upon codepletion of BLM, however, to a level equivalent to that seen in control cells (Fig. 3 C). Rates of nonhomologous end joining (NHEJ) were not significantly different in any of the conditions tested (Fig. 3 D). The differences in cell viability and genomic instability between Brca1Δ11/Δ11 and Brca1Δ11/Δ11;BlmΔ/Δ cells are therefore correlated with the efficiency of HR.
Figure 3.
BLM regulates HR in BRCA1Δ11 cells. (A) Western blot analysis of EJ-DR reporter cells transfected with siRNA oligos and/or BRCA1 expression vector or control empty vector (EV). (B) Representative flow cytometry data in the U2OS EJ-DR reporter assay, in which HR-mediated repair of I-SceI–mediated DSBs produces GFP fluorescence and mutagenic NHEJ (mNHEJ) produces DsRed fluorescence. (C) Analysis of relative HR frequency in the U2OS EJ-DR reporter cell line. (D) Analysis of relative mNHEJ frequency in the U2OS EJ-DR reporter cell line. Graphs represent mean ± SD of three independent experiments.
As a second measure of HR, we assayed the frequency of SCEs, which are formed through crossover HR, in Brca1Δ11/Δ11, BlmΔ/Δ, and Brca1Δ11/Δ11;BlmΔ/Δ metaphase chromosome spreads (Fig. S2, A and B). Whereas WT and Brca1Δ11/Δ11 cells showed an approximately equivalent frequency of SCEs, BlmΔ/Δ cells showed an elevated rate of SCEs, consistent with the known role of BLM in promoting noncrossover resolution of Holliday junctions during the final stages of HR (Chaganti et al., 1974; Wu and Hickson, 2003). Brca1Δ11/Δ11;BlmΔ/Δ chromosomes had fewer SCEs than the high rate seen in BlmΔ/Δ cells, but the rate was nonetheless equivalent to or higher than that seen in WT cells.
BLM modifies HR activity in WT cells, but not BRCA1-nullizygous cells
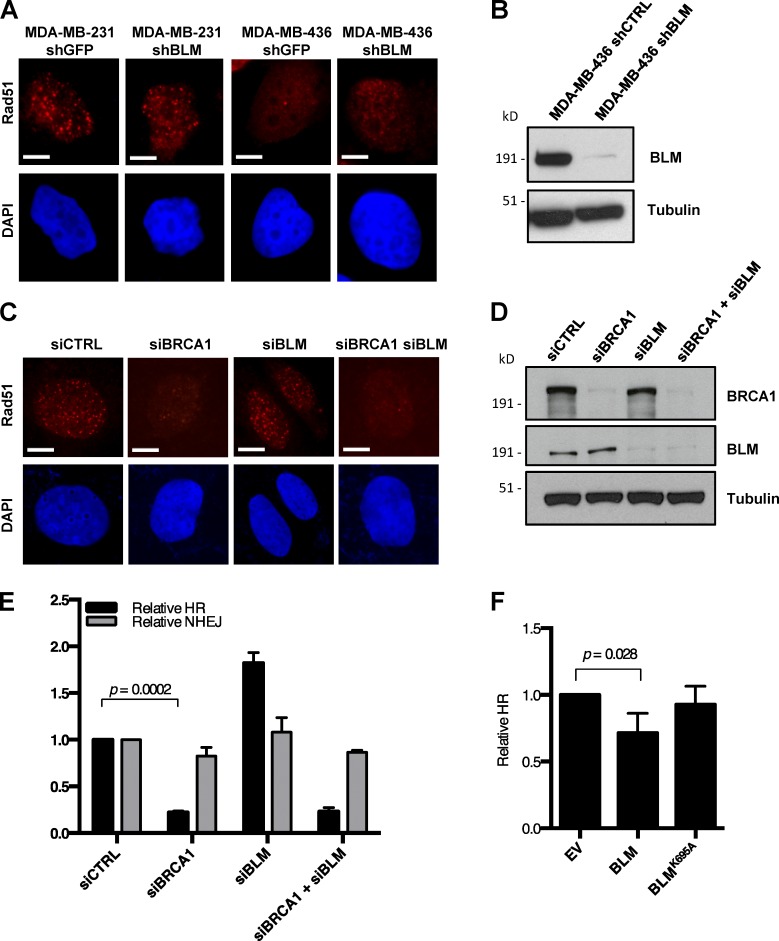
The Brca1Δ11 allele is considered to be hypomorphic (Evers and Jonkers, 2006); hence, we sought to test whether the observed anti-recombinase activity of BLM is also relevant in other BRCA1 mutant cells or in WT cells. Although silencing Blm rescues HR in Brca1Δ11/Δ11 cells, we found that the extent of rescue of RAD51 foci formation was insubstantial in MDA-MB-436 (Elstrodt et al., 2006), a human mammary adenocarcinoma cell line that expresses no BRCA1 protein (Fig. 4, A and B). We also failed to observe rescue of RAD51 foci formation in U2OS osteosarcoma cells after cosilencing of BRCA1 and BLM using siRNA (Fig. 4, C and D; and Fig. S2 C). Silencing of BLM likewise failed to rescue the HR defect of U2OS EJ-DR reporter cells in which BRCA1 was knocked down (Fig. 4 E). Some residual pro-recombinogenic activity provided by the hypomorphic BRCA1Δ11 protein that is expressed in Brca1Δ11/Δ11 cells is therefore necessary for HR even in the absence of Blm. BLM appears to regulate the efficiency of HR, and loss of BLM cannot completely substitute for essential HR factors.
Figure 4.
BLM modifies HR activity in WT cells, but not BRCA1-nullizygous cells. (A) Immunofluorescence analysis of Rad51 IRIF in MDA-MB-231 (BRCA1 wt) and MDA-MB-436 (BRCA1 mutant) cells after control shRNA or shBLM knockdown. (B) Western blot analysis of BLM knockdown efficiency in MDA-MB-436 cells. (C) Immunofluorescence analysis of Rad51 IRIF in U2OS cells with indicated siRNA knockdown. (D) Western blot analysis of BRCA1 and BLM knockdown efficiency in U2OS cells. (E) Analysis of relative HR and mutagenic NHEJ (mNHEJ) frequency in the EJ-DR U2OS reporter cell line with indicated siRNA knockdown. (F) Analysis of relative HR frequency in the EJ-DR U2OS reporter cell line after overexpression of WT BLM and helicase-dead BLM (BLMK695A). Graphs represent mean ± SD of three independent experiments. Bars, 10 µm.
