NuMA has been suggested to function as a Ran-regulated spindle assembly factor that modulates spindle pole assembly. Chang et al. provide structural and biochemical evidence showing that NuMA contains an Importin-β–regulated microtubule-binding region, which allows Ran to regulate the NuMA functions required for the assembly of higher-order microtubule structures.
Abstract
Ran–guanosine triphosphatase orchestrates mitotic spindle assembly by modulation of the interaction between Importin-α/-β and spindle assembly factors (SAFs). The inhibition of SAFs performed by importins needs to be done without much sequestration from abundant nuclear localization signal (NLS) –containing proteins. However, the molecular mechanisms that determine NLS-binding selectivity and that inhibit activity of Importin-β–regulated SAFs (e.g., nuclear mitotic apparatus protein [NuMA]) remain undefined. Here, we present a crystal structure of the Importin-α–NuMA C terminus complex showing a novel binding pattern that accounts for selective NLS recognition. We demonstrate that, in the presence of Importin-α, Importin-β inhibits the microtubule-binding function of NuMA. Further, we have identified a high-affinity microtubule-binding region that lies carboxyl-terminal to the NLS, which is sterically masked by Importin-β on being bound by Importin-α. Our study provides mechanistic evidence of how Importin-α/-β regulates the NuMA functioning required for assembly of higher-order microtubule structures, further illuminating how Ran-governed transport factors regulate diverse SAFs and accommodate various cell demands.
Introduction
In conjunction with transport factors such as Importin-α/-β, the small guanosine triphosphatase Ran plays a crucial role in the delivery of macromolecules between the nucleus and cytoplasm during interphase. For example, in the nuclear import pathway, importins recognize cargoes by binding to the cargoes’ nuclear localization signals (NLSs) in the cytoplasm and then shuttle them into the nucleus. Subsequently, in the nucleus, GTP-bound Ran binds to importins to trigger the release of the imported cargoes (Cook et al., 2007). Importantly, assembly of the mitotic spindle is also orchestrated by the Ran pathway (Nachury et al., 2001; Wiese et al., 2001). Ran functions by modulating the interaction between transport factors and the spindle assembly factors (SAFs) that control many aspects of microtubule behavior, such as microtubule nucleation, stabilization, and bundling. In current models, Ran–GTP binds to importins and liberates SAFs (e.g., nuclear mitotic apparatus protein [NuMA] and targeting protein for Xklp2 [TPX2]), modulating organization of the mitotic spindle (Forbes et al., 2015).
As SAFs only represent a small fraction of the nuclear proteins that contain NLS, they use atypical ways to interact with importins to reduce competition from other NLS-proteins during open mitosis. For example, TPX2, an Importin-α–regulated SAF, adopts a nonclassic NLS binding mode to interact with Importin-α (Schatz et al., 2003; Giesecke and Stewart, 2010). Although a mechanism has been proposed for how Importin-α–regulated SAFs reduce sequestration during mitosis (Giesecke and Stewart, 2010), the molecular basis of molecule selectivity and activity inhibition by Importin-β–regulated SAFs remains to be defined. Microtubule aster assembly in Xenopus laevis egg extracts promoted by NuMA is suppressed by addition of Importin-β, even though NuMA has been shown to pull down both Importin-α and -β (Nachury et al., 2001; Wiese et al., 2001). Further, regulation of NuMA-mediated higher-order microtubule structure assembly (e.g., microtubule asters and mitotic spindles) is facilitated by the NuMA C-terminal tail domain that bears microtubule-binding regions and NLS (Fig. 1 A; Merdes et al., 1996; Haren and Merdes, 2002; Gallini et al., 2016). Here, we investigated how NuMA determines molecule selectivity and the molecular mechanism by which Importin-β inhibits NuMA activity.
Figure 1.
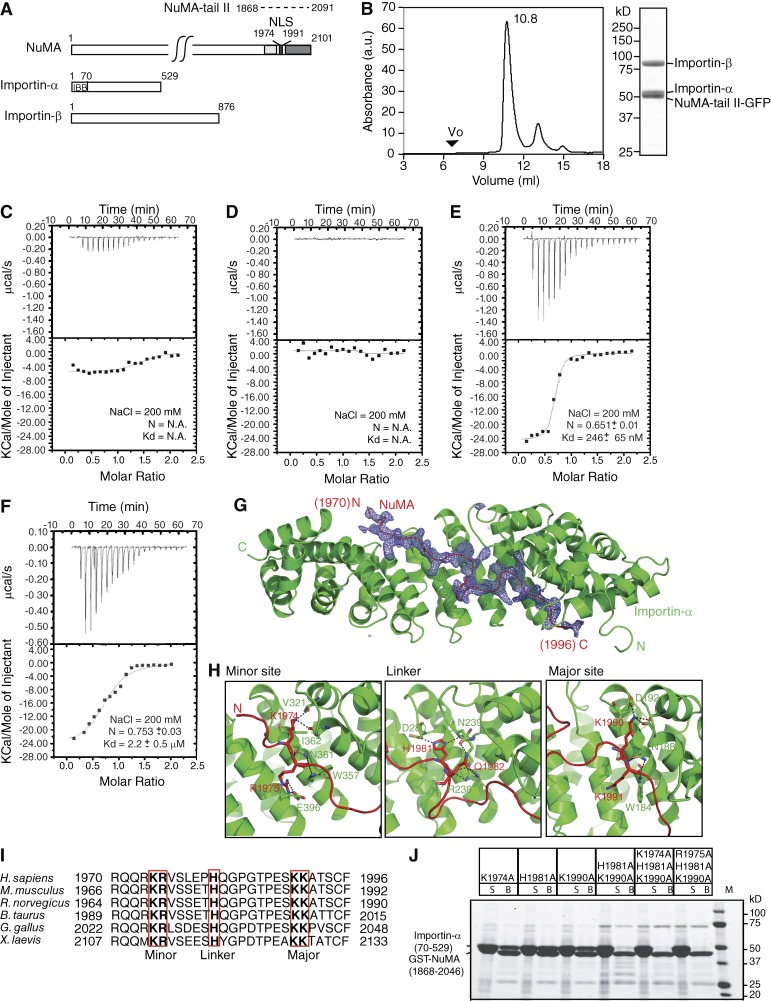
Biochemical and structural characterization of the heterotrimeric NuMA-tail II–Importin-α–Importin-β complex. (A) Domain structures of NuMA, Importin-α, and Importin-β. For NuMA, the microtubule-binding regions suggested by the Merdes and Mapelli groups are shown in light and dark gray, respectively (Haren and Merdes, 2002; Gallini et al., 2016). The NLS is colored black. Residue numbers of NuMA-tail II (dotted line) used in biochemical studies are indicated. For Importin-α, the Importin-β binding domain is indicated. (B) Purified recombinant Importin-α, Importin-β, and NuMA-tail II-GFP were mixed and analyzed by SEC. The peak fraction (10.8 ml) was analyzed by SDS-PAGE and stained with Coomassie blue. SEC (Superdex 200) elution profile for the NuMA-tail II-GFP–Importin-α–Importin-β trimeric complex. The void volume (Vo) of the peak fraction and absorbance (a.u.) at 280 nm for the complex is indicated. (C–F) ITC titration curves (top) and binding isotherms (bottom) of full-length Importin-α (C), full-length Importin-β (D), Importin-α (ΔIBB; E), and 1:1 stoichometric mixture of full-length Importin-α and -β with NuMA-tail II-GFP (F). (G) An omit difference (Fo-Fc) map contoured at 2.5 sigma with a superimposed atomic model of the NuMA (1955–2046)–Importin-α (70–498) complex, showing NuMA in red and Importin-α in green. Residues 1,970 to 1,996 in NuMA are assigned. (H) The three panels highlight the interactions at the minor site, linker region, and major site. (I) Sequence alignment of the NuMA-NLS peptide from different species (numbers represent amino acid positions). The conserved minor-site, linker-region and major-site interacting residues are highlighted by red boxes. (J) GST pull-down assays of Importin-α (70–529) with NuMA-tail mutants. GST-fused NuMA-tail (1,868–2,046) mutants were incubated with recombinant Importin-α. Unbound (S) and bound (B) samples were analyzed by SDS-PAGE and stained with Coomassie blue.
