ABSTRACT
Tumor infiltrating lymphocytes (TIL) in Epstein-Barr virus (EBV)-associated/microsatellite-unstable (MSI) gastric carcinomas (GC) constitute immune-active principal cellular components of tumor microenvironment and contribute to better prognosis. With the remarkable success of cancer immunotherapies, there is an urgent need for a comprehensive understanding of tumor-immune interactions in patients with GC in the context of host immune response. To identify GC subtype-specific immune response gene set, we tested differentially expressed genes for MSI and EBV+ GC subtypes in randomly selected test set (n = 278) in merged ACRG-SMC microarray and TCGA RNA sequencing data set. We identified Host ImmunE Response index (HIERÏ) consisting of 29 immune genes classifying GC patients into robust 3 groups with prognostic significance. Immune-high cluster 1 was enriched with PD-L1High/EBV+/MSI/TILHigh with the best clinical outcome while immune-low cluster 3 displayed worst outcome and exemplified with PD-L1Low/EBV-/MSS. The results were validated in the same cohort (n = 279) and independent cohort (n = 181) with RNA from formalin-fixed paraffin-embedded (FFPE) tissue. Unexpectedly, nearly half of GC in cluster 1 were EBV-/MSS and 10% of cluster 3 GC were EBV+/MSI GC patients, suggesting that in addition to EBV+/MSI GC subtypes, EBV-/MSS subtype also constitutes almost half of high immune cluster and would be a good candidate for immune checkpoint inhibitor therapy. In contrary, almost 10% of EBV+/MSI GC patients may not respond to immune checkpoint inhibitor therapy. Thus, our HIERÏ gene signature demonstrates the potential to subclassify tumor immunity levels, predict prognosis and help immunotherapeutic decisions.
KEYWORDS: biomarker, EBV, gastric cancer, gene signature, immune response, prognosis, PD-L1, prognosis, survival, tumor infiltrating lymphocyte
Abbreviations
- AJCC
American joint committee on cancer
- CA
conventional adenocarcinoma
- CD
clusters of differentiation
- CI
confidence interval
- CLR
carcinoma with Crohn-like reaction
- DFS
disease free survival
- EBER
Epstein-Barr virus-encoded small RNA
- EBV
Epstein-Barr virus
- EBV+ GC
Epstein-Barr virus-associated gastric cancer
- FFPE
formalin-fixed paraffin-embedded
- GC
gastric cancer
- HR
hazard ratio; immunohistochemical staining
- NK cell
natural killer cell
- OS
overall survival
- PD-1
programmed death receptor-1
- PD-L1
programmed death receptor-ligand 1
- TCGA
The Cancer Genome Atlas
- TCR
T cell receptor
- TIL
tumor infiltrating lymphocytes
- CIMP
CpG island methylator phenotype
- ACRG-SMC
Asian Cancer Research Group-Samsung Medical Center
Introduction
Immune infiltrates are heterogeneous between tumor types; variable numbers of tumor-infiltrating lymphocytes (TIL) are found in different tumors of the same type and are very diverse from patient to patient.1 Immune-active or -suppressive tumoral microenvironment (TM) plays critical role in patient's outcome. TIL constitutes the principal cellular component of immune active TM and contributes to improved survival of cancer patients. Therefore, in addition to genomic features of tumors, TM represents promising candidates for predictive and prognostic biomarkers.2
Gene expression profiling of cancer has resulted in the identification of molecular subtypes and the development of models for prediction prognosis and has enriched our knowledge of the molecular pathways of tumorigenesis.3 In gastric cancer (GC), 4 molcular subtypes based on genetic alterations by The Cancer Genome Atlas (TCGA)4 and expression data by Asian Cancer Research Group-Samsung Medical Center (ACRG-SMC)5 have been proposed. Lin et al. analyzed >1600 GCs and found that gene signatures related to inflammation and immunity differ significantly between Asian and non-Asian GCs, with T-cell pathways preferentially associated with non-Asian GC.6 These results suggest that immune signatures also vary geographically within the same tumor.
Epstein-Barr virus (EBV) infection and mutually exclusive microsatellite instability (MSI) are frequently accompanied with TIL and have been known as significant favorable prognostic markers in patients with GC.5,7 Patients with EBV+ GC have more favorable survival than those with EBV- GC possibly caused by immune responses directed against EBV-related proteins expressed by tumor cells. In EBV+ GC, genes involving in cytokine (chemokine) pathways were significantly deregulated8 and programmed cell death ligand 1 (PD-L1) expression is remarkable increased via multiple mechanisms.9 In MSI GC, abundant PD-L1 expression in cases associated with TIL via increased host immune response correlated with favorable survival.11
With the remarkable success of cancer immunotherapies, there is an urgent need for a comprehensive understanding of tumor-immune interactions. Efforts have been made to elucidate the tumor-immune interactions and provide prognostic predictors.10 Recently, Rooney et al.11 quantified the cytolytic activity of the local immune infiltrate across 18 tumor types and identified marked increases in EBV+ GC compared with EBV- GC (possibly due to PDL1 amplifications) and in colorectal tumors with MSI compared with MSS. Given that natural anti-tumor immunity requires cytolytic immune response, increased cytolytic activities observed in EBV (due to virus) and MSI (due to neoantigens) GC may serve as a surrogate marker of cytolytic host immune attack associated with TIL and better prognosis. To identify Host ImmunE Response index (HIERÏ) in GC, we analyzed expression data set of TCGA RNA sequencing and ACRG-SMC microarray results to obtain differentially expressed genes (DEG) in GCs with MSI and EBV with better prognosis.