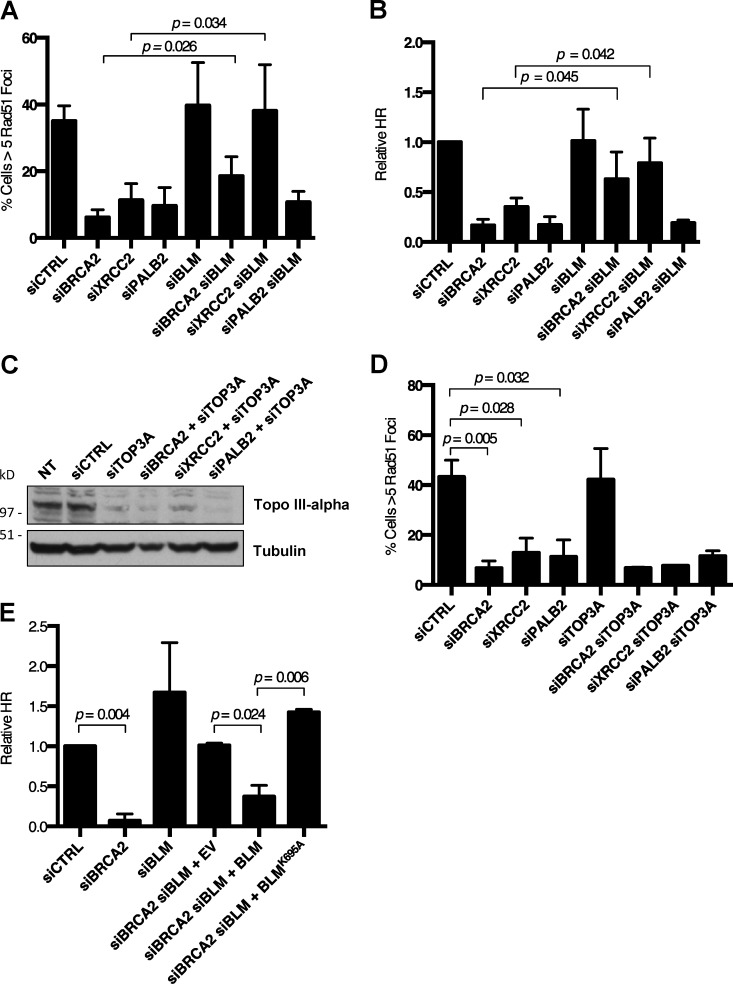
An anti-recombinogenic activity of BLM has previously been inferred from the high rate of SCEs in BS cells (Chaganti et al., 1974; Hu et al., 2001). Using the EJ-DR reporter assay, we tested whether overexpression of BLM could suppress HR in BLM+/+ cells. As has been seen with other mammalian anti-recombinases (Fugger et al., 2009; Lorenz et al., 2009; Schwendener et al., 2010), overexpression of BLM caused a modest but statistically significant reduction in HR efficiency (Figs. 4 F and S2 D). Suppression of HR was not observed, however, upon overexpression of a helicase-dead BLM mutant (BLMK695A; Bugreev et al., 2007). BLM therefore has the potential to disrupt HR in WT and Brca1Δ11/Δ11 cells through its helicase activity. The ability of BLM to dissolve Holliday junctions is further dependent on formation of the multiprotein BTR complex (BLM–topoisomerase III-α–RMI1/2; Manthei and Keck, 2013). To test whether BTR is required for BLM anti-recombinase activity, we measured HR efficiency in cells after siRNA-mediated silencing of BRCA1 and topoisomerase IIIα (TOP3A). In cells expressing mutant BRCA1Δ11, silencing TOP3A did not rescue the low efficiency of HR (Fig. 5 A). The anti-recombinase effect of BLM that is active in this assay therefore appears to be independent of the BTR complex. This represents a difference from BLM-mediated dissolution of Holliday junctions, which is dependent on the BTR complex (Wu and Hickson, 2003). NHEJ was not defective in any of the cells tested upon depletion of BLM or TOP3A (Fig. 5 B).
Figure 5.
Ablation of Blm increases 53BP1 accumulation at DNA break sites in Brca1Δ11/Δ11 cells. (A) Analysis of relative HR frequency in the EJ-DR U2OS reporter cell line after knockdown of BRCA1 and TOP3A and overexpression of BRCA1Δ11. (B) Analysis of relative mutagenic NHEJ (mNHEJ) frequency in the EJ-DR U2OS reporter cell line. (C) Immunofluorescence analysis of 53BP1 IRIF in primary B cells from mice of indicated genotypes after IR. (D) Quantification of 53BP1 IRIF in primary B cells. (E) Immunofluorescence analysis of RPA IRIF in primary B cells from indicated genotypes after IR. (F) Quantification of RPA IRIF in primary B cells. Graphs represent mean ± SD of three independent experiments. Bars, 10 µm.
The presence of 53BP1 at DSB sites regulates the efficiency of DSB resection and entry into the HR pathway (Chapman et al., 2012). 53BP1 limits DSB resection and increases the proportion of DSBs that are repaired by NHEJ. We hypothesized that the different levels of repair seen in Brca1Δ11/Δ11 and Brca1Δ11/Δ11;BlmΔ/Δ cells may be related to differential 53BP1 recruitment to DSB sites in these cells. We therefore measured the number of 53BP1 foci formed after ionizing radiation (IR) treatment (Fig. 5 C). Deletion of Blm was associated with a slight reduction in 53BP1 foci formation relative to WT, as has been reported previously (Grabarz et al., 2013). However, Brca1Δ11/Δ11;BlmΔ/Δ cells showed a significantly greater number of 53BP1 foci after IR compared with BlmΔ/Δ cells (Fig. 5, C and D). As the difference in 53BP1 foci was not of great magnitude, we measured class switch recombination (CSR), which is highly sensitive to 53BP1 levels (Manis et al., 2004; Ward et al., 2004). Although Brca1Δ11/Δ11 and BlmΔ/Δ cells did not show substantial differences in CSR, the rate of switching was markedly reduced in Trp53bp1−/− cells (Fig. S3, A and B). Interestingly, the increased accumulation of 53BP1 in Brca1Δ11/Δ11;BlmΔ/Δ B cells relative to Brca1Δ11/Δ11 cells was sufficient to impact the efficiency of CSR. On the other hand, we did not observe any substantial defects in the accumulation of RPA at sites of IR–induced damage in any of the genotypes used (Fig. 5, E and F; and Fig. S3, C and D). These results show that although deletion of Brca1 or Blm can impact 53BP1 recruitment, this effect does not substantially alter DSB resection, as revealed by RPA accumulation. The difference in HR efficiency between Brca1Δ11/Δ11 and Brca1Δ11/Δ11;BlmΔ/Δ cells is therefore likely to be dependent on a subsequent step in the HR pathway, when RAD51 is loaded.
Rescue of RAD51 foci formation and HR in BRCA2- and XRCC2-deficient cells by silencing of BLM
Efficient HR is dependent on several factors that contribute to accumulation of RAD51 at resected DSBs. These include PALB2 (partner and localizer of BRCA2), BRCA2, and XRCC2 (Johnson et al., 1999; O’Regan et al., 2001; Xia et al., 2006; Jensen et al., 2010). To further dissect the mechanism by which BLM coordinates HR, we knocked down each of these factors individually, and in combination with siBLM oligonucleotides (Fig. S4, A–C). Interestingly, ablation of BLM was sufficient to rescue RAD51 foci formation in cells treated with either siBRCA2 or siXRCC2 oligos (Fig. 6 A). This result suggests that BLM has a general role in reversing RAD51 nucleoprotein filament formation, which can be revealed in cells lacking key HR factors. Loss of RAD51 foci in siPALB2 cells was not rescued by ablation of BLM, suggesting that PALB2 may have a distinct role in RAD51 recruitment. To validate the importance of BLM depletion after silencing of BRCA2 or XRCC2, we used EJ-DR cells to measure HR efficiency. Consistent with the immunofluorescence measurements of RAD51 foci, HR was rescued in both siBRCA2 and siXRCC2 cells after codepletion of BLM (Fig. 6 B).
Figure 6.
