Abstract
Congenital heart disease (CHD) is the leading cause of mortality from birth defects. Exome sequencing of a single cohort of 2,871 CHD probands including 2,645 parent-offspring trios implicated rare inherited mutations in 1.8%, including a recessive founder mutation in GDF1 accounting for ~5% of severe CHD in Ashkenazim, recessive genotypes in MYH6 accounting for ~11% of Shone complex, and dominant FLT4 mutations accounting for 2.3% of Tetralogy of Fallot. De novo mutations (DNMs) accounted for 8% of cases, including ~3% of isolated CHD patients and ~28% with both neurodevelopmental and extra-cardiac congenital anomalies. Seven genes surpassed thresholds for genome-wide significance and 12 genes not previously implicated in CHD had > 70% probability of being disease-related; DNMs in ~440 genes are inferred to contribute to CHD. There was striking overlap between genes with damaging DNMs in probands with CHD and autism.
INTRODUCTION
Congenital heart disease (CHD) affects ~1% of live births and remains the leading cause of mortality from birth defects1. After surgical repair, patients remain at risk of cardiac arrhythmias, heart failure, neurodevelopmental deficits and other congenital anomalies2, 3. While aneuploidies and copy number variations (CNVs) account for ~23% of CHD patients4–6, these have yielded few individual causal genes. While genes causing rare Mendelian syndromic forms of CHD have been identified, genes underlying the large majority of sporadic CHD remain unknown.
To this end, the NHLBI Pediatric Cardiac Genomics Consortium (PCGC) has collected >10,000 CHD probands, including >5,000 parent-offspring trios7. Whole exome sequencing (WES) of 1,213 trios from this cohort showed that ~10% of cases are attributable to de novo mutations (DNMs) in >400 target genes, including dramatic enrichment for damaging mutations in genes encoding chromatin modifiers8, 9. Moreover, these studies demonstrated a striking shared genetic etiology between CHD and neurodevelopmental disorders (NDD)6, 9.
Genetic studies of humans and mice predict a role for inherited variants with large effect10, 11. Analysis of rare multigenerational CHD families has identified mutations in cardiac transcription factors, signaling molecules and structural components12. Inherited heterozygous protein-truncating variants have been implicated in non-syndromic CHD and have suggested distinct genetic architectures for syndromic and non-syndormic CHD9, 13. To date, the roles of recessive inheritance and novel genes operating via dominant transmission have not been systematically studied. Discovery of additional large-effect mutations requires large cohorts, comprehensive genomic data and robust statistical methods.
Here, we analyze the impact of rare inherited recessive and dominant variants, and of DNMs on CHD via WES of a single large CHD cohort.
RESULTS
Cohort Characteristics and Sequencing
We studied 2,871 CHD probands comprising 2,645 parent-offspring trios and 226 singletons recruited to the PCGC and the Pediatric Heart Network (PHN) programs (Supplementary Data Set 1). These include 1,204 previously reported trios9. The ethnicities, gender and clinical features of probands are shown in Supplementary Table 2 and Supplementary Tables 3a–c. Patients with known trisomies and CHD-associated CNVs were prospectively excluded from analysis.
Genomic DNAs underwent WES (see Online Methods). In parallel, WES from 1,789 control trios comprising parents and unaffected siblings of autism probands was analyzed14. Cases and controls showed similar sequencing metrics (Supplementary Table 4). Variants were called and annotated as described in methods.
Recessive Genotypes Enriched in CHD
Principal component analysis (PCA) from WES genotypes showed that CHD cases were more frequently of non-European ancestry than controls. The inbreeding coefficient of probands was higher than controls (Supplementary Figure 1). These differences complicate direct comparison recessive genoytpes (RGs) in cases and controls. Accordingly, we implemented a binomial test to quantify the enrichment of damaging RGs in genes or gene sets in cases, independent of controls. This method compares the observed number of rare damaging RGs to the expected frequency, estimated from from the de novo probability, adjusting for inbreeding, using the polynomial model (see Online Methods and Supplementary Figures 2–6).
We curated a set of 212 human CHD genes (H-CHD genes) from the Online Mendelian Inheritance in Man (OMIM) and published data13, and human orthologs of 61 mouse CHD genes (M-CHD genes) identified in a recessive screen for CHD (Supplementary Data Set 2 and Supplementary Note)11. The H-CHD set comprised 104 dominant genes, 85 recessive genes, 12 X-linked genes, and 11 genes showing both dominant and recessive transmission. Accounting for 20 genes identified in both human and mouse, the combined set comprised 253 human genes (Supplementary Data Set 2).
We identified rare (minor allele frequency [MAF] < 0.001) likely loss-of-function (LoF; frameshift, nonsense, canonical splice site, and start loss), likely damaging missense variants (by MetaSVM; D-Mis), and non-frameshift insertion/deletion variants, and identified homozygous or compound heterozygous genotypes comprising these alleles. This identified 467 damaging RGs in CHD cases (Supplementary Data Set 3) and 165 in controls (Supplementary Data Set 4).
We used the one-tailed binomial test to determine whether damaging RGs were enriched among 96 genes implicated in recessive human CHD (Table 1a). This gene set had 29 damaging RGs vs. 6.7 expected (enrichment = 4.4, P = 8.0×10−11; Table 1, Supplementary Figure 5b, Supplementary Table 5). This set showed zero RGs in controls (Table 1). Adding 41 recessive mouse genes, there were 34 damaging RGs compared to 11.1 expected (enrichment = 3.1, P = 1.4×10−8; Table 1). Adding 116 dominant CHD genes added 17 damaging RGs in 9 genes (cumulative total, observed 51 vs. expected 25.2, enrichment = 2.0, P = 1.8×10−6; Table 1). Similar results were obtained from independently modeling homozygous and compound heterozygous genotypes (see Online Methods, Supplementary Table 6, and Supplementary Figures 7–8) and further corroborated using a burden test-based approach15, 16 that also integrates proband phenotype17 (see Online Methods and Supplementary Figure 9). These findings implicate RGs in known CHD genes in 0.9% of these CHD cases.
Table 1.
