ABSTRACT
The anti-tumor immune response has been shown to be of great prognostic importance in colorectal cancer (CRC) and so has the tumors ability for immune evasion. Our aim of this study was to investigate tumor factors that influence immunity. We used a gene expression array to search for potential mechanisms of tumor immune escape.
One candidate gene identified was TAP1, involved in antigen presentation by MHC class I. TAP1 protein expression was evaluated by immunohistochemistry in 436 CRC patients of the Colorectal Cancer in Umeå Study cohort. We found a significant association between a downregulated expression of TAP1 and low infiltration of various subtypes of lymphocytes as well as macrophages. A downregulated expression of TAP1 was further found to be independent of molecular characteristics, suggesting TAP1 down-regulation to reach beyond the well described highly immunogenic MSI CRCs. A low expression of TAP1 was also significantly associated with poor prognosis in patients with CRC, a result that stayed significant in tumor front of early stage tumors (stage I-II) through multivariable analyses. Furthermore, we found that TAP1 expression was inversely correlated with methylation at sites in close proximity to the promoter region.
In summary, our results show down-regulation of TAP1 to be a general mechanism of tumor immune escape in CRC and a poor prognostic factor in stage I-II CRC patients. We also suggest that methylation of the TAP1 gene may be a putative mechanism for TAP1 downregulation.
KEYWORDS: TAP1, antigen presentation, immune escape, prognosis, colorectal cancer
Introduction
Colorectal cancer (CRC) is the third most common form of cancer among men and women and the second most common cause of cancer death in industrialized countries.1 The basis for curative therapy today is surgical resection, but still almost half of the patients die from metastatic disease. It is imperative to find more efficient treatment strategies to improve patient survival. A better understanding of the molecular basis of this disease is called for.
Several studies, by us and others, have focused on the tumor immune response which has proven critical to prognosis in CRC.2-7 Findings in this area have driven the design of the Immunoscore system,8 which is to be implemented in clinic as a complement to the TNM staging system.
Just as the immune system can play a pivotal role in the control of tumor progression in many cancers, so can the tumors ability to evade it.9-11 Tumors can achieve immune evasion using different tactics including the secretion of immune-inhibiting cytokines and FAS-expression that induces T-lymphocyte apoptosis. Another example is the down-regulation of components of, and thus altered surface expression of, the major histocompatibility complex class 1 (MHC class I).11-13 The MHC or, as it is also called in humans: human leukocyte antigen (HLA), presents peptides and molecules generated by the ubiquitin-proteasome pathway. This pathway involves degradation of cytosolic proteins.
Normal peptides do not elicit a response from the cytotoxic T lymphocytes (CTL/CD8+). However, cells presenting mutated or non-self proteins from intracellular pathogens on the plasma membrane, by MHC class I, trigger an adaptive immune response through binding to the T-cell receptor of CTLs. All healthy nucleated cells express MHC class I molecules and not expressing them induces the cytotoxicity of natural killer (NK) cells of the innate immune system,14,15 this since the MHC class I molecules also serve as ligands of inhibitory killer cell immunoglobulin like receptors (KIRs) on NK cells.16,17
In this study, we investigated possible molecular mechanisms involved in tumor immune escape in CRC. We found that down-regulation of TAP1 of the antigen-presenting machinery (APM) was linked to immune escape and a poor patient prognosis. We further found DNA methylation to be a putative mechanism for regulation of TAP1 expression. A better understanding of the immune evasion mechanisms in CRC may enable improved treatment strategies and prognostic tools for this disease.
Results
Gene expression analyses in tumor tissue for the identification of tumor genes involved in immune regulation
It is still not fully known why some tumors give rise to a strong immunological response while others do not. To identify tumor factors that influence (activate or suppress) immunity we performed a human whole-genome DASL expression array on a selected number of archival tumor specimens (n = 20). Tumor tissue was isolated from FFPE-tumor sections by the use of laser microdissection to avoid contamination by other cell types. Tumor tissue was isolated from 10 tumors highly infiltrated by CD3 positive T-lymphocytes, and 10 tumors with low T-lymphocyte infiltration. All tumors were selected to be of stage II, to be able to capture early changes. Tumors were also selected to be MSS, CIMP negative and BRAF wild type, since MSI, CIMP high, and BRAF mutation have previously been linked to a unique immune signature,18 likely driven by MSI associated hypermutation. Gene expression analysis identified 9 genes that were differentially expressed between the 2 groups (diff P < 0.01) (Supplementary Table 1). The analysis was restricted by poor RNA yield and quality, and data extracted was therefore used mainly as a hypothesis. Of the identified differentially expressed genes, we found several genes involved in antigen presentation and immune regulation. One of the candidate genes identified was TAP1, involved in antigen presentation by MHC class I. Tumors with low infiltration of lymphocytes generally showed a downregulated expression of TAP1 suggesting that TAP1 downregulation might be a mechanism of tumor immune escape in CRC.
Immunohistochemical evaluation of TAP1 expression in a CRC cohort of 436 patients
To verify and further investigate the role of TAP1 in immune regulation, TAP1 expression was evaluated by immunohistochemistry in 436 patients of the CRUMS cohort. The clinicopathological characteristics of the study patients are presented in Table 1. Immunohistochemical staining of TAP1 revealed that TAP1 was expressed at varying levels in CRC (Fig. 1). The expression of TAP1 was evaluated at the tumor front and center according to the IRS score19 and further divided in to groups of low (IRS ≤ 6) and high (IRS > 6) expression according to Kasajima et al.20 More than half of the patients were identified with a low expression of TAP1 (62.2% in tumor front; 71.3% in tumor center). The correlation of TAP1 expression to patient clinicopathological characteristics is shown in Table 2. We found a significant correlation between TAP1 expression and tumor stage (P = 0.001 in tumor front; P = 0.049 in tumor center), where tumors of higher stage more often displayed low TAP1 expression. A low TAP1 expression in tumor front was further correlated with perineural invasion (P = 0.025). No correlation was found to sex, age, localization, lymphovascular invasion, grade, histology type, tumor budding or growth pattern. TAP1 expression was further analyzed in relation to tumor molecular characteristics, e.g. MSI screening and CIMP status, BRAF and KRAS mutation, with no significant findings (Table 3). Since MSI, CIMP-high and BRAF mutated tumors were excluded from the initial gene expression analysis identifying TAP1 as a candidate for immune regulation, this finding is not surprising but instead suggests immune regulatory events that reach beyond the MSI associated hypermutation phenotype.
Table 1.