Results
Host ImmunE Response index (HIERÏ) in gastric cancer
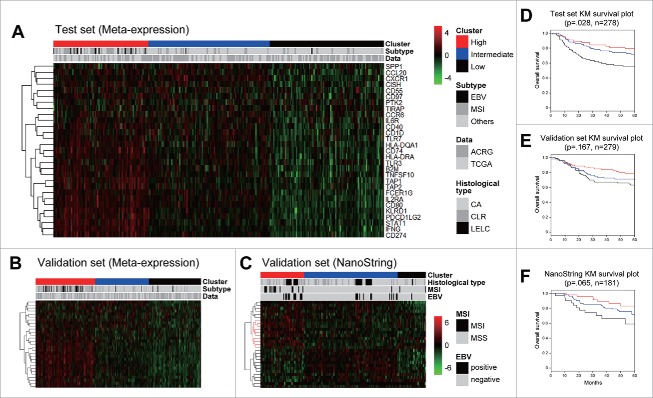
To obtain differentially expressed immune-related genes in GC with MSI and EBV, and with better prognosis, we performed network analysis assigning GC subtype shown in Fig. 1 and analyzed functional modules (significantly differentially expressed subnetwork), and defined HIERÏ consisting of 29 genes [SPP1, CCL20, CXCR1, CISH, CD55, ADGREE5 (CD97), PTK2, TIRAP, CCR6, IL6R, CD40, CD1D, TLR7, HLA-DQA1, CD74, HLA-DRA, TLR3, B2M, TNFSF10, TAP1, TAP2, FCER1G, IL2RA, CD80, KLRD1, PDCD1LG2, STAT1, IFNγ, CD274 (PD-L1)]. All clustering results of meta-expression were absent to platform bias, and microarray and RNA-Sequencing (RNA-Seq) samples were uniformly distributed across clusters.
Figure 1.
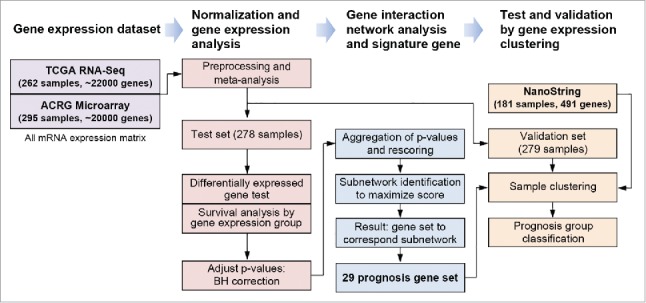
Overall process of gene expression analysis composed of data set preparation, and normalization, gene interaction network-based analysis to identify gene set, test and validation by clustering using signature gene.
To test prognosis classification using HIERÏ, we clustered distinct 3 sample groups for each test set (n = 278) and validation set (n = 279) randomly selected from merged meta-expression data set (n = 557). Additional clustering for validation was performed in an independent validation set with different test platform in which data was obtained from nCounter® gene expression assay using RNA extracted from FFPE tissue (n = 181) (Fig. 2). HIERÏ significantly enriched in multiple immune response pathways of immune system (q-value 0.03), PD-L1 signaling, downstream TCR signaling pathway, and costimulation by the CD28 family (q-value 0.05) (Supplementary Table 1).
Figure 2.
Gene expression clustering and its Kaplan-Meier survival plot. Clustering was performed in 3 data sets: (A) test set of meta-expression data, (B) validation set of meta-expression, and (C) Independent NanoString validation set, and its survival was investigated.
Our HIERÏ classified independent validation cohort with 181 patients into robust 3 groups with prognostic significance and denoted close association with clinical characteristics. All clusters and related clinical features were summarized in Table 1.
Table 1.
Three clusters of HIERÏ immune gene signatures and their characteristics in 181 independent validation cohort.
| Immune-high (n = 48) n (%) | Intermediate (n = 102) n (%) | Immune-low (n = 31) n (%) | p value | total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| gender | .144 | |||||||||
| male | 39 | (81.3) | 67 | (65.7) | 21 | (67.7) | 127 | |||
| female | 9 | (18.8) | 35 | (34.3) | 10 | (32.3) | 54 | |||
| location | .499 | |||||||||
| cardia | 2 | (4.2) | 9 | (8.8) | 3 | (9.7) | 14 | |||
| body | 26 | (54.2) | 62 | (60.8) | 11 | (35.5) | 99 | |||
| antrum | 19 | (39.6) | 19 | (18.6) | 17 | (54.8) | 55 | |||
| whole/multiple | 1 | (2.1) | 12 | (11.8) | 0 | (0.0) | 13 | |||
| histologic type by Lauren | .002 | |||||||||
| intestinal | 23 | (47.9) | 25 | (24.5) | 16 | (51.6) | 64 | |||
| diffuse | 14 | (29.2) | 61 | (59.8) | 12 | (38.7) | 87 | |||
| mixed/indeterminate | 11 | (22.9) | 16 | (15.7) | 3 | (9.7) | 30 | |||
| histologic type by immune response | .043 | |||||||||
| CA | 23 | (47.9) | 65 | (63.7) | 25 | (80.6) | 113 | |||
| CLR | 11 | (22.9) | 14 | (13.7) | 1 | (3.2) | 26 | |||
| LELC | 14 | (29.2) | 23 | (22.5) | 5 | (16.1) | 42 | |||
| EBV | .033 | |||||||||
| negative | 37 | (77.1) | 90 | (88.2) | 30 | (96.8) | 157 | |||
| positive | 11 | (22.9) | 12 | (11.8) | 1 | (3.2) | 24 | |||
| MSI | <.001 | |||||||||
| MSS | 40 | (83.3) | 102 | (100.0) | 29 | (93.5) | 171 | |||
| MSI | 8 | (16.7) | 0 | (0.0) | 2 | (6.5) | 10 | |||
| TNM | .434* | |||||||||
| I | 9 | (18.8) | 27 | (26.5) | 5 | (16.1) | 41 | |||
| II | 19 | (39.6) | 28 | (27.5) | 9 | (29.0) | 56 | |||
| III | 19 | (39.6) | 37 | (36.3) | 16 | (51.6) | 72 | |||
| IV | 1 | (2.1) | 10 | (9.8) | 1 | (3.2) | 12 | |||
CA, conventional adenocarcinoma; CLR, carcinoma with crohn-like reaction; LELC, lymphoepithelioma-like carcinoma; EBV, Ebstein-Barr virus; MSI, microsatellite instability; MSS, microsatellite stable
Chi-square test using linear-by-linear association
Cluster 1 (immune-high, red) with mostly PD-L1High/EBV+/MSI (n = 48) showed the best outcome while cluster 3 (immune-low, black) displayed worst outcome and PD-L1Low/EBV−/MSS (n = 31) in all test set, validation set and independent validation set (Fig. 2B, E). Hierarchical clustering of signature showed that CD274, IFNG, PDCD1LG2, KLRD1, CD80, IL2RA were consistently overexpressed while CXCR1, SPP1, CISH, CD55, CD97, PTK2, TIRAP showed decreased expression in immune-high cluster 1 group. Cluster 2 (blue) was intermediated in overall status. Surprisingly, cluster 1 was significantly associated with histological subtype of lymphoepithelioma-like carcinoma (LELC) (22.9%, odds ratio 2.33, p-value 0.05). Network analyses, p value aggregation analyses and gene set enrichment analyses (GSEA) revealed total 286 functional interactionsamong 65 prognostic marker genes after exclusion of genes with low p values. Genes involved in adaptive immune system (p = .0004), costimulation by the CD28 family (p = .004), PD-1 signaling (p = 0.0023), and generation of second messenger molecules (p = 0.0133) pathways were considered as key determinant (Supplementary Table 1). PD-L1 signaling and additional pathways share core immune genes including CD274 (PD-L1) and binding partner PDCD1LG2 in submodule network shown in Fig. 3.