Damaging recessive genotypes in known CHD genes in cases and controls
| 2,871 CHD cases | |||||||
|
| |||||||
| Gene set (# genes) | Observed | Expected | Enrichment | P-value | |||
|
|
|
||||||
| # homozygotes |
# compound heterozygous |
# unique genes |
# recessive genotypes |
# recessive genotypes |
|||
|
| |||||||
| All genes (18,989) | 265 | 202 | 391 | 467 | - | - | - |
|
| |||||||
| Recessive Known Human (96) | 19 | 10 | 16 | 29 | 6.65 | 4.36 | 8.0×10−11 |
| Recessive Known Mouse or Human (137) | 21 | 13 | 19 | 34 | 11.06 | 3.07 | 1.4×10−8 |
| Known Mouse or Human CHD (253) | 28 | 23 | 28 | 51 | 25.15 | 2.03 | 1.8×10−6 |
|
| |||||||
| 1,789 controls | Observed | Expected | |||||
|
| |||||||
| All genes (18,989) | 22 | 131 | 146 | 165 | - | - | - |
|
| |||||||
| Recessive Known Human (96) | 0 | 0 | 0 | 0 | 2.61 | 0 | 1 |
| Recessive Known Mouse or Human (137) | 1 | 1 | 2 | 2 | 4.47 | 0.45 | 0.94 |
| Known Mouse or Human CHD (253) | 2 | 3 | 5 | 5 | 10.18 | 0.49 | 0.98 |
The expected number of recessive genotypes was determined based on fitted values from the polynomial regression model using the damaging de novo probabilities. P-values were calculated using the one-tailed binomial probability. Values in bold are p-values exceeding the Bonferroni multiple testing cutoff = 0.05/(3×2) = 8.3×10−3.
For previously identified recessive genes, the observed and previously reported cardiac phenotypes were concordant in 22 of 31 cases, suggesting variable expressivity of RGs. For previously identified dominant genes, observed cardiac phenotypes matched those previously reported in only 3 of 17 probands. Of these, phenotypes seen with RGs were more severe than previously described dominant phenotypes (COL1A1, COL5A2, FBN2, MYH6, NSD1, and TSC2), or at the severe end of the described spectrum (CHD7 and NOTCH1; Supplementary Table 5).
We examined the contribution of consanguinity to RGs. 161 probands (5.6%) had homozygous segments implying parental relationships of 3rd cousins or closer (see Supplementary Note). This group included 81 of 84 probands with reported consanguinity. Thirteen (8.1%) of these probands had damaging RGs in recessive H-CHD genes (2.4 expected, 5.4-fold enrichment, P = 1.3×10−6; Supplementary Table 7); all but one genotype was homozygous. Among the remaining 2710 probands, RGs were also enriched (3.9-fold, 16 observed vs. 4.1 expected, P = 5.3×10−6), however RG’s comprised only 0.6% of this group (Supplementary Table 7). Among the seven homozygotes in this group, five probands had inbreeding coefficients between 0.0015 and 0.0035, implying distant parental relatedness, whereas two homozygotes and all nine compound heterozygotes had inbreeding coefficients of zero. Thus, cryptic or overt parental consanguinity was a strong driver of recessive CHD in this cohort. Importantly, 38% of RGs in recessive CHD genes were attributable to a single GDF1 founder mutation (see below). Significant enrichment for RGs in known CHD genes persists after removal of GDF1 homozygotes (Supplementary Table 8).
We observed 44 genes with > 1 damaging RG compared to 26.4 expected (enrichment = 1.7; P = 8.9×10−5 by permutation; see Online Methods); synonymous RGs were not significantly enriched (167 observed, 156.7 expected, P = 0.15 by permutation). This excess persisted after removal of 5 known recessive genes (GDF1, ATIC, DNAH5, DAW1, LRP1; enrichment = 1.6; P = 10−3 by permutation). GO ontology of the novel gene set revealed enrichment of genes involved in muscle cell development (GO:0055001, enrichment = 29.5, FDR = 3.2×10−3), including KEL, MYH6, MYH11, NOTCH1, and RYR1 (Supplementary Data Sets 3,5).
Founder Mutation in GDF1 in Ashkenazim
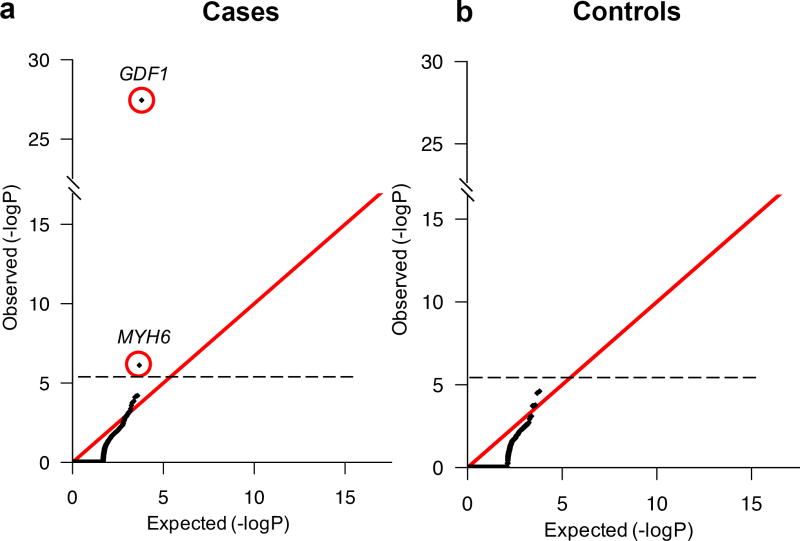
Q-Q plots comparing the observed and expected damaging RGs in each gene using the binomial test showed that two genes, GDF1 and MYH6, had more RGs than expected (genome-wide threshold, P < 2.6×10−6, Figure 1a; Supplementary Table 9); modeling homozygotes and compound heterozygotes separately yielded similar results (Supplementary Table 10). No genes approached genome-wide significance in controls (Figure 1b).
Figure 1. Quantile-quantile plots comparing observed versus expected P-values for recessive genotypes in each gene in cases and controls.
Recessive genotypes (RGs) shown include LoF, D-Mis, and non frameshift insertion/deletions. The expected number of RGs in each gene was calculated from the total number of observed RGs as described in Methods. The significance of the difference between the observed and expected number of RGs was calculated using a one-sided binomial test. (a). Quantile-quantile (Q-Q) plot in cases. (b). Q-Q plot in controls. While the observed values closely conform to expected values in controls, two genes, GDF1 and MYH6, show a significantly increased burden of RGs in cases and survive the multiple-testing correction threshold.
GDF1 had 11 damaging RGs in apparently unrelated subjects compared with 0.016 expected (enrichment = 692.6, one-tailed binomial P = 3.6×10−28; Supplementary Table 9); all were confirmed by Sanger sequencing (Supplementary Figure 10). Ten RGs were homozygous for a p.Met364Thr (c.1091T>C) variant, suggesting a founder mutation; the other was p.Met364del (c.1090_1092delATG)/p.Cys227* (c.681C>A). Consistent with a founder mutation, PCA showed that all p.Met364Thr homozygotes clustered with Ashkenazim (Supplementary Figure 11).