Clinicopathological characteristics of patients included in the study.
| Frequencies, n (%) |
436 (100.0) |
| Sex, n (%) | |
| Male | 241 (55.3) |
| Female | 195 (44.7) |
| Age, n (%) | |
| ≤ 59 years | 80 (18.3) |
| 60–69 years | 106 (24.3) |
| 70–79 years | 160 (36.7) |
| ≥ 80 years | 90 (20.6) |
| TNM stage, n (%) | |
| I | 65 (15.3) |
| II | 164 (38.6) |
| III | 88 (20.7) |
| IV | 108 (25.4) |
| Localization, n (%) | |
| Right colon | 138 (32.0) |
| Left colon | 133 (30.9) |
| Rectum | 160 (37.1) |
| Lymphovascular invasion, n (%) | |
| Yes | 71 (18.7) |
| No | 309 (81.3) |
| Perineural invasion, n (%) | |
| Yes | 83 (20.1) |
| No | 330 (79.9) |
| Grade, n (%) | |
| Low grade | 391 (90.5) |
| High grade | 41 (9.5) |
| Histology type, n (%) | |
| Mucinous | 69 (16.0) |
| Non-mucinous | 362 (84.0) |
| Tumor budding, n (%) | |
| Low grade | 209 (50.5) |
| High grade | 205 (49.5) |
| Growth pattern, n (%) | |
| Pushing | 139 (32.5) |
| Infiltrating | 289 (67.5) |
| Preoperative radiotherapy, n (%)a | |
| No | 357 (82.4) |
| Yes | 76 (17.6) |
| Cancer-specific survival, % | |
| 5y (95% CI) | 61.6 (56.7–66.5) |
Abbreviations: CI, confidence interval.
Preoperative radiation therapy mainly in rectal cancers.
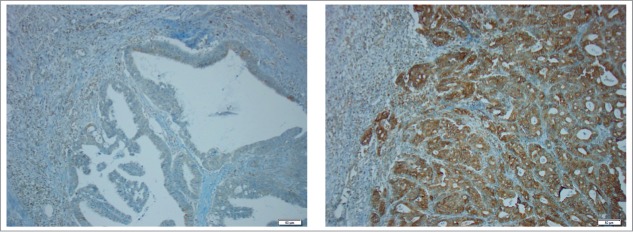
Figure 1.
Immunohistochemical stainings of TAP1 in CRC specimens. Representative light microscopic images (x20 objective magnification) of tumor specimens with low (left) and high (right) TAP1 expression.
Table 2.
TAP1 expression in relation to clinicopathological characteristics in CRC.
| TAP1 expression |
TAP1 expression |
|||||
| |
IRS Front ≤ 6 |
IRS Front > 6 |
P-valuea |
IRS Center ≤ 6 |
IRS Center > 6 |
P-valuea |
| Frequencies, n (%) | 268 (62.2) | 163 (37.8) | 311 (71.3) | 125 (28.7) | ||
| Sex, n (%) | 0.727 | 0.984 | ||||
| Male | 145 (61.4) | 91 (38.6) | 172 (71.4) | 69 (28.6) | ||
| Female | 123 (63.1) | 72 (36.9) | 139 (71.3) | 56 (28.7) | ||
| Age, n (%) | 0.145 | 0.104 | ||||
| ≤ 59 years | 47 (61.0) | 30 (39.0) | 54 (67.5) | 26 (32.5) | ||
| 60–69 years | 69 (65.7) | 36 (34.3) | 83 (78.3) | 23 (21.7) | ||
| 70–79 years | 105 (66.0) | 54 (34.0) | 117 (73.1) | 43 (26.9) | ||
| ≥ 80 years | 47 (52.2) | 43 (47.8) | 57 (63.3) | 33 (36.7) | ||
| TNM stage, n (%) | 0.001 | 0.049 | ||||
| I | 30 (46.9) | 34 (53.1) | 39 (60.0) | 26 (40.0) | ||
| II | 97 (59.9) | 65 (40.1) | 116 (70.7) | 48 (29.3) | ||
| III | 53 (60.2) | 35 (39.8) | 61 (69.3) | 27 (30.7) | ||
| IV | 81 (76.4) | 25 (23.6) | 86 (79.6) | 22 (20.4) | ||
| Localization, n (%) | 0.582 | 0.413 | ||||
| Right colon | 82 (60.7) | 53 (39.3) | 94 (68.1) | 44 (31.9) | ||
| Left colon | 87 (65.4) | 46 (34.6) | 100 (75.2) | 33 (24.8) | ||
| Rectum | 95 (59.7) | 64 (40.3) | 112 (70.0) | 48 (30.0) | ||
| Lymphovascular invasion, n (%) | 0.164 | 0.691 | ||||
| Yes | 48 (69.6) | 21 (30,4) | 52 (73.2) | 19 (26.8) | ||
| No | 186 (60.6) | 121 (39.4) | 219 (70.9) | 90 (29.1) | ||
| Perineural invasion, n (%) | 0.025 | 0.219 | ||||
| Yes | 59 (72.8) | 22 (27.2) | 64 (77.1) | 19 (22.9) | ||
| No | 194 (59.3) | 133 (40.7) | 232 (70.3) | 98 (29.7) | ||
| Grade, n (%) | 0.902 | 0.377 | ||||
| Low grade | 241 (62.4) | 145 (37.6) | 283 (72.4) | 108 (27.6) | ||
| High grade | 26 (63.4) | 15 (36.6) | 27 (65.9) | 14 (34 .1) | ||
| Histology type, n (%) | 0.552 | 0.928 | ||||
| Mucinous | 44 (65.7) | 23 (34.3) | 49 (71.0) | 20 (29.0) | ||
| Non-mucinous | 222 (61.8) | 137 (38.2) | 259 (71.5) | 103 (28.5) | ||
| Tumor budding, n (%) | 0.230 | 0.459 | ||||
| Low grade | 120 (58.3) | 86 (41.7) | 145 (69.4) | 64 (30.6) | ||
| High grade | 130 (64.0) | 73 (36.0) | 149 (72.7) | 56 (27.3) | ||
| Growth pattern, n (%) | 0.243 | 0.331 | ||||
| Pushing | 90 (66.2) | 46 (33.8) | 103 (74.1) | 36 (25.9) | ||
| Infiltrating | 173 (60.3) | 114 (39.7) | 201 (69.6) | 88 (30.4) | ||
| Preoperative radiation therapy, n (%)b | 0.291 | 0.258 | ||||
| No | 222 (63.1) | 130 (36.9) | 258 (72.3) | 99 (27.7) | ||
| Yes | 43 (56.6) | 33 (43.4) | 50 (65.8) | 26 (34.2) | ||
χ2 test.
Preoperative radiation therapy mainly in rectal cancers.
Table 3.
