Cancer-associated fibroblasts (CAFs) in the tumor stroma play a key role in tumor progression. Erdogan et al. show that CAF-mediated alignment of the fibronectin matrix is a key factor promoting directional cancer cell migration.
Abstract
Cancer-associated fibroblasts (CAFs) are major components of the carcinoma microenvironment that promote tumor progression. However, the mechanisms by which CAFs regulate cancer cell migration are poorly understood. In this study, we show that fibronectin (Fn) assembled by CAFs mediates CAF–cancer cell association and directional migration. Compared with normal fibroblasts, CAFs produce an Fn-rich extracellular matrix with anisotropic fiber orientation, which guides the cancer cells to migrate directionally. CAFs align the Fn matrix by increasing nonmuscle myosin II- and platelet-derived growth factor receptor α–mediated contractility and traction forces, which are transduced to Fn through α5β1 integrin. We further show that prostate cancer cells use αv integrin to migrate efficiently and directionally on CAF-derived matrices. We demonstrate that aligned Fn is a prominent feature of invasion sites in human prostatic and pancreatic carcinoma samples. Collectively, we present a new mechanism by which CAFs organize the Fn matrix and promote directional cancer cell migration.
Introduction
Cancer-associated fibroblasts (CAFs) are one of the most abundant cell types in the tumor microenvironment and have the ability to promote tumor growth (Olumi et al., 1999; Orimo et al., 2005). A key function of normal fibroblasts (NFs) is to maintain the homeostasis of the ECM (Kalluri and Zeisberg, 2006). In contrast, CAFs and other activated fibroblasts exhibit changes in this critical process. CAFs secrete high levels of ECM proteins, such as fibronectin (Fn), type I and type II collagen, and express oncofetal isoforms of Fn (Barsky et al., 1984; Tuxhorn et al., 2002; Schor et al., 2003; Clarke et al., 2016; Gopal et al., 2017). In addition, CAFs have been shown to alter the architecture and physical properties of the ECM, influencing cell migration, invasion, and growth (Jolly et al., 2016; Kaukonen et al., 2016). Through force-mediated matrix remodeling, CAFs deform collagen I matrices, generating tracks that cancer cells follow (Gaggioli et al., 2007). CAFs also have been shown to generate aligned matrix fibers in vitro (Amatangelo et al., 2005; Lee et al., 2011; Franco-Barraza et al., 2017). Alignment of ECM fibers has also been observed in tumors and found to be associated with poor patient prognosis (Conklin et al., 2011; Franco-Barraza et al., 2017). However, the mechanisms of ECM alignment and its role in CAF–cancer cell interactions remain poorly understood.
Fn is one of the most abundant ECM proteins and mediates various cellular activities, including adhesion, migration, growth, and differentiation (Pankov and Yamada, 2002). Fn binds to ECM proteins, such as collagen, periostin, fibrillin, and tenascin-C, and facilitates their assembly and organization (Kadler et al., 2008; Kii et al., 2010). Aberrant expression of Fn has also been associated with tumor progression (Insua-Rodríguez and Oskarsson, 2016; Topalovski and Brekken, 2016; Wang and Hielscher, 2017). Hence, there is substantial interest in understanding the function of Fn in the tumor microenvironment.
Fn is assembled into fibers through its binding to transmembrane integrin adhesion receptors (Mao and Schwarzbauer, 2005; Campbell and Humphries, 2011). Integrin α5β1 is the major Fn receptor and facilitates Fn fibrillogenesis by activating cellular contractility and applying traction forces to Fn (Hinz, 2006; Lemmon et al., 2009; Schwarzbauer and DeSimone, 2011). Although the role of α5β1 integrin in the Fn matrix assembly is well known, it is not clear how inside–out signaling in activated fibroblasts is regulated and leads to matrix reorganization.
Growth factor signaling is important in mediating cancer cell–tumor stroma interactions to promote tumor progression. One of the key growth factors connecting cancer and stromal cells is PDGF. PDGF is a potent activator of fibroblasts through its binding to cell-surface PDGF receptors (PDGFRs). PDGFRs are tyrosine kinase receptors composed of homo- or heterodimers of two PDGFR chains, PDGFRα and PDGFRβ (Donovan et al., 2013). Most cancer cells, including prostate carcinomas, express PDGF ligands but not PDGFRs (Sariban et al., 1988; Sitaras et al., 1988). In contrast, CAFs overexpress both PDGFRs compared with NFs (Augsten, 2014). PDGF ligands secreted by cancer cells are known to induce proliferation, migration, and recruitment of stromal fibroblasts (Östman, 2004). A recent study showed that inactivation of PDGFRα in fibroblasts decreases connective tissue remodeling (Horikawa et al., 2015); however, its role in remodeling of other tissues and/or disease states is poorly understood.
In the present study, we demonstrate that Fn fibrillogenesis by CAFs promotes CAF–cancer cell interactions and mediates directional migration of cancer cells in co-culture assays. Fn-rich cell-derived matrices (CDMs) isolated from CAF cultures, but not NF cultures, exhibit aligned fiber organization and promote directional cancer cell migration. Compared with NFs, we find that matrix organization by CAFs is mediated by enhanced myosin-II–driven contractility and increased traction forces, transduced to the ECM via α5β1 integrin. Furthermore, we provide evidence that up-regulated PDGFRα activity in CAFs has a role in contractility and parallel Fn organization. We also identify αv integrin as a regulator of cancer cell migration on CAF matrices. Collectively, we demonstrate a new mechanism driving CAF–cancer cell interaction and directional cancer cell migration.
Results
Fn promotes CAF–cancer cell association and directional cancer cell migration
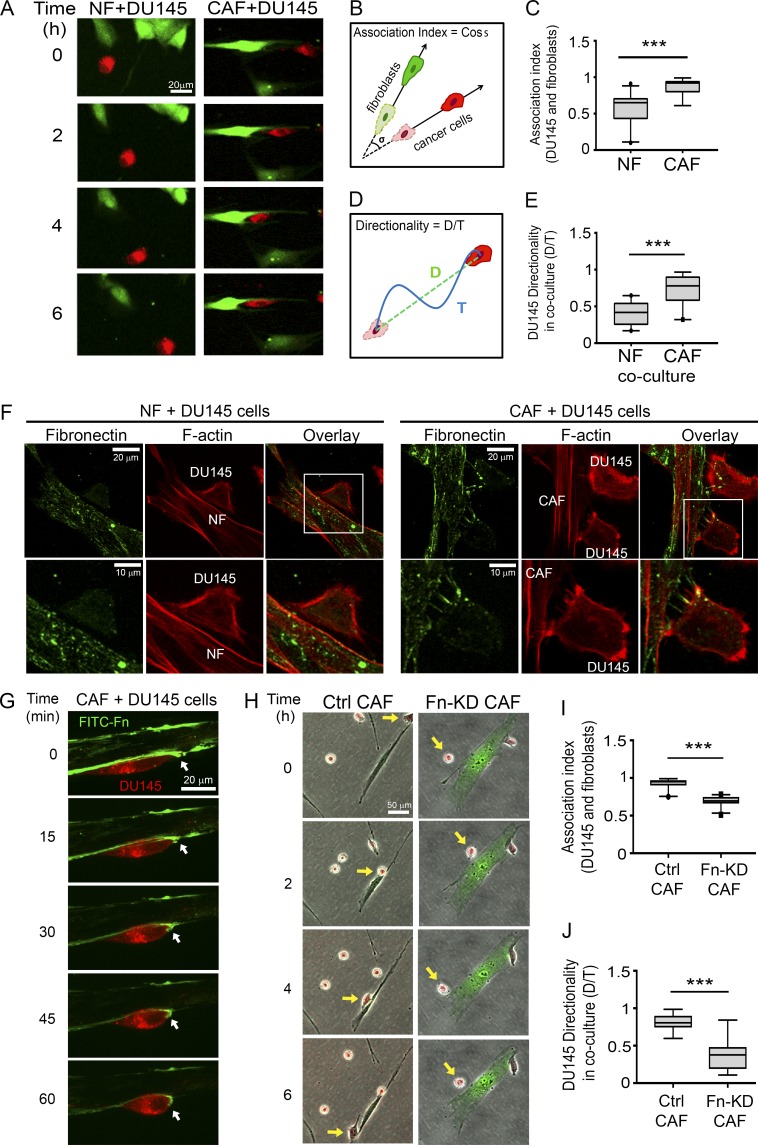
To investigate the effects of CAFs on cancer cell migration, we co-cultured prostatic fibroblasts with DU145 prostate cancer cells. CellTracker green-labeled CAFs or NFs were mixed with CellTracker red-labeled DU145 prostate cancer cells in a 1:1 ratio and loaded into two separate, side-by-side chambers of a microfluidic device to mimic the close interactions within the tumor microenvironment (Fig. S1 A). When DU145 cells were co-cultured with NFs, they exhibited minimal interaction with NFs and migrated randomly (Fig. 1, A–E; and Video 1). In contrast, in co-cultures with CAFs, DU145 cells migrated toward and along the axis of CAFs, resulting in a greater association index with CAFs (Fig. 1, A–E; and Video 2). Interestingly, no difference in the migration speeds of DU-145 cells was found in either co-culture condition (Fig. S1 B). To test whether CAF-promoted directional cancer cell migration was restricted to tissue-matched cancer cells or whether CAFs could induce similar effects on other cancer cell types, we subjected head and neck squamous cell carcinoma (HNSCC) cell lines JHU012 and SCC61 to co-culture with prostate CAFs and NFs. Intriguingly, although derived from a different tissue, HNSCC cell lines also displayed an increased association with fibroblasts and directional migration when co-cultured with prostate CAFs. However, co-culturing with NFs did not affect the migration directionality of HNSCC cells or induce an association between NFs and HNSCC cells (Fig. S1, C–G; and Video 3).
Figure 1.
Fn secreted by CAFs promotes CAF–cancer cell association and directional cancer cell migration. (A) Time-lapse images showing co-culture of DU145 prostate cancer cells (red) with NFs (left; green) or CAFs (right; green) in microfluidic devices. Bar, 20 µm (Videos 1 and 2). (B) Schematic representation of the calculation to determine the association index between fibroblasts and cancer cells. (C) Association index of DU145 cells with NFs or CAFs. (D) Schematic representation for the calculation to determine the directionality ratio. (E) Directionality ratio of DU145 cells in co-cultures with NFs or CAFs. (C and E) The data represent ≥30 cells per condition in four individual experiments. (F) Fn staining of NF + DU145 cells (left) and CAF + DU145 (right) cell co-cultures. Fn, green; F-actin, phalloidin, red. Bars: (F, top) 20 µm; (bottom) 10 µm. White boxes indicate areas of increased magnification. (G) Co-culture of CAFs (unlabeled) with DU145 cells (red), cultured in culture medium supplemented with 5 µg/ml FITC–Fn (green). Bar, 20 µm. Arrows point to the leading edge of the cancer cell, where it binds to the Fn fibers assembled by CAFs (Video 3). (H) Time-lapse images showing co-culture of DU145 cells (red) with CAFs (green) transfected with nontargeting siRNA control (Ctrl CAF, left) or Fn siRNA (Fn-KD CAF, right). Arrows point to the cells of interest. Bar, 50 µm (Video 4). (I) Association index for DU145 cells with control (Ctrl) or Fn–KD CAFs. (J) Directionality ratio of DU145 cells in co-cultures with control (Ctrl) or Fn–KD CAFs. (I and J) The data represent ≥30 cells per condition in three individual experiments (C, E, I, and J) ***, P < 0.001, determined by Mann-Whitney U test. All box plots range from the 25th to the 75th percentiles; the central line indicates the median, and the whiskers range from the 5th to the 95th percentiles.
Prostate CAF-induced directional migration, in both the prostate cell line DU145 and the HNSCC cell lines JHU012 and SCC61, suggests that the mechanism by which CAFs modulate cancer cell migration is not organ specific. Previous studies identified various ways that CAFs alter the ECM composition and architecture (Gaggioli et al., 2007; Jolly et al., 2016). Because ECM is a major factor that regulates cell migration, we hypothesized that increased association and directional cancer cell migration in co-cultures with CAFs are a result of changes in the ECM. Fn is a major ECM protein, which is secreted and assembled into fibers by fibroblasts. Aberrant expression of Fn and its fetal isoforms have been reported in many cancers (Bae et al., 2013; Topalovski and Brekken, 2016; Gopal et al., 2017; Wang and Hielscher, 2017). Therefore, we first studied the expression of Fn and its splice variant (with extra domain A [Fn-EDA]) in prostate CAFs. We found that CAFs expressed 50% more Fn compared with NFs (Fig. S1, H and I). In addition, there was a 3.5-fold increase in the expression of the isoform of Fn in CAFs relative to NFs (Fig. S1, J and K). Next, we studied whether cancer cells interact with Fn in co-cultures. After 24 h of incubation, cells were fixed and stained for Fn and F-actin. In co-cultures with NFs, in the rarer cases, in which cancer cells made physical contact with fibroblasts, we did not observe detectable Fn in the contact area (Fig. 1 F, left). However, when co-cultured with CAFs, cancer cells frequently appeared to be attached to Fn fibers at contact sites with the CAFs (Fig. 1 F, right). To visualize the interaction between CAF-assembled Fn fibrils and cancer cells live, we added FITC-labeled Fn to the co-culture medium at the same time that cells were plated. In the 24 h incubation, CAFs incorporated FITC–Fn into Fn fibrils. The next day, time-lapse microscopy was performed. We observed that DU145 cells actively pull on the Fn fibers on the periphery of CAFs as they migrate (Fig. 1 G and Video 4). Next, we tested whether Fn derived from CAFs was critical for directional cancer cell migration and increased association with CAFs. First, Fn expression was knocked down in CAFs using an siRNA pool, which reduced Fn expression by 80% (Fig. S1, L and M). Then, DU145 cells were co-cultured with control or Fn-knockdown (KD) CAFs for 24 h, and time-lapse microscopy was performed. Knocking down Fn significantly reduced the association and migration directionality of DU145 cells with CAFs (Fig. 1, H–J; and Video 5). Notably, migration speed of DU145 cells was also decreased in co-cultures with Fn-KD CAFs compared with control CAFs (Fig. S1 N).