Additional evidence supports homozygosity for p.Met364Thr in CHD risk among Ashkenazim. p.Met364Thr shows remarkable violation of Hardy Weinberg equilibrium among Ashkenazi CHD cases, with 10 homozygotes and only 1 heterozygote among 204 Ashkenazi cases defined by PCA (P = 5.5×10−38, 1-df chi-square test with Yate’s correction; Supplementary Table 11a). In contrast, among 302 Ashkenazi autism parental controls and 926 additional Ashkenazi adults from an independent cohort without CHD, there were no homozygotes and 12 heterozygotes (carrier frequency = 1.0%), providing strong association of p.Met364Thr homozygosity with CHD among Ashkenazim (two-sided Fisher’s Exact P = 2.8×10−9, Supplementary Table S11b). Moreover, this allele was absent among African, Asian, and Finnish European populations in ExAC.
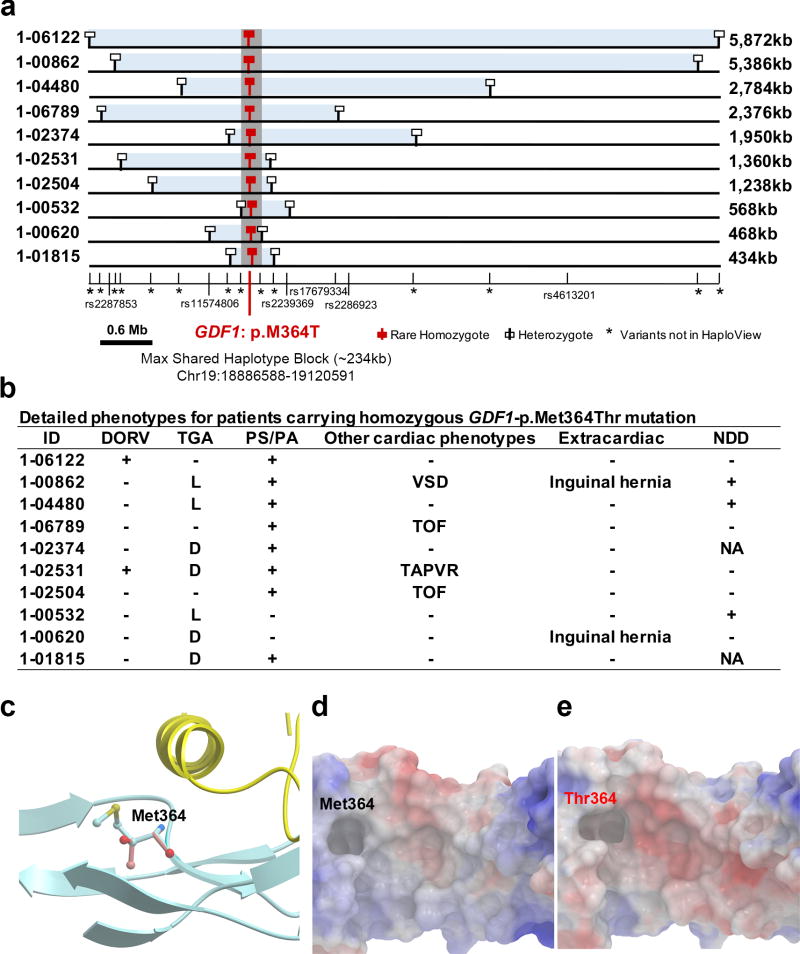
Lastly, all homozygotes shared p.Met364Thr on a common haplotype background, indicating identity by descent (Figure 2a). The length of the shared haplotype varied widely (0.4–5.9 Mb; Figure 2a), indicating remote shared ancestry. The inferred coalescent time for the last shared ancestor, using DMLE+2.3 software18, is 50 generations (95% CI: 45 to 63 generations; Supplementary Figure 12).
Figure 2. Phenotypes and shared haplotypes among homozygotes for GDF1-p.Met364Thr.
(a). Extent of homozygous SNPs flanking homozygous GDF1-p.Met364Thr genotypes. A 5.9 Mb segment of chromosome 19 extending across the location of the homozygous GDF1-p.Met364Thr mutation (denoted by red square) in each unrelated subject is depicted. At the bottom, tick marks indicate location of all SNPs found by exome sequencing among Ashkenazim in cases. Known SNPs are shown via their rs identifiers. Allele frequencies of novel SNPs are indicated by asterisks. The closest heterozygous SNP to either side of the GDF1-p.Met364Thr in each subject is shown as a white square; all SNPs between these two heterozygous SNPs, encompassed by the light blue bar, are homozygous for the same allele seen in other subjects, consistent with the p.Met364Thr variant being identical by descent among all subjects. The length of each homozygous segment is indicated at the right of the panel. The maximum length of the homozygous segment shared by all subjects is 234 kb (shown as grey vertical bar), consistent with the mutation having been introduced into a shared ancestor many generations ago. (b). Cardiac and extracardiac phenotypes of GDF1-p.Met364Thr homozygotes. Present phenotypes are denoted with ‘+’, those absent with ‘−’, and those unavailable for testing with ‘NA’ (c). Ribbon diagram of part of GDF1 homodimer containing p.Met364. The hydrophobic helix from one subunit (yellow) sits above p.Met364 on the other subunit (blue). (d). Space filling model of the segment of GDF1 containing the wild-type p.Met364 showing surface electrostatic charge (blue=positive, red=negative). (e). Surface electrostatic charge of the segment containing mutant p.Thr364. Compared to wild-type, the mutant peptide shows a more negatively charged cavity.
Consistent with this RG causing CHD and not merely being in linkage disequilibrium with the causal variant, the phenotype of p.Met364Thr homozygotes is shared by previously described cases with different recessive GDF1 mutations19. Like prior cases, all GDF1 p.Met364Thr homozygotes had D- or L-transposition of the great arteries, pulmonary stenosis/atresia or both (Figure 2b). GDF1 belongs to the transforming growth factor-beta (TGF-β) superfamily. Studies in mouse implicated Gdf1 in establishment of left-right asymmetry and neural development20–22. GDF1 functions as a homodimer with two-fold inverted symmetry (Figure 2c and Supplementary Figure 13). The interaction surface between monomers comprises a hydrophophic α-helix in one monomer and a hydrophobic cavity in the other; this interaction occurs reciprocally. Met364 lies in the hydrophobic cavity (Figure 2d–e). p.Met364Thr substitutes the polar threonine in the hydrophobic cavity; we infer that this variant impairs dimer formation and downstream signaling (Figure 2c), consistent with recessive transmission.
Homozygosity for GDF1 p.Met364Thr accounts for ~5% of severe CHD among Ashkenazim, including 18% of those with TGA (7 of 38), and 31% with TGA plus PS/PA (5 of 16). This finding has clinical implications for assessing risk of CHD among Ashkenazim.