TAP1 expression in relation to molecular characteristics in CRC.
| TAP1 expression |
TAP1 expression |
|||||
| |
IRS Front ≤ 6 |
IRS Front > 6 |
P-valuea |
IRS Center ≤ 6 |
IRS Center > 6 |
P-valuea |
| Frequencies, n (%) | 268 (62.2) | 163 (37.8) | 311 (71.3) | 125 (28.7) | ||
| MSI screening status, n (%) | 0.784 | 0.219 | ||||
| MSI | 41 (61.2) | 26 (38.8) | 44 (65.7) | 23 (34.3) | ||
| MSS | 221 (63.0) | 130 (37.0) | 260 (73.0) | 96 (27.0) | ||
| CIMP status, n (%)b | 0.880 | 0.266 | ||||
| CIMP-negative | 130 (61.9) | 80 (38.1) | 160 (75.1) | 53 (24.9) | ||
| CIMP-low | 102 (63.4) | 59 (36.6) | 112 (68.7) | 51 (31.3) | ||
| CIMP-high | 34 (59.6) | 23 (40.4) | 38 (66.7) | 19 (33.3) | ||
| BRAFV600E, n (%) | 0.486 | 0.184 | ||||
| wild type | 227 (62.7) | 135 (37.3) | 267 (72.8) | 100 (27.2) | ||
| mutated | 36 (58.1) | 26 (41.9) | 40 (64.5) | 22 (35.5) | ||
| KRAS (codon 12, 13), n (%) | 0.336 | 0.333 | ||||
| wild type | 212 (61.3) | 134 (38.7) | 248 (70.9) | 102 (29.1) | ||
| mutated | 53 (67.1) | 26 (32.9) | 61 (76.3) | 19 (23.8) | ||
Abbreviations: MSI, microsatellite instability; MSS, microsatellite stable; CIMP, CpG island methylator phenotype.
χ2 test.
CIMP negative, no promoter hypermethylation; CIMP low, one to 5 genes methylated; CIMP high, 6 to 8 genes methylated.
TAP 1 expression is linked to immune infiltration and has prognostic value
Since TAP1 is involved in antigen-presentation by MHC class I and was found at lower levels in tumors with low infiltration of CD3 positive cells (Supplementary Table 1), we reasoned that TAP1 down-regulation might be a mechanism of tumor immune escape. Interestingly, we found significant positive correlations between TAP1 protein expression and infiltration of most immune cell subsets analyzed, both at the tumor front and center (Table 4). TAP1 expression was significantly correlated with infiltration of general T cells (CD3+), cytotoxic T cells (CD8+), regulatory T cells (FOXP3+), T helper cells (Tbet+), M1 macrophages (NOS2+) and M2 macrophages (CD163+) (Table 4). However, no correlation was found to neutrophil infiltration (CD66b+) (Table 4). The correlation between TAP1 expression and expression of other APM components was evaluated by immunohistochemical staining of MHC class I (HLA-A, -B, and -C) and TAP2 in tumor tissue from 22 of the CRUMS patients, classified using IRS with either low (n = 12) or high (n = 10) expression of TAP1. Even though the patient material was small, we found significant correlations between TAP1 expression and expression of both MHC class I and TAP2 (Table 5).
Table 4.
The correlation of expression of TAP1 with immune cell markers.
| TAP1 expression |
||||
| Immune | IRS Front |
IRS Center |
||
| markers |
rs |
P-value |
rs |
P-value |
| CD3 | 0.297 | <0.001 | 0.240 | <0.001 |
| CD8 | 0.215 | <0.001 | 0.154 | 0.003 |
| FOXP3 | 0.110 | 0.035 | 0.103 | 0.049 |
| Tbet | 0.277 | <0.001 | 0.233 | <0.001 |
| NOS2 | 0.234 | <0.001 | 0.202 | <0.001 |
| CD163 | 0.298 | <0.001 | 0.262 | <0.001 |
| CD66b | 0.018 | 0.729 | 0.012 | 0.818 |
rs, Spearman´s rank correlation coefficient. Total score 3–4, 5–6, 7–12, was used for correlations of CD3, CD8, FOXP3 and Tbet. For NOS2, CD163 and CD66b, score 1–4 at the tumor front was used.
Table 5.
TAP1 expression in relation to expression of MHC class I (HLA-A, B and C) and TAP2.
| TAP1 front |
||||
| |
|
IRS ≤ 6 |
IRS > 6 |
P-valuea |
| HLA front | IRS ≤ 6 | 4 (100.0) | 0 (0.0) | 0.087 |
| IRS > 6 | 6 (40.0) | 9 (60.0) | ||
| TAP2 front | IRS ≤ 6 | 8 (88.9) | 1 (11.1) | 0.015 |
| IRS > 6 | 2 (22.2) | 7 (77.8) | ||
| TAP1 center |
||||
| |
|
IRS ≤ 6 |
IRS > 6 |
P-valuea |
| HLA center | IRS ≤ 6 | 6 (100.0) | 0 (0.0) | 0.014 |
| IRS > 6 | 5 (35.7) | 9 (64.3) | ||
| TAP2 center | IRS ≤ 6 | 7 (87.5) | 1 (12.5) | 0.020 |
| IRS > 6 | 3 (25.0) | 9 (75.0) | ||
Fischer´s exact test.
Immune infiltration has been positively linked to increased survival in patients with CRC. Next, we therefore investigated the prognostic importance of TAP1 expression in the tumor front and center. We found that TAP1 expression was significantly correlated with prognosis (Fig. 2) (P = 0.001 for tumor front; P = 0.013 for tumor center). Patients with a low expression of TAP1 had a worse prognosis, suggesting that TAP1 down-regulation may facilitate tumor immune escape resulting in poor patient prognosis. The prognostic significance remained significant in multivariable analyses including sex, localization and tumor grade (Table 6). The significance for TAP1 expression was however lost when stage was added to the multivariable analyses. Yet, in multivariable analyses stratified by stage, TAP1 expression in the tumor front was found to be significantly associated with prognosis in CRCs of early stages (stage I-II) (Table 6).
Figure 2.
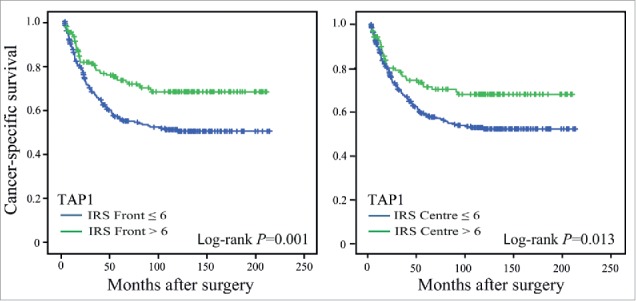
Cancer-specific survival in CRC patients. Kaplan-Meier plots of cases scored for TAP1 expression in the tumor front and tumor center: IRS ≤ 6, low expression; IRS > 6, high expression. Log-rank tests were used to calculate P-values.
Table 6.