The close interaction between the cancer cells and CAFs in our co-culture experiments gave rise to the question of whether cancer cells and CAFs make heterotypic E-cadherin/N-cadherin adhesions, as recently reported (Labernadie et al., 2017). Immunofluorescence (IF) staining of DU145-CAF co-cultures for N-cadherins and E-cadherins revealed that DU145 cells make E-cadherin junctions with other DU145 cancer cells (Fig. S2 A, top). In contrast, CAFs exhibited N-cadherin junctions when contacting other CAFs (Fig. S2 A, bottom). However, we did not observe any N-cadherin/E-cadherin connections at sites at which DU145 cancer cells made contact with CAFs (Fig. S2 A, bottom).
Fn is an essential component of the CAF-derived matrix and promotes directional migration of cancer cells
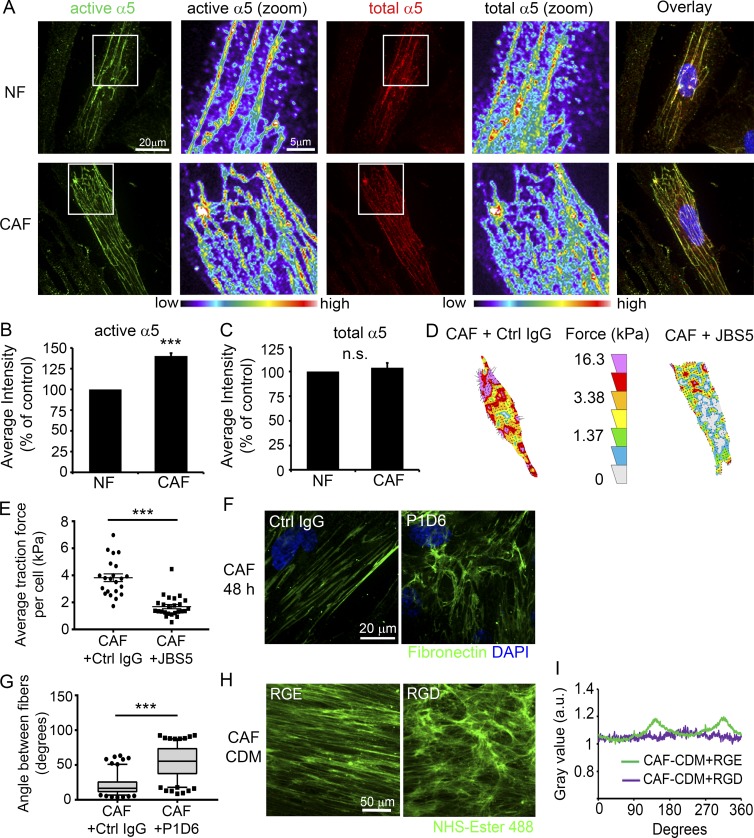
Our results indicated that Fn, secreted by CAFs, is important for regulation of cancer cell migration. In addition to Fn expression, changes in Fn organization can have a role in mediating cancer cell migration. Therefore, we studied the architecture of the Fn matrix in NFs and CAFs. After 48 h of incubation, NFs assembled Fn into an intricate network of fibers resembling a mesh; in contrast, CAFs organized Fn into parallel fibers (Fig. S2 B). The angles between the Fn fibers in the CAF matrix were significantly smaller than those of the NF matrix, indicating a more-aligned fiber organization (Fig. S2 C). Similarly, the peaks observed in Fast-Fourier transform (FFT) analyses indicated that Fn was arranged in a specific direction by CAFs, compared with the unorganized fiber network assembled by NFs (Fig. S2 D).
To better understand the role of CAF-derived Fn in regulating cancer cell migration, we generated CDMs. The CDMs are produced by the CAFs and NFs that were removed from the matrix on d 8, leaving the matrix intact (Franco-Barraza et al., 2016). The matrix was then visualized by labeling with N-hydroxysuccinimide (NHS)–ester-488. The CDMs produced by NFs displayed a random network of fibers; however, ECM fibers in the CAF-derived CDMs presented an anisotropic fiber orientation with prominent peaks 180° apart by FFT analysis (Fig. 2, A–C). Most of the fibers in both NF and CAF CDMs colocalized with Fn, indicating that Fn is an abundant component of those matrices (Fig. 2 A).
Figure 2.
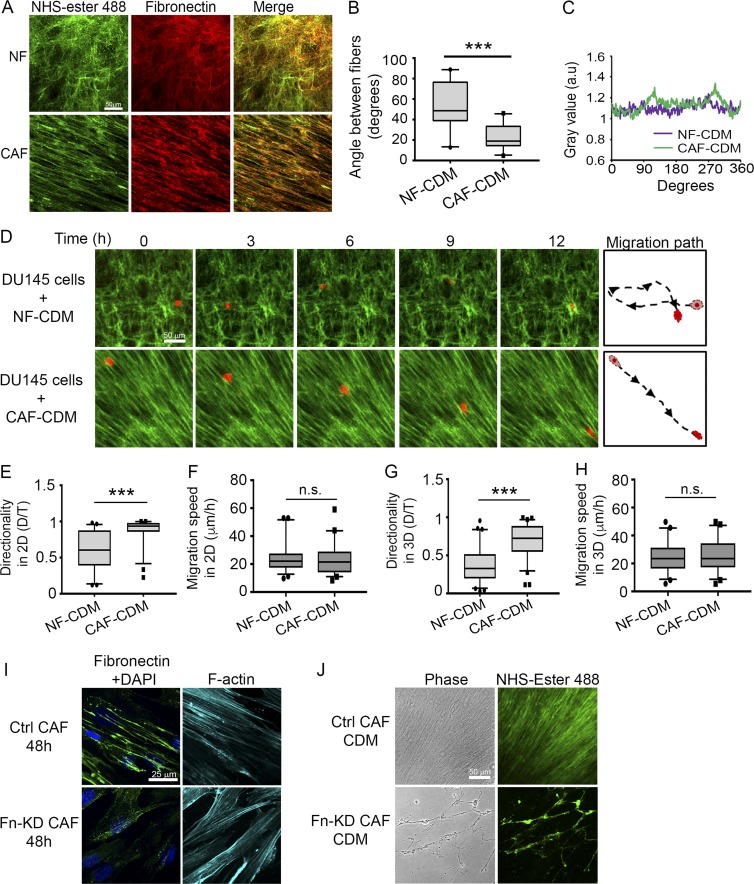
Aligned Fn organization by CAFs mediates directional cancer cell migration. (A) Representative images of NHS–ester-488 (green) and anti–Fn (red) staining in CDMs generated by NFs and CAFs. Bar, 50 µm. (B) Measurements of angles between Fn fibers in NF and CAF CDMs. More than 100 angles per condition were measured from at least 12 images from three independent experiments. ***, P < 0.001, Mann-Whitney U test. (C) FFT analysis of CDMs stained with the Fn antibody shown in A. (D) Time-lapse images showing DU145 cells (red) migrating on NF- and CAF-derived matrices (labeled with NHS–ester-488 dye, green). Bar, 50 µm (Videos 5 and 6). (E–H) Box plots showing DU145 cell migration directionality ratio on NF and CAF CDMs in 2D (E) or 3D (G), and migration speed in 2D (F) and 3D (H). Greater than 70 cells were analyzed per condition from three independent experiments. ***, P < 0.001, analyzed by Mann-Whitney U test. Box plots range from the 25th to the 75th percentiles; the central line indicates the median, and the whiskers range from the 5th to the 95th percentiles. (I) Fn staining of control or Fn-KD CAFs at 48 h. Nucleus, DAPI, blue; F-actin, phalloidin, cyan. Bar, 25 µm. (J) Representative images of CDMs generated by control or Fn-KD CAFs. Bar, 50 µm.
To study cancer cell migration on the CDMs, cells were plated either on top of the CDMs in a 2D format or allowed to invade into the matrix after an overnight incubation (3D format). In both 2D and 3D conditions, directional migration of DU145 cells was enhanced on CAF CDMs compared with NF-CDMs (Fig. 2, D, E, and G; and Videos 6 and 7). Of note, we did not observe a difference in migration speed of DU145 cells on either NF or CAF CDMs (Fig. 2, F and H). Similar to DU145 cells, JHU012 and SCC61 cells also migrated more directionally in CAF CDMs, as opposed to random migration observed in NF-generated CDMs (Fig. S2, E–G, and Video 8, for JHU012 cells; and Fig. S2, J and K, for SCC61 cells). There was no difference in migration speed of JHU012 and SCC61 cells on NF or CAF CDMs in 2D or 3D conditions (Fig. S2, H and I, for JHU012 cells; and Fig. S2, L and M, for SCC61 cells).
Because our data suggested that CAF-secreted Fn mediated migration of cancer cells in co-culture experiments, we sought to determine the role of Fn on cell migration in CDMs. Knocking down Fn in CAFs completely abrogated Fn fibrillogenesis; at 48 h, we observed a minimal number of short Fn fibers in KD cells, with most of the Fn appearing as spots (Fig. 2 I). KD of Fn in CAFs lasted ≥6 d (Fig. S1 L); therefore, we used the control and Fn-KD CAFs to generate CDMs. Although the CDM by control CAFs was abundant and exhibited aligned organization (Fig. 2 J, top), CDM assembled by Fn-KD CAFs exhibited sparse fibers and bare areas in the culture dish (Fig. 2 J, bottom). Because of the defective matrix assembly by Fn-KD CAFs, we could not test cancer cell migration on those CDMs. Nevertheless, these results emphasize the previously reported importance of Fn in matrix assembly and organization, including incorporation of other ECM proteins into the matrix (Singh et al., 2010). Collectively, our data demonstrate that CAFs assemble an Fn-rich, highly organized matrix that promotes directional migration of both prostate cancer and HNSCC cells.
CAFs organize Fn as parallel fibers through increased traction forces and contractility
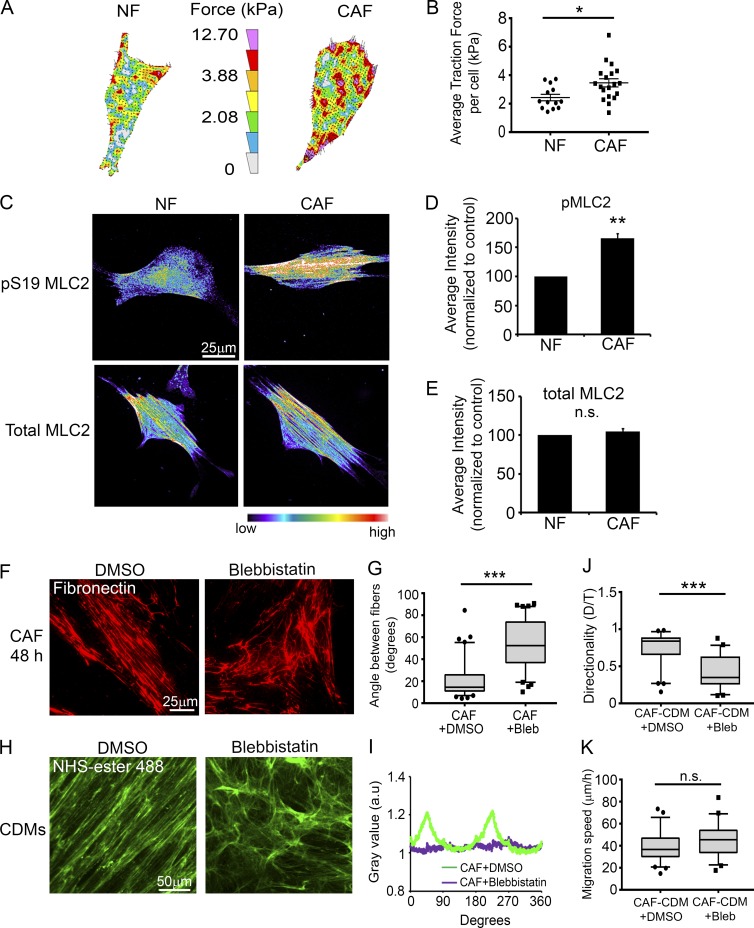
Our data indicate that anisotropic organization of the ECM by CAFs promotes the directional migration of cancer cells, which prompted us to investigate how CAFs mediate Fn organization. Cellular traction forces and nonmuscle myosin II (MyoII)–mediated contractility are critical factors in Fn matrix assembly (Lemmon et al., 2009). In addition, actomyosin contractility has been associated with matrix remodeling in 3D organotypic assays (Calvo et al., 2013). Therefore, we hypothesized that changes in mechanical force by CAFs led to alignment of Fn fibers. We first compared traction stresses generated by CAFs and NFs using traction-force microscopy and observed that CAFs exert ∼50% greater traction force on Fn compared with NFs (Fig. 3, A and B). Next, we performed collagen gel contraction assays to assess contractility of CAFs and NFs. In those assays, CAFs contracted the collagen-I gel to 40% of its original area; however, NFs contracted the gels to 58%, indicating that CAFs are significantly more contractile than NFs are (Fig. S3, A and B). Moreover, IF staining of pS19–myosin light chain 2 (MLC2) revealed that CAFs have 60% more active MyoII than NFs have (Fig. 3, C and D). However, we did not observe a difference in the total amounts of MLC2 expression between NFs and CAFs (Fig. 3, C and E). To further investigate whether MyoII-mediated contractility has a role in alignment of Fn by CAFs, we treated CAFs and NFs with 20 µM blebbistatin, a MyoII-specific inhibitor, for 48 h, and then stained them for Fn. Indeed, blebbistatin treatment disrupted the linear organization of Fn by CAFs and led to a more-random network of fibers compared with vehicle-treated control CAFs (Fig. 3, F and G). No changes were observed in matrix organization when NFs were treated with 20 µM blebbistatin (Fig. S3, C). Because actomyosin contractility is necessary for Fn fibrillogenesis, we treated CAFs with greater concentrations of blebbistatin (50 and 100 µM). Those increased blebbistatin concentrations almost completely abolished Fn fiber formation by CAFs (Fig. S3 D). We also treated CAFs with 20 µM blebbistatin during CDM generation. NHS–ester-488 staining of those matrices showed that the anisotropic fiber orientation by CAFs reverted to a NF-like CDM organization (Fig. 3, H and I). To test whether CAF CDMs generated during blebbistatin treatment affected directional cell migration, DU145 cells were plated onto those CDMs, and time-lapse microscopy was performed. DU145 cells migrated directionally on control CAF CDMs with a directionality ratio of 0.75. In contrast, CAF CDMs treated with blebbistatin did not support the directional migration of cancer cells, decreasing the directionality ratio to 0.42 (Fig. 3 J). We did not observe a significant difference in DU145 migration speed on either of the CDMs (Fig. 3 K).