Rescue of RAD51 IRIF and HR in BRCA2- and XRCC2-deficient cells by silencing of BLM requires BLM helicase activity, but not TOP3A. (A) Quantification of RAD51 IRIF in U2OS cells after indicated knockdowns and IR. (B) Analysis of relative HR frequency in the U2OS EJ-DR reporter cell line after indicated knockdown. (C) Western blot analysis of TOP3A knockdown efficiency in U2OS cells with no treatment (NT) and indicated siRNA-mediated knockdowns. (D) Quantification of RAD51 IRIF in U2OS cells after indicated knockdowns and IR. (E) Analysis of relative HR frequency in the U2OS EJ-DR reporter cell line after indicated knockdowns. Graphs represent mean ± SD of three independent experiments.
In separate experiments, we found that knockdown of TOP3A did not afford a similar rescue phenotype as was seen with siBLM (Fig. 6, C and D), again suggesting that the ability of BLM to regulate RAD51 stability is likely to be independent of the BTR complex. The helicase activity of BLM appears to influence HR in siBRCA2 cells, however. Expression of exogenous BLMWT in siBRCA2 cells reversed the rescue of HR that was achieved by knockdown of BLM, but an equivalent effect was not observed upon expression of the helicase-dead BLMK695A mutant (Fig. 6 E). Collectively with our other data (Fig. 4 F), this result suggests that the helicase activity of BLM is essential for its anti-recombinase effect in cells of several different genetic backgrounds.
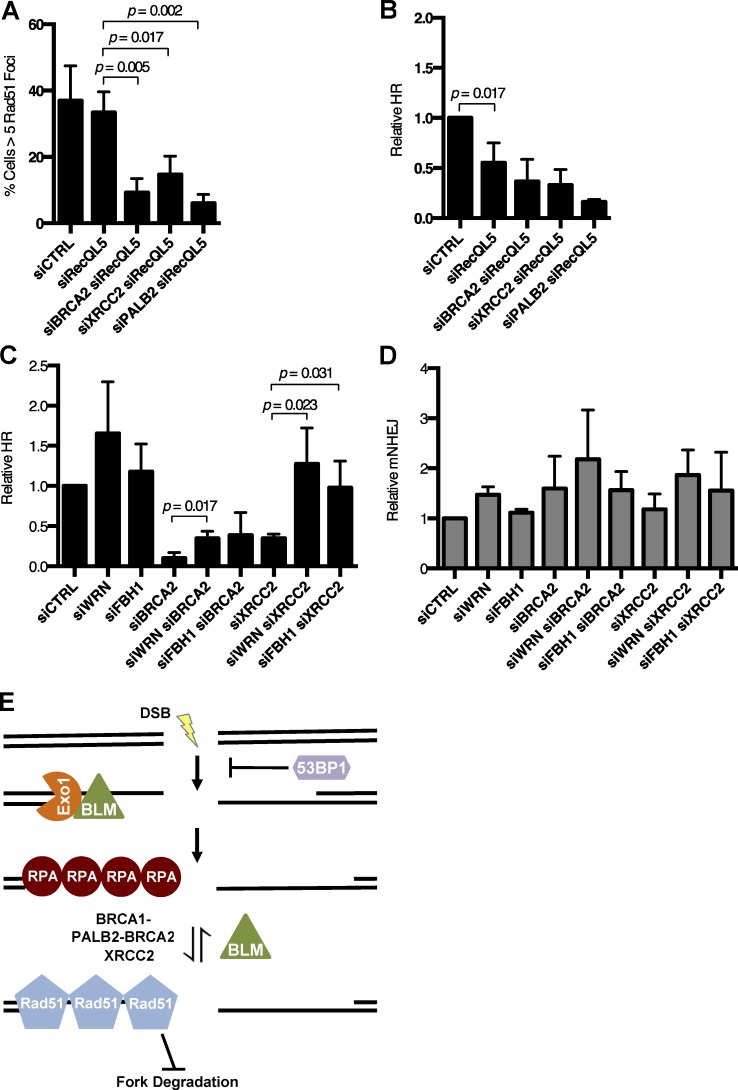
Several enzymes have been reported to exhibit anti-recombinase activity in mammalian cells (Karpenshif and Bernstein, 2012). We performed siRNA knockdown of three of these factors to evaluate if they could rescue HR in cells lacking BRCA2 or XRCC2 (Fig. S5, A–C). First, we tested the RECQ5/RECQL5 helicase, which is reported to have anti-recombinase activity (Hu et al., 2007). Although BLM knockdown can partially rescue the defect in RAD51 foci and HR in cells treated with siBRCA2 or siXRCC2 (Fig. 6, A and B), equivalent knockdown of RECQL5 had little effect (Fig. 7, A and B). FBH1 is a second protein that has been reported to act as a mammalian anti-recombinase, and WRN is a RECQ helicase that has been shown to target RAD51-loaded DNA structures (Constantinou et al., 2000; Fugger et al., 2009; Wang et al., 2015). To evaluate the effect of these factors on recombination efficiency, we repeated our HR assay in siBRCA2 and siXRCC2 cells with combined knockdown of either FBH1 or WRN (Fig. 7 C). Knockdown of WRN or FBH1 afforded a rescue of HR in siXRCC2 cells. Knockdown of WRN also gave a partial rescue of HR in siBRCA2 cells, but no significant difference in HR efficiency was observed in these cells upon knockdown of FBH1. The efficiency of NHEJ was not affected in any case (Fig. 7 D). We conclude that the ability of BLM to suppress HR is of an equivalent magnitude to that seen with other mammalian anti-recombinases, at least in cells that are HR deficient.
Figure 7.
Effect of pro- and anti-recombinogenic proteins on HR efficiency. (A) Quantification of RAD51 IRIF in U2OS cells after indicated knockdowns and IR. (B and C) Analysis of relative HR frequency in the EJ-DR U2OS reporter cell line after indicated knockdowns and I-Sce1 induction. (D) Analysis of relative mutagenic NHEJ (mNHEJ) frequency in the EJ-DR U2OS reporter cell line after indicated knockdowns and I-Sce1 induction. (E) Model for BLM acting as an anti-recombinogenic factor at the level of RAD51 loading to regulate efficient HR. Graphs represent mean ± SD of three independent experiments.
Discussion
Regulation of HR through altered stability of the RAD51 nucleoprotein filament
Our results show that BLM promotes genomic instability in HR-deficient cells, to a level that can cause cell death, by reducing the stability of RAD51 at the presynaptic filament. BLM appears to act by disrupting RAD51 on resected DNA ends. In WT cells, the presence of BRCA1, BRCA2, and factors including XRCC2 stabilizes the RAD51 nucleoprotein filament despite the anti-recombinogenic effect of BLM, but when these factors are absent, there is a failure to retain RAD51 at the break site that leads to insufficient HR, genomic instability, and loss of cell viability (Fig. 7 E). Hyperaccumulation of RAD51 at DNA damage sites in BS cells has been observed previously (Bischof et al., 2001; Wu et al., 2001), and an ability of BLM to displace RAD51 from single-stranded DNA is supported by several lines of cellular and biochemical evidence (Bugreev et al., 2007; Tripathi et al., 2007, 2008). This work is the first to demonstrate that this activity has substantial physiological importance. This ability of BLM is distinct from proposed roles for the protein in DSB resection (Gravel et al., 2008; Nimonkar et al., 2008, 2011), D-loop displacement (van Brabant et al., 2000; Bachrati et al., 2006), or Holliday junction dissolution (Wu and Hickson, 2003).