Recessive MYH6 Genotypes in Shone Complex
MYH6 encodes the cardiac alpha myosin heavy chain, which is highly expressed in embryonic heart. Dominant MYH6 mutations are implicated in atrial septal defect23 and cardiomyopathy24, 25. We identified seven rare damaging RGs in MYH6 versus 0.482 expected (enrichment = 14.5, P = 7.6×10−7; Supplementary Table 9). These included diverse LoF alleles and D-Mis variants, all validated by Sanger sequencing (Table 2, Supplementary Table 9, and Supplementary Figure 14). Five probands had left ventricular obstruction, including four with Shone complex26, having mitral valve and aortic valve obstruction plus aortic arch obstruction (Table 2). Echocardiography revealed abnormal ventricular function in 4 of 7 probands, consistent with a previous report of two patients with RGs in MYH6 who had decreased ventricular function27. RGs in MYH6 accounted for 11% of the 37 sequenced patients with Shone complex (enrichment = 57.45, two-sided Fisher’s exact P = 6.7×10−5).
Table 2.
Recessive MYH6 genotypes associated with Shone complex and valvular disease.
| ID | AA Change (coding DNA Change) |
ExAC Ethnic Specific Freq |
Shone complex |
Detailed Cardiac Phenotype |
Cardiac Function | Extracardiac | NDD | Age at follow-up |
|---|---|---|---|---|---|---|---|---|
| 1-00051 | p.Lys1932*/p.Ala1891Thr (c.5794A>T/c.5671G>A) | 3.0×10−5/0 | + | LSVC, abn MV, sub AS, valve AS, CoA | LV diastolic dysfunction | − | + (LD) | 22 |
| 1-01407 | p.Glu98Lys (c.292G>A) | 3.0×10−4 | − | mitral atresia, DORV, CoA | mild RV systolic dysfunction | Hypothyroid | + (LD) | 16 |
| 1-04847 | p.Arg1899His/p.Asn598Lysfs*38 (c.5696G>A/c.1793dupA) | 0/0 | + | parachute MV, BAV, CoA | NL | − | − | 16 |
| 1-05009 | p.Ala1327Val/p.Leu388Phe(c.3980C>T/c.1162C>T) | 2.7×10−3/0 | − | TA, PA | dilated, hyper-trabeculated LV | − | NA | 0 |
| 1-06399 | p.Gly585Ser/p.Ile512Thr(c.1753G>A/c.1535T>C) | 2.0×10−4/3.0×10−5 | + | mitral stenosis, VSD, BAV, hypoplastic transv. Ao | NL | − | NA | 0.08 |
| 1-06876 | p.Ile1068Thr/Splice site(c.3203T>c.3979-2A>C) | 1.5×10−5/2.0×10−5 | + | LSVC, abn mitral valve, valve AS, CoA | dilated LV | − | − | 22 |
| 1-07343 | p.Arg1610Cys (c.4828C>T) | 3.0×10−5 | − | ASD/VSD | NA | − | NA | NA |
Abbreviations: ASD- Atrial septal defect, AS- Aortic stenosis, BAV- Bicuspid aortic valve, CoA- Coarctation of the aorta, DORV- Double outlet right ventricle. MV-mitral valve, PA-Pulmonary atresia, TA-Tricuspid atresia, VSD-Ventricular septal defect. Extracardiac manifestations refer to CHD probands displaying additional abnormalities not pertaining to the heart. NDD-neurodevelopmental disabilities, LD-Learning Disability, NA-NDD status not attained as proband < age 1. “+”:Present; “ −”:Not present.
Recessive Genotypes Enriched in Patients with Laterality Defects
Among the major CHD subgroups (laterality defects, left ventricular obstruction, conotruncal defects and others; Supplementary Table 3a), only laterality defects (heterotaxy and D-TGA) were significantly enriched for damaging RGs in known CHD genes (21 damaging RGs in 13 genes vs. 4.8 expected; enrichment = 4.4, P = 8.5×10−9; Supplementary Table 12). Significant enrichment persisted after removing GDF1 RGs (enrichment = 3.2, P = 1.2×10−4). These RGs occurred in eight genes previously implicated in laterality defects (ARMC4, BBS10, DAW1, DNAAF1, DNAH5, DYNC2H1, GDF1, and PKD1L1) and five not previously implicated (ATIC, COL1A1, COL5A2, DGCR2, and MYH6).
We also performed GO ontology analysis of all 82 genes with LoF RGs. This identified significant terms related to cilia structure/regulation, a predominant mechanism in laterality determination (Supplementary Data Set 6). Genes in these GO terms included DNAI2, ARMC4, DNAH5, and DNAAF1 (proband phenotypes in Supplementary Data Set 3). Although all these genes have been associated with human primary ciliary dyskinesia and situs inversus totalis, only DNAH5 has been previously associated with human CHD28.
Heterozygous LoF Mutations in FLT4 in Tetralogy of Fallot
We compared the observed and expected frequency of rare (MAF ≤ 10−5) heterozygous LoF variants in 115 known dominant CHD genes in cases and controls using the binomial test and found no significant enrichment in either group (Supplementary Data Sets 7–8; Supplementary Table 13a,b). Analysis of heterozygous LoF variants in all 212 known human CHD genes also showed no enrichment.
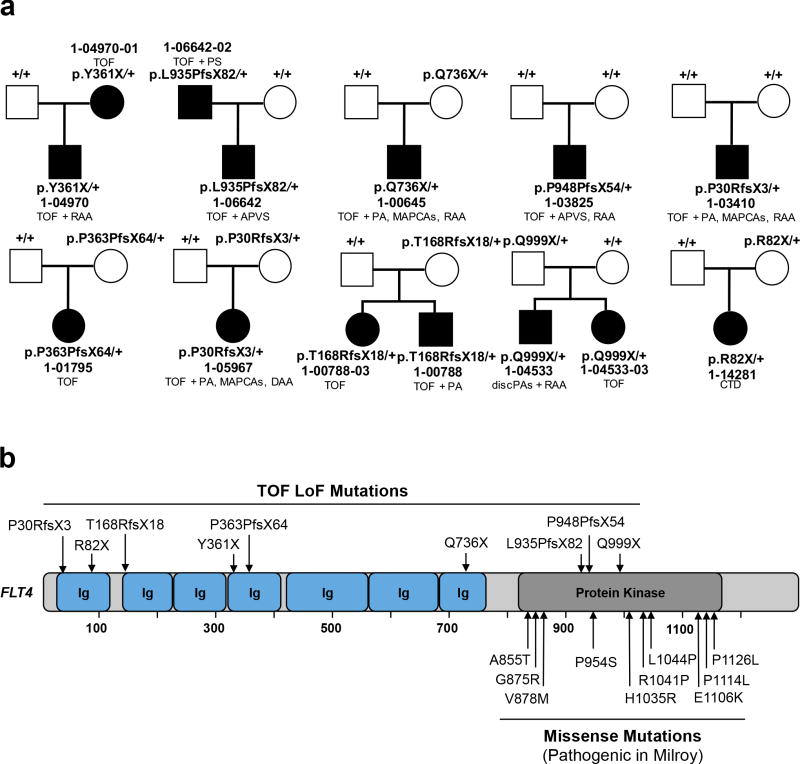
To search for novel haploinsufficient CHD genes, we compared the observed and expected distribution of rare heterozygous LoFs in each gene (see Online Methods). Q-Q plots (Supplementary Figure 15) showed that FLT4, with eight different inherited LoFs, significantly departed from expectation (enrichment = 15.5, P = 7.6×10−8, Supplementary Table 14). Moreover, there were two de novo FLT4 LoF mutations, yielding a combined p-value of 9.8×10−10 (p-values combined by Fisher’s method, Figure 3). LoF variants were distributed throughout the encoded protein; all were confirmed by Sanger sequencing (Supplementary Figure 16).