Figure 3.
Myosin-II-driven traction force and contractility mediate parallel Fn organization by CAFs. (A) Representative traction force vector maps of a NF and a CAF. Warmer colors indicate areas with high traction forces. (B) Dot plot shows mean traction forces in NFs and CAFs. Line indicates the mean; error bars indicate SEM. A total of 13 NFs and 19 CAFs were analyzed in three independent experiments. *, P < 0.02, determined by Student’s t test. (C) Immunostaining for pS19-MLC2 or total MLC2 in NFs and CAFs. Images are shown in pseudocolor; warmer colors indicating high intensity, whereas cooler colors indicating low intensity. (D and E) Quantification of mean fluorescent intensity of pS19-MLC2 (D) and total MLC2 (E) in NFs and CAFs, normalized to NFs. Error bars indicate SEM from three individual experiments. More than 77 cells were analyzed per condition. **, P < 0.01; n.s., not significant, as determined by Student’s t test. (F) Fn staining of CAFs after 48 h treatment with DMSO (left) or 20 μM blebbistatin (Bleb; right). Bar, 25 µm. (G) Measurements of angles between Fn fibers in CAFs treated with DMSO or blebbistatin. More than 80 angles measured per condition from at least 12 images from three independent experiments. ***, P < 0.001, analyzed by Mann-Whitney U test. (H) NHS–ester-488 staining of CAF CDMs generated during DMSO (left) or blebbistatin (right) treatment. Bar, 50 µm. (I) FFT analysis of CDM images shown in H. (J and K) Box plots showing directionality ratio (J) and migration speed (K) of DU145 cell migration on CAF CDMs generated during DMSO or blebbistatin treatment. ***, P < 0.001; n.s., not significant, analyzed by Mann-Whitney U test. Greater than 50 cells per condition from three independent experiments were analyzed. The box plots range from the 25th to the 75th percentiles; the central line indicates the median, and the whiskers range from the 5th to the 95th percentiles.
CAFs form larger adhesions with slower turnover rates compared to NFs
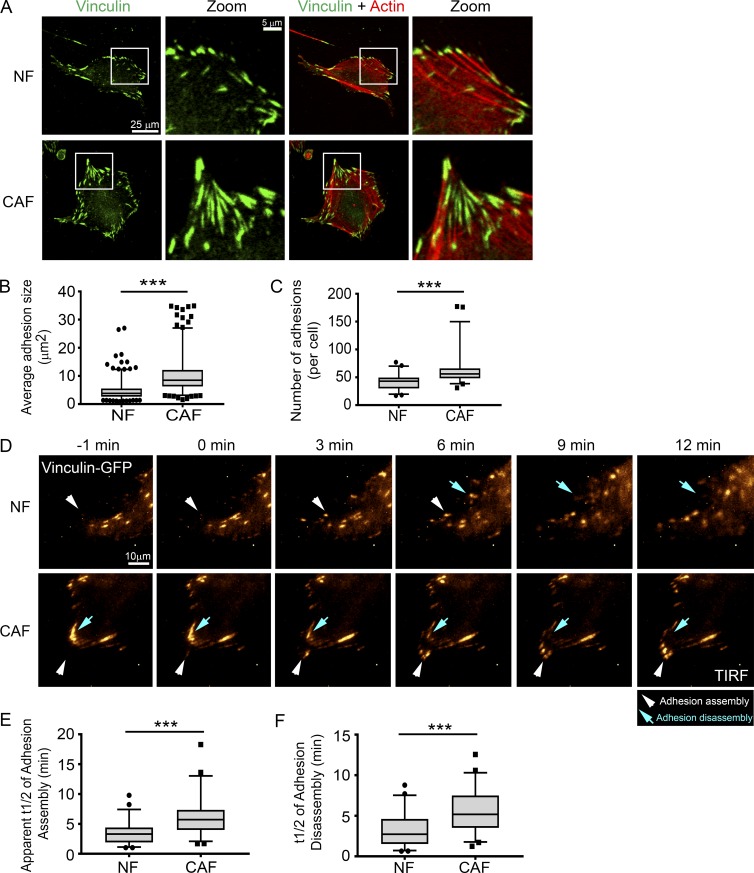
Adhesions are attachment points in cells that link the actin cytoskeleton and transmit MyoII-mediated mechanical force to the ECM (Burridge and Fath, 1989). The adhesion protein vinculin is mechanosensitive, and the increased size of the vinculin-containing adhesions has been correlated with increased mechanical force (Galbraith et al., 2002; Grashoff et al., 2010). Because we observed significant changes in the traction forces and contractility of CAFs, we investigated the size and number of vinculin-positive adhesions in CAFs and NFs. CAFs displayed significantly larger adhesions, relative to NFs (Fig. 4, A and B). Moreover, vinculin adhesions were more abundant in CAFs than they were in NFs (Fig. 4 C).
Figure 4.
CAFs form larger adhesions that turn over more slowly than NFs do. (A) IF staining of vinculin (green) and actin (red) in NFs and CAFs. White boxes indicate selected areas of interest in the zoom boxes. Bars: (original images) 25 µm; (zoomed images) 5 µm. (B and C) Box plots show mean adhesion size (μm2; B) and adhesion number (C) in NFs and CAFs. More than 600 adhesions (B) and than 50 cells (C) per condition were analyzed from three independent experiments. ***, P < 0.001, determined by Mann Whitney U test. (D) Time-lapse images show adhesion assembly and disassembly. NFs and CAFs were transfected with vinculin–GFP (pseudo-colored in gold; white arrowheads, adhesion assembly; blue arrows, adhesion disassembly; Videos 9 and 10). Bar, 10 µm. (E and F) Quantification of the apparent t1/2 of adhesion assembly (E) and disassembly (F) for NFs and CAFs. 50–60 adhesions were analyzed per condition from four independent experiments. ***, P < 0.001, determined by Mann Whitney U test. All box plots range from the 25th to the 75th percentiles; the central line indicates the median, and the whiskers range from the 5th to the 95th percentiles.
Larger adhesions in CAFs could be a result of altered adhesion dynamics in those cells. Therefore, we assessed adhesion turnover in CAFs and NFs by live-cell imaging. Vinculin-GFP–transfected fibroblasts were plated onto Fn-coated, glass-bottom dishes and imaged for 20 min using total internal reflection fluorescence (TIRF) microscopy. Analysis of individual adhesions revealed that CAFs assemble and disassemble adhesions at approximately one-half the speed of NFs (Fig. 4, D–F; and Videos 9 and 10).
Increased α5β1 integrin activity in CAFs transduce mechanical forces to Fn, leading to its alignment
α5β1 Integrin is the major Fn-binding integrin responsible for Fn matrix assembly in fibroblasts. To test whether changes in Fn organization by CAFs might be mediated by α5β1 activity, we assessed active and total α5 integrin levels in CAFs and NFs. IF analysis of active α5 integrin, using the SNAKA51 antibody, which recognizes the active form of α5 integrin in fibrillar adhesions (Clark et al., 2005), revealed that CAFs display an ∼30% increase in active α5 integrin relative to NFs (Fig. 5, A and B). However, no difference was observed in the mean fluorescent intensities of total α5 integrin staining between CAFs and NFs (Fig. 4, A and C). Western blot (WB) analysis of α5 integrin protein levels also did not show a difference between CAFs and NFs (Fig. S4, A and B). Assessment of active and total β1 integrin levels similarly showed a higher level of active β1 integrin in CAFs compared with NFs without a change in total β1 levels (Fig. S4, C–F).
Figure 5.
α5β1 integrin in CAFs transduce mechanical forces to Fn. (A) IF staining of NFs and CAFs for active α5 integrin (green, left) and total α5 integrin (red, middle). Bar, 20 µm. Zoomed images have been pseudocolored to show differences in fluorescence intensity, with warmer colors indicating higher intensity. Bar, 5 µm. (B and C) Quantification of mean fluorescent intensity of active (B) and total (C) α5 integrin. 80–95 cells per condition from five independent experiments were analyzed. Error bars indicate SEM for five experiments. ***, P < 0.001; n.s., not significant as determined by Student’s t test. (D) Representative traction-force vector maps of CAFs treated with 5 µg/ml control IgG (left) or α5 integrin function blocking antibody JBS5 (right). Warmer colors indicate areas with high traction forces. (E) The dot plot shows mean traction forces in control and JBS5-treated CAFs. Line indicates mean; error bars indicate SEM. 22 control CAFs and 25 JBS5-treated CAFs were analyzed in four independent experiments. ***, P < 0.001 as determined by Mann Whitney U test. (F) Fn staining of CAFs after 48 h treatment with 5 µg/ml control IgG or α5 integrin function-blocking antibody P1D6. Bar, 20 µm. (G) Measurements of angles between Fn fibers in CAFs treated with IgG or JBS5. Greater than160 angles measured per condition from ≥16 images from three independent experiments. ***, P < 0.001 as determined by Mann-Whitney U test. (H) NHS–ester-488 staining of CAF CDMs generated during 10 μM RGE or RGD treatment. Bar, 50 µm. (I) FFT analysis of CAF CDMs shown in Fig. 4 H.
Mechanical force can lead to activation of integrins, and integrins act as transducers of the force generated within the cell to the ECM (Ross et al., 2013). Because we observed that CAFs exert higher traction stresses on Fn, we tested whether that force was transmitted via α5β1 integrin to the Fn. CAFs were plated on Fn-coated polyacrylamide (PAA) gels and treated with 5 µg/ml of either anti–integrin α5 function-blocking antibody (clone JBS5) or control IgG and subjected to traction force microscopy. Treatment with JBS5 led to a significant decrease in the mean traction forces of the CAFs, which was similar to the mean traction forces observed with NFs (Fig. 5, D and E), indicating that α5β1 integrin has a central role in force transmission to Fn in CAFs. We then tested whether blocking α5β1 integrin affects Fn organization by CAFs. A synthetic arginine-glycine-aspartic acid (RGD) peptide was used at 10 µM concentration to block α5β1 integrin in CAFs during matrix formation, and an equal concentration of the arginine-glycine-glutamic acid (RGE) peptide was used as a control because of its decreased affinity for integrins. RGE peptide–treated CAFs displayed aligned Fn fiber organization; however, treatment with the RGD peptide disrupted fiber assembly and alignment by CAFs, resulting in a more NF-like fiber organization, which was quantified by measuring the angles among Fn fibers (Fig. S4, G and H). Notably, the RGD peptide not only blocks α5β1 integrin but also affects other integrins that bind to the RGD sequence. Thus, we also evaluated the Fn matrix organization during treatment of the CAFs with an anti–α5 integrin blocking antibody (clone P1D6) or control IgG. Control IgG-treated CAFs aligned Fn fibers, similar to our previous observations, whereas treatment with 5 µg/ml of P1D6 antibody perturbed the fiber assembly and alignment, resulting in fewer fibers that were randomly organized (Fig. 5, F and G).
Next, we investigated whether changes in Fn organization by RGD treatment of CAFs affected cancer cell migration by treating the CAFs with RGD or RGE peptides during CDM generation. RGD treatment of CAFs resulted in CDMs with a meshwork-like fiber organization in the CDMs, compared with anisotropic fiber orientation in control CDMs generated by RGE-treated CAFs (Fig. 5, H and I). DU145 cells were plated onto those CDMs and subjected to time-lapse microscopy. DU145 cells exhibited enhanced directional migration on control CAF CDMs; however, the directionality ratio was significantly reduced on CAF CDMs that were generated during RGD treatment (Fig. S4 I). Interestingly, we observed a slight, but discernible, increase in the migration speed of DU145 cells on RGD-treated CAF CDMs in comparison to the control (Fig. S4 J).
Aligned matrix organization by CAFs is mediated by PDGFRα
Fn–α5β1 integrin binding has been shown to activate PDGFRα in mesenchymal stem cells (Veevers-Lowe et al., 2011). Furthermore, PDGFRα has been associated with connective tissue remodeling by fibroblasts (Horikawa et al., 2015), and overexpression of PDGFRs in tumor stroma is correlated with poor prognosis in several types of cancer (Heldin, 2013). To dissect the role of PDGFRα in CAF-mediated ECM organization, we studied PDGFRα expression and function in prostate CAFs and NFs. IF staining and WB analysis both showed that CAFs express approximately threefold more PDGFRα compared with NFs (Fig. 6, A–D). In addition, CAFs exhibited a 60% increase in PDGFRα activity, as assessed by tyrosine-762 phosphorylation, upon stimulation with growth medium containing serum (an abundant source of PDGFs; Fig. 6, E and F).
Figure 6.