Results and discussion
Biochemical reconstitution of a heterotrimeric NuMA-tail II–Importin-α–Importin-β complex
Coimmunoprecipitation studies from Xenopus egg extracts have shown that NuMA-tail II interacts with Importin-α and Importin-β (Nachury et al., 2001). Hence, we first sought to recapitulate these interactions biochemically using recombinant GFP-tagged WT NuMA-tail II (aa 1868–2091; hereafter NuMA-tail II-GFP), Importin-α and Importin-β (Fig. 1 A). Purified recombinant NuMA-tail II-GFP, Importin-α and Importin-β form a stable trimeric complex, as they comigrated in a sharp mono-dispersed peak in size exclusion chromatography (SEC; Fig. 1 B and Fig. S1 A).
Next, Importin-β, but not Importin-α, has been shown to inhibit the microtubule aster formation induced by NuMA-tail II (Nachury et al., 2001; Wiese et al., 2001). We used isothermal titration calorimetry (ITC) to examine if Importin-β interacts with NuMA-tail II directly or if interaction is mediated by Importin-α.Whereas NuMA-tail II-GFP showed no measurable interaction with either full-length Importin-α or full-length Importin-β under our experimental conditions, it bound to Importin-α lacking the Importin-β binding (IBB) domain (aa 70–524; hereafter Importin-α (ΔIBB)) and an equal stoichiometric mixture of full-length Importin-α and -β (Fig. 1, C–F; and Fig. S1 B). These results indicate that Importin-α but not Importin-β directly contacts NuMA-tail II, as the Importin-α and -β mixture did not show a lower dissociation constant to NuMA-tail II compared with that of Importin-α (ΔIBB; ∼2.2 µM vs. ∼250 nM). However, the interaction between full-length Importin-α and NuMA-tail II required the presence of Importin-β.
An atypical NLS interaction between NuMA and Importin-α
As Importin-α directly binds to NuMA-tail II, we wanted to determine the crystal structure of the NuMA-tail II and Importin-α (ΔIBB) complex to understand the basis of the NLS binding mode of NuMA, which could determine the selectivity of the Importin-α interaction. To identify the minimum region in NuMA-tail II that was required for Importin-α (ΔIBB) binding and suitable for crystallization, NuMA-tail II (aa 1868–2101) and Importin-α (ΔIBB) complex was purified (Fig. S1 C) and then subjected to limited proteolysis using either trypsin or elastase over different time courses (Fig. S1 D). Two stable peptides that appeared after 1-h elastase treatment and showed as a monodispersed peak were examined to determine the boundaries of the N and C termini using Edman sequencing and mass spectrometry (Fig. S1 E). Although only ∼30 residues in the C terminus of Importin-α were removed by elastase, less than half of NuMA-tail II (91 residues, aa 1955–2046) was protected by Importin-α. Next, the NuMA-tail peptide (aa 1955–2046)–Importin-α (aa 70–498) complex was purified and yielded crystals of the P212121 orthorhombic space group that diffracted to 2.4 Å (Table S1). The structure was determined by molecular replacement using Importin-α as a search model. After initial refinement, a continuous electron density of NuMA-tail peptide was clear only from residues 1970 to 1996, so that only this portion could be assigned and built unambiguously (Fig. 1 G).
The NuMA-NLS fragment (aa 1970–1996) comprised two stretches of basic residues that were bound to the minor and major NLS-binding sites on Importin-α (Fig. 1 H). Interestingly, residues in the linker region that connected the two basic stretches and that are normally disordered in conventional bipartite NLS were also involved in Importin-α recognition and binding (Fig. 1 H). Among these residues, a histidine (aa 1981) that is evolutionarily conserved from Xenopus to human interacts with residues Asp-280 and Asn-239 on Importin-α (Fig. 1, H and I). Thus, NuMA-NLS and Importin-α display an atypical NLS interaction pattern that contains three binding patches (Fig. S2, A and B).
Next, a GST pull-down assay was performed to biochemically verify the interaction between Importin-α and GST-NuMA-tail (aa 1868–2046) that contains variant NLS point mutations. Whereas single point mutations in each binding patch of the NuMA-tail (aa 1868–2046) fragment were not sufficient to disrupt its interaction with Importin-α (ΔIBB), a double mutation (K1990A/H1981A) caused a substantial reduction of Importin-α binding (Fig. 1 J). Furthermore, Importin-α was essentially not detectable in the pull-down assay using a GST-NuMA-tail (aa 1868–2046) fragment containing triple mutations (K1974A or R1975A/H1981A/K1990A; Fig. 1 J), suggesting that all three patches on NuMA are crucial for Importin-α binding. Consistently, a dissociation constant value was not detectable by ITC analysis for Importin-α (ΔIBB) and a NuMA-tail II-GFP construct that contained the triple mutation (R1975A/H1981A/K1990A; hereafter RHK-NuMA-tail II-GFP; Fig. S2 C). Notably, the RHK mutant also showed no measurable interaction with the Importin-α/-β mixture compared with WT (Fig. 1 F and Fig. S2 D), further suggesting that NuMA-tail II mainly binds Importin-α via its NLS in the presence of Importin-β.