Figure 3. FLT4 loss-of-function mutations in Tetralogy of Fallot.
(a). Pedigrees of 10 CHD kindreds with rare FLT4 loss-of-function (LoF) mutations are shown. Subjects with and without CHD are shown as filled and unfilled symbols, respectively. Each kindred ID number is shown along with the FLT4 genotype of each subject and CHD phenotype of affected subjects. (b) Diagram of FLT4 protein is shown with seven immunoglobulin domains (Ig) and a kinase domain. The top panel shows LoF mutations associated with Tetralogy-type CHD, whereas the bottom panel displays missense mutations associated with the Milroy disease (Hereditary Lymphedema).
FLT4 was highly intolerant to LoF variation in ExAC (pLI = 1) and only one LoF allele was identified among 3,578 parental controls. Pedigrees of FLT4 probands revealed four family members with CHD; all shared the proband’s FLT4 mutation (Figure 3a). However, only 4 of 10 FLT4 mutation carriers reported CHD, indicating incomplete penetrance.
Strongly supporting a pathogenic role for the FLT4 LoFs, the phenotype of 9 of 10 probands and 3 of 4 affected relatives was tetralogy of Fallot (TOF) (Figure 3a); mutation carriers had no extracardiac malformations, growth abnormalities or NDD. Among 426 probands with TOF in our cohort, 2.3% had FLT4 LoF mutations (95.2-fold enrichment, P = 1.9×10−12; Supplementary Table 15).
FLT4 encodes a VEGF receptor expressed in lymphatics and the vasculature. Interestingly, diverse missense mutations that cluster in the kinase domain and impair enzymatic activity cause hereditary lymphedema (Figure 3b)29.
De Novo Damaging Mutations Enriched in Isolated CHD Cases
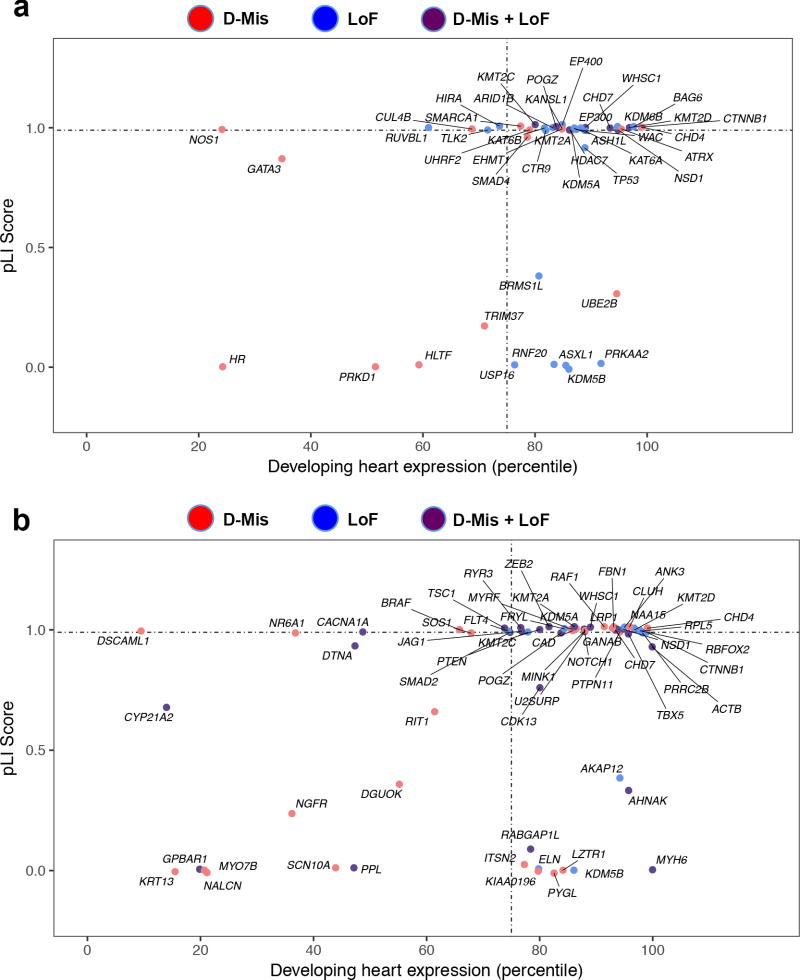
The number of observed DNMs in cases and controls closely fit the Poisson distribution (Supplementary Figure 17; Supplementary Data Sets 9–10). Damaging DNMs were enriched in cases (1.4-fold, P = 2.4×10−17, Supplementary Table 16) but not controls. We inferred that damaging DNMs contribute to ~8.3% of cases. Additionally, we found 89 damaging DNMs in 46 chromatin modifiers accounting for 2.3% of cases (enrichment = 3.1, P = 8.7×10−20; Figure 4a; Supplementary Tables 17–18), including seventeen chromatin modifier genes not previously implicated in CHD.
Figure 4. Chromatin modification genes and genes with multiple damaging de novo mutations are enriched for high expression in developing heart and intolerance to loss-of-function mutation.
(a) Enrichment of damaging mutations in chromatin modifiers in genes highly expressed in developing heart and intolerant to loss-of-function (LoF) mutation. X axis (0–100) denotes the percentile rank of heart expression in developing mouse heart at E14.5, and y axis (0–1.0) denotes intolerance to LoF mutation (pLI) in the ExAC database. (b) 66 genes with 2 or more damaging de novo mutations are plotted. Multihit genes are highly enriched (N=31) for genes that are highly expressed in developing heart and intolerant to LoF mutation (pLI ≥ 0.99).