Figure 3.
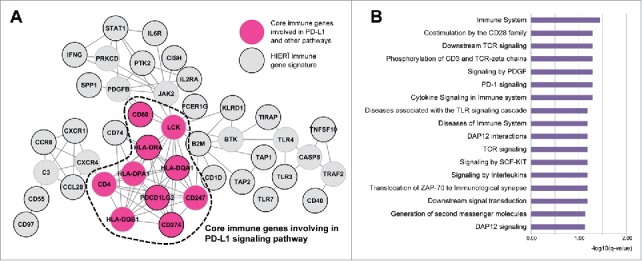
HIERÏ immune gene signature network including PD-L1 signaling pathway and associated gene functions by gene set enrichment analysis.
Table 2 summarizes the HIERÏ clustering in all 738 patients. High immune cluster 1 was closely associated with high PD-L1 mRNA expression and high TIL characterized with LELC histology, suggesting high host cellular immune response. In this cluster (229 patients; 31.3%), although 59.8% of them were EBV+ or MSI, it is noteworthy that nearly half of the patients are EBV-/MSS subtype, and they also would be a good candidate for immune checkpoint inhibitor therapy. In contrary, our results also suggest that about 10% of EBV+/MSI GC patients are not associated with immune response (cluster 3) and would not be a good candidate for immune checkpoint inhibitor therapy.
Table 2.
Three clusters of HIERÏ immune gene signatures and their characteristics.
| HIERÏ Cluster |
Immune-high |
Intermediate |
Immune-low |
|---|---|---|---|
| Total (n = 738) | 229 (31.0%) | 294 (39.8%) | 215 (29.1%) |
| GC subtype | EBV+ or MSI | EBV- and MSS | EBV- and MSS |
| 59.80% | 84.50% | 89.30% | |
| Prognosis | Good | Intermediate | Poor |
| (event observed, expected) | (ME 47, 64.5) | (ME 73, 72.6) | (ME 77, 59.9) |
| (NS 7, 11.8) | (NS 25, 24.9) | (NS 11, 6.3) | |
| PD-L1 mRNA expression | High | Intermediate | Low |
| (mean, CI 95%) | (ME 6.4, 6.3 – 6.5) | (ME 5.6, 5.5 – 5.7) | (ME 5.0, 4.9 – 5.1) |
| (NS 8.3, 8.0 – 8.6) | (NS 7.3, 7.2 – 7.5) | (NS 7.1, 6.8 – 7.4) | |
| Histological type | LELC | CA or CLR | CA or CLR |
| (LELC %, odds ratio) | (22.9%, 2.33) | (13.7%, 0.89) | (3.2%, 0.17) |
ME, merged meta-expression set; NS, nanostring; GC, gastric cancer, CI, confidence interval; CA, conventional adenocarcinoma, CLR, Carcinoma with crohn-like reaction; LELC, lymphoepithelioma-like carcinoma
We tested applicability of our newly developed HIERÏ gene signature in gene expression data of 272 TCGA12 colorectal adenocarcinomas (Supplementary Fig. 1A). All cases were also classified into 3 immune groups: high (22.8%, n = 62), intermediate (48.9%, n = 133) and low (28.3%, n = 77). We identified that immune-high group included 78.9% of 38 MSI-high cases (p < 2.2e-16), and 69.4% of 36 CIMP-high cases (p < 1.96e-07) (Supplementary Figure 1B). Finally, we revealed that our HIERÏ gene signature classified MSI/CIMP expression subtype in different cancer subtype.
PD-L1 mRNA expression and clinicopathologic variables in 181 independent validation cohort
As PD-L1 (CD274) was a main gene contributing immunological pathway and a major constituent in high-immune cluster 1 group by HIERÏ, we tested discriminating power of PD-L1 mRNA expression as a biomarker to predict clinical outcomes. PD-L1 was divided into 2 groups as overexpression (PD-L1High, n = 41 22.6%) and under expression (PD-L1Low, n = 140 77.3%) by optimizing expression cut-off using log-rank test (Supplementary Fig. 2). Overall survival (OS) difference of PD-L1High group (p< 0.001) is more significant than immune-high cluster group using HIERÏ (p = 0.082), and disease free survival (DFS) also shown significant survival difference in PD-L1High group compared toPD-L1High group (p = 0.001). PD-L1High was significantly associated with MSI GC (p = 0.034), EBV+ GC (p = 0.004), and LELC (p = 0.025), however, was not association with gender, Lauren histologic type, location of tumor, and pTNM stages (Table 3).
Table 3.