Importin-α/-β modulate NuMA-tail II–mediated microtubule assembly
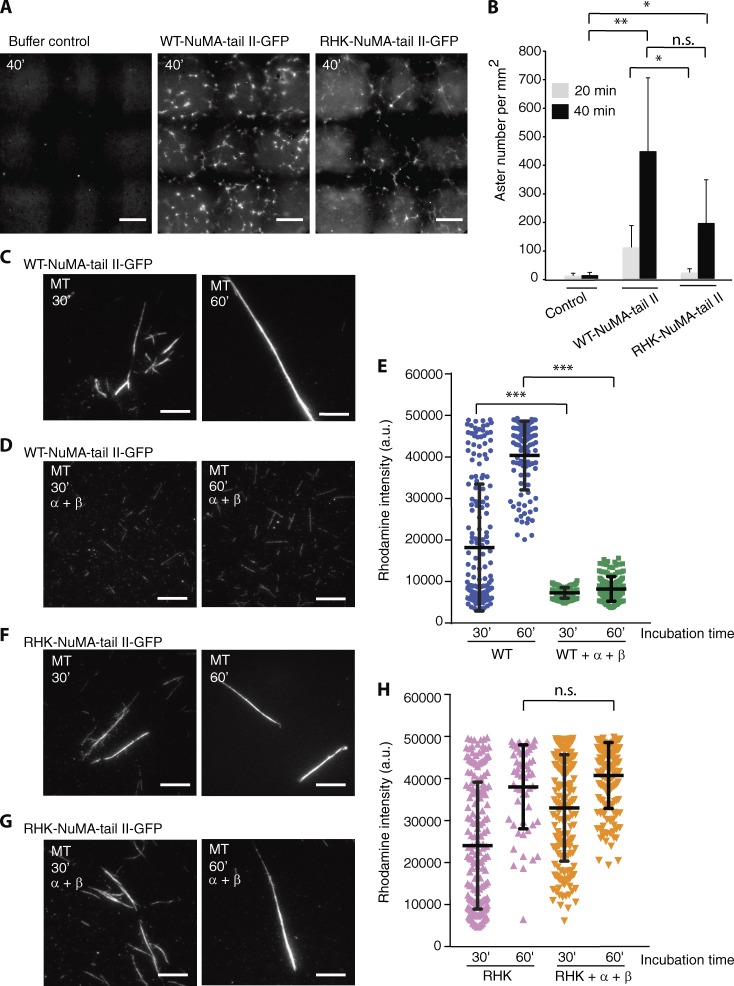
Addition of NuMA-tail II to Xenopus egg extracts induces microtubule aster assembly (Merdes et al., 1996; Nachury et al., 2001; Wiese et al., 2001), which could be caused by direct (e.g., bundling of microtubules) or indirect effects (e.g., activating endogenous SAFs by sequestering importins). To verify if NuMA-tail II directly promotes microtubule aster assembly, we performed a microtubule aster assembly assay in Xenopus egg extracts using the Importin-α–insensitive NuMA-tail II mutant (RHK-NuMA-tail II-GFP). We found that addition of NuMA-tail II-GFP (WT, 5 µM) led to substantial microtubule aster assembly compared with control (Fig. 2, A and B). Interestingly, aster formation was also promoted by addition of the RHK-NuMA-tail II-GFP mutant (5 µM), albeit with reduced efficiency compared with the WT NuMA-tail II-GFP (Fig. 2, A and B). This result suggests that NuMA-tail II may directly promote microtubule aster formation, because the RHK mutant that does not bind Importin-α cannot activate SAFs by sequestering endogenous importins.
Figure 2.
Importin-α/-β regulates NuMA-tail II-mediated higher-order microtubule structure assembly. (A) Microtubule aster assembly in Xenopus egg extracts, examined with buffer control (left), WT NuMA-tail II-GFP (5 µM, center), and RHK-NuMA-tail II-GFP (5 µM, right) after 40-min incubation at 18°C. Microtubules were imaged using X-rhodamine tubulin (100 nM). Large representative images were created with 3×3 raster scan images. Bars, 100 µm. (B) Dependency of aster assembly activity on NuMA. The number of microtubule asters was counted in microtubule images at indicated incubation times. Data are mean ± SD from n = 6 trials, performed using three independent extract preparations (10 ± 9, 109 ± 76, and 21 ± 14 at 20 min; 12 ± 10, 445 ± 256, and 194 ± 152 at 40 min; for control, WT, and RHK mutant, respectively). (C–H) GMPCPP-stabilized microtubules (X-rhodamine–labeled) were incubated separately with equal concentrations (400 nM) of WT-tail II-GFP (C and D) and RHK-tail II-GFP (F and G) in the presence or absence of Importin-α and Importin-β (4 µM each). Samples were fixed and processed to determine rhodamine fluorescence intensity after 30 and 60 min incubation. Bars, 10 µm. (E and H) Rhodamine intensity of microtubules for each condition was analyzed and plotted. SD was determined from data pooled from three independent experiments (E: WT, 30′, n = 180; WT+α+β, 30′, n = 153; WT, 60′, n = 100; WT+α+β, 60′, n = 147. H: RHK, 30′, n = 180; RHK+α+β, 30′, n = 180; RHK, 60′, n = 55; RHK+α+β, 60′, n = 147). Two-tailed Student t test for comparison of 30 min incubation; Mann-Whitney U test for comparison of 60 min incubation. Statistical differences: *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
NuMA-tail II does not directly promote microtubule growth (Wiese et al., 2001), so it may induce microtubule aster assembly by directly cross-linking microtubules. Next, NuMA-tail II-GFP (WT, 400 nM) was incubated with Taxol-stabilized, X-rhodamine–labeled microtubules for 30 and 60 min. After 60-min incubation, both the mean and high (i.e., >30,000 a.u.) intensities of rhodamine were increased compared with 30 min of incubation, suggesting that NuMA-tail II can directly stimulate the formation of microtubule bundles (Fig. 2, C and E). However, in the presence of Importin-α/-β (4 µM of each), mean rhodamine signals were comparable for both 30- and 60-min incubation and, essentially, only a low intensity of rhodamine (∼10,000 a.u.) could be observed at these two time points (Fig. 2, D and E). These results suggest that the presence of Importin-α/-β prevents formation of NuMA-tail II–mediated microtubule bundles. Furthermore, the RHK-NuMA-tail II-GFP (Importin-α insensitive; 400 nM) construct bundled microtubules both in the absence and presence of Importin-α/-β, further suggesting that Importin-α/-β modulate NuMA-mediated microtubule cross-linking (Fig. 2, F–H).