Several cellular factors have been shown to maintain replication fork integrity by stabilizing RAD51 filaments (Schlacher et al., 2012; Leuzzi et al., 2016; Sato et al., 2016), and we find an equivalent role for BLM. Deletion of Blm largely rescues the fork degradation that is normally seen in Brca1Δ11/Δ11 cells (Fig. 2, F and G), suggesting that BLM influences both HR efficiency and fork protection by regulating RAD51 filament stability. Interestingly, silencing of BLM has been reported to help protect replication forks and maintain genomic integrity in cells lacking the replication-associated protein BOD1L (Higgs et al., 2015). Targeting the anti-recombinase activity of BLM may therefore rescue genomic integrity in an even greater range of genotypes than those that we have tested in this study.
BLM as an Srs2-like anti-recombinase
Like BLM, the DNA damage response factor 53BP1 also regulates the efficiency of HR, albeit by a different mechanism (Bouwman et al., 2010; Bunting et al., 2010). 53BP1 limits DSB resection, thereby inhibiting HR and promoting use of alternative pathways for DSB repair such as NHEJ. The effect of BLM appears to be exerted at the level of stabilization of RAD51 on the resected 3′ single-stranded DNA region. This direct effect on RAD51 is reminiscent of the anti-recombinogenic effect of Srs2 during HR in Saccharomyces cerevisiae. Diploid yeast lacking Srs2 are hyperrecombinogenic and exhibit hypersensitivity to UV and IR, which can be rescued by mutations in RAD51 (Aboussekhra et al., 1989, 1992). Srs2 stimulates the ATPase activity of RAD51 at resected DSBs, promoting release of RAD51, and limiting the formation of an active presynaptic filament (Krejci et al., 2003; Veaute et al., 2003). Previous biochemical work has shown that the K695A mutant of BLM, which lacks DNA translocase and helicase activity, is not able to displace RAD51 from DNA templates in vitro (Bugreev et al., 2007). Our cell biological assays support the idea that the helicase activity of BLM is critical for its anti-recombinase function. Our work additionally shows that the association of BLM with topoisomerase IIIα, RMI-1, and RMI-2 as the BTR complex, which is essential for Holliday junction dissolution, is likely dispensable for its anti-recombinase activity.
BLM is one of a group of mammalian anti-recombinases
Although no precise mammalian orthologue of Srs2 has been identified, several proteins have been suggested to have an analogous function. These include FBH1, RECQ5, PARI, and POLQ (Hu et al., 2007; Fugger et al., 2009; Schwendener et al., 2010; Moldovan et al., 2012; Simandlova et al., 2013; Ceccaldi et al., 2015). It is not clear which of these proteins is of greatest importance for regulation of mammalian HR (Kowalczykowski, 2015). In our work, we measured the effect of ablation of BLM and the putative anti-recombinases, RECQ5, WRN, and FBH1 on the efficiency of HR in cells lacking key HR factors. We found that depletion of BLM has at least as significant an effect as depletion of these other putative anti-recombinases (Fig. 7, A–C). These results raise an important question of whether these factors act redundantly or are regulated to work in specific instances. Notably, BLM and RECQ5 act in a nonredundant fashion to suppress crossover recombination (Wang et al., 2003; Hu et al., 2005).
Effect of BLM on HR in BRCA2-, XRCC2-, and PALB2-deficient cells
Whereas our overexpression analysis suggests that BLM works as an active anti-recombinase in normal cells, the effects of the absence of the anti-recombinase activity of BLM was most easily observed in cells with deficiencies in HR. In addition to its effect in Brca1Δ11/Δ11 cells, deletion of BLM afforded some degree of rescue of the HR defect normally seen in siBRCA2 and siXRCC2 cells. Deletion of BLM did not, however, afford any substantial rescue of HR in PALB2-deficient cells. This result was unexpected, because PALB2 interacts with both BRCA1 and BRCA2 to allow stable association of BRCA2 at DSB sites and loading of RAD51 (Xia et al., 2006; Sy et al., 2009b; Zhang et al., 2009). As PALB2 deficiency is not rescued by BLM deletion, it is possible that PALB2 plays a role separate from BRCA1 and BRCA2 in the control of genomic integrity or RAD51 loading. Notably, PALB2 has been shown to directly interact with D-loop structures, where it is able to interact with and stabilize RAD51 (Buisson et al., 2010). PALB2 also binds MRG15, a chromodomain protein that modifies the efficiency of HR (Sy et al., 2009a). Clearly, the full range of activities that regulate the stability of the RAD51 nucleoprotein filament warrants further study.
Our results support a hypothesis in which a failure to constrain mutagenic RAD51-dependent processes such as nonallelic HR may lead to the chromosome instability seen in BS cells (Bhargava et al., 2016; Carvalho and Lupski, 2016). SGS1, the yeast homologue of BLM, suppresses recombination between homologous sequences and can partially substitute for Srs2 in srs2 mutant yeast (Myung et al., 2001; Mankouri et al., 2002; Ira et al., 2003). Deletion of Blm in mouse spermatocytes also leads to a block in meiosis associated with mispairing of homologous chromosomes and increased chiasmata (Holloway et al., 2010). We propose that BLM deserves further consideration as an anti-recombinase with substantial importance in regulating the HR pathway for repair of mammalian DSBs.
Materials and methods
Cell culture
Resting primary B cells were isolated from mouse spleen by ACK lysis of erythrocytes and negative selection with anti-CD43 MACS microbeads (Miltenyi). B cells were resuspended at 0.5–1 × 106 cells/ml and activated with 25 µg/ml lipopolysaccharide (L2630; Sigma Aldrich), 50 U/ml IL-4 (I1020; Sigma Aldrich), and 1:1,000 RP105 antibody (55128; BD) for 48–72 h. MEFs as well as U2OS, MDA-MB-231, and MDA-MB-436 cells were cultured in DMEM supplemented with 15% FBS and 1% penicillin/streptomycin.
DNA repair assay
The EJ-DR assay was performed as previously described (Bindra et al., 2013). EJ-DR reporter cells were plated at 200,000–500,000 cells per well in 10% charcoal-stripped FBS (100–119; Gemini), antibiotic/antimycotic, and DMEM and transfected the next day with siRNA and/or plasmid DNA using Lipofectamine 2000 (11668; Invitrogen) or Lipofectamine RNAiMAX (13778; Invitrogen). After knockdown and expression (24 h), cells were grown in 10% Tet-free FBS (100-800; Gemini), antibiotic/antimycotic, and DMEM. Incorporated I-Sce1 was induced with Shield1 (632189; Clontech) and triamcinolone (T6510; Sigma Aldrich) ligands for 24 h. NHEJ and HR repair activity was assessed 48 h postinduction by quantification of DsRed- and GFP-positive cells on BD FACS Calibur system and analyzed on FlowJo (Tree Star).