Cox regression analyses of TAP1 expression in predicting survival of CRC patients.
| TAP1 IRS Front (≤ 6; > 6) |
TAP1 IRS Center (≤ 6; > 6) |
|||||
| Gene symbol |
HR |
95% CI |
P-value |
HR |
95% CI |
P-value |
| Univariable | 0.57 | 0.40–0.80 | 0.001 | 0.62 | 0.43–0.91 | 0.014 |
| Multivariablea | 0.54 | 0.38–0.77 | 0.001 | 0.59 | 0.40–0.86 | 0.006 |
| Multivariableb | 0.79 | 0.55–1.14 | 0.213 | 0.74 | 0.50–1.09 | 0.131 |
| Multivariablea (stage I-II) | 0.43 | 0.21–0.89 | 0.022 | 0.48 | 0.22–1.06 | 0.069 |
| Multivariablea (stage III) | 0.69 | 0.42–1.78 | 0.688 | 0.60 | 0.26–1.37 | 0.306 |
| Multivariablea (stage IV) | 1.04 | 0.61–1.77 | 0.892 | 0.95 | 0.55–1.65 | 0.853 |
including sex, age, localization and tumor grade.
including sex, age, localization, tumor grade and TNM stage.
Abbreviations: HR, hazard ratio; CI, confidence interval
TAP1 expression is inversely correlated with DNA methylation
Since a decreased expression of the TAP1 gene was linked to a weaker immune response and a worse prognosis in patients with I-II CRC, we initiated a search for possible mechanisms of TAP1 down-regulation. Methylation is a known epigenetic alteration that can control gene expression.21,22 To investigate the relationship between DNA methylation and gene expression of the TAP1 gene, we used the TCGA colon adenocarcinoma (COAD) data set. The data set includes methylation status for 88 CpG sites associated with the TAP1 gene and limiting analyses to samples for which both gene expression and methylation data were available, we analyzed 298 samples. For 8 samples multiple methylation data sets were available and the final data was therefore generated to include all combinations of TAP1 expression and methylation data, resulting in 312 such combinations. The relationship between methylation status for these CpG sites and expression of the TAP1 gene was modeled using OPLS, resulting in 1 predictive and 3 orthogonal statistically significant components with cumulative R2(X), R2(Y), and Q2 values of 0.823, 0.592, and 0.505 respectively. The model was further validated using a permutation test and by splitting the data into training and test sets. We found that most TAP1-associated CpG sites with the highest VIP scores in the OPLS analysis clustered in 3 distinct regions along the gene; close to the promoter region upstream of exon 1, in a region spanning exon 2 and 3, and in a region spanning exon 11 (Fig. 3A).
Figure 3.
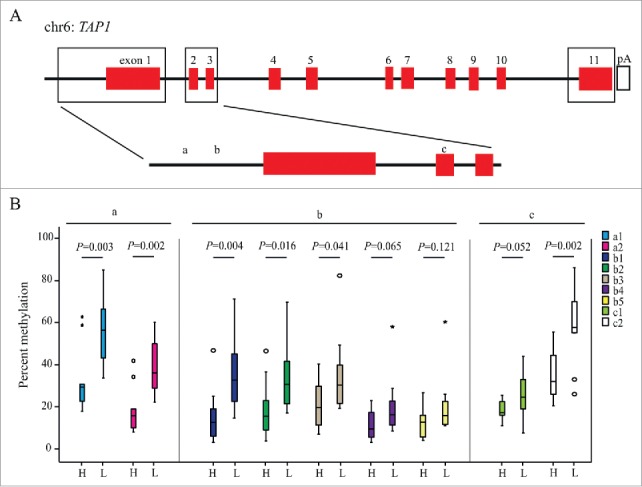
TAP1 methylation and expression in CRC. (A) A schematic illustration of the TAP1 gene. Methylation of putative CpG regions correlated with TAP1 expression, as identified from the TCGA data set, are highlighted by boxes. (B) Percent methylation of specific CpG-sites (a-c) (described in Supplementary Table 3) as analyzed by pyrosequencing and illustrated by Boxplots. Outlier values (o) and far-out values (*) are indicated. The analyses include patients with high (H, IRS > 6, n = 10) and low (L, IRS ≤ 6, n = 12) expression of TAP.
Focusing on the CpG sites closest to the TAP1 promoter, with the strongest correlation to expression of TAP1, we designed 3 pyrosequencing assays including a total of 9 CpG-sites (Fig. 3). Specific methylation assays were performed on tumor tissue from 22 CRUMS patients, classified using IRS with either low (n = 12) or high (n = 10) expression of TAP1. Six out of the 9 CpG sites tested exhibited a statistically significant difference regarding methylation status between these groups, and all CpG sites displayed a higher level of methylation in the group of tumors with low TAP1 expression (Fig. 3B).
Discussion
Tumors can achieve immune evasion using different strategies of which one is the down-regulation of components of the APM and thus altered surface expression of the MHC class I molecules.12,13 MHC class I play a crucial role in the elimination by CD8+ lymphocytes of both virally infected as well as tumor cells. In this study, we searched for differences between tumors that were highly infiltrated by immune cells and tumors that had escaped the immune response. We found that tumors with low infiltration of lymphocytes generally showed a down-regulated expression of TAP1, a component of the APM, suggesting that TAP1 down-regulation might be a mechanism of tumor immune escape in CRC. We further found that expression of TAP1 may be regulated by methylation.
We initiated the study performing a whole genome expression analysis comparing CD3 high and CD3 low CRC tumors. Among the genes that were identified as differentially expressed between the 2 groups, we found TAP1. By using LMPC, where one can selectively cut out tumor tissue, we decreased the risk of contamination from stromal cells, not the least surrounding immune cells which potentially could blur the outcome. However, the number of detected genes (P < 0.01) was low, leading to a low number of differentially expressed genes even without FDR correction. The reasons are probably low RNA input levels and possibly poor RNA quality since the FFPE tissue was of older date. Considering this we chose not to systematically analyze all of the significant factors but instead used this as the hypothesis generator we once set out for it to be.
The finding that a decreased expression of TAP1 was associated with poor immune infiltration was verified in a large cohort of CRC patients. TAP1 was stained using immunohistochemistry on whole-tissue sections of CRC tumors from a total of 436 patients. Using IRS scoring, a semi-quantitative assessment of expression, we divided the tumors into either TAP1 high or TAP1 low. The semi-quantitative method has the built-in risk of inter-observer variability but it also enables the assessment of possibly heterogenic expression levels in a tumor section. The observer can thus consider variations and also avoid necrotic areas in the evaluation, a forte in comparison to the use of a purely quantitative computerised method where this is not as easily done. Here we sought to avoid the possible pitfalls of a semi-quantitative method by letting 2 observers independently score all the tumors and thereafter, together, reach a conclusive judgement for discordant cases.
We found significant associations between TAP1 expression and the number of different subtypes of tumor infiltrating lymphocytes as well as macrophages. No significant correlation was seen with neutrophil infiltration which complies well with the APM mainly activating the adaptive immune system. Studying the different clinical characteristics of the tumors, a decreased expression of TAP1 was more often found in later tumor stages, suggesting a link between immune escape and tumor progression. A decreased expression of TAP1 in tumor front was also correlated with increased perineural invasion. Sex, age, localization, lymphovascular invasion, grade, histology type, tumor budding and growth pattern showed no significant association to TAP1 expression. Neither did MSI, CIMP-status or BRAF or KRAS mutational status. This could in one way be considered a surprise since MSI is associated with a higher immune cell infiltration. Our findings thereby suggest that TAP1 down-regulation is not restricted to the well described highly immunogenic right sided, MSI, CIMP-high, BRAF mutated subgroup of CRC. In summary, TAP1 down-regulation appears to be a more general mean of immune evasion in colorectal carcinomas, independent of different molecular characteristics.