Importin-β regulates microtubule binding of NuMA-tail II
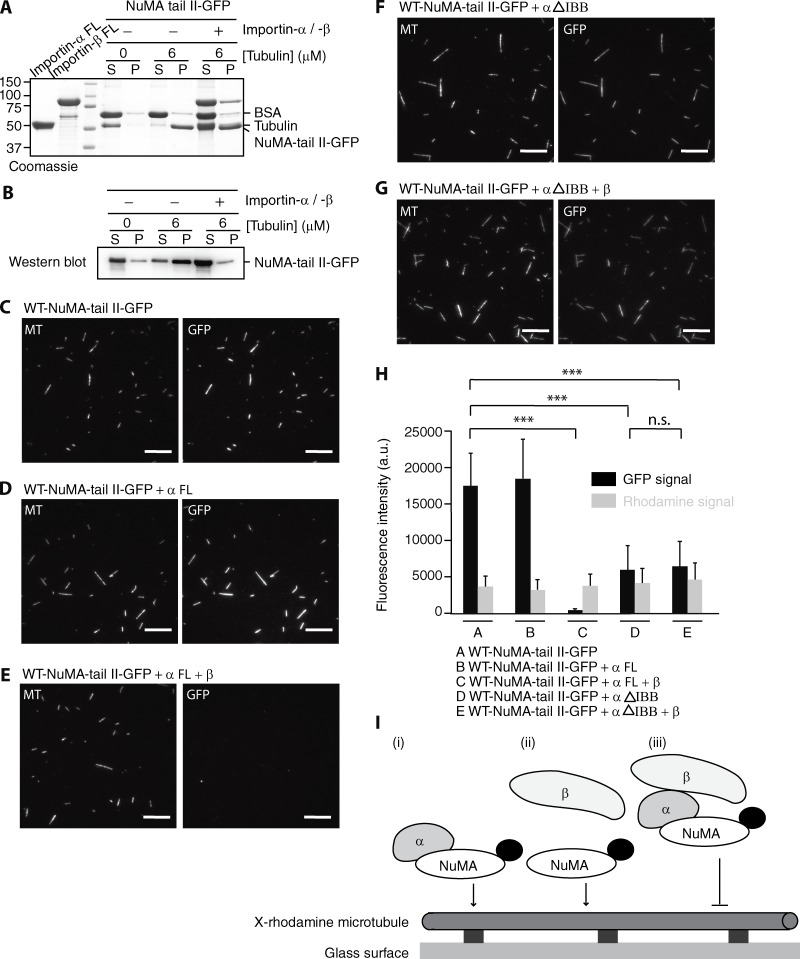
We next performed microtubule cosedimentation assays to examine whether Importin-α/-β regulates the microtubule-binding activity of NuMA-tail II. NuMA-tail II-GFP (20 nM) in the absence or presence of Importin-α/-β (200 nΜ of each) was incubated with Taxol-stabilized microtubules, and the mixture was then pelleted by centrifugation. Proteins in the pellet and supernatant fractions were analyzed by Coomassie blue staining and Western blotting. Whereas NuMA-tail II-GFP was found almost exclusively in the pellet fraction in the absence of importins, the majority of NuMA-tail II-GFP was in the supernatant fraction in the presence of Importin-α/-β, suggesting that Importin-α/-β blocks interaction of NuMA-tail II with microtubules (Fig. 3, A and B).
Figure 3.
Importin-β regulates interaction of NuMA-tail II with microtubules. (A) SDS-PAGE analysis of microtubule cosedimentation assays for NuMA-tail II-GFP in the presence of Importin-α or Importin-α/-β. BSA (final, 0.25 mg/ml), which was used to suppress nonspecific interactions, and tubulin concentrations are indicated. Supernatant and pellet fractions are indicated as S and P, respectively. FL, full length. (B) The NuMA-tail II-GFP bands in the gel shown in A were detected by Western blot using anti-GFP antibody as the positions of NuMA-tail II-GFP and tubulin overlap. (C–G) GMPCPP-stabilized microtubules (X-rhodamine– and biotin-labeled), immobilized on a glass surface, were incubated with WT-NuMA-tail II-GFP (C) in the presence of full-length Importin-α (D), full-length Importin-α and Importin-β (E), Importin-α (ΔIBB; F), and Importin-α (ΔIBB) and full-length Importin-β (G). Bars, 10 µm. (H) Analysis of NuMA (GFP) and microtubule (X-rhodamine) fluorescence signals. Mean fluorescence signals under the different conditions shown in C–G were measured and plotted. SD was determined from data pooled from three independent experiments (n = 200 microtubules for each condition). Two-tailed Student t test; statistical differences: ***, P < 0.001; n.s., not significant. (I) A summary of microtubule binding of NuMA-tail II in the presence of Importin-α/-β. X-rhodamine–labeled microtubules were immobilized on the glass surface and NuMA-tail II-GFP (indicated as NuMA; GFP is shown as a black dot) in the presence of Importin-α and Importin-β was imaged using TIRF microscopy.
To further dissect the detailed molecular mechanism of how Importin-α/-β regulates microtubule binding of NuMA-tail II, we used total internal reflection fluorescence (TIRF) microscopy to analyze the interaction of NuMA-tail II with microtubules in the presence or absence of Importin-α or Importin-β (Fig. 3 I). Although NuMA-tail II-GFP could bind to microtubules immobilized on coverslips in the absence or presence of full-length Importin-α, there was a substantial reduction (∼50-fold) in GFP signal in the presence of full-length Importin-α/-β (Fig. 3, C–E and H). However, GFP intensities of RHK-NuMA-tail II-GFP along microtubules were comparable regardless of the presence or absence of Importin-α/-β (∼1.7-fold difference in GFP intensity in the absence and presence of Importin-α/-β) (Fig. S3, A–C). These results indicate that full-length Importin-α/-β complex suppresses the microtubule-binding activity of NuMA-tail II. Notably, in the presence of Importin-α (ΔIBB), as well as Importin-α (ΔIBB) together with full-length Importin-β, GFP signal along the microtubules was only slightly reduced (∼3-fold) compared with NuMA-tail II-GFP alone (Fig. 3, F–H). These results suggest that Importin-β, but not Importin-α, inhibits the interaction of NuMA-tail II with microtubules (Fig. 3 I).