The association of PD-L1 mRNA expression and clinicopathologic variables in 181 independent validation cohort.
| PD-L1 High (n = 41) n (%) | PD-L1 Low (n = 140) n (%) | p value | total | |||||
|---|---|---|---|---|---|---|---|---|
| gender | .928 | |||||||
| male | 29 | (70.7) | 98 | (70.0) | 127 | |||
| female | 12 | (29.3) | 42 | (30.0) | 54 | |||
| location | .499 | |||||||
| cardia | 2 | (4.9) | 12 | (8.6) | 14 | |||
| body | 21 | (51.2) | 78 | (55.7) | 99 | |||
| antrum | 16 | (39.0) | 39 | (27.9) | 55 | |||
| whole/multiple | 2 | (4.9) | 11 | (7.9) | 13 | |||
| histologic type by Lauren | .664 | |||||||
| intestinal | 16 | (39.0) | 48 | (34.3) | 64 | |||
| diffuse | 20 | (48.8) | 67 | (47.9) | 87 | |||
| mixed/indeterminate | 5 | (12.2) | 25 | (17.9) | 30 | |||
| histologic type by immune response | .025 | |||||||
| CA | 20 | (48.8) | 93 | (66.4) | 113 | |||
| CLR | 10 | (24.4) | 15 | (10.7) | 26 | |||
| LELC | 11 | (26.8) | 32 | (22.9) | 42 | |||
| EBV | .004 | |||||||
| negative | 30 | (73.2) | 127 | (90.7) | 157 | |||
| positive | 11 | (26.8) | 13 | (9.3) | 24 | |||
| MSI | .034 | |||||||
| MSS | 36 | (87.8) | 135 | (96.4) | 171 | |||
| MSI | 5 | (12.2) | 5 | (3.6) | 10 | |||
| PD-L1 tumor cells | <.001 | |||||||
| negative | 16 | (39.0) | 106 | (75.7) | 122 | |||
| positive | 25 | (61.0) | 34 | (24.3) | 59 | |||
| PD-L1 immune cells (%) | .003 | |||||||
| negative | 29 | (70.7) | 125 | (89.3) | 154 | |||
| positive | 12 | (29.3) | 15 | (10.7) | 27 | |||
| HIERÏ cluster | <.001 | |||||||
| high | 24 | (58.5) | 24 | (17.1) | 48 | |||
| intermediate | 16 | (39.0) | 86 | (61.4) | 102 | |||
| low | 1 | (2.4) | 30 | (21.4) | 31 | |||
| TNM | .772* | |||||||
| I | 7 | (17.1) | 34 | (24.3) | 41 | |||
| II | 18 | (43.9) | 38 | (27.1) | 56 | |||
| III | 14 | (34.1) | 58 | (41.4) | 72 | |||
| IV | 2 | (4.9) | 10 | (7.1) | 12 | |||
CA, conventional adenocarcinoma; CLR, carcinoma with crohn-like reaction; LELC, lymphoepithelioma-like carcinoma; EBV, Ebstein-Barr virus; MSI, microsatellite instability; MSS, microsatellite stable
Chi-square test using linear-by-linear association
Out of 41 PD-L1High cases, immunohistochemical staining (IHC) on tumor cells showed 3+ in 8, 2+ in 14, and 1+ in 3, and negative in 16 cases, in which 9 cases showed positive in stromal immune cells. Of the 140 PD-L1Low cases, IHC staining on tumor cells showed 3+ in 1, 2+ in 23, and 1+ in 10, and negative in 106 cases, in which 47 cases showed positive in stromal immune cells. The PD-L1 IHC-positive group had a mean mRNA expression level of 6.80 (standard deviation 1.02), and the PD-L1 IHC-negative group had a mean mRNA expression level of 5.58 (standard deviation 0.66). The rate of concordance between mRNA expression and IHC for tumor cells was 81.8% (148/181) and was significantly correlated (p< 0.001). The proportion of PD-L1 IHC-positive immune cells did not correlate significantly with mRNA levels (p>.05). The IHC protein expression of PD-L1 on tumor cells (p< .001) were strongly associated with mRNA expression of PD-L1. Moreover, PD-L1 IHC on stromal immune cells were also correlated with mRNA expression (p = .003) (Table 3).
Immune cell-related gene expression by histologic subtypes and PD-L1 status
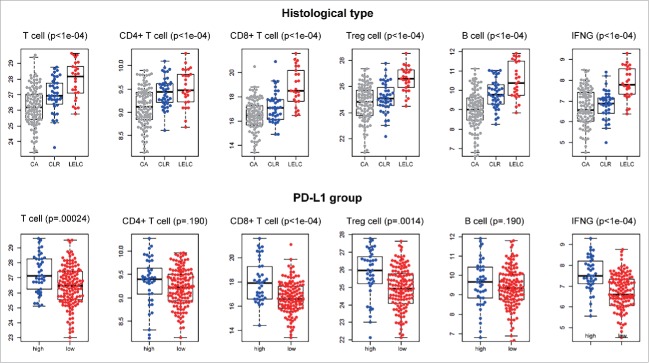
The differences in mRNA expression levels of genes (n = 491) related to representative inflammatory cells between the immune response-related histologic subtypes and PD-L1 categories are shown in Supplementary Figures 3 and 4, respectively. Between the different histologic subtypes, mRNA expression of both T cell (p< 0.001) and B cell (p< 0.001) genes were highest in LELC followed by carcinoma with crohn-like reaction (CLR) and conventional adenocarcinoma (CA). Among the T cells, CD4+, CD8+ and Treg cells (CD4+, FOXP3+, CD25[IL2RA]+) also showed significantly different expression levels according to immune histologic subtypes (p< 0.001). Additionally, LELC showed higher expression of IFNγ compared with other histologic subtypes (Fig. 4, upper lane). Based on expression of PD-L1 subgroups, mRNA levels of T cell (p< 0.001), IFNγ (p< 0.001), CD8+ T cell (p< 0.001), and Treg (p = 0.0013) were significantly higher in PD-L1High group compared with PD-L1Low group (Fig. 4, lower lane).
Figure 4.
mRNA expression difference of immune cells divided by immune histologic subtypes (upper lane) and PD-L1 expression subgroups (lower lane).
Discussion
Understanding the interactions between tumor and the host immune system is critical to finding prognostic biomarkers, reducing drug resistance, and developing new therapies.10,11 Here, we analyzed expression data set of TCGA RNA sequencing and ACRG-SMC microarray results and found HIERÏ associated with host immune response and predicting prognosis. Additionally, high mRNA of PD-L1 or tumor cell PD-L1 IHC+ group predicted improved survival in GC patients.