In univariate survival analysis, we found that a low expression of TAP1 significantly correlated with a poorer patient prognosis, a result that stayed significant in multivariable analyses in the lower tumor stages (stage I-II). This finding implies that TAP1 down-regulation is a potential marker for poor prognosis in patients with stage I-II CRC, indicating that more vigorous efforts are called for in the treatment of these patients. The association between TAP1 down-regulation and immune infiltration, suggests that part of the poor prognosis seen in TAP1 down-regulated tumors may be linked to immune regulation. Interferons (both α and gamma) are expressed by activated T-lymphocytes and have been shown to upregulate the expression of various components of the antigen presenting machinery, including TAP1.23-25 In this study, tumors with intact TAP1 expression were more infiltrated by immune cells posing the question of whether TAP1 expression may be supported by interferons secreted by the infiltrating immune cells? Then it would rather be that immune infiltration would support an intact APM, thus conserving the immunogenicity of the tumor, than vice versa. Interferons have shown promising therapeutic effects in melanoma patients,26 and a link has been suggested to an upregulation of APM-molecules.23 Identifying patients with tumors lacking TAP1 expression could therefore be of relevance in the future of personalized therapy.
In the APM, peptides from the cytoplasmic proteins degraded by the proteasome are translocated across the endoplasmic reticulum (ER) membrane via the antigen-processing heterodimer of TAP1 and TAP2. The TAP-associated peptides are then loaded into MHC class I complexes which are transported to the surface. Earlier studies have shown TAP1 and TAP2 to be down-regulated in various kinds of cancers with a correlation to immune cell infiltration and even prognosis.20,27-32 In CRC, Kasajima et al. have previously shown a link between downregulation of TAP1 and loss of inflammatory response.20 The authors further showed that TAP1 and TAP2 expression was strongly correlated with expression of MHC class I. However, no correlation was found between TAP1 expression and prognosis in their study. The reasons for these discrepancies are not clear since we have used the same IRS scoring system. Down-regulation of TAP2 has further been suggested by Cabrera et al. to be a mechanism for loss of MHC class I expression in MSS tumors.33 MHC class I expression has previously been linked to prognosis in CRC, but the results so far are inconclusive.34-37 In this study, we found down-regulation of TAP1 expression to be correlated with decreased levels of both MHC class I (HLA-A, -B and -C) and TAP2, suggesting that immune escape in tumors with down-regulated TAP1 may at least partly be linked to loss of other APM components. However, we also found tumors with down-regulated TAP1 expression but intact expression of MHC class I, suggesting that there may also be other possible scenarios. Leone et al. suggests that TAP down-regulation might lead to the presentation of empty HLA-molecules.14 If that would be the case, tumor cells could evade recognition by NK cells by still expressing HLA molecules on their surface but without presenting the immune instigating antigens. In a study by Lampen et al., a discrepancy was found between TAP blockade in cell lines and MHC class I expression at the cell surface, leading to the conclusion that TAP-inhibited cells still presented peptides on their surface but from a different TAP-independent peptide repertoire.38 Many questions remain and further studies are required to understand the importance of TAP1 and other components of the APM in immune regulation and CRC prognosis.
Searching for possible mechanisms of TAP1 down-regulation, we performed methylation assays on tumor tissue from a subset of patients from the CRUMS cohort. Epigenetic regulation, including DNA hypermethylation, of expression of TAP1 and other components of the APM has been found in a variety of cancers.21,39,40 Here we focused on the CpG sites closest to the TAP1 promoter, holding the strongest correlation to expression of TAP1 (highest VIP scores). Specific methylation assays were performed on tumor tissue with low and high TAP1 expression, respectively. In the group of tumors with a low TAP1 expression we found that all CpG sites displayed higher levels of methylation compared with the group with a high expression of TAP1. The results from these analyses, although performed on a small number of patients, suggest hypermethylation of TAP1 to be a possible tumor strategy for APM-alterations and hence immune evasion in CRC. Future studies are required to address the impact of methylation on expression on the different APM components in immune regulation and cancer.
From this study, we conclude that down-regulation of TAP1 is a potential mechanism of tumor immune escape in CRC and could be an additional prognostic factor in CRC patients selecting stage I and II patients who might be in need of more vigorous treatments. We further found that hypermethylation of TAP1 could be a possible mechanism for down-regulation of TAP1 and tumor immune escape. In the future, in the context of personalized medicine, reactivation of a down-regulated TAP1 expression might be a way to regenerate tumor-specific immunity in CRC patients.
Patients and methods
Study population
Patients included in the study were from the Colorectal Cancer in Umeå Study (CRUMS), described previously in full detail.41 The clinicopathological characteristics of the study patients can be found in Table 1. In brief, tissue specimens were collected from patients surgically resected for CRC between 1995 and 2003 at the Department of Surgery, Umeå University Hospital, Sweden. Formalin-fixed paraffin-embedded (FFPE) tissue was sampled from all patients and clinicopathological and molecular variables have been characterized and previously published.6,41 Lymphovascular invasion was recorded as present or absent (yes or no). Tumor budding at the invasive front was quantified on pancytokeratin-stained slides according to Prall et al.42 and scored as low grade (0–9 buds per microscopic field) or high grade (10 or more), adapted from Ueno et al.43
Infiltration of different subsets of lymphocytes and macrophages has also previously been assessed in the CRUMS cohort,5,6,41,44 along with infiltration of neutrophils (CD66b+) (unpublished evaluation). After exclusions due to unavailable or insufficient tumor sample, and/or lack of clinical information, 436 patients were included in the study. For survival analysis, another 37 patients were excluded, due to incomplete follow-up data or to death from perioperative complications (death within 30 d of surgery). An informed consent was obtained from the patients and the handling of tissue samples and patient data was approved by the Regional Ethical Review Board of Umeå, Sweden, and in accordance with the Declaration of Helsinki.
External reference data
Reference methylation and expression data was obtained for CRC patients from The Cancer Genome Atlas (TCGA) Research Network,45 representing 291 primary colon adenocarcinoma samples for which both RNA-seq expression data and array methylation data in the form of β values was available. Accession numbers for reference data sets used in this study are provided in Supplementary Table 2.