There were 66 genes with two or more damaging DNMs compared to 21 previously8, 9 (Figure 4b, Supplementary Tables 19–20). Interestingly, 108 damaging DNMs affecting 39 of 104 known dominant H-CHD genes accounted for 3.7% of cases (enrichment = 9.3, P = 5.5×10−65; Supplementary Table 21). An orthogonal analytic approach yielded similar results (see Supplementary Note and Supplementary Figure 18).
Unlike prior studies8, 9, 13, we found that damaging DNMs were enriched in isolated CHD cases (CHD without extracardiac congenital anomaly, clinically diagnosed syndrome or neurodevelopmental abnormality, and limited to patients over age 1 at enrollment); these mutations contributed to ~3.1% of cases (1.5-fold enrichment, P = 8.5×10−4; Supplementary Table 22a). Damaging DNMs in known CHD genes accounted for ~50% (13/26) of the excess mutation burden in isolated CHD. DNMs contributed to 6%–8% of probands with any extracardiac features (EA alone or NDD alone), and to 28% of cases with both EA and NDD (Supplementary Tables 22a–d and 23).
De novo mutations are Enriched in Autism-Associated Genes
We previously showed unexpected overlap of genes harboring damaging DNMs in CHD and neurodevelopmental disorders8, 9. We compared the genes harboring damaging DNMs in our CHD cohort and in 4,778 probands with autism30, 31, focusing on genes in the upper quartile of brain and heart expression. Nineteen such genes had de novo LoF mutations in both cohorts (enrichment 5.2, P < 10−6) and 48 had damaging mutations in both (enrichment 2.8, P < 10−6; Supplementary Table 24). Notably, among CHD patients with neurodevelopmental phenotyping, 67% (21/31) of those with LoF DNMs in the overlapping gene set had NDD, compared to 32.8% in the total cohort with neurodevelopmental phenotyping (OR = 4.3; two-sided Fisher’s P = 1.4 ×10−4; Supplementary Table 25). Notably, 14/35 of all genes with LoF DNMs in both the CHD and autism cohorts are chromatin modifiers (enrichment = 14.7, P < 10−6 by permutation; Supplementary Table 25). Most strikingly, 87% of patients who had LoF DNMs in chromatin modifiers had NDD at enrollment.
Meta-Analysis of Damaging De Novo and Loss-of-function Heterozygous Variants
We tested each gene for an excess of de novo and rare inherited heterozygous variants. Seven genes (CHD7, KMT2D, PTPN11, RBFOX2, FLT4, SMAD6, and NOTCH1) surpassed genome wide significance (Table 3) compared to four previously9, 13. Among the remaining top 25 genes, KDM5B had strong prior statistical support, ELN, NSD1, NODAL, RPL5, and SOS1 have previously been found associated with syndromic CHD; GATA6, FRYL, and TBX18 were identified in case reports with a phenotype that included CHD. Our findings strengthen the evidence supporting a role for these genes.
Table 3.
Top 25 genes in the meta-analysis of damaging de novo mutations and loss-of-function heterozygous mutations in probands
| Gene | Damaging de novo | LoF heterozygotes | Meta P-value | pLI | HHE Rank | Gene Set | ||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| # Damaging | P-value | # LoF | P-value | |||||
| CHD7 | 14 | 1.6×10−20 | 0 | 1 | 7.5×10−19 | 1 | 93.4 | H-CHD/Chromatin |
| KMT2D | 16 | 2.1×10−20 | 1* | 0.86 | 8.5×10−19 | 1 | 96.8 | H-CHD/Chromatin |
| PTPN11 | 9 | 4.6×10−17 | 0 | 1 | 1.8×10−15 | 1 | 94.2 | H-CHD |
| FLT4 | 2 | 5.2×10−4 | 8 | 7.6×10−8 | 9.8×10−10 | 1 | 74.4 | NA |
| NOTCH1 | 5 | 2.7×10−5 | 6* | 1.8×10−4 | 9.4×10−8 | 1 | 87.9 | H-CHD |
| RBFOX2 | 3 | 3.4×10−7 | 1* | 0.18 | 1.1×10−6 | 0.99 | 97.8 | NA |
| SMAD6 | 1 | 0.012 | 8 | 6.0×10−6 | 1.3×10−6 | 0 | 78.3 | M-CHD |
| GATA6 | 4 | 2.4×10−7 | 0 | 1 | 3.8×10−6 | N/A | 94.8 | H-CHD |
| ELN | 2 | 1.3×10−4 | 5* | 8.7×10−3 | 1.7×10−5 | 0 | 79.8 | H-CHD |
| CCDC154 | 0 | 1 | 7* | 5.5×10−6 | 7.2×10−5 | 0.31 | 18.4 | NA |
| SLCO1B3 | 0 | 1 | 9 | 6.6×10−6 | 8.5×10−5 | 0 | 11.7 | NA |
| GPBAR1 | 2 | 2.6×10−5 | 1 | 0.27 | 9.1×10−5 | 0 | 19.9 | NA |
| PTEN | 2 | 6.0×10−5 | 1 | 0.16 | 1.2×10−4 | 0.98 | 77.9 | H-CHD |
| RPL5 | 2 | 6.2×10−5 | 1 | 0.16 | 1.3×10−4 | 0.99 | 97.9 | H-CHD |
| NSD1 | 5 | 1.0×10−5 | 0 | 1 | 1.3×10−4 | 1 | 94.8 | H-CHD/Chromatin |
| SAMD11 | 2 | 1.8×10−4 | 4* | 0.06 | 1.4×10−4 | 0 | N/A | NA |
| C21ORF2 | 0 | 1 | 5 | 1.2×10−5 | 1.5×10−4 | 0.01 | 46.7 | NA |
| NODAL | 0 | 1 | 4 | 1.2×10−5 | 1.5×10−4 | 0.95 | 16.4 | H-CHD |
| SMAD2 | 3 | 5.5×10−5 | 1 | 0.24 | 1.6×10−4 | 0.99 | 74.7 | NA |
| H1FOO | 0 | 1 | 4 | 1.6×10−5 | 1.9×10−4 | 0.4 | 10.3 | NA |
| FRYL | 2 | 2.8×10−3 | 5* | 8.3×10−3 | 2.8×10−4 | 1 | 84.4 | NA |
| KDM5B | 3 | 2.9×10−5 | 2* | 0.86 | 2.9×10−4 | 0 | 86 | Chromatin |
| POGZ | 3 | 2.5×10−5 | 0 | 1 | 2.9×10−4 | 1 | 83.8 | Chromatin |
| SOS1 | 3 | 2.6×10−5 | 0 | 1 | 3.0×10−4 | 1 | 67.9 | H-CHD |
| TBX18 | 1 | 0.02 | 3 | 1.8×10−3 | 3.0×10−4 | 1 | 72.6 | NA |
Meta-analysis was performed by combining the p-values from damaging de novo mutations and loss-of-function (LoF) heterozygous mutations using the Fisher's method with 4 degrees of freedom. The top 25 genes are shown. Genes which are bolded surpass the Bonferroni multiple testing correction (2.6×10−6, 0.05/18,989) for p-values tabulated by either de novo, heterozygous, or meta-analysis. H-CHD: Known human CHD genes. M-CHD: Known mouse CHD genes. Chromatin: Chromatin modification genes consists of 546 genes in GO:0016569.
denotes that at least one of the carriers has unknown transmission.