Recently, Li et al., analyzed tumor-infiltrating immune cells in over 10,000 RNA-Seq samples across 23 cancer types from TCGA by computational approach and developed a web-accessible resource, TIMER (Tumor Immune Estimation Resource), to enable further explorations of the disease-specific clinical impact of different immune infiltrates in the tumor microenvironment. Gentles et al.,2 also developed CIBERSORT (http://precog.stanford.edu) by analyzing ∼18,000 human tumors with overall survival outcomes across 39 malignancies and identified a FOXM1 regulatory network as a major predictor of adverse outcomes, and favorably prognostic genes related with tumor-associated leukocytes. However, the clinical impact of immune cells in many cancers remains poorly understood and controversial. Moreover, for reasons that remain unclear, immune prognostic value is known to vary according to tumor site and histology.13 Therefore, new diagnostic strategies that comprehensively and simultaneously assess the tumor immunity levels and could enhance patients' prognosis and inform immunotherapeutic decisions for cancer patients are urgently needed.
Here, we identified a HIERÏ composed of 29 immune-related genes associated with cytokine activity and extracellular matrix binding (SPP1), cytokine activity and chemokine activity (CCL20, CCR6), a receptor for interleukin 8 (CXCR1), negative regulation of cytokines (CISH), lipid binding and virus receptor activity (CD55), leukocyte migration (ADGREE5), transferase activity (PTK2), cytokine secretion and inflammatory response (TIRAP), interleukin-6 receptor binding (IL6R), induction of immunoglobulin secretion (CD40), immunoregulatory interactions (CD1 d, B2M, KLRD1), transmembrane signaling receptor activity (TLR7, TLR3, FCER1G), MHC class II receptor activity (HLA-DQA1, CD74, HLA-DRA), IFNγ signaling (B2M, TAP1, STAT1, IFNγ), receptor binding (TNFSF10), transporter activity and ATPase activity (TAP2), interleukin-2 binding (IL2RA), T-lymphocyte activation (CD80), and immune escape for tumor cells (CD274). Hierarchical clustering of signature showed that CD274, IFNG, PDCD1LG2, KLRD1, CD80, IL2RA were consistently overexpressed in both test and validation sets, suggesting activation of T cell-inflamed phenotype consisting of infiltrating T cells, a broad chemokine profile and a type I interferon signature indicative of innate immune activation in EBV/MSI GC subtypes. In these tumors, tumor cells resist immune attack through the dominant inhibitory effects of immune system-suppressive pathways.14
Cluster 1 (immune-high) was significantly associated with MSI/EBV+ GC, LELC histology (high TIL) and best prognosis. In a recent clinical trial for PD-1 blockade with pembrolizumab (MK-3475) in patients with advanced solid tumors, immune-related gene expression signatures composed of genes associated with T cell cytotoxic function, antigen presentation machinery, and IFNγ signaling showed reproducible and sensitive tools that define common features of the immune microenvironment associated with response to pembrolizumab across multiple tumor types. Based on these observations, our HIERÏ immune gene signatures would be helpful to select patients for PD-1 blockade therapy. Our results are consistent with recent data from GC patients suggesting that patients with EBV+ and MSI GC may have greater likelihood of response to PD-1 blockade and that EBV and MSI status should be evaluated as variables in clinical trials of these emerging inhibitors.9 However, in the present study, we first identified that in addition to EBV+/MSI GC subtypes, EBV-/MSS subtype also constitute almost half of high immune cluster and would be a good candidate for immune checkpoint inhibitor therapy. In contrary, almost 10% of EBV+/MSI GC patients may not respond to immune checkpoint inhibitor therapy. Our observations warrant prospective clinical investigation in the near future.
Links between cancer and inflammation have long been debated. The PD-1/PD-L1 interaction inhibits T lymphocyte proliferation, survival, and effector functions (cytotoxicity, cytokine release), induces apoptosis of tumor-specific T cells, promotes the differentiation of CD4+ T cells into Foxp3+ T-reg cells, and increases the resistance of tumor cells to cytotoxic T lymphocyte attacks.15 Although the precise mechanisms explaining better outcome in GC patients with upregulation of PD-L1 are not well established, the relationship between PD-L1 expression and TILs is a promising association and needs to be studied in more depth. In this study, PD-L1 expression was highest in LELC followed by CLR and CA and closely associated with MSI, EBV infection, increased expression of IFNγ, total T cells, CD8+ T cells, CD4+ T cells, and Treg cells. These results are consistent with previous reports that elevated PD-L1 is significantly associated with high CD8+ T-lymphocyte infiltration induced by TH1/IFNγ signaling.16 This observation indicates that PD-L1 upregulation and increased TILs seen in some GC may be in part due to changing antigen expression in constantly evolving tumors as a consequence of genetic instability as the carcinogenic process progresses.17 Better prognosis of GC patients with high TILs has also been reported in other studies.18-20 In certain subtype of GC, both TILs (host immune response) and PD-L1 upregulation (tolerance induced by the tumor itself) may impart its anti-tumorigenic properties by helping to foster a microenvironment which induces an anti-tumoral immune reaction around the tumor by recruiting particular inflammatory cells. Based on these observations, we hypothesize that this tumor-induced tolerance occurs due to 1) The immune cells ignoring the tumor (through tumor PD-L1 expression); 2) The tumor inducing anergy among tumor specific T cells (through upregulated CTLA4); 3) Exhaustion of TIL (through persistent viral antigen exposure).17 Overall, tumor PD-L1 associated with dense TIL supports survival benefit of GC patients.
In our study cohorts, no patients have received immunotherapy because anti-PD-1/PD-L1 therapy is still in ongoing clinical trial in GC patients. Recently, it has been reported that PD-L1 expression levels evaluated using the tumor proportion score (TPS) can predict patients who may respond to immunotherapy in solid tumors.21-23 Based on these observations, although its predictive role cannot be directly implicated at the present, our immune-high HIERÏ cluster based on differentially expressed genes in EBV+/MSI GCs, which are characterized by abundant PD-L1 expression, can be used to identify the patients who may likely to respond to anti PD-1/PD-L1 blockade therapy.
