Rowe and colleagues preview work from Cuellar et al. that identifies SETDB1 as a repressor of immunostimulatory retrotransposons in leukemia.
Abstract
Cancer cells thrive on genetic and epigenetic changes that confer a selective advantage but also need strategies to avoid immune recognition. In this issue, Cuellar et al. (2017. J. Cell Biol. https://doi.org/10.1083/jcb.201612160) find that the histone methyltransferase SETDB1 enables acute myeloid leukemia cells to evade sensing of retrotransposons by innate immune receptors.
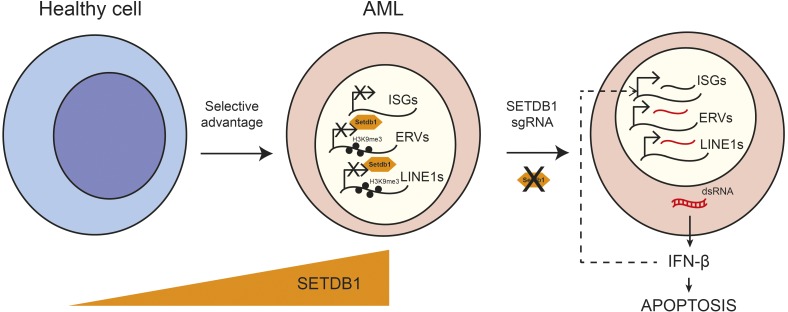
Cancer cells frequently exhibit dysregulation of the enzymes that control epigenetic modifications of the genome. For example, three recent studies have suggested that DNA methyltransferases might play a key role in enabling cancer cells to evade recognition by the immune system by limiting the expression of endogenous retrotransposons (Chiappinelli et al., 2015; Roulois et al., 2015; Goel et al., 2017). Pharmacological inhibition of DNA methylation could induce the expression of retrotransposons in cancer cells, which trigger an antiviral response that leads to tumor cell death. However, the exact mechanisms used by cancer cells to prevent the production of these retrotransposons are unknown. In this issue, Cuellar et al. report that SETDB1 is a gatekeeper of tumor survival that enables acute myeloid leukemia (AML) cells to evade innate immune sensing of retrotransposons. An elevated level of SETDB1 in cancer cells promotes the formation of heterochromatin at retrotransposons. This in turn prevents transcription of retrotransposon-derived double-stranded RNA (dsRNA), its downstream recognition by cytosolic RNA sensors, and the activation of a type I IFN response (Fig. 1).
Figure 1.
SETDB1 enables AML cells to evade innate immune sensing of retrotransposons. Cancer cells elevate their levels of SETDB1, which promotes the formation of H3K9me3-based heterochromatin at retrotransposons. This in turn prevents transcription of retrotransposon-derived dsRNA and its downstream recognition by cytosolic RNA sensors. Genetic ablation of SETDB1 leads to a type I IFN response, the subsequent activation of ISGs, and apoptosis. LINE1, long interspersed nuclear element 1; sgRNA, single-guide RNA.
Retrotransposons are a class of transposable elements that includes endogenous retroviruses (ERVs) and long interspersed nuclear elements (LINEs). Because of their viral origin, these elements can produce nucleic acids that can be distinguished as “nonself” rather than “self” by the immune system, which recognizes pathogen-specific molecules (pathogen-associated molecular patterns [PAMPs]). For example, LINEs have bidirectional promoters and can produce dsRNA, and ERVs produce cytoplasmic DNA during reverse transcription. Both of these PAMPs activate IFN by binding to distinct pattern-recognition receptors (PRRs; Zeng et al., 2014). Retrotransposon-derived nucleic acids have been proposed to modulate cancer survival and tumor immune responses. Indeed, several pioneering papers have recently revealed ERVs to be central to antitumor immunity through the sensing of dsRNA produced from them and the subsequent triggering of innate and adaptive immune responses (Chiappinelli et al., 2015; Roulois et al., 2015; Goel et al., 2017). These papers show that immunostimulatory ERVs are usually under epigenetic control that includes DNA methylation and that they can be reactivated with standard cancer drugs, such as those based on 5-azacytidine, which block DNA methylation, or by CDK4/6 inhibitors, which indirectly block DNA methylation (Chiappinelli et al., 2015; Roulois et al., 2015; Goel et al., 2017). However, although it is known that ERVs are controlled at the chromatin level in cancer cells, the epigenetic factors responsible for this immune masking of retrotransposons remain obscure.