Aligned matrix organization by CAFs is mediated by PDGFRα. (A) IF staining of PDGFRα in NFs and CAFs. Images are shown pseudocolor, with warmer colors indicating high intensity, and cooler colors indicating low intensity. Bar, 25 µm. (B) Quantification of mean fluorescent intensity of PDGFRα. More than 60 cells per condition from three independent experiments were analyzed. Error bars indicate SEM for three experiments. **, P < 0.01 as determined by Student’s t test. (C and E) WB analysis of PDGFRα (C) and pY762-PDGFRα (E) in NFs and CAFs. β-actin was used as a loading control. (D and F) Quantification of PDGFRα (D) and pY762-PDGFRα (E) mean intensity in NFs and CAFs, normalized to β-actin. Error bars represent the SEM from three independent experiments. *, P < 0.05 as determined by Student’s t test. (G) Collagen gel contraction of CAFs treated with either 10 µg/ml of control (Ctrl) antibody or PDGFRα neutralizing antibody (AF-307). Dashed lines circle the gels after 24 h treatment. (H) Quantification of collagen gel contraction at 24 h. Three gels from three independent experiments were analyzed. Error bars indicate SEM for three experiments. *, P < 0.05 as determined by Student’s t test. (I) Representative traction-force vector maps of CAFs treated with 10 µg/ml control IgG (left) or AF307 (right). Warmer colors indicate areas with high traction forces. (J) Scatter dot plot shows mean traction forces in control and AF307-treated CAFs. Line indicates means, whereas error bars indicate SEM. 17 control CAFs and 21 AF307-treated CAFs were analyzed in three independent experiments. ***, P < 0.001 as determined by Mann Whitney U test. (K) Fn staining of CAFs after 48 h treatment with 10 µg/ml control IgG or AF307. Bar, 25 µm. (L) Measurements of angles between Fn fibers in CAFs treated with control IgG or AF307. Greater than100 angles measured per condition from ≥12 images from three independent experiments. ***, P < 0.001 as analyzed by Mann-Whitney U test. The box plots range from the 25th to the 75th percentiles; the central line indicates the median, and the whiskers range from the 5th to the 95th percentiles. (M) Active α5 integrin staining of CAFs after 48 h treatment with 10 µg/ml control IgG or AF307. Bar, 25 µm. (N) Quantification of mean fluorescent intensity of active α5 integrin in control IgG or AF307-treated CAFs. Greater than 70 cells per condition were analyzed in three independent experiments. *, P < 0.05 as determined by Student’s t test.
To determine whether up-regulated PDGFRα expression and activity in CAFs affected contractility, we performed collagen-gel contraction assays. Blocking PDGFRα activity with a neutralizing antibody, AF307, significantly reduced the contractility of CAFs (Fig. 6, G and H). Furthermore, we found that inhibiting PDGFRα in CAFs decreased the traction forces applied to Fn (Fig. 6, I and J). Likewise, addition of PDGFRα blocking antibody, AF307 significantly changed the matrix organization by CAFs, from aligned fibers to a more-random organization (Fig. 6, K and L). Because previous studies have reported that α5β1 integrin and PDGFRs can crosstalk on the cell membrane and modulate each other’s activity (Eliceiri, 2001; Zemskov et al., 2009), we also tested whether PDGFRα has a similar function in our system. Indeed, we observed a decrease in active α5β1 levels when CAFs were treated with AF307 (Fig. 6, M and N). These data suggest that PDGFRα collaborates with α5β1 integrin to promote cellular contractility and to organize the ECM.
Fn fibers are aligned at sites of invasion in human prostate cancer tissues
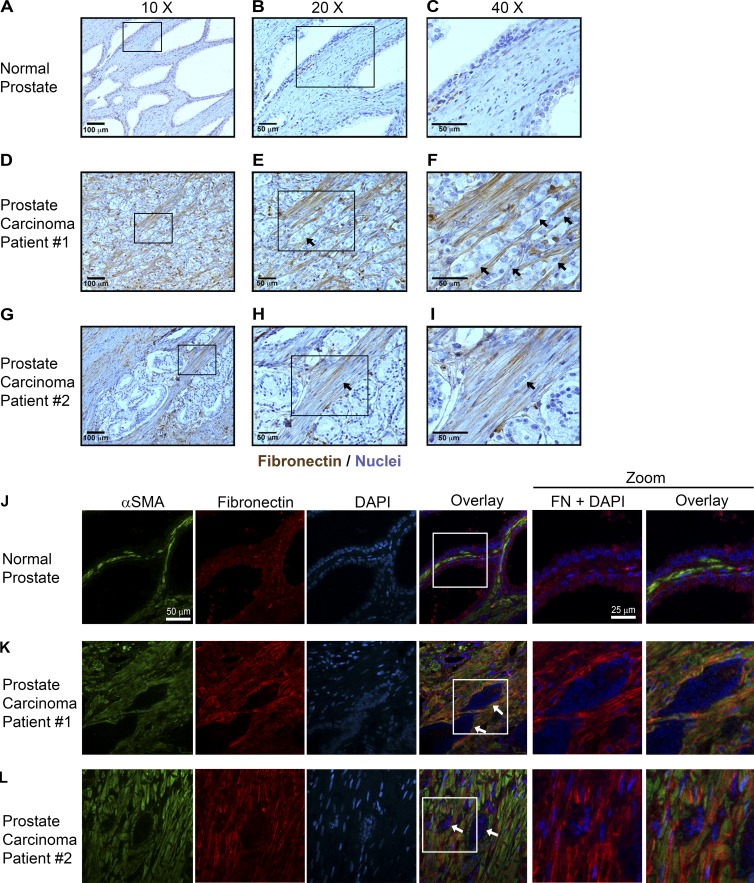
To determine whether Fn alignment in vivo is regulated differently by CAFs and NFs, we examined Fn in four prostate cancer cases (Fig. 7 and Fig. S5, A–I), comparing regions of normal adjacent prostate tissue to regions of invasive carcinoma. Regions of normal adjacent prostate tissue contained low levels of Fn (Fig. 7, A–C). In regions of benign prostatic hyperplasia, Fn was more abundant than in normal prostate but was largely disorganized (Fig. S5, A–C). However, in regions of invasion, Fn was present at high levels and formed well-organized, parallel fibers contacting many invading cancer cells (Fig. 7, D–I; and Fig. S5, D–I). By double IF, α smooth muscle actin–positive (αSMA+) fibroblasts adjacent to normal prostatic epithelium (NFs) were surrounded by disorganized Fn (Fig. 7 J), whereas αSMA+ fibroblasts around cancer cells (CAFs) were surrounded by well-organized, linear Fn fascicles (Fig. 7, K and L). Within the tumor, expansive regions of confluent epithelial growth with minimal stroma exhibited little to no Fn (not depicted), supporting the idea that Fn fibers are produced by surrounding CAFs. Consistent with the prostate cancer results, pancreatic cancer samples showed a similar rearrangement of Fn around benign and malignant lesions. Fn was largely disorganized surrounding acinar-to-ductal metaplasia and pancreatic intraepithelial neoplasm (benign precursor) areas within pancreatic ductal adenocarcinoma (PDAC) samples (Fig. S5, J and K). However, invading clusters of cancer cells were arranged in parallel with numerous, well-organized Fn fibers in PDAC (Fig. S5, L and M).
Figure 7.
Fn structure differs around normal versus malignant prostate epithelium. (A–C) A minimal staining for Fn (brown) is observed around healthy (normal) prostate epithelium adjacent to cancer. Cells were counterstained with hematoxylin (blue). (D–I) Fn forms long parallel fascicles in invasive regions of two different cancer cases (D–F and G–I). E is an enlargement of D, and F is an enlargement of E. Similarly, H is an enlargement of G, and I is and enlargement of H. (D–I) Arrows indicate invading tumor cells. (J–L) Immunofluorescent labeling of αSMA (green; left), Fn (red; middle), and nuclear counterstain with DAPI (blue; right) indicate that myofibroblasts surround both benign (J) and malignant prostate (K and L) epithelium but only form long, parallel Fn fascicles around invading cells. Bars: (J–L, αSMA, Fibronectin, DAPI, and Overlay) 50 µm; (J–L, Zoom) 25 µm.
Prostate cancer cells use αv integrin to migrate on CAF CDMs
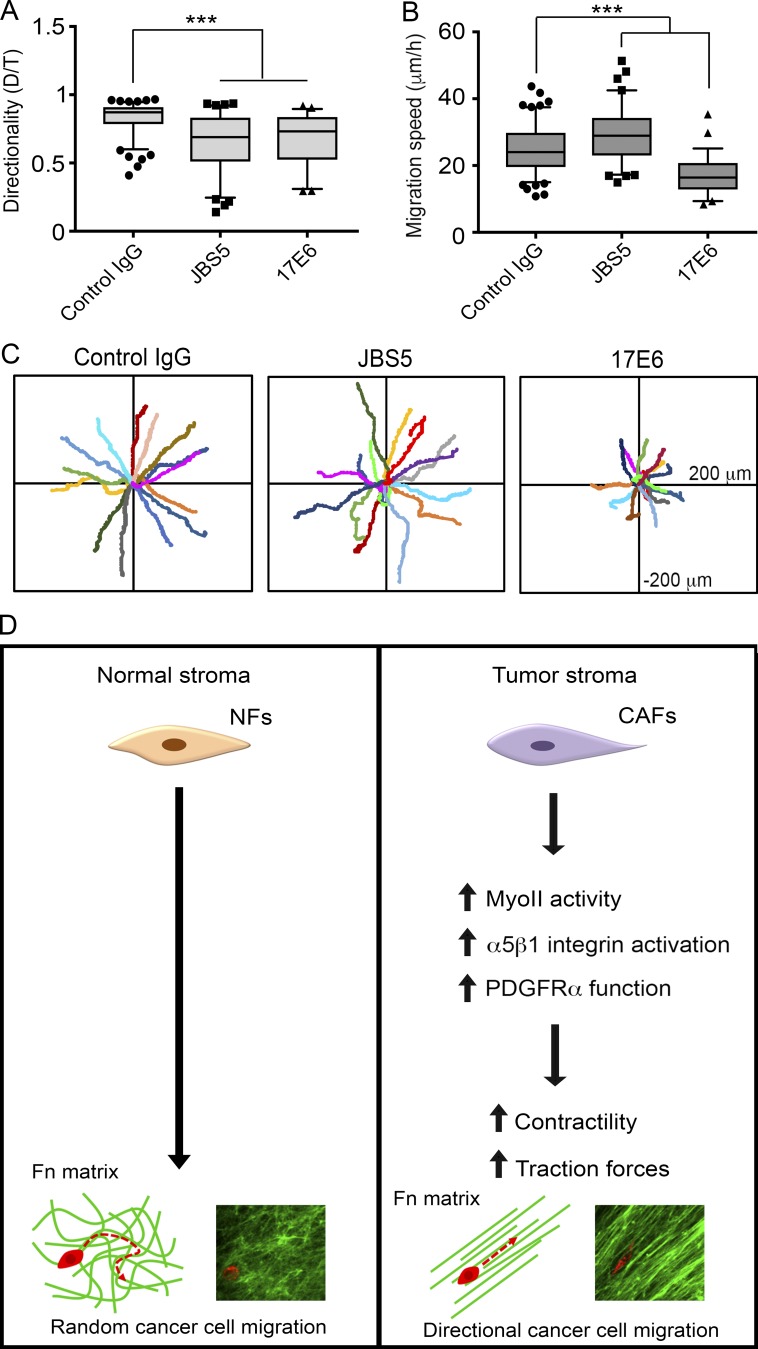
Our data show that the alignment of matrix fibers by CAFs promotes directional cancer cell migration. Interestingly, a recent study identified integrins αvβ6 and α9β1 as responsible for efficient and directional cell migration on HNSCC CAF CDMs (Gopal et al., 2017). Therefore, we tested integrins known to be expressed by prostate cancer cells to determine which class of integrins is responsible for directional cancer cell migration on prostatic CAF CDMs. The expression of α5 and αv integrins are deregulated in prostate cancers and changes in their expression and activity have been linked to cancer cell migration and invasion (Sutherland et al., 2012). Therefore, we assessed DU145 cell migration in 2D cell migration assays on CAF CDMs using respective α5 and αv function blocking antibodies JBS5 and 17E6. When α5 integrin was blocked, DU145 cells migrated with decreased directionality but increased migration speed compared with control IgG–treated cells (Fig. 8, A–C). In contrast, when αv integrins were inhibited, both migration directionality and speed was significantly impaired compared with control cells (Fig. 8, A–C). These results suggest that αv integrin mediates both directional and efficient cell migration on CAF CDMs.
Figure 8.
αv integrins are critical for cancer cell migration on CAF CDMs. (A and B) Quantifications of migration directionality (A) and speed (B) of DU145 cells on CAF CDMs when treated with control IgG, α5 integrin function-blocking antibody JBS5, or αv function-blocking antibody 17E6. Greater than 80 cells per condition were analyzed in four independent experiments. ***, P < 0.001 as determined by one-way ANOVA and Tukey’s post hoc test. (A and B) The box plots range from the 25th to the 75th percentile, the middle line indicates the median, and the whiskers range from the 5th to the 95th percentile. (C) Rose plots showing migration trajectories of 14 representative cells treated with control IgG, JBS5, or 17E6 antibodies. (D) Proposed model of CAF-mediated, directional cancer cell migration. The healthy (normal) fibroblastic matrix resembles a random meshwork of fibers that do not induce migration directionality. However, CAFs organize the Fn matrix into aligned fibers through increased MyoII, α5β1 integrin, and PDGFRα-mediated contractility and traction forces. Anisotropic organization of matrix fibers by CAFs promotes directional migration of cancer cells.
Discussion
Evidence has accumulated to show that changes in the tumor microenvironment support cancer progression (Miles and Sikes, 2014). CAFs are a key component of the tumor microenvironment with tumor-supportive roles (Mezawa and Orimo, 2016). Cancer cell migration and invasion are critical initial steps in metastasis; however, the mechanisms by which tumor–stroma interactions regulate those processes are not well understood. In this study, we identified a new mechanism by which CAFs promote cancer cell migration (Fig. 8 D). Using a co-culture system, we demonstrated that cancer cells associate with primary human prostate CAFs and migrate directionally along them. We provided evidence that CAF–cancer cell association is promoted by the Fn fibrils assembled by CAFs, and cancer cells pull on the CAF-assembled Fn to migrate along CAFs. Furthermore, we show that the CAF–cancer cell association is blocked when Fn is knocked down in CAFs. Interestingly, prostate CAFs were also able to promote an increased association with HNSCC cells and induce their directional migration. This finding suggests a ubiquitous mechanism by which CAFs from different tumor microenvironments can modulate cancer cell migration, which was not, to our knowledge, previously known.