In conclusion, we developed HIERÏ immune gene signatures that comprehensively and simultaneously assess the tumor immunity levels, predict prognosis and inform immunotherapeutic decisions. This study also confirmed the association between PD-L1 with MSI/EBV+ GC and specific tumoral immune responses. Our findings suggest that the definition of PD-L1-mediated immune suppression could include or expand to PD-L1-mediated immune balance between an offense (by T cell) and the defense (by PD-L1-armed shield). Further elucidation of the underlying mechanisms may guide to tailored immunotherapy of patients of cancer with wide spectra of tumor heterogeneity and complex immune cells.
Materials and methods
HIERÏ inference from gene-gene interaction network
We downloaded whole genome (20639 genes) mRNA microarray data of ACRG-SMC (GSE62254) and RNA sequencing data from TCGA gastric adenocarcinomas. To adjust gene expression scale from different platforms, we merged expression data set of TCGA (n = 262) and ACRG-SMC (n = 295) from meta-analysis to integrate microarray and RNA-Seq expression (n = 557). Samples of meta-expression data were divided by 2 sets for test (n = 278) and validation (n = 279) (Fig. 1). Platform independent validation (n = 181) was performed in different gene expression data collected from NanoString Human Immunology Panel.
To identify GC subtype-specific immune response gene set, we tested DEG for MSI and EBV subtypes based on ANOVA in 278 randomly selected test set. Prognosis markers were calculated from survival analysis p-value from 3 sample groups by gene expression: high, intermediate, and low. All p-values were adjusted by Benjamini-Hochberg correction. Immune genes as a backgroup set to investigate activity were referred from NanoString Human Immuneology Panal Signature gene set. Gene interaction network-based analysis method, BioNet assigning p-values calculated from GC subtype DEG test and survival analysis were adopted.24 The reference gene interaction network was composed from Reactome.25 Assigned p-values for each node were aggregated into one p-value and it was converted to score, then we suppose to a significant subnetwork to maximize score using FastHeinz algorithm. Genes of subnetwork were filtered by cutoff (node score > 1, EBV DEG p-value < 0.05 and MSI DEG p-value <0.05). Finally, 29 genes were listed up as HIERÏ. The pathways involving in 29 genes were investigated by GSEA using Reactome pathway. To remove background effect, we defined background gene set as 491 all test immune response genes before GSEA. We confirmed prognosis ability and subtype prediction performance of HIERÏ by K-means clustering method. All samples of test (n = 278) and validation set from the same cohort (n = 279) were divided by 3 groups. The overall workflow of gene set identification and validation are depicted in Fig. 1.
To test applicability of our gene signatures in colorectal carcinoma where prevalence of MSI is high, we performed sample clustering method using expression matrix of our 29 signature genes in TCGA colorectal adenocarcinoma (n = 216).12
Patients in an independent validation set (n = 181)
Out of patients who underwent surgery for primary GC from September 2004 to September 2011 at Samsung Medical Center, we randomly selected cases from EBVaCG study (n = 44),20 the ARTIST trial (n = 98),26 and a recent prospective immune cell study (n = 39).27 In all cases, enough archival tissue was available for RNA extraction.
The clinicopathological characteristics included age, sex, tumor location, Lauren classification, histologic subtypes by host cellular immune responses,20 EBV and MSI status, pTNM stage (AJCC 7th edition), DFS, and OS.
A total of 181 patients with GC included 44 EBV+ GC, 16 MSI GC and 121 EBV- GC/MSS GC cases. The mean age of the patients was 56.6 y (range, 28 to 88 years). The male-to-female ratio was 2.4:1. The mean follow-up period was 44.8 ± 15.2 months. 55 (29.7%) patients experienced a recurrence during follow up and 42 (22.7%) patients died of disease.
Histologic subtypes based on cellular immune response patterns consisted of 27 LELC, 42 CLR, and 112 CA. EBV was positive in 44 GCs and included 27 LELC (61.4%), 11 CLR (25%), and 6 CA (13.6%) by histologic subtypes based on cellular immune responses. In contrary, 137 EBV- GC consisted of 0 LELC, 11 CLRs (8.3%) and 126 CA (91.7%). Of the 44 EBV+ GCs, 19 cases (43.2%) were diagnosed as stage I, 14 cases (31.8%) as stage II, 10 cases (22.7%) as stage III, and 1 case (2.3%) as stage IV. Of note, more than half the cases classified as LELC were stage I (51.9%) followed by stage II (29.6%), stage III (14.8%) and stage IV (3.7%). Of the 137 EBV- GCs, 21 cases (15.3%) were diagnosed as stage I, 42 cases (30.7%) as stage II, 64 cases (46.7%) as stage III, and 10 cases (7.3%) as stage IV. Kaplan-Meier survival curves of pT stage showed the expected association between AJCC stages and survival for both DFS (p< 0.001) and OS (p = 0.0032).
NanoString nCounter assay
Total RNA was extracted from 3 to 4 sections of 4 μm thick FFPE tissue sections from representative primary tumor blocks using the High Pure RNA Paraffin kit (Roche Diagnostic, Mannheim, Germany). For NanoString nCounter assay, we used nCounter® Gene Expression Human Immunology Panel 491 human immune signature genes and 15 housekeeping genes (NanoString Technologies, Seattle, WA, USA). For all cases selected, archival tissue was available for RNA extraction with the estimated tumor cell percentage of > 60% after microdissection of tumor areas. RNAs (200 ng) were hybridized to target sequence-specific capture probes and fluorescent-labeled reporter probes. The mRNA-probe complexes were washed, immobilized, and quantified by fluorescence imaging as described previously.28,29
To remove batch effect of nCounter gene expression, we did 2 step normalizations for gene expression matrix. First, within-normalization using NanoStringNorm R package with options (CodeCount+Sum, Bacground = mean, and SampleContent = total.sum) was performed, and outliers were adjusted to median value with outlier R package.30 Next, gene expression matrixes spanning 2 batch were rescaled by between-normalization using edgeR R package, and log2 transformed expression was considered as a final gene expression matrix (Supplementary Table 2).31 To validate classification performance of HIERÏ, gene expression matrix of these validation data set was also classified by 3 prognostic groups using K-mean clustering, and survival analysis of these 3 groups was tested.