Gene expression analysis
Gene expression analysis was performed on isolated tumor tissue from 20 patients of the CRUMS cohort. Patients included were selected to be stage II, microsatellite stable (MSS), CpG island methylator phenotype (CIMP) negative, and BRAF wild-type. Patients were further selected with respect to tumor infiltration of CD3 positive lymphocytes. CD3 was previously assessed in the CRUMS cohort, and recorded as a CD3 totalscore.41 According to this total score, 10 patients were selected with low CD3 infiltration (total score 3–4) and 10 patients with high CD3 infiltration (total score 7–12). FFPE-embedded tumor specimens were cut into 4 µM sections and stained with hematoxylin. Tumor cells were isolated from the sections by laser microdissection and pressure catapulting (LMPC) using the PALM MicroBeam Laser Capture Microdissection System (Zeiss) in the Auto Laser Pressure Catapulting (AutoLPC) mode. Cells were collected into one AdhesiveCap (Zeiss). RNA was extracted from the isolated tumor cells using the High Pure RNA Paraffin Kit (Roche, Stockholm, Sweden). Gene expression was analyzed by the Whole-Genome Gene Expression DASL HT Assay (Illumina). Labeled cRNA was hybridized to the human HT-12 v4 expression BeadChip. Microarrays were scanned using the Illumina HiScan® System and the data was analyzed using the Genome Studio. 6 microarrays (CD3 low (n = 3) and CD3 high (n = 3)) failed the scanning process due to low signal average and were excluded from the analyses. The number of detected genes (P < 0.01) was relatively low (between 6000–8000), due to low RNA input levels and poor RNA quality. Data was analyzed by the differential expression analysis using cubic spline normalization and the Illumina custom error model without FDR correction.
Immunohistochemical evaluations
Four μm FFPE tissue sections were dried, de-waxed, and rehydrated. The anti-TAP1 antibody (11114–1-AP; Nordic Biosite) and the anti-TAP2 antibody (K0137–3; MBL) were used at a dilution of 1:50, and the anti-HLA class I ABC antibody (ab70328; Abcam) at a dilution of 1/400 on an automated Ventana Benchmark Ultra staining machine with the ultraVIEW DAB Detection Kit for visualization (Ventana Medical Systems, Inc.). Expression of TAP1 was evaluated in the CRUMS cohort by 2 observers, of whom one was an experienced gastro pathologist, using light microscopy. The kappa coefficient for inter-observer agreement was 0.538 and 0.574 for front and center, respectively. In cases of discrepant scoring, a third estimation was made by both observers followed by a conclusive judgement. TAP1 expression was evaluated at the most representative area of the invasive tumor front and the tumor center according to the Immunoreactive score (IRS).19 IRS is a semi-quantitative assessment of expression determined by multiplying the staining intensity in 4 gradations (0 = no, 1 = weak, 2 = moderate, and 3 = strong intensity) with the percentage of positive cells in 5 gradations (0 = no, 1 = < 10%, 2 = 10–50%, 3 = 51–80%, and 4 = > 80%). Lymphocytes were used as internal positive control. Using the resulting IRS score (1–12), patients were further divided into groups of low (IRS ≤ 6) and high (IRS > 6) expression according to Kasajima, A et al.20 Expression of MHC class I (HLA-A, -B and -C) and TAP2 were evaluated by the same procedure in tumor tissue from a subset of 22 CRUMS patients, selected to have either low (IRS ≤ 6, n = 12) or high (IRS > 6, n = 10) expression of TAP1.
Methylation analyses
Specific methylation assays were performed on DNA isolated from FFPE tumor tissue of 22 CRUMS patients, selected to have either low (IRS ≤ 6, n = 12) or high (IRS > 6, n = 10) expression of TAP1. The patients in the 2 groups were selected to have tumors with a similar distribution of tumor stage, localization, MMR status, CIMP status, and KRAS and BRAF mutations. The positions for the specific TAP1 methylation sites are presented in Supplementary Table 3. DNA was extracted using the Illustra Nucleon Genomic Extraction Kit (GE Healthcare) and bisulfate treated with the Epitec Fast DNA Bisulfite Kit (Qiagen), according to the manufacturer's instructions. The methylation-specific PCRs were performed using the PyroMark® PCR Kit (Qiagen). The PCR was run in an ABI Veriti thermal cycler (ABI) using following cycling conditions: denaturation at 95°C for 15min, followed by 45 cycles each comprising of 30s at 95°C, 30s at 61°C and 30s at 72°C, with an extended last step of 72°C for 10 minutes. The primers used for the TAP1 assays are available in Supplementary Table 4. DNA methylation in the TAP1 region was determined by pyrosequencing using the PyroMark Q24 Advanced System (Qiagen). Data was analyzed using PyroMark Q24 1.0.10 Software (Qiagen) and methylation percentage (mC/mC+C) of each CpG site was calculated.
Statistics
Statistical analyses were performed using the PASW Statistics 24 (SPSS Inc., Chicago, IL, USA). χ2 tests were used for cross-tabulations and the exact linear-by-linear association test was used to test linear relationships. The Spearman´s rank correlation test was used to test correlations between categorical variables and the Mann–Whitney U-test was used for differences in continuous variables between groups. Cancer-specific survival was assessed using the Kaplan-Meier survival analysis, and comparisons between outcomes in different groups were performed with the log-rank test. Cancer-specific survival was defined as death with known disseminated or recurrent disease. Multivariable survival analyses were performed using Cox proportional hazard models. P < 0.05 was considered statistically significant.
Multivariate modeling of the reference data was performed using Simca-P v. 14.1 (Umetrics, Sweden). The relationship between DNA methylation (β-values) and gene expression was modeled using orthogonal partial least squares (OPLS) regression, using default settings for limiting the number of extracted components. Methylation β values were centered but not scaled and gene expression values were centered and scaled to unit variance. Validity of calculated models were assessed using 7-fold cross-validation (default settings), permutation tests using 50 iterations, and by random division of data into training and test sets of equal size. Variable Importance in the Projection (VIP) was used to identify the relative influence of individual markers in obtained models.
Statement of author contribution
Study concept and design: AL, ALB, MLW, SE, RP; acquisition of data: AL, ALB, PL, XL, ÅÖ, RS, SE; data analyses: AL, ALB, PL, SE, RP; drafting of the manuscript: AL, SE, ALB; critical revision of the manuscript for important intellectual content: PL, XL, MLW, ÅÖ, RS, RP. All authors approved the final version of the manuscript.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Kerstin Näslund and Dr Åsa Stenberg for expert technical assistance, and Dr Anna Dahlin, Maria Jacobsson and Vincy Eklöf for previous evaluations in CRUMS. All from the department of Medical Biosciences, Pathology, Umeå University, Umeå.