Metaphase spreads
Telomere DNA FISH analysis was performed and analyzed as previously described (Misenko and Bunting, 2014). B cells were activated for 24 h and treated with 250 nM mitomycin C (Sigma Aldrich), 2 µM olaparib (KU0059436; Selleckchem), or 4 µM cisplatin (Sigma Aldrich) for 16 h. Cells were arrested in metaphase with 100 ng/ml colcemid (Sigma Aldrich) for 1 h. Cells were collected, incubated in hypotonic solution (0.075 M KCl) for 15 min at 37°C, and fixed in a 3:1 methanol/acetic acid solution (three washes) and stored overnight at −20°C. Cells were dropped onto glass microscope slides in a Thermotron at 52% humidity at 22.9°C and dried for 30 min to 1 h and stored in 37°C chamber. Telomere DNA FISH analysis was performed by first preparing the telomeric Cy-3 PNA probe (Cy3-00-CCCTAACCCTAACCCTAA, F1002; PNA Bio Inc.). The probe was incubated in deionized formamide, pH 7.0, at 37°C for 1 h, followed by addition of FISH Master Mix (4× SSC, 20% dextran sulfate) and incubation in at 37°C for 1 h. Probe was denatured at 80°C followed by preannealing at 37°C for 1 h.
Concurrently, chromosome slides were pretreated with pepsin in 0.01 M HCl acidic solution for 90 s at 37°C, followed by washes in 1× PBS and 1× PBS/50 mM MgCl2, and fixed in 1% formaldehyde/1× PBS/50 mM MgCl2 for 10 min. Slides were then washed in 1× PBS, dehydrated in a series of ethanol incubations (70%, 90%, and 100%), and air dried. Slides were denatured by incubation at 80°C in 70% deionized formamide/2× SSC for 90 s, followed by dehydration in a series of ethanol incubations (70%, 90%, and 100%), and air dried. In a humidity chamber, the preannealed probe mix was applied to the slide, covered with 18 × 18 mm coverslip, and incubated for 1 h at 37°C. The probe was cross-linked in 50% formamide/2× SSC (three 5-min washes), followed by washes in 1× SSC (3 × 5 min washes) and 1× SSC/0.1% Tween 20 (three 5-min washes). Slides were counterstained in DAPI solution, washed in 3× SSC, and mounted in Mowiol antifade solution. For each experiment, 50 metaphases per genotype and treatment were analyzed.
Animal husbandry
All animal experiments were approved by the Institutional Animal Care and Use Committee at Rutgers University (protocol 12–024).
Flow cytometry
Primary T cells were isolated from thymi of 2- to 3-mo-old mice in 1× HBSS, 1% FBS/1× penicillin/streptomycin and strained through a 70-μm mesh. Blocking was with anti–CD16/CD32 antibody (rat, 553142; BD Biosciences) and stained with APC-CD8a (rat; 553035; BD Biosciences) and/or FITC-CD4 (rat, 553046; BD Biosciences) antibodies. For analysis of CSR, B cells were stained with 1:100 dilutions of anti-B220-FITC (rat; 553088, BD Biosciences) and biotin anti–mouse IgG1 (rat, 553441; BD) followed by incubation in streptavidin Alexa Fluor 647 (S32357; Thermo Fisher). Cells were washed twice in 1× HBSS, 1% FBS, and 1× penicillin/streptomycin. For analysis of RPA staining, B cells treated with 10 Gy IR followed by 2 h of recovery were extracted with PBS with Tween 20, fixed in 4% PFA, permeabilized, and stained with RPA32 antibody (rat, 1:500, 2208; Cell Signaling). After secondary staining with Alexa Flour 488 anti-rat (1:200; A11006; Life Technologies), cells were stained for DNA content using propidium iodide (Forment et al., 2012). Flow cytometry analysis was performed on BD FACSCalibur system and analyzed on FlowJo (Tree Star).
Sister chromatid exchange assay
B cells were activated in the presence of 10 μm BrdU for 48 h followed by arrest in metaphase with 100 ng/ml colcemid (Sigma Aldrich). Cells were fixed and dropped onto glass slides for metaphase spreads (see Materials and Methods). Slides were stained with Hoechst 33258 (Thermo Fisher), incubated in McIlvaine’s solution (164 mM Na2HPO4, 16 mM citric acid, pH 7.0), and exposed to UV light for 45 min. Slides were incubated in a 1:12 Giemsa staining solution in 3% methanol, dehydrated with xylenes, and mounted with Permount (Fisher Scientific).
DNA combing
Splenic B cells were grown in vitro for 48 h under activating conditions. Cells were incubated with 25 µM 5-iododeoxyuridine (I7125; Sigma Aldrich) for 20 min followed by a 20 min incubation with 250 µM 5-chlorodeoxyuridine (C6891; Sigma Aldrich). Cells were subsequently treated with 4mM hydroxyurea for five hours (H8627; Sigma Aldrich). DNA fiber spreads were prepared and analyzed by immunofluorescence as previously described (Li et al., 2016). Cell suspension was collected in 2-µl volume and dried on glass slide. Cells were lysed in 7 μl lysis buffer consisting of 50 mM EDTA and 0.5% SDS in 200 mM Tris-HCl, pH 7.5, for 2 min. Slides were tilted at 15° angle to dry and create spread of DNA fibers. Slides were fixed in methanol/acetic acid solution (3:1) for 10 min followed by incubation in 2.5M HCl for 80 min. Slides were washed with PBS, blocked in 5% BSA for 20 min, and stained with 1:25 anti-BrdU (mouse) and 1:400 anti-BrdU (rat) in 5% BSA for 2 h in a humidity chamber. Slides were washed with PBS and stained with 1:500 sheep anti–mouse Cy3 and 1:400 goat anti–rat Alexa Fluor 488 secondary antibody for 1 h. Slides were again washed in PBS and mounted using Vectashield mounting medium with glass coverslips. Approximately 300 fiber lengths were analyzed per genotype in three independent experiments using ImageJ software.
Western blotting and immunofluorescence
Primary antibodies were used at the following dilutions for Western blotting: anti-tubulin (mouse, 1:50,000; Sigma Aldrich), mouse anti-BLM (rabbit, 1:1,000; A300-110A; Bethyl), human anti-BLM (rabbit, 1:1,000; ab2179; Abcam), anti-BRCA1 (mouse, 1:1,000; MABC199; EMD Millipore), anti-RECQ5 (rabbit, 1:1,000; ab31609; Abcam), anti-TOP3A (rabbit, 1:2,000; 14525-1-AP; Proteintech), anti-XRCC2 (mouse, 1:1,000; ab20253; Abcam), anti-BRCA2 (rabbit, 1:1,000; sc28235; Santa Cruz), anti-WRN (mouse, 1:500; ab66606; Abcam), anti-FBH1 (mouse, 1:1,000; sc-81563; Santa Cruz), and anti-PALB2 (1:1,000, M11, raised in rabbits against amino acids 601–880 of human PALB2). Secondary HRP-conjugated anti–mouse (sheep; 1:2,000; NA931V; GE Healthcare) and anti–mouse (donkey; 1:2,000; NA9340V; GE Healthcare) antibodies were used.
For immunofluorescence, U2OS, MEF, and MDA-231/436 cells were grown on 18 × 18-mm coverslips overnight before 10 Gy IR treatment and 2-h recovery. B lymphocyte cells were activated and treated with 10 Gy IR and 4 h recovery before being dropped onto slides coated with Cell-tak (BD). Cells were preextracted in 0.5% Triton X-100 in PBS, fixed in 2% paraformaldehyde in PBS, and permeabilized in 0.5% Triton X-100 in PBS and incubated in antibody. Primary antibodies were used at the following dilutions: anti-RAD51 (rabbit, 1:100; H-92; Santa Cruz), anti-53BP1 (rabbit, 1:2,000; NB100-304; Novus), and anti-RPA32 (rat, 1:100; 2208; Cell Signaling) and detected with anti–rabbit Alexa Fluor 546 or Alexa Fluor 488 antibody (1:200; Thermo Fisher) or anti–rat Alexa Fluor 488 (1:200; Life Technologies). Cells were counterstained with DAPI and mounted in Mowiol solution. Approximately 200–500 cells per treatment were analyzed for each experiment.