Histologic examinations and EBV in situ hybridization
Histologic examinations were performed with H&E slides from entire tumor tissue. EBV in situ hybridization was performed as described previously.20 Based on host cellular immune responses, we classified histologic subtypes into LELC, CLR and CA as described previously.20
Microsatellite instability
For the screening of MSI, we performed IHC for DNA mismatch reHIERÏ protein expression (MLH1, MSH2, and MSH6) on 4 µm thick paraffin sections using mouse monoclonal antibodies specific for each MLH1 (clone G168–15, 1:200; BD Pharmingen, San Diego, CA, USA), MSH2 (clone FE11, 1:400; Calbiochem, La Jolla, CA, USA), and MSH6 (clone 44, 1:400; BD Transduction Laboratories, San Diego, CA, USA). MMR protein expression was described as negative for absent or <10% nuclear staining, and positive for ≥ 10% nuclear staining. Normal gastric epithelium or lymphocytes adjacent to the tumor served as positive controls. In cases with loss of MMR proteins, we performed MSI test. Samples with no allelic size variations in any of the 5 quasimonomorphic mononucleotide repeat markers were classified as microsatellite stable as described previously.32
PD-L1 Immunohistochemistry
As we observed significant DEG in both EBV (p = 9.31E-05) and MSI (p = 2.93E-10) GCs and showed significant survival difference (p = 2.93E-01), we performed IHC for PD-L1 also known as cluster of differentiation CD274 or B7 homolog 1 (B7-H1). Staining for PD-L1 in FFPE tissue sections was conducted with a rabbit monoclonal antibody (clone SP142; Spring Bioscience, Pleasanton, CA) on an automated staining platform (Benchmark; Ventana, Tucson, AZ, USA) with the representative tumor paraffin blocks. The SP142 was developed for Roche/Genentech's anti-PD-L1 (MPDL3280 A) immunotherapy development program and included in research of various tumor types, and has been proven excellent performance in staining both tumor and immune cells with superior quality as described previously.33 The percentages of tumor cells and peritumoral immune cells that stained positive for PD-L1 were reviewed independently by 2 pathologists (J.C. and K.M.K.).
As we used representative tumor blocks for PD-L1 staining, tumor cells stained in greater than 1% was considered positive/high expression; IHC intensity on tumors cells was arbitrary scored as 0 (negative), 1+ (weak), 2+ (intermediate) and 3+ (strong) where among tissues with PD-L1 IHC positive in more than 1% on tumor cells score 0 was considered IHC negative or low while score 1+ or greater was considered IHC positive or high. For intratumoral (immune cells within cancer cells) and peritumoral (immune cells apart from cancer cells) immune cells, staining of < 1%, 1–5%, 5–10% or ≥ 10% of cells per area was scored as 0, 1, 2, or 3, respectively33,34; grades ≥ 2 were considered overexpression in immune cells. The cases of discrepancy between both pathologists were re-reviewed for a consensus using a double-headed microscope. At all times, the pathologists were blinded to mRNA status of PD-L1.
Supplementary Material
Grant support
This work was supported by Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (NRF-S20140364000 and NRF-2015R1A2A2A01006459), the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (HI14C3418, HI16C1990), and 20 by 20 project of Samsung Medical Center (GF01140111).
References
- 1.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nature Reviews Cancer. 2012;12:298-306. doi: 10.1038/nrc3245. PMID:22419253 [DOI] [PubMed] [Google Scholar]
- 2.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al.. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nature Medicine. 2015;21:938-45. doi: 10.1038/nm.3909. PMID:26193342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, et al.. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature Communications. 2013;4:2612. doi: 10.1038/ncomms3612. PMID:24113773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research N Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-9. doi: 10.1038/nature13480. PMID:25079317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al.. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nature Medicine. 2015;21:449-56. doi: 10.1038/nm.3850. PMID:25894828 [DOI] [PubMed] [Google Scholar]
- 6.Lin SJ, Gagnon-Bartsch JA, Tan IB, Earle S, Ruff L, Pettinger K, Ylstra B, van Grieken N, Rha SY, Chung HC, et al.. Signatures of tumour immunity distinguish Asian and non-Asian gastric adenocarcinomas. Gut 2015;64:1721-31. doi: 10.1136/gutjnl-2014-308252. PMID:25385008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camargo MC, Kim WH, Chiaravalli AM, Kim KM, Corvalan AH, Matsuo K, Yu J, Sung JJ, Herrera-Goepfert R, Meneses-Gonzalez F, et al.. Improved survival of gastric cancer with tumour Epstein-Barr virus positivity: an international pooled analysis. Gut. 2014;63:236-43. doi: 10.1136/gutjnl-2013-304531. PMID:23580779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SY, Park C, Kim HJ, Park J, Hwang J, Kim JI, Choi MG, Kim S, Kim KM, Kang MS. Deregulation of immune response genes in patients with Epstein-Barr virus-associated gastric cancer and outcomes. Gastroenterology. 2015;148:137-47 e9. [DOI] [PubMed] [Google Scholar]