Identification of an Importin-β–regulated microtubule-binding region in NuMA-tail II
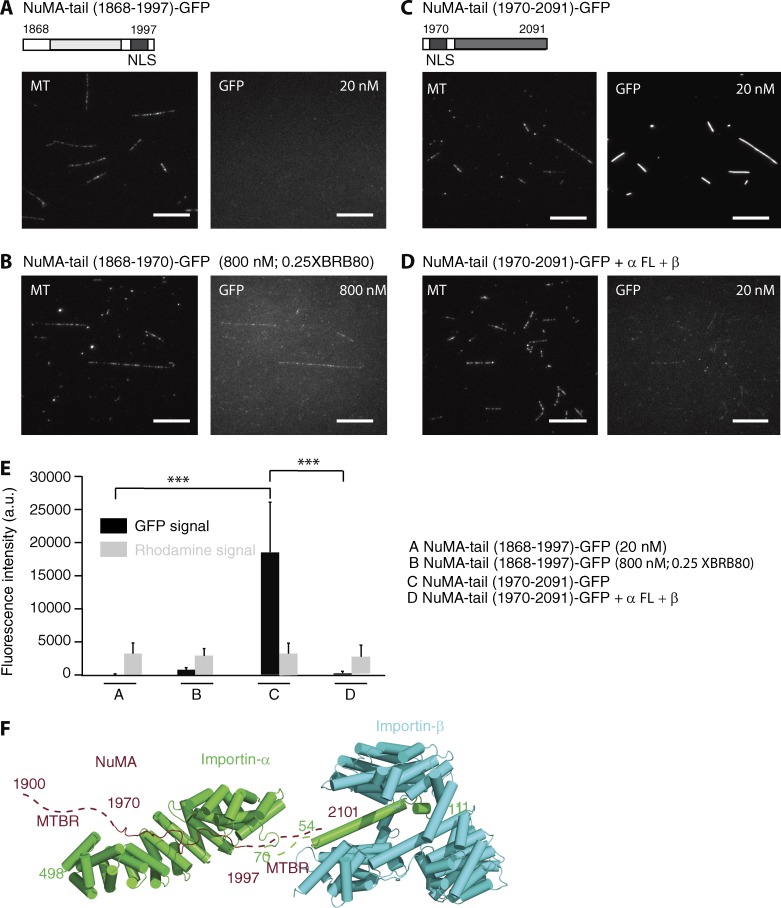
Our structural model showed that NuMA-NLS and Importin-α exhibit an antiparallel configuration (Fig. 1 G). Because Importin-β binds to the IBB domain of the N terminus of Importin-α, Importin-β may sterically hinder the microtubule-binding site existing in the C terminus of the NLS, thereby dramatically reducing the microtubule-binding affinity of NuMA. To examine this hypothesis, microtubule binding of two NLS-containing constructs (NuMA-tail [1868–1997] and NuMA-tail [1970–2091]) was examined by TIRF-based analysis (Fig. 4, A and C; and Fig. S3 D). Although NuMA-tail (1868–1997) showed no (20 nM) or weak (up to 800 nM) binding along microtubules (Fig. 4, A, B, and E), 20 nM of NuMA-tail (1970–2091) displayed strong GFP fluorescence signal along microtubules (Fig. 4, C and E). These results suggest that NuMA-tail (1970–2091) has a higher affinity for microtubules compared with that of NuMA-tail (1868–1997). Furthermore, the microtubule-binding activity of NuMA-tail (1970–2091) was reduced significantly (∼80-fold) in the presence of Importin-α/-β (Fig. 4, D and E). Together, these results suggest that a microtubule-binding region with relatively higher affinity for microtubules exists in the C terminus of the NuMA NLS and is regulated by Ran-governed Importin-β (Fig. 4 F).
Figure 4.
Importin-β regulates the microtubule-binding region that lies C-terminal to the NuMA NLS. GMPCPP-stabilized microtubules (X-rhodamine– and biotin-labeled) were incubated with NuMA-tail (1868–1997) and NuMA-tail (1970–2091) that contain microtubule-binding regions (light and dark gray) and NLS (black). (A and B) 20 nM and 800 nM of NuMA-tail (1868–1997) —GFP were examined under conditions of 1XBRB80 (A) and 0.25XBRB80 (B), respectively. (C and D) 20 nM of NuMA-tail (1970–2091)—GFP in the absence (C) or presence (D) of Importin-α/-β was analyzed. Bars, 10 µm. (E) Mean fluorescence signals under the different conditions shown in A–D were measured and plotted (n = 200 microtubules for each condition). SD was determined from data pooled from three independent experiments. Two-tailed Student t test; statistical differences: ***, P < 0.001. (F) The microtubule-binding region that lies C-terminal to the NLS (aa 1997–2101; brown dotted line) in NuMA is sterically blocked by Importin-β (light blue) that binds to the IBB domain of Importin-α (green; PDB code: 1QGK). Two microtubule-binding regions (MTBRs) are indicated.
In summary, we have determined a crystal structure of the Importin-α and NuMA-NLS dimer, showing that there is an antiparallel binding configuration and that three patches of the NuMA-NLS are involved in binding Importin-α. Although the interaction pattern between the NuMA-NLS and the minor NLS-binding site on Importin-α is similar to that of classic NLSs, the NuMA-NLS retains only two positively charged residues interacting with positions P2 and P3 of the major NLS-binding site and, as a result, it has a diminished interaction network compared with that of the classic NLS (a cluster of four positively charged Lys and Arg residues; Fig. S3, E and F). Interestingly, in addition to the two clusters of basic residues that bind to major and minor NLS-binding sites, His-1981 and Gln-1982 in the linker region form salt bridges with residues on Importin-α, resulting in an overall much more extensive interaction interface between NuMA-NLS and Importin-α compared with others (e.g., nucleoplasmin; Fontes et al., 2000; Fig. S3, E and F). Thus, NuMA-NLS exhibits a novel nonclassic interaction mode with Importin-α, which could increase selectivity for Importin-α.