In addition to its influence on CAF–cancer cell association, we show that Fn is critical for ECM synthesis and organization by CAFs. Fn is a major component of the CDMs generated by CAFs and NFs and knocking down Fn expression in CAFs completely disrupts ECM synthesis and organization. Moreover, CAFs organize Fn into parallel fibers, whereas the Fn matrix assembled by NFs resembles a mesh. ECM architecture can guide directional migration of cells through physical cues because migrating cells use the ECM as attachment points during migration (Petrie et al., 2009). CAF-mediated parallel organization of CAF CDMs promotes directional migration of both prostate cancer and HNSCC cells, where cell migration is in the same direction as the orientation of the fibers. In clinical prostate carcinoma samples, we observed aligned Fn fibers at the sites of invasion, which were adjacent to invading cancer cells. Interestingly, parallel-organized Fn was also present in PDACs, indicating that alignment of Fn fibers is a clinical feature of both carcinomas and may contribute to cancer cell dissemination.
Although overexpression of Fn and its EDA isoform was reported to be a feature of CAFs more than a decade ago (Kalluri and Zeisberg, 2006), the effect of changes in the Fn matrix on cancer cell migration are just now emerging. A recent article by Gopal et al. (2017) studied the CAF matrisome and identified Fn–EDA as a marker of poor survival in patients with HNSCC. Interestingly, this study reports directional migration of HNSCC cells on CAF CDMs, in a collective manner. We did not observe collective cell migration of either prostate cancer or HNSCC cells on CAF CDMs; however, this could be a result of differences in epithelial properties of the cancer cell lines that were selected or differences in cell density between the two studies. Nevertheless, both studies identify Fn as a critical component of CAF CDMs that regulates directional cell migration.
A few studies have identified factors that can lead to ECM alignment, including the serine proteinase fibroblast activation protein (Lee et al., 2011), and the transcription factors Snail1 and Twist1, which may act downstream of TGF-β to induce the CAF phenotype (Stanisavljevic et al., 2015; García-Palmero et al., 2016). However, the mechanism by which CAFs organize the ECM remains largely unclear. Here, we demonstrated that mechanical force is an important factor that enables CAFs to generate an aligned ECM. MyoII-mediated contractility is a prominent feature of CAFs (Calvo et al., 2013). We found that prostate CAFs have elevated MyoII activity, are highly contractile, and apply high-traction stresses on Fn. Remarkably, treatment with low doses of MyoII inhibitor perturbed the aligned Fn organization by CAFs, giving rise to a more-random network of fibers similar to that assembled by NFs. Because DU145 cells did not migrate directionally when plated onto CAF CDMs generated during blebbistatin treatment, these results suggest that matrix organization is a major driver of cancer cell migration directionality and is mediated by MyoII-driven contractility and high traction force generated by CAFs.
α5β1 integrin has a major role in Fn fibrillogenesis and can be activated by mechanical force from within the cell, through actomyosin contractility (Friedland et al., 2009). Consistent with previous studies identifying α5β1 integrin as a mechanotransducer (Schwartz and DeSimone, 2008; Roca-Cusachs et al., 2012), we found that high traction forces produced by CAFs were transduced by α5β1 integrin to Fn. Although there were no differences in the expression of α5 and β1 integrin subunits between CAFs and NFs, we found increased activation of α5 and β1 integrins in CAFs in comparison to NFs. The increased Fn expression and contractility of CAFs explain the enhancement observed in α5β1 integrin activation (Lin et al., 2013). The overexpression and activation of PDGFRα may also enhance α5β1 integrin activity, as indicated by our blocking-antibody results. Because many signals converge on integrins to induce inside–out signaling, it seems likely that there are additional mechanisms that could lead to enhanced activation of α5β1 integrin in CAFs, such as any deregulations in the cell metabolism sensor AMP-activated protein kinase, which was recently reported to be a negative regulator of β1 integrin activity and Fn fibrillogenesis in fibroblasts (Georgiadou et al., 2017) or deregulation of the integrin inhibitor Sharpin protein, which was shown to control collagen remodeling and traction forces (Peuhu et al., 2017). Other integrins can also contribute to changes in CDM organization, including αvβ5, as recently reported by Franco-Barraza et al. (2017).
As part of the mechanism by which CAFs promote ECM organization, we found that prostate CAFs overexpress PDGFRα and exhibit increased Y762 phosphorylation. Inhibition of PDGFRα significantly abrogated collagen gel contraction and traction stresses generated by CAFs, as well as α5β1 integrin activity and Fn organization. These data are consistent with previous demonstrations of cross talk between α5β1 integrin and PDGFRα in mesenchymal stem cells (Veevers-Lowe et al., 2011). Interestingly, α5β1 integrin and PDGFRs have been shown to be in complex with tissue transglutaminase, which modulates the activity of both receptors, and potentially converges and amplifies their downstream signaling (Akimov and Belkin, 2001; Zemskov et al., 2009). However, it is also possible that PDGFRα signaling can activate contractility directly, which would lead to indirect activation of α5β1 integrin. For example, PDGFRα signaling can activate RhoA–ROCK pathway in mesenchymal stem cells, leading to increased polymerization of αSMA in actin filaments (Ball et al., 2007), which is also a characteristic feature of CAFs. Therefore, integrin α5β1 and PDGFRα signaling may converge on activation of RhoA-mediated contractility (Danen et al., 2002). PDGFR signaling is a promising target in many cancers (Heldin, 2013); thus, understanding the regulation of CAFs and stromal ECM by PDGFRs may provide valuable information for targeting the tumor stroma in carcinomas.
In response to the profound changes in the tumor microenvironment, cancer cells express and activate different integrins to regulate processes, such as cell attachment and migration. Gopal et al. (2017) reported that HNSCC cells up-regulate α5β1, αvβ5, and αvβ6 integrins when cultured on Fn-EDA–rich CAF CDMs. They further identified that the collective migration of HNSCC cells on CAF CDMs was mediated by αvβ6 and Fn-EDA–binding α9β1 integrins (Gopal et al., 2017). These findings prompted us to investigate the integrins involved in directional migration of prostate cancer cells on CAF CDMs. Similar to Gopal et al. (2017), we found that blocking α5β1 integrin in prostate cancer cells induced faster migration with decreased directionality, and blocking αv integrin activity decreased both directionality and speed. These results suggest that α5β1 integrins may be responsible for forming stronger attachments to matrix, as previously reported (Roca-Cusachs et al., 2009), whereas αv integrins are critical for cell migration. Aberrant expression of RGD-binding integrins α5 and αv have been reported in prostate cancers and are being explored as potential targets for therapy (Goel et al., 2008; Sutherland et al., 2012). Gopal et al. (2017) also showed that EDA-binding integrin α9β1 regulates cell migration on CAF CDMs. Although, CAFs in our study also express high levels of EDA-Fn (Fig. S1, J and K), we did not find any studies that report α9 integrin expression in prostate cancers. ITGA9 expression was not detected in 11 prostate cancer tissues examined in the Human Protein Atlas; therefore, we did not study that integrin. Other EDA-binding integrins, α4β1 and α4β7, were also not investigated because there are multiple studies showing that α4 integrin is not expressed in prostate cancer and DU145 cells (Rokhlin and Cohen, 1995; Barthel et al., 2013; Chen et al., 2015). Collectively, our results indicate differing roles for integrins in CAFs and cancer cells in mediating matrix assembly and cell migration, thus, highlighting the complexity of integrin signaling in tumors.
In summary, our study shows that CAFs organize the Fn matrix through increased contractility and traction forces, which are mediated by MyoII, α5β1 integrin, and PDGFRα. This matrix organization leads cancer cells to migrate directionally using αv integrins (Fig. 8 D). Alignment of the Fn fibers is a prominent feature of both prostatic and pancreatic cancer stromata both in vivo and in vitro and is likely to guide the invasion of cancer cells. Surprisingly, our data suggest that CAFs in the tumor microenvironment are not tissue type specific in their ability to regulate cancer cell migration. For example, CAFs from prostate can regulate the migration of HNSCC cells. This indicates a commonly used mechanism for modulating the migration of cancer cells, which has far reaching implications in the development of tumor metastases. Furthermore, biochemical targeting of this pathway could prove beneficial in limiting stromal support during the metastasis of cancer cells.
Materials and methods
Antibodies and reagents
Primary antibodies were diluted 1:300 for IF, unless otherwise noted. The primary antibodies used were as follows: mouse anti–Fn (610077, diluted 1:300 for IF, 1:1,000 for WB; BD), rabbit anti-Fn (clone F14, 1:1,000 for immunohistochemistry and 1:100 for IF; BioGenex), mouse anti–integrin α5 (clone SNAKA51; MilliporeSigma), rabbit anti–integrin α5 (1:750 for IF; MilliporeSigma; AB1949), mouse anti–integrin α5 (clone 6F4, 1:1,000 for WB; was a gift from R. Horwitz, Allen Institute for Cell Science, Seattle, WA), mouse anti–integrin β1 (clone 12G10; Abcam), rabbit anti–integrin β1 (1:1,000 for WB; AB1952; MilliporeSigma), rabbit anti–MLC2 (3672S; Cell Signaling Technology), rabbit anti–phospho-S19-MLC2 (3671S; Cell Signaling Technology), rabbit anti–PDGFRα (clone D1E1E; Cell Signaling Technology), rabbit anti–p-PDGFRα-Y762 (clone D9B1N; 1:300 for WB; Cell Signaling Technology), mouse anti-vinculin (clone hVIN-1; 1:1,000 for IF; Sigma-Aldrich), mouse anti–E-cadherin (clone 36; BD), mouse anti–N-cadherin (clone 8C11; Santa Cruz Biotechnology, Inc.), mouse anti–α tubulin (clone DM1A, 1:5,000 for WB; Sigma-Aldrich), and mouse anti–β-actin (clone AC15; 1:5,000 for WB; Sigma-Aldrich; or clone 1A4, 1:1,000 for tissue IF; BioGenex). For function-blocking experiments, mouse anti–integrin α5 (clone JBS5; MilliporeSigma), mouse anti–integrin α5 (clone P1D6; MilliporeSigma), mouse anti–integrin αv (clone 272-17E6; Abcam), and goat anti–PDGFRα (clone AF307; R&D Systems) were used. Mouse IgG (0107-01) and rabbit IgG (0111-01) were purchased from SouthernBiotech and were used as controls in function-blocking experiments. For IF staining, Alexa Fluor 488 goat anti–mouse, Alexa Fluor 488 donkey anti–rabbit, Alexa Fluor 488 goat anti–mouse IgG1, Alexa Fluor 555 goat anti–rabbit, and Alexa Fluor 555 goat anti–mouse IgG2a secondary antibodies were used at 1:600 dilution (Thermo Fisher Scientific). For WB, Alexa Fluor 680 donkey anti–mouse, Alexa Fluor 680 goat anti–rabbit (Thermo Fisher Scientific), IRDye800 donkey anti–mouse (Rockland Immunochemicals) and anti–rabbit HRP-conjugated IgG (Promega) secondary antibodies were used. siGENOME siRNA SMARTpool for FN1 (M-009853-01-005) and nontargeting control (D-001206-14-05) were ordered from Dharmacon (Horizon Discovery) and 50 nM of either siRNA pool was transfected into CAFs using DharmaFECT 1 transfection reagent (Horizon Discovery), following the manufacturers’ protocols. Cells were kept in Fn-depleted medium, which was prepared using gelatin–agarose beads (G5384; MilliporeSigma). Fn (F2006) and fluorescein isothiocyanate isomer (F7250) were purchased from MilliporeSigma. RGD and RGE peptides were purchased from Bachem. Rat tail type I collagen was purchased from BD. Blebbistatin was purchased from MilliporeSigma. GFP–vinculin plasmid was a gift from S. Craig (Johns Hopkins University School of Medicine, Baltimore, MD).
Fibroblast isolation and cell maintenance
Human prostatic CAFs were isolated from prostate cancers and NFs were from benign prostate hyperplasia tissues. Fibroblasts were isolated from six separate patients’ tissue samples (three patients with prostate cancer and three patients with benign prostate hyperplasia). CAFs and NFs were prepared as previously described (Olumi et al., 1999). Cells were verified using a tissue-recombination bioassay to confirm that CAFs induced tumor formation from BPH1 cells and that NFs did not elicit tumorigenesis. CAFs and NFs were used between passages four and eight to ensure proper function in ECM production and communication with cancer cells.
Prostatic cancer DU145 cells and fibroblasts were maintained in RPMI 1640 with 10% FBS and penicillin–streptomycin, as described previously (Ao et al., 2007). Human head and neck cancer cells SCC61 and JHU012 were maintained in DMEM/F12 (Thermo Fisher Scientific), supplemented with 10% FBS and penicillin–streptomycin (Ao et al., 2015).
Fabrication of two-channel co-culture devices
The co-culture microfluidic device was prepared using standard soft-lithography techniques, as previously described (Xia and Whitesides, 1998; Jean et al., 2014). First, a master mold was fabricated using photolithography, patterning a layer of photoresist SU8 by UV exposure through a 20,000-dots per inch photomask. Second, polydimethylsiloxane (PDMS; Ellsworth Adhesives) prepolymer was mixed with a curing agent at a mass ratio of 10:1 and then poured over the mold. After degassing for 30 min and curing in a 70°C oven for 2 h, the PDMS was fully polymerized. The resulting PDMS component was then cut and removed from the mold. Inlet and outlet holes were punched through the PDMS layer using a 3.5-mm-diam punch. Third, the PDMS layer was bonded to a 100-µm-thick glass coverslip (No. 1; VWR VistaVision, VWR International) after both components were exposed to oxygen plasma. Fourth, Pyrex cloning cylinders of 8 mm diam and 8 mm tall (Thermo Fisher Scientific) were attached to the inlet and outlet holes using uncured, liquid PDMS for loading and removing cells and medium. Fifth, the liquid PDMS “glues” were subsequently allowed to cure at 70°C for 2 h. Sterilized water was loaded into the device to keep the walls of the microfluidic channels hydrophilic. Sixth, the assembled device was sterilized under UV light for 1 h.