Cuellar et al. (2017) uncovered a role for SETDB1 in preventing retrotransposon recognition through an unbiased CRISPR/Cas9 screen that focused on 350 epigenetic modifiers to identify factors essential for the survival and propagation of a human AML cell line. They show that SETDB1 is overexpressed in a broad range of cancers, including solid tumors, and that it is required for survival in five out of seven tested human AML cell lines. A recent study also shows that colorectal cancer cells are dependent on SETDB1 to survive a lethal dose of kinase inhibitors (Guler et al., 2017). Genetic ablation of SETDB1 leads to apoptosis, which is preceded by a striking up-regulation of IFN-stimulated genes (ISGs) in multiple cancer cell lines. Cuellar et al. (2017) show that this induction of IFN production by depletion of SETDB1 correlates with increased expression of some ERVs, LINEs, and satellite repeats and a slight decrease in global H3K9 trimethylation (H3K9me3) levels (including at ERVs and LINEs), as well as at some zinc finger genes that are known targets of SETDB1. Of note, not all retrotransposons are affected because there is no change in H3K9me3 at HERVH integrants, for example. Interestingly, Cuellar et al. (2017) find that the loss of H3K9me3 after SETDB1 ablation occurs only in concert with neighboring unmodified, rather than acetylated, H3K14. This supports a function for SETDB1 in inactive chromatin and indicates its potential collaboration with other cellular factors to target specific loci. Cuellar et al. (2017) verify that at key ISGs (of the IFN-induced protein with tetratricopeptide repeats or IFIT family), H3K9me3 is not affected by SETDB1 knockout, suggesting that ISGs are activated after a bona fide immune response to the expression of retrotransposons. Most remarkably, Cuellar et al. (2017) show that SETDB1 knockout cells produce dsRNA, which is readily detected by antibody staining, and that ERVs and LINEs exhibit the bidirectional transcription required for dsRNA formation. Finally, the authors demonstrate that SETDB1-mediated ISG induction and cell death depend on RNA sensing by the cytosolic PRRs MDA5 and RIG-I.
Although Cuellar et al. (2017) provide evidence that SETDB1-regulated retrotransposons are involved in this mechanism of innate sensing, it is unclear which ones are most relevant in producing immunostimulatory nucleic acids. With ERVs and LINEs together comprising at least 25% of the human genome, this is a difficult yet important question to pursue in future work. Another interesting possibility raised by this work is that SETDB1 may control natural immune responses at the chromatin level through the regulated reactivation of retrotransposons in certain contexts, as ERVs can enhance T-independent B cell responses (Zeng et al., 2014) and SETDB1 prevents ERV expression by B cells (Collins et al., 2015). Further work will also be necessary to determine whether SETDB1, like TREX1, is required to prevent downstream sensing of DNA produced from retrotransposons that are competent for reverse transcription through the cGAS–STING pathway (Zeng et al., 2014; Thomas et al., 2017), as well as its role defined here in preventing RNA sensing (Cuellar et al., 2017).