Funding
This work was supported by the Swedish Cancer Society under Grant CAN 2014/858; Lion's Cancer Research Foundation under Grant LP 15–2083; the Cancer Research Foundation in Northern Sweden under Grant AMP 16–832; and the County Council of Vasterbotten under Grant VLL-463871. The study sponsors had no role in study design, data collection, analysis and interpretation of the data.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. PMID:25651787 [DOI] [PubMed] [Google Scholar]
- 2.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, et al. . Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. PMID:17008531 [DOI] [PubMed] [Google Scholar]
- 3.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: Impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. PMID:22419253 [DOI] [PubMed] [Google Scholar]
- 4.Mlecnik B, Tosolini M, Kirilovsky A, Berger A, Bindea G, Meatchi T, Bruneval P, Trajanoski Z, Fridman WH, Pagès F, et al. . Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol. 2011;29:610–8. doi: 10.1200/JCO.2010.30.5425. PMID:21245428 [DOI] [PubMed] [Google Scholar]
- 5.Ling A, Lundberg IV, Eklof V, Wikberg ML, Oberg A, Edin S, Palmqvist R. The infiltration, and prognostic importance, of Th1 lymphocytes vary in molecular subgroups of colorectal cancer. J Pathol Clin Res. 2016;2:21–31. doi: 10.1002/cjp2.31. PMID:27499912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling A, Edin S, Wikberg ML, Oberg A, Palmqvist R. The intratumoural subsite and relation of CD8(+) and FOXP3(+) T lymphocytes in colorectal cancer provide important prognostic clues. Br J Cancer. 2014;110:2551–9. doi: 10.1038/bjc.2014.161. PMID:24675384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deschoolmeester V, Baay M, Lardon F, Pauwels P, Peeters M. Immune cells in colorectal cancer: Prognostic relevance and role of MSI. Cancer Microenviron. 2011;4:377–92. doi: 10.1007/s12307-011-0068-5. PMID:21618031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galon J, Mlecnik B, Bindea G, Angell HK, Berger A, Lagorce C, Lugli A, Zlobec I, Hartmann A, Bifulco C, et al. . Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J Pathol. 2014;232:199–209. doi: 10.1002/path.4287. PMID:24122236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. PMID:21376230 [DOI] [PubMed] [Google Scholar]
- 10.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: From immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–8. doi: 10.1038/ni1102-991. PMID:12407406 [DOI] [PubMed] [Google Scholar]
- 11.Pernot S, Terme M, Voron T, Colussi O, Marcheteau E, Tartour E, Taieb J. Colorectal cancer and immunity: What we know and perspectives. World J Gastroenterol. 2014;20:3738–50. doi: 10.3748/wjg.v20.i14.3738. PMID:24833840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seliger B. Different regulation of MHC class I antigen processing components in human tumors. J Immunotoxicol. 2008;5:361–7. doi: 10.1080/15476910802482870. PMID:19404870 [DOI] [PubMed] [Google Scholar]
- 13.Algarra I, Garcia-Lora A, Cabrera T, Ruiz-Cabello F, Garrido F. The selection of tumor variants with altered expression of classical and nonclassical MHC class I molecules: Implications for tumor immune escape. Cancer Immunol Immunother. 2004;53:904–10. doi: 10.1007/s00262-004-0517-9. PMID:15069585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leone P, Shin EC, Perosa F, Vacca A, Dammacco F, Racanelli V. MHC class I antigen processing and presenting machinery: Organization, function, and defects in tumor cells. J Natl Cancer Inst. 2013;105:1172–87. doi: 10.1093/jnci/djt184. PMID:23852952 [DOI] [PubMed] [Google Scholar]
- 15.Liao NS, Bix M, Zijlstra M, Jaenisch R, Raulet D. MHC class I deficiency: Susceptibility to natural killer (NK) cells and impaired NK activity. Science. 1991;253:199–202. doi: 10.1126/science.1853205. PMID:1853205 [DOI] [PubMed] [Google Scholar]
- 16.Malmberg KJ, Bryceson YT, Carlsten M, Andersson S, Bjorklund A, Bjorkstrom NK, Baumann BC, Fauriat C, Alici E, Dilber MS, et al. . NK cell-mediated targeting of human cancer and possibilities for new means of immunotherapy. Cancer Immunol Immunother. 2008;57:1541–52. doi: 10.1007/s00262-008-0492-7. PMID:18317755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colonna M, Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–8. doi: 10.1126/science.7716543. PMID:7716543 [DOI] [PubMed] [Google Scholar]
- 18.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, et al. . The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–6. doi: 10.1038/nm.3967. PMID:26457759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe. 1987;8:138–40. PMID:3303008 [PubMed] [Google Scholar]
- 20.Kasajima A, Sers C, Sasano H, Johrens K, Stenzinger A, Noske A, Buckendahl AC, Darb-Esfahani S, Müller BM, Budczies J, et al. . Down-regulation of the antigen processing machinery is linked to a loss of inflammatory response in colorectal cancer. Hum Pathol. 2010;41:1758–69. doi: 10.1016/j.humpath.2010.05.014. PMID:20869097 [DOI] [PubMed] [Google Scholar]
- 21.Poage GM, Butler RA, Houseman EA, McClean MD, Nelson HH, Christensen BC, Marsit CJ, Kelsey KT. Identification of an epigenetic profile classifier that is associated with survival in head and neck cancer. Cancer Res. 2012;72:2728–37. doi: 10.1158/0008-5472.CAN-11-4121-T. PMID:22507853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sigalotti L, Fratta E, Coral S, Tanzarella S, Danielli R, Colizzi F, Fonsatti E, Traversari C, Altomonte M, Maio M. Intratumor heterogeneity of cancer/testis antigens expression in human cutaneous melanoma is methylation-regulated and functionally reverted by 5-aza-2'-deoxycytidine. Cancer Res. 2004;64:9167–71. doi: 10.1158/0008-5472.CAN-04-1442. PMID:15604288 [DOI] [PubMed] [Google Scholar]
- 23.Heise R, Amann PM, Ensslen S, Marquardt Y, Czaja K, Joussen S, Beer D, Abele R, Plewnia G, Tampé R, et al. . Interferon alpha signalling and its relevance for the upregulatory effect of transporter proteins associated with antigen processing (TAP) in patients with malignant melanoma. PLoS One. 2016;11:e0146325. doi: 10.1371/journal.pone.0146325. PMID:26735690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma W, Lehner PJ, Cresswell P, Pober JS, Johnson DR. Interferon-gamma rapidly increases peptide transporter (TAP) subunit expression and peptide transport capacity in endothelial cells. J Biol Chem. 1997;272:16585–90. doi: 10.1074/jbc.272.26.16585. PMID:9195970 [DOI] [PubMed] [Google Scholar]