SMAD6, an inhibitor of BMP signaling, had 8 inherited and one de novo LoF mutation (Meta P = 1.3×10−6; Table 3). Phenotypes included TOF, hypoplastic left heart syndrome, coarctation and D-TGA. Only two probands had extracardiac abnormalities. Zero LoFs were found among 7,156 parental control alleles, and LoFs were markedly enriched among European probands compared to non-Finnish European controls in ExAC (OR = 20.5, two-sided Fisher’s P = 2.7×10−6). SMAD6 missense variants, but not LoFs, have been previously identified in three sporadic cases with bicuspid aortic valve and mitral valve disease32. Among parents transmitting SMAD6 LOFs, only one had a CHD diagnosis, BAV. Interestingly, SMAD6 LoFs showing incomplete penetrance have also been implicated in midline craniosynostosis, with a common variant near BMP2 modifying penetrance33. Our findings suggest that SMAD6 LoFs produce variable phenotypes, dependent on additional genetic or environmental factors.
DISCUSSION
This study represents the largest genetic investigation of a single CHD cohort, and the first comprehensive analysis of recessive and dominant inherited variants in CHD. Our search for disease-associated transmitted variants and pathways was enhanced by comparing observed and expected numbers of recessive or dominant genotypes independent of control subjects, accommodating for variation in inbreeding and ethnic background. While extension of the expected frequency of DNMs to standing variation is confounded by the impact of selection and drift on allele frequencies over subsequent generations, our analysis demonstrates that this approach is robust for estimating the expected frequency of rare inherited variants, which are likely to be recently introduced into the population. We anticipate this approach will be broadly relevant.
Rare inherited genotypes in known CHD genes, and genome-wide significant new CHD candidate genes accounted for 1.8% of CHD in this cohort. The excess of genes with RGs suggests that more genes await discovery. A recessive founder mutation in GDF1 accounted a large fraction of severe CHD among Ashkenazim. Genotyping this specific variant, which has a minor allele frequency of ~0.5% in Ashkenazim, can immediately be used for diagnosis and population-based risk assessment.
Enrichment of damaging RGs was particularly marked in probands with laterality defects. This is consistent with epidemiology showing that laterality defects have the highest recurrence risk of any CHD10, are more prevalent in populations with high consanguinity34, and conversely show no enrichment for damaging DNMs8, 9.
We also found new phenotypes arising from recessive mutations in genes previously implicated in CHD caused by monoallelic mutations, including RGs in MYH6 in Shone complex, a disease of previously unknown cause. The finding of abnormal ventricular function in several of these patients, as well as in other patients with monoallelic MYH6 mutation, suggests that patients with Shone complex and biallelic MYH6 mutations may be at particular risk for ventricular dysfunction, potentially allowing early identification and intervention. Other genes without previously described recessive phenotypes included CHD7, COL1A1, COL5A2, FBN2, NOTCH1, NSD1, and TSC2, as well as genes previously implicated only in mouse CHD (DGCR2, and DAW1, LRP1, and MYH10).
Ten probands had rare LoFs in FLT4 and predominantly had TOF. None had NDD and only 1 had EA, unlike 25% of all TOF probands in this study. FLT4 LoFs resulted in phenotypes distinct from heterozygous missense mutations in the kinase domain that cause defective lymphatic development35. Further studies of the expression and role of FLT4 in the developing heart will be of interest.
Doubling the size of our sequenced cohort more than doubled the identified CHD risk genes. The current data set includes 66 genes with two or more damaging DNMs compared to 21 previously, and 19 with two or more LoF DNMs compared to five previously9. Highly enriched gene sets, in which 72%–85% of genes are expected to confer risk, include 12 genes (AKAP12, ANK3, CLUH, CTNNB1, KDM5A, KMT2C, MINK1, MYRF, PRRC2B, RYR3, U2SURP, and WHSC1) not previously implicated in CHD9, and have increased the strength supporting a role for 6 additional genes which as yet do not reach thresholds for significance (CAD, FRYL, GANAB, KDM5B, NAA15, and POGZ). DNMs are highly enriched in cases with neurodevelopmental abnormalities or extra-cardiac structural manifestations, or both. Importantly, we report for the first time a significant contribution of DNMs to 3.1% of isolated CHD. From the distribution of genes with multiple damaging DNMs, the estimated number of genes in which DNMs contribute to CHD in this cohort is 443 (95% CI = [154.1, 731.9]; Supplementary Figure 19; see Supplementary Note).
Pathway analysis identifies DNMs, predominantly LoFs, in chromatin modifiers as a major contributor to CHD, accounting for 2.3% of probands (Figure 4). Eleven chromatin modifiers have two or more damaging DNMs, and we estimate that mutations in at least ~38 (95% CI = [7, 69]) chromatin modifier genes contribute to CHD using a maximum likelihood approach (Supplementary Figure 20). The implication of LoF DNMs in writers, erasers and readers of many different specific chromatin marks in CHD underscores the dosage sensitivity of these genes, which is supported by their general intolerance to LoF mutation. Together these findings suggest that heart development depends on precise control of transcription mediated by changes in chromatin state in response to developmental signals36–38.
After removing chromatin modifiers from GO term enrichment analysis (for GO enrichment analysis with chromatin modifiers see Supplementary Data Set 11), several terms involved in developmental processes show enrichment (Supplementary Data Set 12). Extension of pathway analysis to genes with damaging RGs demonstrated enrichment of genes involved in cilia formation and function. These genes have long been known to play a critical role in establishment of the left-right body axis, and cilia gene mutations frequently contribute to heterotaxy. Understanding the mechanisms underlying the effects of these mutations will be of great interest in determining mechanisms of normal and abnormal human development.