Microscopy
A WaveFX spinning disk confocal system (Quorum Technologies) equipped with an Eclipse Ti microscope (Nikon) and an ImagEM-CCD camera (Hamamatsu Photonics) was used for imaging the IF-stained coverslips and for recording cell migration when multiple channel acquisition was required. A Plan Fluor 40× objective (NA 1.3) was used for imaging the IF-stained coverslips. A Nikon 10× Ph1 ADL objective (NA 0.25) and a Plan Fluor 20× objective (NA 0.75) were used to image cell migration. DAPI, Alexa Fluor 488, Alexa Fluor 555, and acti-stain 670 were excited by laser at 405, 491, 561, and 642 nm, respectively (Semrock). Emission filters for those fluorophores were 460/50, 525/50, 593/40 or 620/60, and 700/75, respectively (Semrock). Images were acquired and analyzed with MetaMorph software (Molecular Devices).
TIRF microscopy and cell migration movies in phase contrast were performed using an inverted IX71 microscope (Olympus) with a Retiga EXi CCD camera (QImaging). An Olympus UPlanFl N 10× objective (NA 0.30) was used for cell-migration assays. TIRF images were taken with an Olympus PlanApo 60× oTIRFM objective (NA 1.45) and a 488-nm laser line from a HeNe laser (Prairie Technologies). MetaMorph software was used for image acquisition and analysis.
Co-culture in microfluidic devices
Microfluidic devices were incubated with culture medium at 37°C and subconfluent fibroblasts and cancer cells were labeled with CellTracker Green or CellTracker Red (C2925 and C34552; Thermo Fisher Scientific), respectively. After a gentle, but thorough, wash, cells were detached and mixed at 1:1 ratio in RPMI-1640 complete growth medium. A total of 104 cells, suspended in 20 µl medium, were loaded into each inlet reservoir of a microfluidic cell-culture chamber. The cell density was kept low to be able to observe interactions among individual cells. After incubating for ∼2 h at 37°C to allow cells to attach, 200–300 µl of RPMI-1640 full-growth medium was added to the inlet reservoir, and cells were incubated overnight. The next day, the cell culture medium was replaced with phenol-red–free RPMI 1640, supplemented with 5% FBS and 50 mM Hepes, and time-lapse imaging was performed at 37°C in a temperature-controlled chamber (Live Cell Instrument). Areas of interest were chosen where a fibroblast–cancer cell pair could be identified, and their movement with regard to each other was analyzed. Images were taken every 10 min for 12 h using the spinning disk confocal system described in the Microscopy section. To visualize Fn in co-culture experiments, 5 µg/ml FITC–Fn was added to the culture medium. For those experiments, CAFs and DU145 cells were mixed in 1:1 ratio to yield a total of 4 × 104 cells, which were seeded to a 35-mm, glass-bottomed dish and incubated overnight. During that incubation, CAFs incorporated FITC–Fn as they assembled Fn fibers, which enabled us to visualize the Fn matrix. The next day, the cells were imaged using the 40× objective in the spinning disk confocal system. Images were taken every 30 s for 1 h. Cell migration association index was determined by calculating the angle (σ) between the axis of migrating cancer cells and fibroblasts. The association index was defined as the cosine of the angle σ; an index of 0 indicated perpendicular migration of cells in relation to each other, whereas an index of 1 signifies cells migrating parallel to each other. The migration directionality ratio was calculated by dividing the net distance (D) by the actual path length traveled by the cell (T).
Preparation of an FITC-labeled Fn
Fn was diluted to 0.5 mg/ml in borate buffer (170 mM Na2B4O7, pH 9.3, and 40 mM NaCl) and 6 mg FITC was dissolved in 200 ml borate buffer. 1 ml of 0.5 mg/ml Fn solution was loaded to a dialysis cassette and placed in the FITC-containing borate buffer and dialyzed at RT for 1.5 h in the dark. Next, the Fn cassette was dialyzed extensively against 1× PBS, pH 7.4, for 2 d, changing the PBS buffer four to five times. Protein concentration was determined using the following formula: FITC-protein (mg/ml) = [OD280 − (0.36 × OD493)]/1.4. Fn was then dialyzed against 50% glycerol and stored at −20°C.
Generation of 3D CDMs and migration assays
3D CDMs were generated as previously described (Beacham et al., 2006). In brief, 35-mm-diam, glass-bottom dishes or 6-well plates were coated with a 0.2% (wt/vol) gelatin solution for 1 h at 37°C. The gelatin-coated dishes were treated with 1% (vol/vol) glutaraldehyde in PBS for 30 min, followed by a 30-min treatment of 1M ethanolamine at RT. The dishes were then washed extensively using PBS. NFs or CAFs were plated on dishes as a confluent layer, and they were cultured for 8 d in complete growth medium (RPMI-1640 with 10% FBS and penicillin–streptomycin). For CDM generation with CAFs transfected with Fn siRNA, an RPMI-1640 medium with 10% Fn-depleted FBS and antibiotics was used. In all experiments, the cell culture medium was replenished and supplemented with freshly prepared 50 µg/ml of ascorbic acid on alternate days. On d 8, cells were rinsed with PBS, then extracted from the matrix using an alkaline detergent (0.5% [vol/vol] Triton X-100 and 20 mM NH4OH in PBS), leaving the 3D matrix intact and attached to the culture dish.
To visualize the matrix, the CDMs were labeled using 2 µg/ml NHS-ester Alexa Fluor 488 dye (A20000; Thermo Fisher Scientific), which was dissolved in 50 mM sodium bicarbonate buffer, pH 9, by incubation for 15 min in the dark. The matrices were washed with PBS and treated with 200 mM Tris buffer ,pH 7.5, for 10 min to deactivate the NHS esters. The labeled 3D CDMs were blocked with 1% BSA solution and stored at 4°C until ready to use. This protocol was provided by A. Doyle (National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD).
For the cell-migration assays with the CDMs, cancer cells were labeled with CellTracker Red CMTPX dye (C34552; Thermo Fisher Scientific), following the manufacturer’s protocol. For 2D migration assays, cells were plated 2 h before imaging. For 3D migration assays, cells were plated 24 h before imaging to allow for invasion of the cells into the 3D matrix. The cells were maintained in phenol red-free RPMI-1640 medium supplemented with 5% FBS and 50 mm Hepes in a 37°C temperature-controlled chamber (Live Cell Instrument) during acquisition. Images were acquired every 5–10 min for 6 h using the spinning disk confocal microscope. MetaMorph software was used to track migrating cells and to measure the net distance from the first time point to the last time point. The migration directionality ratio was calculated as described in Co-culture in microfluidic devices. Migration speed was calculated by dividing the total distance traveled (in micrometers) by total time (in hours). In experiments testing integrin function in DU145 cell migration on CAF-CDMs, 10 µg/ml of either the function-blocking antibody (JBS5 or 17E6) or the control IgG was added to culture medium 30 min before imaging. Function-blocking properties of each antibody were confirmed by cell-attachment assays using crystal violet staining.
IF
For most experiments, cells were plated onto glass coverslips coated with 5 µg/ml Fn (F0895; Sigma-Aldrich) and allowed to attach for 3 h. However, to allow for Fn matrix deposition and formation of fibrillar adhesions, Fn and integrin staining was performed 48 h after the cells were plated onto uncoated glass coverslips. Cells were fixed using 4% PFA supplemented with 0.12 M sucrose in PBS for 15 min at RT. After fixation, cells were permeabilized with 0.2% (vol/vol) Triton X-100 for 3 min in most experiments. Cells were not permeabilized for Fn staining to observe the extracellular Fn matrix organization. Blocking was performed for 1 h with 20% goat serum in PBS. Primary and secondary antibodies were diluted in 5% goat serum and were incubated with the cells at 4°C overnight and 45 min at RT, respectively. After each antibody step, coverslips were washed with PBS extensively. DAPI (AS-83210; AnaSpec) and phalloidin (acti-stain 670, PHDN1; Cytoskeleton, Inc.) costains were performed at the same time as the secondary antibodies. Coverslips were mounted on the glass slides using Aqua Poly/Mount (Polysciences, Inc.).
The mean fluorescence intensity was quantified by dividing the background-corrected, integrated fluorescence intensity in individual cells by the cell unit area with MetaMorph software. The size and number of vinculin adhesions were calculated by tracing individual adhesions and using the measure tool within MetaMorph.
Calculation of angles between the fibers and FFT analysis
Fn images were opened in MetaMorph software, and a template of nine dots, which was formed by selecting three dots 120° apart on three concentric circles, was placed on the image. The angle between a fiber that intersected with one of the reference dots and its closest intersecting fiber was quantified using ImageJ (http://rsb.info.nih.gov/ij). A minimum of 50 angles were measured for each experimental condition in each experiment. This procedure was adapted from a previous protocol for quantifying ECM alignment (Yang et al., 2011).
To characterize fiber orientation, FFT was also used as previously reported (Ayres et al., 2006). In brief, the FFT function was performed on fluorescence images of Fn and CDMs using ImageJ. Then, a 512-pixel-diam circle was overlaid on the FFT output image (2048 × 2048 pixels) in the center using the Oval Profile plug-in (W. O’Connell, University of California, San Diego, San Diego, CA). A radial summation of gray value intensities over the circle was conducted and normalized by dividing it by the total intensity. Peak intensities observed 180° apart from each other indicated an aligned fiber orientation, whereas no noticeable peaks were observed when fibers were unorganized.
Traction-force microscopy
Traction forces of fibroblasts were measured as described previously (Sabass et al., 2008; Jean et al., 2013). In brief, rectangular glass coverslips were mounted with PAA gels embedded with 0.2-µm FluoSpheres crimson (625:645) fluorescent beads (Thermo Fisher Scientific). Immediately after gels solidified, the PAA gel surface was activated using 1 mg/ml Sulfo-SANPAH (ProteoChem) solution in double-distilled H2O under UV light for 5 min on ice. The PAA gels were then washed with double-distilled H2O and incubated with 25 µg/ml Fn overnight at 4°C. The Young’s modulus of the PAA gels was 15.6 kPa as previously calculated (Yeung et al., 2005; Sabass et al., 2008). This PAA gel compliancy was chosen based on previous studies of prostate cancer tissue stiffness (Hoyt et al., 2008; Zhai et al., 2010). A total of 2 × 103 CellTracker green-labeled fibroblasts were incubated on top of the coated coverslip for 3 h at 37°C to allow cells to adhere before being subjected to imaging. For each cell of interest, a differential interference contrast and a fluorescent image of the cell and a fluorescence image of the FluoSpheres beads beneath the attached cell were taken. Then, trypsin was added to dissociate the cells from the PAA gel, and another fluorescence image of the FluoSpheres beads was acquired from the same field. The spinning-disk confocal microscope was used to acquire all images. The images were analyzed using the LIBTRC software developed by M. Dembo (Boston University, Boston, MA) to determine the mean traction forces by cells, normalized to the cell area. Traction-force maps were then generated using the software (Dembo and Wang, 1999).
Cell-contraction assay
CAFs and NFs were suspended in full-growth medium at a density of 6 × 105 cells/ml. Then, rat tail type I collagen was neutralized and diluted to a concentration of 3 mg/ml. The cell suspension and prepared collagen were mixed on ice at 1:2 ratio, to get a final mixture with 2 × 105 cells/ml and 2 mg/ml of collagen. 600 µl of that mixture was loaded into each well of a 12-well plate and incubated at 37°C until polymerization was complete. Gels were then covered with 1 ml of medium and detached from plates using a pipette tip to circle around the inside wall of each well. The plates were scanned at the beginning of the assay and after 24 h incubation (end of assay) at 37°C. The gel area was measured at those times using MetaMorph software. The percentage of contraction by the gels was calculated by dividing the gel area at 24 h by the gel area at 0 h, and multiplying by 100%.
WB
Cells were lysed using radioimmunoprecipitation assay buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% SDS) with protease inhibitor cocktail (Sigma-Aldrich). The protein concentration in the cell lysates was measured with a bicinchoninic acid assay (Bio-Rad Laboratories). 30 µg of each cell lysate was run in an SDS-PAGE gel and transferred to a nitrocellulose membrane. After blocking in 4% nonfat dry milk in TBS-T solution for 1 h, the membrane was first incubated with the primary antibody at 4°C overnight, then incubated with the secondary antibody for 1 h at RT. The membranes were imaged using an Odyssey CLx imaging system (LI-COR Biosciences). For phosphorylated PDGFRα WB, CAFs and NFs were starved overnight and, the next morning, were stimulated with complete culture medium for 2 h. Cell lysates were then prepared with addition of PhosSTOP phosphatase inhibitor cocktail (MilliporeSigma) to the lysis buffer. HRP-conjugated secondary antibodies were used and detected with SuperSignal West Femto maximum sensitivity substrate kit (Thermo Fisher Scientific) via an Amersham Imager 600 (GE Healthcare). The normalized band intensities were measured with Image Studio Lite Software version 4 (LI-COR Biosciences), which were further normalized to loading controls, either β-actin or α-tubulin.