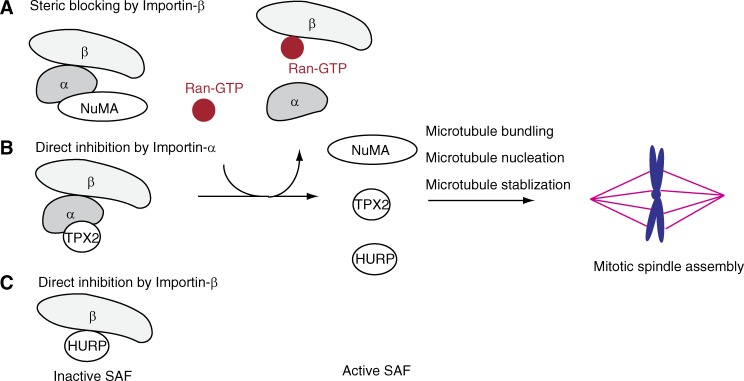
Furthermore, Importin-β significantly suppressed interactions of NuMA-tail II with microtubules in an Importin-α–dependent manner. We have demonstrated that the microtubule-binding region that lies C-terminal to the NLS has a greater microtubule affinity and is regulated by Importin-β. Although Gallini et al. (2016) also suggested the existence of a microtubule-binding region in the C terminus of NuMA-tail II NLS, our study further suggests that this microtubule-binding region could be Ran-pathway–regulated. Steric blockage of this microtubule-binding region by Importin-β could further prevent microtubule bundling induced by NuMA-tail II. NuMA-tail II has been suggested to induce microtubule aster formation in Xenopus egg extracts by mediating microtubule bundling (Merdes et al., 1996; Nachury et al., 2001; Wiese et al., 2001). Hence, Importin-β that suppresses NuMA-mediated microtubule bundles most likely inhibits microtubule aster assembly. Collectively, we provide mechanistic evidence for how the Ran-governed transport factors, Importin-α/-β, regulate interaction of NuMA and microtubules, which is required for the assembly of microtubule asters and mitotic spindles. Therefore, regulation of SAF activities by Importin-α/-β can be (1) by direct binding of Importin-α (e.g., TPX2; Schatz et al., 2003; Giesecke and Stewart, 2010), (2) by direct binding of Importin-β (e.g., HURP; Silljé et al., 2006), and (3) by steric blockage of importin-β mediated by Importin-α (e.g., NuMA; Fig. 5).
Figure 5.
Schematic for how Ran–GTP regulates SAF activities required for mitotic spindle assembly. SAFs (e.g., NuMA, TPX2, and HURP) are suppressed by the Ran-governed transport factors Importin-α and -β. Ran–GTP modulates interaction between Importins and SAFs. Free SAFs that regulate distinct microtubule behaviors control organization of the mitotic spindle.
Materials and methods
Protein expression and purification
NuMA-tail II (aa 1868–2091) was amplified and inserted intoa modified pET-DUET expression vectorthat contains TEV cleavage site and GFP sequence, allowing for expression of a C-terminal-GFP–tagged NuMA-tail II fragment. The RHK-NuMA-tail II-GFP mutant was obtained by PCR-based mutagenesis and verified by sequencing. Human full-length Importin-α and Importin-β were amplified by PCR and cloned into the pGEX-6p1 expression vector (GE Healthcare) to produce the N-terminal GST-fused proteins. Mouse Importin-α (ΔIBB) was cloned into pRSF-DUET with an N-terminal His tag. Whereas all NuMA-tail II, full-length Importin-α and full-length Importin-β were human proteins, mouse Importin-α (ΔIBB) proteins were used for biochemical and structural studies. Mouse Importin-α shares ∼95% identity with human Importin-α over the full protein sequence.
NuMA-tail II-GFP, RHK-NuMA-tail II-GFP, and Importin-α (ΔIBB) proteins were expressed in BL21(DE3) Rosetta (Novagen) Escherichia coli overnight at 18°C after induction with 0.5 mM IPTG. The culture was harvested by centrifugation and lysed according to the following procedure. The cell pellets were resuspended in lysis buffer (50 mM KH2PO4, 50 mM Na2HPO4, 150 mM NaCl, and 2 mM β-mercaptoethanol), disrupted by French press, and centrifuged at 15,000 g at 4°C for 20 min. The supernatant was incubated with Ni resin (Sigma) for 30 min, followed by a prewash with 25 mM imidazole. Proteins were eluted with the buffer containing 250 mM imidazole and dialyzed against low salt buffer (20 mM Hepes, pH 7.4, 50 mM NaCl, and 3 mM DTT) overnight. After dialysis, protein samples were loaded into a HiTrap Q HP column and eluted by a salt gradient. Then the samples were further purified by SEC (Superdex 200 16/60) and analyzed by SDS-PAGE. The protein fraction was collected, concentrated, and stored in a buffer consisting of 1× BRB80, 100 mM KCl, and 10% sucrose.
The same bacterial strain and protein overexpression strategy used to express NuMA-tail II was applied to express full-length Importin-α and Importin-β. Cells were harvested and suspended in buffer A (20 mM Hepes pH 7.4, 150 mM NaCl, and 3 mM DTT). After lysis of the cells, the clarified lysate was loaded onto a GST column (GE Healthcare). The GST-fused protein was then eluted by a buffer containing 20 mM Hepes, pH 7.4, 150 mM NaCl, 50 mM of reduced glutathione, and 3 mM DTT. The GST tag was removed by overnight digestion with PreScission protease at 4°C. The protein without the GST tag was further purified by HiTrap Q HP column and SEC (Superdex 200 16/60). Protein quality was analyzed and confirmed by SDS-PAGE.
GST-tagged NuMA-tail II (aa 1955–2046) and His-tagged Importin-α (ΔIBB) (aa 70–498) were coexpressed in BL21(DE3) Rosetta (Novagen) E. coli overnight at 18°C after induction with 0.5 mM IPTG. The cell pellet was disrupted by French press and clarified by centrifugation at 15,000 g at 4°C for 20 min. After cell lysis, the protein complex was purified using a series of chromatographic methods, including GST resins, HiTrap S column, and SEC (Superdex 200). Both GST and His tags were removed by overnight incubation with PreScission protease at 4°C. Purified protein fractions were collected and concentrated to 25 mg/ml using Amicon Ultra 15 (Millipore) for crystallization.
Protein crystallization and structure determination
The NuMA (aa 1955–2046)–Importin-α (aa 70–498) complex was purified and screened for crystallization. Crystals were grown in a 25-mg/ml protein concentration at 20°C in hanging drops containing 1 µl of the protein and 1 µl of a reservoir solution consisting of 1 M KCl, 0.8 M ammonium sulfate, and 100 mM Hepes, pH 7.0. Crystals grew to their maximum size in 1 wk. Crystal form was in the orthorhombic space group P212121 and contained a heterodimer in the asymmetric unit. For cryoprotection, crystals were stabilized in 1 M KCl, 0.8 M ammonium sulfate, 100 mM Hepes pH 7.0, and 20% (vol/vol) glycerol. Data were collected at the Structural Biology Resource Center at The Rockefeller University and beamline BL13B1 and BL13C1 at the National Synchotron Radiation Research Center (Taiwan). X-ray intensities were processed using HKL2000 (Otwinowski and Minor, 1997), and molecular replacement was performed by Phaser in the Phenix software using Importin-α (PDB code: 1IQ1) as a research model (McCoy et al., 2007).