Plasmids
The BRCA1 expression constructs were based on pcDNA-3xMyc-BRCA1, which was provided by B. Xia (Rutgers Cancer Institute of New Jersey, New Brunswick, NJ) and has been previously described (Chen et al., 1998; Anantha et al., 2017). The BRCA1Δ11 construct was generated through site-directed mutagenesis following the QuikChange protocol (Agilent Technologies).
The full-length hBLM was PCR amplified from the pGFP-C1-BLM (80070; Addgene), provided by N. Ellis (The University of Arizona Cancer Center, Tuscon, AZ; Hu et al., 2001), using the iPROOF High-Fidelity PCR kit (Bio-Rad) and a 3′ primer (5′-TGCT TTAATTAATTACTTGTCGTCATCGTCCTTGTAGTCTGAGAATGCATATGAAGGCTTAAGAAACGGTCTATT-3′) containing codons for the FLAG epitope tag (DYKDDDDK). The amplified product was ligated into the pMx-GFP vector by restriction digest with Xho1 and Pac1 to generate the pMx-GFP-hBLMFLAG construct. The BLMK695A construct was generated through site-directed mutagenesis following the QuikChange protocol (Agilent Technologies).
siRNA and shRNA
For depletion of BLM by shRNA, a pLKO.1 plasmid containing the targeting sequence (5′-CCGGCGAAGGAAACTCACGTCAATACTCGAGTATTGACGTGAGTTTCCTTCGTTTTTG-3′; TRCN0000070996; Sigma Aldrich) for MEFs, a pLKO.1 plasmid containing the targeting sequence (5′-GCCTTTATTCAATACCCATTT-3′; TRCN0000004904; Sigma Aldrich) for human cells, or control vector was transfected into 293T cells with pMD2.G and pVSVG plasmids to produce lentivirus. MEF cells were infected with lentivirus and selected in medium containing 2 µg/ml puromycin for 1 wk and maintained in 1 µg/ml puromycin thereafter. For transient knockdown via siRNA, siGENOME SMARTpool constructs targeting BLM, BRCA2, XRCC2, PALB2, TOP3A, RecQ5, FBH1, and WRN were acquired commercially (Dharmacon; Table S1) and transfected into U2OS and U2OS EJ-DR reporter cell lines. Custom siRNAs targeting the 3′-UTR region of BRCA1 and BLM were used for knockdown and mutant expression experiments (Table S2). Knockdown efficiency was confirmed via Western blot analysis.
Microscopy
Metaphase spreads were mounted in Mowiol solution and imaged at room temperature using an AxioImager.Z2 microscope (Zeiss) with a Plan-Apo 60×/1.4 oil objective lens (Nikon). The microscope was fitted with a CoolCube 1m camera system and MetaSystems Metafer automatic slide platform. Acquisition of images was performed using Metafer4 v3.9.6 software and images were viewed in Photoshop CS6 (Adobe). Immunofluorescence slides were mounted in Mowiol solution and imaged at room temperature using an E800 microscope (Nikon) with anApochromat 63×/1.4 oil objective lens (Zeiss). The microscope was fitted with a DXM1200F camera system (Nikon). Acquisition of images was performed using ACT-1 software (Nikon) and viewed and overlaid in PhotoShop CS6.
Statistical analysis
All experiments were performed with a minimum of three independent experiments. Data and statistical analysis was performed on GraphPad Prism 6. Error bars represent standard deviation between experiments. P-values were calculated using a Student’s t test.
Online supplemental material
Fig. S1 shows that ablation of Blm rescues genomic instability in Brca1Δ11/Δ11 cells. Fig. S2 shows an analysis of SCEs in Brca1Δ11/Δ11;BlmΔ/Δ cells and BLM overexpression and Rad51 foci in U2OS cells. Fig. S3 shows that ablation of Blm increases CSR in Brca1Δ11/Δ11 cells but has no effect on RPA intensity. Fig. S4 shows knockdown of PALB2, BRCA2, and XRCC2 in U2OS cells. Fig. S5 shows knockdown of RECQ5, FBH1, and WRN anti-recombination factors in U2OS cells. Table S1 lists the Dharmacon siRNA oligonucleotides used. Table S2 lists the custom siRNA oligos used.
Supplementary Material
Acknowledgments
The authors thank Dr. Bing Xia for providing the pcDNA-3xMyc-BRCA1 plasmid and Dr. Steve Brill and Dr. Bing Xia for critical review of the manuscript.
This work was funded by National Institutes of Health grant R01CA190858 (to S.F. Bunting). D.S. Patel and S.M. Misenko are supported by New Jersey Commission on Cancer Research predoctoral fellowships and the National Institutes of Health Ruth L. Kirschstein National Research service award T32 GM8339.
The authors declare no competing financial interests.
Author contributions: D.S. Patel and S.F. Bunting designed experiments. D.S. Patel performed the experimental procedures. S.M. Misenko performed analysis of SCEs. J. Her performed the DNA fiber assay.