Adhesion turnover assay
24 h before the assay, fibroblasts were transfected with 1.5 µg of GFP–vinculin with the TransIT-X2 dynamic delivery system according to the manufacturer’s instructions (Mirus Bio LLC). Before the assay, 35-mm-diam, glass-bottom dishes were coated with 5 µg/ml Fn overnight at 4°C. The next day, cells were plated on Fn-coated dishes and allowed to adhere for 1 h at 37°C. Time-lapse images of GFP–vinculin were acquired at 15-s intervals for 20 min with an IX71 microscope and PlanApo 60× OTIRFM objective (NA 1.45) as described in the Microscopy section. Rate constants for adhesion assembly and disassembly were determined as described previously (Webb et al., 2004) with MetaMorph software.
Histology
Deidentified, formalin-fixed, paraffin-embedded tissue was procured from four cases of prostate cancer and 10 cases of PDAC through the Cooperative Human Tissue Network with approval from the Vanderbilt Institutional Review Board. 5-μm sections were processed as described (Shi et al., 2014) and labeled with antibodies to Fn for colorimetric analysis. Detection was performed using Vectastain Elite ABC kit (Vector Laboratories) after a reaction with 3,3′-diaminobenzidine (Vector Laboratories). For fluorescent analysis, antibodies to smooth muscle α-actin (αSMA) were detected using the Vectastain Elite ABC kit, followed by tyramide signal amplification with the TSA Plus Cyanine 3 kit (PerkinElmer), followed by heat inactivation. Binding of anti–Fn antibodies was detected with Cy2–anti-rabbit antibodies (Jackson ImmunoResearch Laboratories, Inc.). Slides were counterstained with hematoxylin (Sigma Aldrich) or Toto3 (Thermo Fisher Scientific) and DAPI (Sigma-Aldrich). Colorimetric images were obtained on an Axioskop 40 microscope (ZEISS), and fluorescent images were captured on the Quorum WaveFX spinning disk confocal system with Nikon Eclipse Ti microscope as described in Microscopy.
Data analysis and statistics
Statistical analyses were performed with SPSS software (version 24; IBM Analytics), and the Shapiro-Wilk test was performed to assess data normality. p-Values were determined using either a Student’s t test (if data were normally distributed) or Mann-Whitney U test (if data were not normally distributed). In figures, *, P < 0.05; **, P < 0.01; and ***, P < 0.001, as indicated, and these were designated as statistically significant. The bar graphs were generated using Microsoft Excel and presented as mean ± SEM from at least three independent experiments. The box and whisker plots were generated using GraphPad Prism version 7.02 (GraphPad Software), in which the box ranges from 25–75th percentile, with the middle line indicating the median, and the whiskers indicating 5–95th percentile.
Online supplemental material
Fig. S1 shows the schematic of microfluidic co-culture device, DU145 migration speed, and co-culture results from JHU012 and SCC61 HNSCC cells with fibroblasts and Fn WB. Fig. S2 shows images of N-cadherin and E-cadherin staining in co-cultures, Fn staining in NFs and CAFs, and JHU012 and SCC61 cell migration data on CDMs generated by NFs and CAFs. Fig. S3 shows collagen gel contraction assays and Fn images after blebbistatin treatments of NFs and CAFs. Fig. S4 shows WB for α5 and β1 integrin subunits, images of Fn organization after RGD treatment of CAFs, and DU145 cell migration on CDMs generated during treatment with RGD peptide. Fig. S5 shows immunohistochemistry images of Fn organization in tissue from patients with prostate and pancreatic cancer. Video 1 shows three fields of DU145–NF co-culture migration. Video 2 shows three fields of DU145–CAF co-culture migration. Video 3 shows JHU012-NF (left) and JHU012-CAF (right) co-culture migration. Video 4 shows a DU145 cell migrating along a CAF and interacting with FITC–Fn fiber (green) assembled by the CAF. Video 5 shows co-culture migration of DU145 cells with control (left) or Fn–KD (right) CAFs. Video 6 shows a DU145 cell migrating on NF–CDM. Video 7 shows a DU145 cell migrating on CAF–CDM. Video 8 shows JHU012 cell migrating on NF (left) and CAF (right) CDM. Video 9 shows adhesion turnover of a NF transfected with vinculin-GFP. Video 10 shows adhesion turnover of a CAF transfected with vinculin-GFP.
Supplementary Material
Acknowledgments
This paper is dedicated to Dr. D.J. Webb who passed away on May 15, 2017, while this manuscript was under review.
We thank Susan Craig and Andrew Doyle for reagents and protocols. We also thank Bong Hwan Sung for his help in video processing.
This study was supported by National Institutes of Health grants GM117916 (to A.M. Weaver and D.J. Webb), CA155572 (to D. Li and D.J. Webb), R01CA206458 (to A.M. Weaver), R01CA163592 (to A.M. Weaver), 5P30 DK058404-13 and GISPORE 5P50 CA095103-13 (to M.K. Washington and C. Shi), and 5U01 CA151924 (to S.W. Hayward); National Center for Research Resources grant S10RR025524 (to D.J. Webb); and Vanderbilt University. B.M. Brewer was supported by the National Science Foundation Graduate Research Fellowship grant DGE-0909667.
The authors declare no competing financial interests.
Author contributions: B. Erdogan and M. Ao designed the study, performed the experiments, and analyzed the data. L.M. White and A.L. Means conducted experiments. B. Brewer and L. Yang designed and prepared the microfluidic devices. M.K. Washington and C. Shi identified prostate and pancreatic tumor samples. O.E. Franko prepared the primary cells. A.M. Weaver, S.W. Hayward, D. Li, and D.J. Webb supervised the project. B. Erdogan prepared the manuscript, and M. Ao, A.M. Weaver, D. Li, and D.J. Webb edited and revised the manuscript.
Footnotes
Abbreviations used:
- αSMA
- smooth muscle α-actin
- CAF
- cancer-associated fibroblast
- CDM
- cell-derived matrix
- FFT
- fast-Fourier transform
- Fn
- fibronectin
- HNSCC
- head and neck squamous cell carcinoma
- IF
- immunofluorescence
- KD
- knockdown
- MLC2
- myosin light chain 2
- NF
- normal fibroblast
- NHS
- N-hydroxysuccinimide
- PAA
- polyacrylamide
- PDAC
- pancreatic ductal adenocarcinoma
- PDGFR
- PDGF receptor
- PDMS
- polydimethylsiloxane
- RGD
- arginine-glycine-aspartic acid
- RGE
- arginine-glycine-glutamic acid
- TIRF
- total internal reflection fluorescence
- WB
- Western blot
References
- Akimov S.S., and Belkin A.M.. 2001. Cell-surface transglutaminase promotes fibronectin assembly via interaction with the gelatin-binding domain of fibronectin: a role in TGFbeta-dependent matrix deposition. J. Cell Sci. 114:2989–3000. [DOI] [PubMed] [Google Scholar]
- Amatangelo M.D., Bassi D.E., Klein-Szanto A.J.P., and Cukierman E.. 2005. Stroma-derived three-dimensional matrices are necessary and sufficient to promote desmoplastic differentiation of normal fibroblasts. Am. J. Pathol. 167:475–488. 10.1016/S0002-9440(10)62991-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao M., Franco O.E., Park D., Raman D., Williams K., and Hayward S.W.. 2007. Cross-talk between paracrine-acting cytokine and chemokine pathways promotes malignancy in benign human prostatic epithelium. Cancer Res. 67:4244–4253. 10.1158/0008-5472.CAN-06-3946 [DOI] [PubMed] [Google Scholar]
- Ao M., Brewer B.M., Yang L., Franco Coronel O.E., Hayward S.W., Webb D.J., and Li D.. 2015. Stretching fibroblasts remodels fibronectin and alters cancer cell migration. Sci. Rep. 5:8334 10.1038/srep08334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augsten M. 2014. Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front. Oncol. 4:62 10.3389/fonc.2014.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres C., Bowlin G.L., Henderson S.C., Taylor L., Shultz J., Alexander J., Telemeco T.A., and Simpson D.G.. 2006. Modulation of anisotropy in electrospun tissue-engineering scaffolds: Analysis of fiber alignment by the fast Fourier transform. Biomaterials. 27:5524–5534. 10.1016/j.biomaterials.2006.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae Y.K., Kim A., Kim M.K., Choi J.E., Kang S.H., and Lee S.J.. 2013. Fibronectin expression in carcinoma cells correlates with tumor aggressiveness and poor clinical outcome in patients with invasive breast cancer. Hum. Pathol. 44:2028–2037. 10.1016/j.humpath.2013.03.006 [DOI] [PubMed] [Google Scholar]
- Ball S.G., Shuttleworth C.A., and Kielty C.M.. 2007. Platelet-derived growth factor receptor-alpha is a key determinant of smooth muscle alpha-actin filaments in bone marrow-derived mesenchymal stem cells. Int. J. Biochem. Cell Biol. 39:379–391. 10.1016/j.biocel.2006.09.005 [DOI] [PubMed] [Google Scholar]
- Barsky S.H., Green W.R., Grotendorst G.R., and Liotta L.A.. 1984. Desmoplastic breast carcinoma as a source of human myofibroblasts. Am. J. Pathol. 115:329–333. [PMC free article] [PubMed] [Google Scholar]
- Barthel S.R., Hays D.L., Yazawa E.M., Opperman M., Walley K.C., Nimrichter L., Burdick M.M., Gillard B.M., Moser M.T., Pantel K., et al. 2013. Definition of molecular determinants of prostate cancer cell bone extravasation. Cancer Res. 73:942–952. 10.1158/0008-5472.CAN-12-3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beacham D., Amatangelo M., and Cukierman E.. 2006. Preparation of Extracellular Matrices. Curr. Protoc. Cell Biol. Chapter 10:Unit 10.9:1–21. [DOI] [PubMed] [Google Scholar]
- Burridge K., and Fath K.. 1989. Focal contacts: transmembrane links between the extracellular matrix and the cytoskeleton. BioEssays. 10:104–108. 10.1002/bies.950100403 [DOI] [PubMed] [Google Scholar]
- Calvo F., Ege N., Grande-Garcia A., Hooper S., Jenkins R.P., Chaudhry S.I., Harrington K., Williamson P., Moeendarbary E., Charras G., and Sahai E.. 2013. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 15:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell I.D., and Humphries M.J.. 2011. Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 3:1–14. 10.1101/cshperspect.a004994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Zhang Q., Liu S., Parajuli K.R., Qu Y., Mei J., Chen Z., Zhang H., Khismatullin D.B., and You Z.. 2015. IL-17 and insulin/IGF1 enhance adhesion of prostate cancer cells to vascular endothelial cells through CD44-VCAM-1 interaction. Prostate. 75:883–895. 10.1002/pros.22971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K., Pankov R., Travis M.A., Askari J.A., Mould A.P., Craig S.E., Newham P., Yamada K.M., and Humphries M.J.. 2005. A specific alpha5beta1-integrin conformation promotes directional integrin translocation and fibronectin matrix formation. J. Cell Sci. 118:291–300. 10.1242/jcs.01623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C.J., Berg T.J., Birch J., Ennis D., Mitchell L., Cloix C., Campbell A., Sumpton D., Nixon C., Campbell K., et al. 2016. The initiator methionine trna drives secretion of type II collagen from stromal fibroblasts to promote tumor growth and angiogenesis. Curr. Biol. 26:755–765. 10.1016/j.cub.2016.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin M.W., Eickhoff J.C., Riching K.M., Pehlke C.A., Eliceiri K.W., Provenzano P.P., Friedl A., and Keely P.J.. 2011. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 178:1221–1232. 10.1016/j.ajpath.2010.11.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danen E.H.J., Sonneveld P., Brakebusch C., Fässler R., and Sonnenberg A.. 2002. The fibronectin-binding integrins α5β1 and αvβ3 differentially modulate RhoA-GTP loading, organization of cell matrix adhesions, and fibronectin fibrillogenesis. J. Cell Biol. 159:1071–1086. 10.1083/jcb.200205014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M., and Wang Y.L.. 1999. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 76:2307–2316. 10.1016/S0006-3495(99)77386-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J., Abraham D., and Norman J.. 2013. Platelet-derived growth factor signaling in mesenchymal cells. Front. Biosci. (Landmark Ed.). 18:106–119. 10.2741/4090 [DOI] [PubMed] [Google Scholar]
- Eliceiri B.P. 2001. Integrin and growth factor receptor crosstalk. Circ. Res. 89:1104–1110. 10.1161/hh2401.101084 [DOI] [PubMed] [Google Scholar]
- Franco-Barraza J., Beacham D.A., Amatangelo M.D., and Cukierman E.. 2016. Preparation of Extracellular Matrices Produced by Cultured and Primary Fibroblasts. Curr. Protoc. Cell Biol. 71:10.9.1–10.9.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Barraza J., Francescone R., Luong T., Shah N., Madhani R., Cukierman G., Dulaimi E., Devarajan K., Egleston B.L., Nicolas E., et al. 2017. Matrix-regulated integrin αvβ5 maintains α5β1-dependent desmoplastic traits prognostic of neoplastic recurrence. eLife. 6:e20600 10.7554/eLife.20600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland J.C., Lee M.H., and Boettiger D.. 2009. Mechanically activated integrin switch controls alpha5beta1 function. Science. 323:642–644. 10.1126/science.1168441 [DOI] [PubMed] [Google Scholar]
- Gaggioli C., Hooper S., Hidalgo-Carcedo C., Grosse R., Marshall J.F., Harrington K., and Sahai E.. 2007. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 9:1392–1400. 10.1038/ncb1658 [DOI] [PubMed] [Google Scholar]
- Galbraith C.G., Yamada K.M., and Sheetz M.P.. 2002. The relationship between force and focal complex development. J. Cell Biol. 159:695–705. 10.1083/jcb.200204153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Palmero I., Torres S., Bartolomé R.A., Peláez-Garcia A., Larriba M.J., Lopez-Lucendo M., Peña C., Escudero-Paniagua B., Muñoz A., and Casal J.I.. 2016. Twist1-induced activation of human fibroblasts promotes matrix stiffness by upregulating palladin and collagen α1(VI). Oncogene. 35:5224–5236. [DOI] [PubMed] [Google Scholar]