- 25.Trowsdale J, Hanson I, Mockridge I, Beck S, Townsend A, Kelly A. Sequences encoded in the class II region of the MHC related to the ‘ABC’ superfamily of transporters. Nature. 1990;348:741–4. doi: 10.1038/348741a0. PMID:2259383 [DOI] [PubMed] [Google Scholar]
- 26.Tarhini AA, Gogas H, Kirkwood JM. IFN-alpha in the treatment of melanoma. J Immunol. 2012;189:3789–93. doi: 10.4049/jimmunol.1290060. PMID:23042723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leibowitz MS, Andrade Filho PA, Ferrone S, Ferris RL. Deficiency of activated STAT1 in head and neck cancer cells mediates TAP1-dependent escape from cytotoxic T lymphocytes. Cancer Immunol Immunother. 2011;60:525–35. doi: 10.1007/s00262-010-0961-7. PMID:21207025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandoh N, Ogino T, Katayama A, Takahara M, Katada A, Hayashi T, Harabuchi Y. HLA class I antigen and transporter associated with antigen processing downregulation in metastatic lesions of head and neck squamous cell carcinoma as a marker of poor prognosis. Oncol Rep. 2010;23:933–9. doi: 10.3892/or_00000717. PMID:20204276 [DOI] [PubMed] [Google Scholar]
- 29.Lou Y, Vitalis TZ, Basha G, Cai B, Chen SS, Choi KB, Jeffries AP, Elliott WM, Atkins D, Seliger B, et al. . Restoration of the expression of transporters associated with antigen processing in lung carcinoma increases tumor-specific immune responses and survival. Cancer Res. 2005;65:7926–33. doi: 10.1158/0008-5472.CAN-04-3977. PMID:16140964 [DOI] [PubMed] [Google Scholar]
- 30.Kageshita T, Hirai S, Ono T, Hicklin DJ, Ferrone S. Down-regulation of HLA class I antigen-processing molecules in malignant melanoma: Association with disease progression. Am J Pathol. 1999;154:745–54. doi: 10.1016/S0002-9440(10)65321-7. PMID:10079252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mehta AM, Jordanova ES, Kenter GG, Ferrone S, Fleuren GJ. Association of antigen processing machinery and HLA class I defects with clinicopathological outcome in cervical carcinoma. Cancer Immunol Immunother. 2008;57:197–206. doi: 10.1007/s00262-007-0362-8. PMID:17622526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ylitalo EB, Thysell E, Jernberg E, Lundholm M, Crnalic S, Egevad L, Stattin P, Widmark A, Bergh A, Wikström P. Subgroups of castration-resistant prostate cancer bone metastases defined through an inverse relationship between androgen receptor activity and immune response. Eur Urol. 2016;71(5):776–87. doi: 10.1016/j.eururo.2016.07.033. PMID:27497761 [DOI] [PubMed] [Google Scholar]
- 33.Cabrera CM, Jimenez P, Cabrera T, Esparza C, Ruiz-Cabello F, Garrido F. Total loss of MHC class I in colorectal tumors can be explained by two molecular pathways: Beta2-microglobulin inactivation in MSI-positive tumors and LMP7/TAP2 downregulation in MSI-negative tumors. Tissue Antigens. 2003;61:211–9. doi: 10.1034/j.1399-0039.2003.00020.x. PMID:12694570 [DOI] [PubMed] [Google Scholar]
- 34.Benevolo M, Mottolese M, Piperno G, Sperduti I, Cione A, Sibilio L, Martayan A, Donnorso RP, Cosimelli M, Giacomini P. HLA-A, -B, -C expression in colon carcinoma mimics that of the normal colonic mucosa and is prognostically relevant. Am J Surg Pathol. 2007;31:76–84. doi: 10.1097/01.pas.0000213343.55605.b9. PMID:17197922 [DOI] [PubMed] [Google Scholar]
- 35.Iwayama Y, Tsuruma T, Mizuguchi T, Furuhata T, Toyota N, Matsumura M, Torigoe T, Sato N, Hirata K. Prognostic value of HLA class I expression in patients with colorectal cancer. World J Surg Oncol. 2015;13:36. doi: 10.1186/s12957-015-0456-2. PMID:25889416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menon AG, Morreau H, Tollenaar RA, Alphenaar E, Van Puijenbroek M, Putter H, Janssen-Van Rhijn CM, Van De Velde CJ, Fleuren GJ, Kuppen PJ. Down-regulation of HLA-A expression correlates with a better prognosis in colorectal cancer patients. Lab Invest. 2002;82:1725–33. doi: 10.1097/01.LAB.0000043124.75633.ED. PMID:12480922 [DOI] [PubMed] [Google Scholar]
- 37.Watson NF, Ramage JM, Madjd Z, Spendlove I, Ellis IO, Scholefield JH, Durrant LG. Immunosurveillance is active in colorectal cancer as downregulation but not complete loss of MHC class I expression correlates with a poor prognosis. Int J Cancer. 2006;118:6–10. doi: 10.1002/ijc.21303. PMID:16003753 [DOI] [PubMed] [Google Scholar]
- 38.Lampen MH, Verweij MC, Querido B, van der Burg SH, Wiertz EJ, van Hall T. CD8+ T cell responses against TAP-inhibited cells are readily detected in the human population. J Immunol. 2010;185:6508–17. doi: 10.4049/jimmunol.1001774. PMID:20980626 [DOI] [PubMed] [Google Scholar]
- 39.Hasim A, Abudula M, Aimiduo R, Ma JQ, Jiao Z, Akula G, Wang T, Abudula A. Post-transcriptional and epigenetic regulation of antigen processing machinery (APM) components and HLA-I in cervical cancers from Uighur women. PLoS One. 2012;7:e44952. doi: 10.1371/journal.pone.0044952. PMID:23024775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khan AN, Gregorie CJ, Tomasi TB. Histone deacetylase inhibitors induce TAP, LMP, Tapasin genes and MHC class I antigen presentation by melanoma cells. Cancer Immunol Immunother. 2008;57:647–54. doi: 10.1007/s00262-007-0402-4. PMID:18046553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dahlin AM, Henriksson ML, Van Guelpen B Stenling R, Oberg A, Rutegard J, Palmqvist R. Colorectal cancer prognosis depends on T-cell infiltration and molecular characteristics of the tumor. Mod Pathol. 2011;24:671–82. doi: 10.1038/modpathol.2010.234. PMID:21240258 [DOI] [PubMed] [Google Scholar]
- 42.Prall F, Nizze H, Barten M. Tumour budding as prognostic factor in stage I/II colorectal carcinoma. Histopathology. 2005;47:17–24. doi: 10.1111/j.1365-2559.2005.02161.x. PMID:15982319 [DOI] [PubMed] [Google Scholar]
- 43.Ueno H, Murphy J, Jass JR, Mochizuki H, Talbot IC. Tumour ‘budding’ as an index to estimate the potential of aggressiveness in rectal cancer. Histopathology. 2002;40:127–32. doi: 10.1046/j.1365-2559.2002.01324.x. PMID:11952856 [DOI] [PubMed] [Google Scholar]
- 44.Edin S, Wikberg ML, Dahlin AM, Rutegard J, Oberg A, Oldenborg PA, Palmqvist R. The distribution of macrophages with a M1 or M2 phenotype in relation to prognosis and the molecular characteristics of colorectal cancer. PLoS One. 2012;7:e47045. doi: 10.1371/journal.pone.0047045. PMID:23077543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. PMID:22810696 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.