The regulation of immunity by retrotransposons is an emerging field in which the identification by Cuellar et al. (2017) of SETDB1 as a key regulator of ERVs in cancer cells paves the way for further research on this topic. Importantly, it builds on previous landmark work that has defined SETDB1 as a director of the epigenetic silencing of ERVs in embryonic stem cells, murine embryonic fibroblasts, and B cells (Matsui et al., 2010; Collins et al., 2015; Tie and Rowe, 2017). However, this ability of SETDB1 to repress ERVs in these noncancerous cell types raises the important question of whether SETDB1 can be selectively targeted by novel drugs without off-target effects on the genome and epigenome. New SETDB1-targeting cancer drugs are an important area of development that could prove valuable in a range of cancers, all bearing the hallmark of SETDB1 dependence. Future work into the mechanism of SETDB1 repression of retrotransposons in cancer cells will reveal whether it involves the same components that are required for ERV silencing early in development (Tie and Rowe, 2017), or perhaps components of the RNA interference machinery as proposed by Cuellar et al. (2017), or both. An understanding of all the epigenetic and related factors necessary for this mechanism should lead to the identification of additional, perhaps more specific, druggable therapeutic targets.
Acknowledgments
We regret not being able to cite more articles because of reference limitations. We thank Pierre V. Maillard for reading through the text.
Research in the Rowe laboratory is funded by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and Royal Society grant 101200/Z/13/Z and a European Research Council starting grant (678350) awarded to H.M. Rowe.
The authors declare no competing financial interests.
References
- Chiappinelli K.B., Strissel P.L., Desrichard A., Li H., Henke C., Akman B., Hein A., Rote N.S., Cope L.M., Snyder A., et al. . 2015. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 162:974–986. 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P.L., Kyle K.E., Egawa T., Shinkai Y., and Oltz E.M.. 2015. The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc. Natl. Acad. Sci. USA. 112:8367–8372. 10.1073/pnas.1422187112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar T.L., Herzner A.-M., Zhang X., Goyal Y., Watanabe C., Friedman B.A., Janakiraman V., Durinck S., Stinson J., Arnott D., et al. . 2017 Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia. J. Cell Biol. 10.1083/jcb.201612160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel S., DeCristo M.J., Watt A.C., BrinJones H., Sceneay J., Li B.B., Khan N., Ubellacker J.M., Xie S., Metzger-Filho O., et al. . 2017. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 548:471–475. 10.1038/nature23465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler G.D., Tindell C.A., Pitti R., Wilson C., Nichols K., KaiWai Cheung T., Kim H.J., Wongchenko M., Yan Y., Haley B., et al. . 2017. Repression of Stress-Induced LINE-1 Expression Protects Cancer Cell Subpopulations from Lethal Drug Exposure. Cancer Cell. 32:221–237.e13. 10.1016/j.ccell.2017.07.002 [DOI] [PubMed] [Google Scholar]
- Matsui T., Leung D., Miyashita H., Maksakova I.A., Miyachi H., Kimura H., Tachibana M., Lorincz M.C., and Shinkai Y.. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 464:927–931. 10.1038/nature08858 [DOI] [PubMed] [Google Scholar]
- Roulois D., Loo Yau H., Singhania R., Wang Y., Danesh A., Shen S.Y., Han H., Liang G., Jones P.A., Pugh T.J., et al. . 2015. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell. 162:961–973. 10.1016/j.cell.2015.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C.A., Tejwani L., Trujillo C.A., Negraes P.D., Herai R.H., Mesci P., Macia A., Crow Y.J., and Muotri A.R.. 2017. Modeling of TREX1-Dependent Autoimmune Disease using Human Stem Cells Highlights L1 Accumulation as a Source of Neuroinflammation. Cell Stem Cell. 21:319–331.e8. 10.1016/j.stem.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie C.H., and Rowe H.M.. 2017. Epigenetic control of retrotransposons in adult tissues: implications for immune regulation. Curr. Opin. Virol. 25:28–33. 10.1016/j.coviro.2017.06.007 [DOI] [PubMed] [Google Scholar]
- Zeng M., Hu Z., Shi X., Li X., Zhan X., Li X.D., Wang J., Choi J.H., Wang K.W., Purrington T., et al. . 2014. MAVS, cGAS, and endogenous retroviruses in T-independent B cell responses. Science. 346:1486–1492. 10.1126/science.346.6216.1486 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]