- 9.Derks S, Liao X, Chiaravalli AM, Xu X, Camargo MC, Solcia E, Sessa F, Fleitas T, Freeman GJ, Rodig SJ, et al.. Abundant PD-L1 expression in Epstein-Barr Virus-infected gastric cancers. Oncotarget. 2016;7:32925-32. doi: 10.18632/oncotarget.9076. PMID:27147580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, et al.. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biology. 2016;17:174. doi: 10.1186/s13059-016-1028-7. PMID:27549193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48-61. doi: 10.1016/j.cell.2014.12.033. PMID:25594174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-7. doi: 10.1038/nature11252. PMID:22810696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chifman J, Pullikuth A, Chou JW, Bedognetti D, Miller LD. Conservation of immune gene signatures in solid tumors and prognostic implications. BMC Cancer. 2016;16:911. doi: 10.1186/s12885-016-2948-z. PMID:27871313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nature Immunology. 2013;14:1014-22. doi: 10.1038/ni.2703. PMID:24048123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zitvogel L, Kroemer G. Targeting PD-1/PD-L1 interactions for cancer immunotherapy. Oncoimmunology. 2012;1:1223-5. doi: 10.4161/onci.21335. PMID:23243584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schalper KA, Carvajal-Hausdorf D, McLaughlin J, Altan M, Velcheti V, Gaule P, Sanmamed MF, Chen L, Herbst RS, Rimm DL. Differential expression and significance of PD-L1, IDO-1, and B7-H4 in human lung cancer. Clinical Cancer Research: an official journal of the American Association for Cancer Research. 2017;23:370-8. doi: 10.1158/1078-0432.CCR-16-0150. PMID:27440266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drake CG, Jaffee E, Pardoll DM. Mechanisms of immune evasion by tumors. Advances in Immunology. 2006;90:51-81. doi: 10.1016/S0065-2776(06)90002-9. PMID:16730261 [DOI] [PubMed] [Google Scholar]
- 18.Lee HE, Chae SW, Lee YJ, Kim MA, Lee HS, Lee BL, Kim WH. Prognostic implications of type and density of tumour-infiltrating lymphocytes in gastric cancer. British Journal Of Cancer. 2008;99:1704-11. doi: 10.1038/sj.bjc.6604738. PMID:18941457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizukami Y, Kono K, Kawaguchi Y, Akaike H, Kamimura K, Sugai H, Fujii H. Localisation pattern of Foxp3+ regulatory T cells is associated with clinical behaviour in gastric cancer. British Journal Of Cancer. 2008;98:148-53. doi: 10.1038/sj.bjc.6604149. PMID:18087278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song HJ, Srivastava A, Lee J, Kim YS, Kim KM, Ki Kang W, Kim M, Kim S, Park CK, Kim S. Host inflammatory response predicts survival of patients with Epstein-Barr virus-associated gastric carcinoma. Gastroenterology. 2010;139:84-92.e2. doi: 10.1053/j.gastro.2010.04.002. PMID:20398662 [DOI] [PubMed] [Google Scholar]
- 21.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al.. Pembrolizumab for the treatment of non-small-cell lung cancer. The New England Journal Of Medicine. 2015;372:2018-28. doi: 10.1056/NEJMoa1501824. PMID:25891174 [DOI] [PubMed] [Google Scholar]
- 22.Kulangara K, Hanks DA, Waldroup S, Peltz L, Shah S, Roach C, et al.. Development of the combined positive score (CPS) for the evaluation of PD-L1 in solid tumors with the immunohistochemistry assay PD-L1 IHC 22C3 pharmDx. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2017;35:(suppl; abstr e14589). doi: 10.1200/JCO.2017.35.15_suppl.e14589 (published online before print). [DOI] [Google Scholar]
- 23.Roach C, Zhang N, Corigliano E, Jansson M, Toland G, Ponto G, Dolled-Filhart M, Emancipator K, Stanforth D, Kulangara K, et al.. Development of a companion diagnostic PD-L1 immunohistochemistry assay for pembrolizumab therapy in non-small-cell lung cancer. Applied Immunohistochemistry & Molecular Morphology: AIMM. 2016;24:392-7. doi: 10.1097/PAI.0000000000000408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel CL, Owens CC 3rd, Murali TM. Network-based functional enrichment. BMC bioinformatics. 2011;12 Suppl 13:S14. doi: 10.1186/1471-2105-12-S13-S14. PMID:22479706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al.. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Research 2011;39:D691-7. doi: 10.1093/nar/gkq1018. PMID:21067998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J, Lim DH, Kim S, Park SH, Park JO, Park YS, Lim HY, Choi MG, Sohn TS, Noh JH, et al.. Phase III trial comparing capecitabine plus cisplatin versus capecitabine plus cisplatin with concurrent capecitabine radiotherapy in completely resected gastric cancer with D2 lymph node dissection: the ARTIST trial. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology 2012;30:268-73. doi: 10.1200/JCO.2011.39.1953. PMID:22184384 [DOI] [PubMed] [Google Scholar]
- 27.Choi HS, Ha SY, Kim HM, Ahn SM, Kang MS, Kim KM, Choi MG, Lee JH, Sohn TS, Bae JM, et al.. The prognostic effects of tumor infiltrating regulatory T cells and myeloid derived suppressor cells assessed by multicolor flow cytometry in gastric cancer patients. Oncotarget 2016;7:7940-51. doi: 10.18632/oncotarget.6958. PMID:26799288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J, Sohn I, Do IG, Kim KM, Park SH, Park JO, Park YS, Lim HY, Sohn TS, Bae JM, et al.. Nanostring-based multigene assay to predict recurrence for gastric cancer patients after surgery. PloS One. 2014;9:e90133. doi: 10.1371/journal.pone.0090133. PMID:24598828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin SJ, Gagnon-Bartsch JA, Tan IB, Earle S, Ruff L, Pettinger K, et al.. Signatures of tumour immunity distinguish Asian and non-Asian gastric adenocarcinomas. Gut 2015;64:1721-31. doi: 10.1136/gutjnl-2014-308252. PMID:25385008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waggott D, Chu K, Yin S, Wouters BG, Liu FF, Boutros PC. NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics. 2012;28:1546-8. doi: 10.1093/bioinformatics/bts188. PMID:22513995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-40. doi: 10.1093/bioinformatics/btp616. PMID:19910308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang SY, Park CK, Chang DK, Kim JW, Son HJ, Cho YB, Yun SH, Kim HC, Kwon M, Kim KM. Lynch-like syndrome: characterization and comparison with EPCAM deletion carriers. International Journal of Cancer. 2015;136:1568-78. doi: 10.1002/ijc.29133. PMID:25110875 [DOI] [PubMed] [Google Scholar]
- 33.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, et al.. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280 A in cancer patients. Nature. 2014;515:563-7. doi: 10.1038/nature14011. PMID:25428504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho J, Lee J, Bang H, Kim ST, Park SH, An JY, Choi MG, Lee JH, Sohn TS, Bae JM, et al.. Programmed cell death-ligand 1 expression predicts survival in patients with gastric carcinoma with microsatellite instability. Oncotarget. 2017;8(8):13320-13328. PMID:28076847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.