Footnotes
Abbreviations used:
- BS
- Bloom syndrome
- CSR
- class switch recombination
- DSB
- double-strand break
- HR
- homologous recombination
- IR
- ionizing radiation
- IRIF
- ionizing radiation–induced foci
- MEF
- mouse embryonic fibroblast
- NHEJ
- nonhomologous end joining
- RPA
- replication protein A
- SCE
- sister chromatid exchange
References
- Aboussekhra A., Chanet R., Zgaga Z., Cassier-Chauvat C., Heude M., and Fabre F.. 1989. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 17:7211–7219. 10.1093/nar/17.18.7211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboussekhra A., Chanet R., Adjiri A., and Fabre F.. 1992. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol. 12:3224–3234. 10.1128/MCB.12.7.3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantha R.W., Simhadri S., Foo T.K., Miao S., Liu J., Shen Z., Ganesan S., and Xia B.. 2017. Functional and mutational landscapes of BRCA1 for homology-directed repair and therapy resistance. eLife. 6: e21350 10.7554/eLife.21350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati C.Z., Borts R.H., and Hickson I.D.. 2006. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 34:2269–2279. 10.1093/nar/gkl258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava R., Onyango D.O., and Stark J.M.. 2016. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 32:566–575. 10.1016/j.tig.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A., Ear U.S., Koller B.H., Weichselbaum R.R., and Bishop D.K.. 2000. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275:23899–23903. 10.1074/jbc.C000276200 [DOI] [PubMed] [Google Scholar]
- Bindra R.S., Goglia A.G., Jasin M., and Powell S.N.. 2013. Development of an assay to measure mutagenic non-homologous end-joining repair activity in mammalian cells. Nucleic Acids Res. 41:e115 10.1093/nar/gkt255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof O., Kim S.H., Irving J., Beresten S., Ellis N.A., and Campisi J.. 2001. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 153:367–380. 10.1083/jcb.153.2.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P., Aly A., Escandell J.M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., et al. . 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17:688–695. 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev D.V., Yu X., Egelman E.H., and Mazin A.V.. 2007. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 21:3085–3094. 10.1101/gad.1609007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson R., Dion-Côté A.M., Coulombe Y., Launay H., Cai H., Stasiak A.Z., Stasiak A., Xia B., and Masson J.Y.. 2010. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 17:1247–1254. 10.1038/nsmb.1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting S.F., Callén E., Wong N., Chen H.T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., et al. . 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 141:243–254. 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho C.M., and Lupski J.R.. 2016. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 17:224–238. 10.1038/nrg.2015.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R., Liu J.C., Amunugama R., Hajdu I., Primack B., Petalcorin M.I., O’Connor K.W., Konstantinopoulos P.A., Elledge S.J., Boulton S.J., et al. . 2015. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature. 518:258–262. 10.1038/nature14184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti R.S., Schonberg S., and German J.. 1974. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc. Natl. Acad. Sci. USA. 71:4508–4512. 10.1073/pnas.71.11.4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J.R., Taylor M.R., and Boulton S.J.. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 47:497–510. 10.1016/j.molcel.2012.07.029 [DOI] [PubMed] [Google Scholar]
- Chen J., Silver D.P., Walpita D., Cantor S.B., Gazdar A.F., Tomlinson G., Couch F.J., Weber B.L., Ashley T., Livingston D.M., and Scully R.. 1998. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell. 2:317–328. 10.1016/S1097-2765(00)80276-2 [DOI] [PubMed] [Google Scholar]
- Chester N., Babbe H., Pinkas J., Manning C., and Leder P.. 2006. Mutation of the murine Bloom’s syndrome gene produces global genome destabilization. Mol. Cell. Biol. 26:6713–6726. 10.1128/MCB.00296-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W.K., Hanada K., Kanaar R., and Hickson I.D.. 2010. BLM has early and late functions in homologous recombination repair in mouse embryonic stem cells. Oncogene. 29:4705–4714. 10.1038/onc.2010.214 [DOI] [PubMed] [Google Scholar]
- Constantinou A., Tarsounas M., Karow J.K., Brosh R.M., Bohr V.A., Hickson I.D., and West S.C.. 2000. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 1:80–84. 10.1093/embo-reports/kvd004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., and German J.. 1995. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 83:655–666. 10.1016/0092-8674(95)90105-1 [DOI] [PubMed] [Google Scholar]
- Elstrodt F., Hollestelle A., Nagel J.H., Gorin M., Wasielewski M., van den Ouweland A., Merajver S.D., Ethier S.P., and Schutte M.. 2006. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 66:41–45. 10.1158/0008-5472.CAN-05-2853 [DOI] [PubMed] [Google Scholar]
- Evers B., and Jonkers J.. 2006. Mouse models of BRCA1 and BRCA2 deficiency: Past lessons, current understanding and future prospects. Oncogene. 25:5885–5897. 10.1038/sj.onc.1209871 [DOI] [PubMed] [Google Scholar]
- Forment J.V., Walker R.V., and Jackson S.P.. 2012. A high-throughput, flow cytometry-based method to quantify DNA-end resection in mammalian cells. Cytometry A. 81:922–928. 10.1002/cyto.a.22155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugger K., Mistrik M., Danielsen J.R., Dinant C., Falck J., Bartek J., Lukas J., and Mailand N.. 2009. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J. Cell Biol. 186:655–663. 10.1083/jcb.200812138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarz A., Guirouilh-Barbat J., Barascu A., Pennarun G., Genet D., Rass E., Germann S.M., Bertrand P., Hickson I.D., and Lopez B.S.. 2013. A role for BLM in double-strand break repair pathway choice: Prevention of CtIP/Mre11-mediated alternative nonhomologous end-joining. Cell Reports. 5:21–28. 10.1016/j.celrep.2013.08.034 [DOI] [PubMed] [Google Scholar]
- Gravel S., Chapman J.R., Magill C., and Jackson S.P.. 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22:2767–2772. 10.1101/gad.503108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O., Resch W., Yamane A., Klein I., Kieffer-Kwon K.R., Jankovic M., Oliveira T., Bothmer A., Voss T.C., Ansarah-Sobrinho C., et al. . 2012. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature. 484:69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennet T., Hagen F.K., Tabak L.A., and Marth J.D.. 1995. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc. Natl. Acad. Sci. USA. 92:12070–12074. 10.1073/pnas.92.26.12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs M.R., Reynolds J.J., Winczura A., Blackford A.N., Borel V., Miller E.S., Zlatanou A., Nieminuszczy J., Ryan E.L., Davies N.J., et al. . 2015. BOD1L is required to suppress deleterious resection of stressed replication forks. Mol. Cell. 59:462–477. 10.1016/j.molcel.2015.06.007 [DOI] [PubMed] [Google Scholar]
- Holloway J.K., Morelli M.A., Borst P.L., and Cohen P.E.. 2010. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J. Cell Biol. 188:779–789. 10.1083/jcb.200909048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P., Beresten S.F., van Brabant A.J., Ye T.Z., Pandolfi P.P., Johnson F.B., Guarente L., and Ellis N.A.. 2001. Evidence for BLM and Topoisomerase IIIalpha interaction in genomic stability. Hum. Mol. Genet. 10:1287–1298. 10.1093/hmg/10.12.1287 [DOI] [PubMed] [Google Scholar]
- Hu Y., Lu X., Barnes E., Yan M., Lou H., and Luo G.. 2005. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell. Biol. 25:3431–3442. 10.1128/MCB.25.9.3431-3442.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Raynard S., Sehorn M.G., Lu X., Bussen W., Zheng L., Stark J.M., Barnes E.L., Chi P., Janscak P., et al. . 2007. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 21:3073–3084. 10.1101/gad.1609107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber L.J., Yang T.W., Sarkisian C.J., Master S.R., Deng C.X., and Chodosh L.A.. 2001. Impaired DNA damage response in cells expressing an exon 11-deleted murine Brca1 variant that localizes to nuclear foci. Mol. Cell. Biol. 21:4005–4015. 10.1128/MCB.21.12.4005-4015.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., and Haber J.E.. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 115:401–411. 10.1016/S0092-8674(03)00886-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.B., Carreira A., and Kowalczykowski S.C.. 2010. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 467:678–683. 10.1038/nature09399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.D., Liu N., and Jasin M.. 1999. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 401:397–399. 10.1038/43932 [DOI] [PubMed] [Google Scholar]