The initial model was built into the electron density map of the orthorhombic crystal form O (Jones et al., 1991) and refined using the Crystallography and NMR System (Brünger et al., 1998). The final model was refined to a resolution of 2.4 Å with an Rwork of 19.8% and an Rfree of 22.5%. An electron density for NuMA NLS could be observed and assigned unambiguously from residues 1970 to 1996. No clear electron density outside this region could be identified. The residues outside the aa 1970–1996 region are presumed to be disordered and so have been omitted from the final model. The stereochemical quality of the final model was assessed with PROCHECK (Laskowski et al., 1993). There are no residues in the disallowed region of the Ramachandran plot. Data collection and refinement statistics are shown in Table S1. Coordinates and structure factors have been deposited in the Protein Data Bank (PDB code: 5GXW).
ITC
Binding affinities of full-length Importin-α, full-length Importin-β, and Importin-α (ΔIBB) to WT or mutant NuMA-tail II-GFP were measured by ITC (MicroCal iTC200). All proteins were dialyzed against ITC buffer (20 mM Hepes pH 7.4, 200 mM NaCl, 1 mM DTT). Protein concentrations were determined by measuring absorbance at 280 nm using a spectrophotometer. NuMA-tail II-GFP was stored in a sample cell, and importins were injected into the cell by syringe. ITC was performed at 25°C. To remove the heat effects produced in the reference titration (Importin-α/Importin-β interaction), heat changes of WT and the RHK mutant in the presence of Importin-α and -β (Fig. 1 F and Fig. S2 D) were subtracted from the reference titration data (buffer control; Fig. S1 B) before curve fitting. The heat changes were fitted using a one-sites binding model.
Microtubule aster assembly in Xenopus egg extracts
Cell-free extracts from Xenopus eggs, arrested in metaphase of meiosis II, were prepared essentially as described in Desai et al., 1999. In brief, for each extract preparation, two to three female frogs were primed using pregnant mare serum gonadtropin (hor-272; Prospec) and induced to ovulate using human chorionic gonadotropin (CG-10; Sigma). Eggs were collected ∼16 h after the last hormone injection, washed with MMR (5 mM Na-Hepes, pH 7.85, 0.1 mM EDTA, 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, and 2 mM CaCl2), treated with 2% cysteine solution for dejellying, and then rinsed with buffers in the following order: XB (10 mM K-Hepes, 100 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, and 50 mM sucrose, pH 7.7), CSF-XB (XB plus 1 mM MgCl2, and 5 mM EGTA), and CSF-XB + PI (CSF-XB plus 10 µg/ml each of the protease inhibitors leupeptin, pepstatin A, and chymostatin). The rinsed eggs were supplemented with 10 µg/ml cytochalasin D, packed in centrifuge tubes (344057; Beckman) using a tabletop centrifuge (5702R; Eppendorf), and then spin-crushed using a SW-55 rotor at 10,000 g for 15 min at 16°C (Optima XE-90; Beckman). After centrifugation, a cytoplasmic fraction of the crushed eggs was extracted using a 16-G needle and supplemented with Energy mix (75 mM creatine phosphate, 1 mM ATP, and 1 mM MgCl2). Protease inhibitors (leupeptin, pepstatin A, and chymostatin) and cytochalasin D (each 10 µg/ml) were also added to the fraction. The entire procedure was performed in a temperature-controlled room at 18°C. The prepared extracts were stored in 1.5-ml test tubes on ice until required and used within 6 h.
For microtubule aster assembly, each reaction was prepared using 20 µl of extracts supplemented with X-rhodamine-labeled tubulin (10 µg/ml). NuMA-tail II proteins (5 µM) were then added to reactions and incubated at 18°C. Negative and positive control samples were prepared with BRB80 and with 20 µM Taxol in BRB80 plus 0.5% DMSO, respectively. At 20 min and 40 min after reagent addition, 4 µl of each extract reaction was squashed between a clean slide and a coverslip (Matsunami), sealed using VaLaP (Desai et al., 1999), and then immediately imaged using an epi-fluorescence microscope (Ti; Nikon) equipped with a 60× objective lens (1.20 NA, Plan Apo WI; Nikon), an excitation light illuminator (Intensilight; Nikon), a fluorescence filter set (TxRed-4040C-000; Semrock), a motorized XY stage (MS-2000; Applied Scientific Instruments), and a sCMOS camera (Neo; Andor). Image acquisition was performed by raster-scanning ∼500 × 500-µm areas of each squashed extract and then stitching them into single composite images using image acquisition software (NIS-Elements, ver. 4.2; Nikon). The number of microtubule asters was counted using an object detection algorithm in the software, after calibrating the detection parameter values using positive control samples (i.e., Taxol asters).
Statistical analysis
Statistical analyses were calculated using Prism (version 6.0c). Figure legends detail the n values and error bars for each experiment. Statistical differences for aster formation assays (Fig. 2 B), microtubule bundling assays (Fig. 2, E and H; 30-min incubation), and microtubule binding assays (Figs. 3 H, 4 E, and S3 C) were evaluated with two-tailed Student t tests. For the t test, data distribution was assumed to be normal, but this was not formally tested. When data distribution was not normal, Mann-Whitney U tests were performed for microtubule bundling assays (Fig. 2, E and H; 60-min incubation). Notation for p-values is as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Microtubule cosedimentation assay
Taxol-stabilized microtubules were polymerized from purified precleared bovine tubulin. Microtubules (6 µM) were incubated with ∼2 µM NuMA-tail II-GFP in the absence or presence of full-length Importin-α and Importin-β (4 µM) for 20 min at room temperature in a buffer containing 0.5 × BRB80, 40 mM KCl, 40 µM Taxol, 2 mM DTT, and 0.25 mg/ml BSA. Subsequently, reaction solutions were subjected to sedimentation in a TLA 120.1 rotor (Beckman Coulter) at 75,000 rpm for 10 min at 27°C. The proteins in the pellet and supernatant were analyzed by SDS-PAGE, followed by Coomassie blue staining. GFP bands were detected by Western blotting using anti-GFP antibody (a gift from T.M. Kapoor, Rockefeller University, New York, NY, used at 1:2,500).
Microtubule bundling visualized by TIRF microscopy
Microtubules were polymerized from a solution in the presence of X-rhodamine–labeled tubulin and guanalyl-(α,β)-methylene diphosphate (GMPCPP). Microtubule solution (1 µl) was incubated with 400 nM WT or RHK-NuMA-tail II-GFP in the presence or absence of 4 µM full-length Importin-α and Importin-β for 30 and 60 min at room temperature. Samples (2 µl) were spotted onto glass slides and fixed for 5 min with 1% formaldehyde. Microtubules were visualized by a TIRF illumination system, and X-rhodamine intensities were determined using ImageJ.