- Georgiadou M., Lilja J., Jacquemet G., Guzmán C., Rafaeva M., Alibert C., Yan Y., Sahgal P., Lerche M., Manneville J.-B., et al. 2017. AMPK negatively regulates tensin-dependent integrin activity. J. Cell Biol. 216:1107–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel H.L., Li J., Kogan S., and Languino L.R.. 2008. Integrins in prostate cancer progression. Endocr. Relat. Cancer. 15:657–664. 10.1677/ERC-08-0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal S., Veracini L., Grall D., Butori C., Schaub S., Audebert S., Camoin L., Baudelet E., Radwanska A., Beghelli-de la Forest Divonne S., et al. 2017. Fibronectin-guided migration of carcinoma collectives. Nat. Commun. 8:14105 10.1038/ncomms14105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grashoff C., Hoffman B.D., Brenner M.D., Zhou R., Parsons M., Yang M.T., McLean M.A., Sligar S.G., Chen C.S., Ha T., and Schwartz M.A.. 2010. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 466:263–266. 10.1038/nature09198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin C.-H. 2013. Targeting the PDGF signaling pathway in tumor treatment. Cell Commun. Signal. 11:97 10.1186/1478-811X-11-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B. 2006. Masters and servants of the force: the role of matrix adhesions in myofibroblast force perception and transmission. Eur. J. Cell Biol. 85:175–181. 10.1016/j.ejcb.2005.09.004 [DOI] [PubMed] [Google Scholar]
- Horikawa S., Ishii Y., Hamashima T., Yamamoto S., Mori H., Fujimori T., Shen J., Inoue R., Nishizono H., Itoh H., et al. 2015. PDGFRα plays a crucial role in connective tissue remodeling. Sci. Rep. 5:17948 10.1038/srep17948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt K., Castaneda B., Zhang M., Nigwekar P., di Sant’agnese P.A., Joseph J.V., Strang J., Rubens D.J., and Parker K.J.. 2008. Tissue elasticity properties as biomarkers for prostate cancer. Cancer Biomark. 4:213–225. 10.3233/CBM-2008-44-505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insua-Rodríguez J., and Oskarsson T.. 2016. The extracellular matrix in breast cancer. Adv. Drug Deliv. Rev. 97:41–55. 10.1016/j.addr.2015.12.017 [DOI] [PubMed] [Google Scholar]
- Jean L., Majumdar D., Shi M., Hinkle L.E., Diggins N.L., Ao M., Broussard J.A., Evans J.C., Choma D.P., and Webb D.J.. 2013. Activation of Rac by Asef2 promotes myosin II-dependent contractility to inhibit cell migration on type I collagen. J. Cell Sci. 126:5585–5597. 10.1242/jcs.131060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean L., Yang L., Majumdar D., Gao Y., Shi M., Brewer B.M., Li D., and Webb D.J.. 2014. The Rho family GEF Asef2 regulates cell migration in three dimensional (3D) collagen matrices through myosin II. Cell Adhes. Migr. 8:460–467. 10.4161/19336918.2014.983778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly L.A., Novitskiy S., Owens P., Massoll N., Cheng N., Fang W., Moses H.L., and Franco A.T.. 2016. Fibroblast-mediated collagen remodeling within the tumor microenvironment facilitates progression of thyroid cancers driven by brafv600e and pten loss. Cancer Res. 76:1804–1813. 10.1158/0008-5472.CAN-15-2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadler K.E., Hill A., and Canty-Laird E.G.. 2008. Collagen fibrillogenesis: fibronectin, integrins, and minor collagens as organizers and nucleators. Curr. Opin. Cell Biol. 20:495–501. 10.1016/j.ceb.2008.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R., and Zeisberg M.. 2006. Fibroblasts in cancer. Nat. Rev. Cancer. 6:392–401. 10.1038/nrc1877 [DOI] [PubMed] [Google Scholar]
- Kaukonen R., Mai A., Georgiadou M., Saari M., De Franceschi N., Betz T., Sihto H., Ventelä S., Elo L., Jokitalo E., et al. 2016. Normal stroma suppresses cancer cell proliferation via mechanosensitive regulation of JMJD1a-mediated transcription. Nat. Commun. 7:12237 10.1038/ncomms12237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kii I., Nishiyama T., Li M., Matsumoto K., Saito M., Amizuka N., and Kudo A.. 2010. Incorporation of tenascin-C into the extracellular matrix by periostin underlies an extracellular meshwork architecture. J. Biol. Chem. 285:2028–2039. 10.1074/jbc.M109.051961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labernadie A., Kato T., Brugués A., Serra-Picamal X., Derzsi S., Arwert E., Weston A., González-Tarragó V., Elosegui-Artola A., Albertazzi L., et al. 2017. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat. Cell Biol. 19:224–237. 10.1038/ncb3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-O., Mullins S.R., Franco-Barraza J., Valianou M., Cukierman E., and Cheng J.D.. 2011. FAP-overexpressing fibroblasts produce an extracellular matrix that enhances invasive velocity and directionality of pancreatic cancer cells. BMC Cancer. 11:245 10.1186/1471-2407-11-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon C.A., Chen C.S., and Romer L.H.. 2009. Cell traction forces direct fibronectin matrix assembly. Biophys. J. 96:729–738. 10.1016/j.bpj.2008.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G.L., Cohen D.M., Desai R.A., Breckenridge M.T., Gao L., Humphries M.J., and Chen C.S.. 2013. Activation of beta 1 but not beta 3 integrin increases cell traction forces. FEBS Lett. 587:763–769. 10.1016/j.febslet.2013.01.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., and Schwarzbauer J.E.. 2005. Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol. 24:389–399. 10.1016/j.matbio.2005.06.008 [DOI] [PubMed] [Google Scholar]
- Mezawa Y., and Orimo A.. 2016. The roles of tumor- and metastasis-promoting carcinoma-associated fibroblasts in human carcinomas. Cell Tissue Res. 365:675–689. 10.1007/s00441-016-2471-1 [DOI] [PubMed] [Google Scholar]
- Miles F.L., and Sikes R.A.. 2014. Insidious changes in stromal matrix fuel cancer progression. Mol. Cancer Res. 12:297–312. 10.1158/1541-7786.MCR-13-0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olumi A.F., Grossfeld G.D., Hayward S.W., Epithelium P., Olumi A.F., Grossfeld G.D., Hayward S.W., Carroll P.R., Tlsty T.D., and Cunha G.R.. 1999. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium carcinoma-associated fibroblasts direct tumor progression of initiated human. Cancer Res. 59:5002–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orimo A., Gupta P.B., Sgroi D.C., Arenzana-Seisdedos F., Delaunay T., Naeem R., Carey V.J., Richardson A.L., and Weinberg R.A.. 2005. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 121:335–348. 10.1016/j.cell.2005.02.034 [DOI] [PubMed] [Google Scholar]
- Östman A. 2004. PDGF receptors-mediators of autocrine tumor growth and regulators of tumor vasculature and stroma. Cytokine Growth Factor Rev. 15:275–286. 10.1016/j.cytogfr.2004.03.002 [DOI] [PubMed] [Google Scholar]
- Pankov R., and Yamada K.M.. 2002. Fibronectin at a glance. J. Cell Sci. 115:3861–3863. 10.1242/jcs.00059 [DOI] [PubMed] [Google Scholar]
- Petrie R.J., Doyle A.D., and Yamada K.M.. 2009. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 10:538–549. 10.1038/nrm2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peuhu E., Kaukonen R., Lerche M., Saari M., Guzmán C., Rantakari P., De Franceschi N., Wärri A., Georgiadou M., Jacquemet G., et al. 2017. SHARPIN regulates collagen architecture and ductal outgrowth in the developing mouse mammary gland. EMBO J. 36:165–182. 10.15252/embj.201694387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P., Gauthier N.C., Del Rio A., and Sheetz M.P.. 2009. Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v)beta(3) and talin enable mechanotransduction. Proc. Natl. Acad. Sci. USA. 106:16245–16250. 10.1073/pnas.0902818106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Cusachs P., Iskratsch T., and Sheetz M.P.M.P.. 2012. Finding the weakest link: exploring integrin-mediated mechanical molecular pathways. J. Cell Sci. 125:3025–3038. 10.1242/jcs.095794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokhlin O.W., and Cohen M.B.. 1995. Expression of cellular adhesion molecules on human prostate tumor cell lines. Prostate. 26:205–212. 10.1002/pros.2990260406 [DOI] [PubMed] [Google Scholar]
- Ross T.D., Coon B.G., Yun S., Baeyens N., Tanaka K., Ouyang M., and Schwartz M.A.. 2013. Integrins in mechanotransduction. Curr. Opin. Cell Biol. 25:613–618. 10.1016/j.ceb.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabass B., Gardel M.L.M.L., Waterman C.M.C.M., and Schwarz U.S.U.S.. 2008. High resolution traction force microscopy based on experimental and computational advances. Biophys. J. 94:207–220. 10.1529/biophysj.107.113670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariban E., Sitaras N.M., Antoniades H.N., Kufe D.W., and Pantazist P.. 1988. Expression of platelet-derived growth factor (PDGF)-related transcripts and synthesis of biologically active PDGF-like proteins by human malignant epithelial cell lines. J. Clin. Invest. 82:1157–1164. 10.1172/JCI113712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor S.L., Ellis I.R., Jones S.J., Baillie R., Seneviratne K., Clausen J., Motegi K., Vojtesek B., Kankova K., Furrie E., et al. 2003. Migration-stimulating factor: a genetically truncated onco-fetal fibronectin isoform expressed by carcinoma and tumor-associated stromal cells. Cancer Res. 63:8827–8836. [PubMed] [Google Scholar]
- Schwartz M.A., and DeSimone D.W.. 2008. Cell adhesion receptors in mechanotransduction. Curr. Opin. Cell Biol. 20:551–556. 10.1016/j.ceb.2008.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer J.E., and DeSimone D.W.. 2011. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb. Perspect. Biol. 3:1–19. 10.1101/cshperspect.a005041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Washington M.K., Chaturvedi R., Drosos Y., Revetta F.L., Weaver C.J., Buzhardt E., Yull F.E., Blackwell T.S., Sosa-Pineda B., et al. 2014. Fibrogenesis in pancreatic cancer is a dynamic process regulated by macrophage-stellate cell interaction. Lab. Invest. 94:409–421. 10.1038/labinvest.2014.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Carraher C., and Schwarzbauer J.E.. 2010. Assembly of fibronectin extracellular matrix. Annu. Rev. Cell Dev. Biol. 26:397–419. 10.1146/annurev-cellbio-100109-104020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaras N.M., Sariban E., Bravo M., Pantazis P., and Antoniades H.N.. 1988. Constitutive production of platelet-derived growth factor-like proteins by human prostate carcinoma cell lines. Cancer Res. 48:1930–1935. [PubMed] [Google Scholar]
- Stanisavljevic J., Loubat-Casanovas J., Herrera M., Luque T., Peña R., Lluch A., Albanell J., Bonilla F., Rovira A., Peña C., et al. 2015. Snail1-expressing fibroblasts in the tumor microenvironment display mechanical properties that support metastasis. Cancer Res. 75:284–295. 10.1158/0008-5472.CAN-14-1903 [DOI] [PubMed] [Google Scholar]
- Sutherland M., Gordon A., Shnyder S.D., Patterson L.H., and Sheldrake H.M.. 2012. RGD-binding integrins in prostate cancer: Expression patterns and therapeutic prospects against bone metastasis. Cancers (Basel). 4:1106–1145. 10.3390/cancers4041106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalovski M., and Brekken R.A.. 2016. Matrix control of pancreatic cancer: New insights into fibronectin signaling. Cancer Lett. 381:252–258. 10.1016/j.canlet.2015.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxhorn J.A., Ayala G.E., Smith M.J., Smith V.C., Dang T.D., and Rowley D.R.. 2002. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin. Cancer Res. 8:2912–2923. [PubMed] [Google Scholar]
- Veevers-Lowe J., Ball S.G., Shuttleworth A., and Kielty C.M.. 2011. Mesenchymal stem cell migration is regulated by fibronectin through α5β1-integrin-mediated activation of PDGFR-β and potentiation of growth factor signals. J. Cell Sci. 124:1288–1300. 10.1242/jcs.076935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.P., and Hielscher A.. 2017. Fibronectin: How its aberrant expression in tumors may improve therapeutic targeting. J. Cancer. 8:674–682. 10.7150/jca.16901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb D.J., Donais K., Whitmore L.A., Thomas S.M., Turner C.E., Parsons J.T., and Horwitz A.F.. 2004. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6:154–161. 10.1038/ncb1094 [DOI] [PubMed] [Google Scholar]
- Xia Y., and Whitesides G.M.. 1998. Soft Lithography. Annu. Rev. Mater. Sci. 28:153–184. 10.1146/annurev.matsci.28.1.153 [DOI] [Google Scholar]
- Yang N., Mosher R., Seo S., Beebe D., and Friedl A.. 2011. Syndecan-1 in breast cancer stroma fibroblasts regulates extracellular matrix fiber organization and carcinoma cell motility. Am. J. Pathol. 178:325–335. 10.1016/j.ajpath.2010.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T., Georges P.C., Flanagan L.A., Marg B., Ortiz M., Funaki M., Zahir N., Ming W., Weaver V., and Janmey P.A.. 2005. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 60:24–34. 10.1002/cm.20041 [DOI] [PubMed] [Google Scholar]
- Zemskov E.A., Loukinova E., Mikhailenko I., Coleman R.A., Strickland D.K., and Belkin A.M.. 2009. Regulation of platelet-derived growth factor receptor function by integrin-associated cell surface transglutaminase. J. Biol. Chem. 284:16693–16703. 10.1074/jbc.M109.010769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L., Madden J., Foo W.-C., Mouraviev V., Polascik T.J., Palmeri M.L., and Nightingale K.R.. 2010. Characterizing stiffness of human prostates using acoustic radiation force. Ultrason. Imaging. 32:201–213. 10.1177/016173461003200401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.