- Karpenshif Y., and Bernstein K.A.. 2012. From yeast to mammals: Recent advances in genetic control of homologous recombination. DNA Repair (Amst.). 11:781–788. 10.1016/j.dnarep.2012.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski S.C. 2015. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7: a016410 10.1101/cshperspect.a016410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., and Sung P.. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 423:305–309. 10.1038/nature01577 [DOI] [PubMed] [Google Scholar]
- Larsen N.B., and Hickson I.D.. 2013. RecQ helicases: Conserved guardians of genomic integrity. Adv. Exp. Med. Biol. 767:161–184. 10.1007/978-1-4614-5037-5_8 [DOI] [PubMed] [Google Scholar]
- Leuzzi G., Marabitti V., Pichierri P., and Franchitto A.. 2016. WRNIP1 protects stalled forks from degradation and promotes fork restart after replication stress. EMBO J. 35:1437–1451. 10.15252/embj.201593265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Cole F., Patel D.S., Misenko S.M., Her J., Malhowski A., Alhamza A., Zheng H., Baer R., Ludwig T., et al. . 2016. 53BP1 ablation rescues genomic instability in mice expressing ‘RING-less’ BRCA1. EMBO Rep. 17:1532–1541. 10.15252/embr.201642497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A., Osman F., Folkyte V., Sofueva S., and Whitby M.C.. 2009. Fbh1 limits Rad51-dependent recombination at blocked replication forks. Mol. Cell. Biol. 29:4742–4756. 10.1128/MCB.00471-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Santoro I.M., McDaniel L.D., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R.A., and Bradley A.. 2000. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26:424–429. 10.1038/82548 [DOI] [PubMed] [Google Scholar]
- Mak T.W., Hakem A., McPherson J.P., Shehabeldin A., Zablocki E., Migon E., Duncan G.S., Bouchard D., Wakeham A., Cheung A., et al. . 2000. Brca1 required for T cell lineage development but not TCR loci rearrangement. Nat. Immunol. 1:77–82. 10.1038/76950 [DOI] [PubMed] [Google Scholar]
- Manis J.P., Morales J.C., Xia Z., Kutok J.L., Alt F.W., and Carpenter P.B.. 2004. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 5:481–487. 10.1038/ni1067 [DOI] [PubMed] [Google Scholar]
- Mankouri H.W., Craig T.J., and Morgan A.. 2002. SGS1 is a multicopy suppressor of srs2: Functional overlap between DNA helicases. Nucleic Acids Res. 30:1103–1113. 10.1093/nar/30.5.1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthei K.A., and Keck J.L.. 2013. The BLM dissolvasome in DNA replication and repair. Cell. Mol. Life Sci. 70:4067–4084. 10.1007/s00018-013-1325-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misenko S.M., and Bunting S.F.. 2014. Rapid analysis of chromosome aberrations in mouse B lymphocytes by PNA-FISH. J. Vis. Exp. (90). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan G.L., Dejsuphong D., Petalcorin M.I., Hofmann K., Takeda S., Boulton S.J., and D’Andrea A.D.. 2012. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell. 45:75–86. 10.1016/j.molcel.2011.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan M.E., Chiu J.W., Koller B.H., and Jasin M.. 1999. Brca1 controls homology-directed DNA repair. Mol. Cell. 4:511–518. 10.1016/S1097-2765(00)80202-6 [DOI] [PubMed] [Google Scholar]
- Myung K., Datta A., Chen C., and Kolodner R.D.. 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27:113–116. 10.1038/83673 [DOI] [PubMed] [Google Scholar]
- Nimonkar A.V., Ozsoy A.Z., Genschel J., Modrich P., and Kowalczykowski S.C.. 2008. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. USA. 105:16906–16911. 10.1073/pnas.0809380105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar A.V., Genschel J., Kinoshita E., Polaczek P., Campbell J.L., Wyman C., Modrich P., and Kowalczykowski S.C.. 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25:350–362. 10.1101/gad.2003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan P., Wilson C., Townsend S., and Thacker J.. 2001. XRCC2 is a nuclear RAD51-like protein required for damage-dependent RAD51 focus formation without the need for ATP binding. J. Biol. Chem. 276:22148–22153. 10.1074/jbc.M102396200 [DOI] [PubMed] [Google Scholar]
- Ray Chaudhuri A., Callen E., Ding X., Gogola E., Duarte A.A., Lee J.E., Wong N., Lafarga V., Calvo J.A., Panzarino N.J., et al. . 2016. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 535:382–387. 10.1038/nature18325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert R.C., Roes J., and Rajewsky K.. 1997. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 25:1317–1318. 10.1093/nar/25.6.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Shimomuki M., Katsuki Y., Takahashi D., Kobayashi W., Ishiai M., Miyoshi H., Takata M., and Kurumizaka H.. 2016. FANCI-FANCD2 stabilizes the RAD51-DNA complex by binding RAD51 and protects the 5′-DNA end. Nucleic Acids Res. 44:10758–10771. 10.1093/nar/gkw876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K., Wu H., and Jasin M.. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 22:106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendener S., Raynard S., Paliwal S., Cheng A., Kanagaraj R., Shevelev I., Stark J.M., Sung P., and Janscak P.. 2010. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J. Biol. Chem. 285:15739–15745. 10.1074/jbc.M110.110478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simandlova J., Zagelbaum J., Payne M.J., Chu W.K., Shevelev I., Hanada K., Chatterjee S., Reid D.A., Liu Y., Janscak P., et al. . 2013. FBH1 helicase disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells. J. Biol. Chem. 288:34168–34180. 10.1074/jbc.M113.484493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy S.M., Huen M.S., and Chen J.. 2009a MRG15 is a novel PALB2-interacting factor involved in homologous recombination. J. Biol. Chem. 284:21127–21131. 10.1074/jbc.C109.023937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy S.M., Huen M.S., and Chen J.. 2009b PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. USA. 106:7155–7160. 10.1073/pnas.0811159106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V., Nagarjuna T., and Sengupta S.. 2007. BLM helicase-dependent and -independent roles of 53BP1 during replication stress-mediated homologous recombination. J. Cell Biol. 178:9–14. 10.1083/jcb.200610051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V., Kaur S., and Sengupta S.. 2008. Phosphorylation-dependent interactions of BLM and 53BP1 are required for their anti-recombinogenic roles during homologous recombination. Carcinogenesis. 29:52–61. 10.1093/carcin/bgm238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brabant A.J., Ye T., Sanz M., German J.L. III, Ellis N.A., and Holloman W.K.. 2000. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 39:14617–14625. 10.1021/bi0018640 [DOI] [PubMed] [Google Scholar]
- Veaute X., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., and Fabre F.. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 423:309–312. 10.1038/nature01585 [DOI] [PubMed] [Google Scholar]
- Wang A.T., Kim T., Wagner J.E., Conti B.A., Lach F.P., Huang A.L., Molina H., Sanborn E.M., Zierhut H., Cornes B.K., et al. . 2015. A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol. Cell. 59:478–490. 10.1016/j.molcel.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Seki M., Narita Y., Nakagawa T., Yoshimura A., Otsuki M., Kawabe Y., Tada S., Yagi H., Ishii Y., and Enomoto T.. 2003. Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol. Cell. Biol. 23:3527–3535. 10.1128/MCB.23.10.3527-3535.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I.M., Reina-San-Martin B., Olaru A., Minn K., Tamada K., Lau J.S., Cascalho M., Chen L., Nussenzweig A., Livak F., et al. . 2004. 53BP1 is required for class switch recombination. J. Cell Biol. 165:459–464. 10.1083/jcb.200403021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., and Hickson I.D.. 2003. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 426:870–874. 10.1038/nature02253 [DOI] [PubMed] [Google Scholar]
- Wu L., Davies S.L., Levitt N.C., and Hickson I.D.. 2001. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 276:19375–19381. 10.1074/jbc.M009471200 [DOI] [PubMed] [Google Scholar]
- Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F.J., and Livingston D.M.. 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 22:719–729. 10.1016/j.molcel.2006.05.022 [DOI] [PubMed] [Google Scholar]
- Xu X., Weaver Z., Linke S.P., Li C., Gotay J., Wang X.W., Harris C.C., Ried T., and Deng C.X.. 1999. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol. Cell. 3:389–395. 10.1016/S1097-2765(00)80466-9 [DOI] [PubMed] [Google Scholar]
- Zhang F., Ma J., Wu J., Ye L., Cai H., Xia B., and Yu X.. 2009. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr. Biol. 19:524–529. 10.1016/j.cub.2009.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.