Microtubule-binding TIRF analysis of GFP-tagged NuMA-tail II
TIRF fluorescence images were collected using an Eclipse microscope with a Ti-E TIRF module (Nikon) and an iXON Ultra 897 EMCCD camera (Andor Technology Ltd.). The microscope was equipped with a two-color, near-simultaneous TIRF illumination system. Samples were visualized using a 100× 1.45 oil immersion objective lens. GFP was excited using a 488-nm laser (Coherent) and X-rhodamine with a 561-nm laser (Cobolt).
Microtubules were polymerized from a solution that contained unlabeled, X-rhodamine–labeled, and biotin-labeled tubulin in a ratio of 10:1:0.7 at 37°C in the presence of GMPCPP. Flow chambers were assembled with biotin–polyethylene glycol–coated coverslips. The chamber was sequentially filled with 0.5 mg/ml α-casein, 0.2 mg/ml NeutrAvidin, and biotinylated microtubules in 1× BRB80, and 20 µM Taxol. Next, GFP-labeled WT or RHK-NuMA-tail II (20 nM) was added to the chamber in 1× BRB80 supplemented with 20 µM Taxol, 0.5 mg/ml α-casein, 10% sucrose, 2 mM DTT, 200 µg/ml glucose oxidase, 35 µg/ml catalase, and 4.5 µg/ml glucose. To examine whether or not importins inhibit the microtubule binding of NuMA-tail II, 200 nM importins was preincubated for 10 min with GFP-labeled NuMA-tail II under the same experimental conditions that were used for NuMA-tail II alone. The chamber was then filled with the solution and sealed. X-rhodamine and GFP intensities were determined using ImageJ.
Online supplemental material
Fig. S1 shows biochemical characterization of the regions of the Importin-α–NuMA-tail II complex suitable for crystallographic studies. Fig. S2 shows the comparison of NuMA-NLS with other classic NLSs, and a triple mutation (RHK-NuMA-tail II) abolishes interaction with Importin-α analyzed by ITC. Fig. S3 demonstrates RHK mutant/microtubule interaction is not regulated by importins, and illustrates the interaction network of Importin-α with NuMA. Table S1 shows crystallographic data collection and refinement statistics for the structure of Importin-α–NuMA-tail II complex.
Supplementary Material
Acknowledgments
We thank the technical services provided by the Synchrotron Radiation Protein Crystallography Facility of the National Core Facility Program for Biotechnology, Ministry of Science and Technology and the National Synchrotron Radiation Research Center, a national user facility supported by the Ministry of Science and Technology of Taiwan, China.
This work was supported by the Ministry of Science and Technology, Taiwan (grants 106-2311-B-001-038-MY3 and 105-2311-B-001-043-MY2), and the Japan Society for the Promotion of Science KAKENHI (grant JP16748896).
The authors declare no competing financial interests.
Author contributions: C.-C. Chang and T.-L. Huang performed biochemical experiments. Y. Shimamoto carried out experiments involving Xenopus egg extracts. C.-C. Chang, T.-L. Huang, Y. Shimamoto, S.-Y. Tsai, and K.-C. Hsia analyzed data. All authors discussed the results and helped write the manuscript. K.-C. Hsia directed the project and prepared the manuscript.
Footnotes
Abbreviations used:
- GMPCPP
- guanalyl-(α,β)-methylene diphosphate
- IBB
- importin-β binding
- ITC
- isothermal titration calorimetry
- NuMA
- nuclear mitotic apparatus protein
- RHK
- R1975A/H1981A/K1990A
- SAF
- spindle assembly factor
- SEC
- size exclusion chromatography
- TIRF
- total internal reflection fluorescence
- TPX2
- targeting protein for Xklp2
References
- Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. . 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921. 10.1107/S0907444998003254 [DOI] [PubMed] [Google Scholar]
- Cook A., Bono F., Jinek M., and Conti E.. 2007. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76:647–671. 10.1146/annurev.biochem.76.052705.161529 [DOI] [PubMed] [Google Scholar]
- Desai A., Murray A., Mitchison T.J., and Walczak C.E.. 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412. 10.1016/S0091-679X(08)61991-3 [DOI] [PubMed] [Google Scholar]
- Fontes M.R., Teh T., and Kobe B.. 2000. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol. 297:1183–1194. 10.1006/jmbi.2000.3642 [DOI] [PubMed] [Google Scholar]
- Forbes D.J., Travesa A., Nord M.S., and Bernis C.. 2015. Nuclear transport factors: global regulation of mitosis. Curr. Opin. Cell Biol. 35:78–90. 10.1016/j.ceb.2015.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallini S., Carminati M., De Mattia F., Pirovano L., Martini E., Oldani A., Asteriti I.A., Guarguaglini G., and Mapelli M.. 2016. NuMA phosphorylation by Aurora-A orchestrates spindle orientation. Curr. Biol. 26:458–469. 10.1016/j.cub.2015.12.051 [DOI] [PubMed] [Google Scholar]
- Giesecke A., and Stewart M.. 2010. Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-alpha. J. Biol. Chem. 285:17628–17635. 10.1074/jbc.M110.102343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L., and Merdes A.. 2002. Direct binding of NuMA to tubulin is mediated by a novel sequence motif in the tail domain that bundles and stabilizes microtubules. J. Cell Sci. 115:1815–1824. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W., and Kjeldgaard M.. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 47:110–119. 10.1107/S0108767390010224 [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., and Thornton J.M.. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291. 10.1107/S0021889892009944 [DOI] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., and Read R.J.. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J.D., and Cleveland D.W.. 1996. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 87:447–458. 10.1016/S0092-8674(00)81365-3 [DOI] [PubMed] [Google Scholar]
- Nachury M.V., Maresca T.J., Salmon W.C., Waterman-Storer C.M., Heald R., and Weis K.. 2001. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 104:95–106. 10.1016/S0092-8674(01)00194-5 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., and Minor W.. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326. 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Schatz C.A., Santarella R., Hoenger A., Karsenti E., Mattaj I.W., Gruss O.J., and Carazo-Salas R.E.. 2003. Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J. 22:2060–2070. 10.1093/emboj/cdg195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silljé H.H., Nagel S., Körner R., and Nigg E.A.. 2006. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 16:731–742. 10.1016/j.cub.2006.02.070 [DOI] [PubMed] [Google Scholar]
- Wiese C., Wilde A., Moore M.S., Adam S.A., Merdes A., and Zheng Y.. 2001. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 291:653–656. 10.1126/science.1057661 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.