It is important to link the genetic causes of CHD to patient outcomes. There is striking overlap of genes mutated in CHD and autism. In particular, patients in our cohort with LoF mutations in chromatin modifiers are at very high risk of NDD (87%). Conversely, virtually all patients with LoF mutations in chromatin modifiers who have been ascertained for autism studies in the Simons Collection do not have CHD31, indicating variable expressivity of CHD. We have noted previously that patients with DNMs in chromatin modifiers have high risk of NDD9, suggesting that mutations in these genes may identify CHD patients at high risk of autism and intellectual disability who may benefit from early neurodevelopmental intervention39.
By combining inherited and de novo variant analysis, we identified a genetic contribution to 10.1% of CHD. Despite these advances, the pathogenesis of a large fraction of CHD cases remains unknown. Potential explanations include contributions from more common variants, structural variants that have eluded detection by WES, variants in non-coding regions, polygenic inheritance, epistasis and gene-environment interactions6, 33, 40, 41.
A recent study estimated that WES of 10,000 trios will yield 80% saturation for identifying genes contributing to syndromic CHD cases13. Our Monte Carlo simulations suggest that two or more damaging DNMs have now been identified in ~10.5% of risk loci, and that sequencing 10,000 trios will yield 170.1 risk genes, predicting 38% saturation of all CHD risk genes, comprising both syndromic and non-syndromic CHD acting via DNMs (Supplementary Figure 21). It is clear that loci suggested from human studies can be further substantiated at low cost by orthogonal approaches engineering mutations into model organisms and cells42. This study indicates that continued sequencing of large, well-phenotyped cohorts will provide an increasingly complete picture of the genetic underpinnings of CHD, allowing new insight into mechanisms governing human development, improved prediction of clinical outcome, and the opportunity to mitigate these risks.
ONLINE METHODS
Patient Subjects
Pediatric Cardiac Genomics Consortium (PCGC)
CHD subjects were recruited to the Congenital Heart Disease Network Study of the Pediatric Cardiac Genomics Consortium (CHD GENES: ClinicalTrials.gov identifier NCT01196182)7. The institutional Review Boards of Boston’s Children’s Hospital, Brigham and Women’s Hospital, Great Ormond Street Hospital, Children’s Hospital of Los Angeles, Children’s Hospital of Philadelphia, Columbia University Medical Center, Icahn School of Medicine at Mount Sinai, Rochester School of Medicine and Dentistry, Steven and Alexandra Cohen Children’s Medical Center of New York, and Yale School of Medicine approved the protocols. All subjects or their parents provided informed consent. Subjects were selected for structural CHD (excluding PDA associated with prematurity, and pulmonic stenosis associated with twin-twin transfusion). Individuals with either an identified chromosomal aneuploidy or a CNV that is known to be associated with CHD were not included. For all subjects, cardiac diagnoses were obtained from review of all imaging and operative reports and entered as Fyler codes based on the International Paediatric and Congenital Cardiac Codes (http://www.ipccc.net/). All patients were evaluated at study entry using a standardized protocol consisting of an interview that includes maternal, paternal and birth history and whether the patient has been examined by a geneticist. A comprehensive review of the proband’s medical record was performed that included height and weight data, along with presence or absence of a broad range of reported extracardiac malformations, the availability and results of genetic testing and the presence or absence of a clinical genetic diagnosis. For probands under age 1, specialty (other than cardiology) services obtained in the course of clinical care were documented. For probands over age 1, parents were asked if their child was diagnosed with developmental delay and whether educational supports were obtained. Each patient has a 3-generation pedigree. For the current study, assessment of neurodevelopmental outcome was based on parental report when the subject was at least 12 months old and classified as having NDD if they answered “Yes” to the presence of at least one of the following conditions: developmental delay, learning disability, mental retardation, or autism. A total of 1,027 cases could not be evaluated for neurodevelopmental outcome because the age at interview was < 1 year.
Pediatric Heart Network (PHN)
CHD subjects were chosen from the DNA biorepository of the Single Ventricle Reconstruction trial43. Subjects underwent in-person neurodevelopment evaluation at 14 months old with the Psychomotor Developmental Index (PDI) and Mental Development Index (MDI) of the Bayley Scales of Infant Development-II44. Subjects were further assessed with the Ages and Stages Questionnaire (ASQ) from which the scores at 3 year of age were analyzed. Subjects were classified as having NDD if PDI or MDI score < 70 or a risk score in at least one of the five domains of the ASQ at 3 year of age. DNA from blood or sputum was collected from trios follow-up visits at or after 3 years.
Controls
Controls included 1,789 previously analyzed families which include one offspring with autism, one unaffected sibling, and unaffected parents14. The permission to access to the genomic data in the Simons Simplex Collection (SSC) on the National Institute of Mental Health Data Repository was obtained. Written informed consent for all participants was provided by the Simons Foundation Autism Research Initiative45. Only the unaffected sibling and parents were analyzed in this study. Controls were designated as unaffected by the SSC14.
Cardiac Phenotyping
Cardiac phenotypes were divided into 5 major categories (Supplementary Table 3a) on the basis of the major cardiac lesion: conotruncal defects (CTD, N=872), D-transposition of the great arteries (D-TGA, N=251), heterotaxy (HTX, N=272), left ventricular outflow tract obstruction (LVO, N=797), or Other (N=679). CTD phenotypes include Tetralogy of Fallot (TOF), double-outlet right ventricle (DORV), truncus arteriosus, membranous ventricular septal defects (VSD), and aortic arch abnormalities. LVO phenotypes include hypoplastic left heart syndrome (HLHS), coarctation of the aorta (CoA), and aortic stenosis/bicuspid aortic valve (AS/BAV). HTX syndromes include situs abnormalities such as dextrocardia, left or right isomerism (LAI, RAI) as the major malformation, and may include other defects such as L-transposition of the great arteries (L-TGA), atrioventricular canal defects (AVC), anomalous pulmonary venous drainage (TAPVR, PAPVR), and double outlet right ventricle. Isomerism of other organs was not considered a separate extra-cardiac malformation for this study. Lesions in the “Other” category include pulmonary valve abnormalities, anomalous pulmonary venous drainage, atrial septal defects (ASD), atrioventricular canal defects, double inlet left ventricle (DILV), and tricuspid valve atresia (TA). Any structural anomaly that was not acquired was called an extracardiac malformation.
Exome sequencing
Samples were sequenced at the Yale Center for Genome Analysis following the same protocol. Genomic DNA from venous blood or saliva was captured using the Nimblegen v.2 exome capture reagent (Roche) or Nimblegen SeqxCap EZ MedExome Target Enrichment Kit (Roche) followed by Illumina DNA sequencing as previously described8. WES data were processed using two independent analysis pipelines at Yale University School of Medicine and Harvard Medical School (HMS). At each site sequence reads were independently mapped to the reference genome (hg19) with BWA-MEM (Yale) and Novoalign (HMS) and further processed using the GATK Best Practices workflows46–48, which include duplication marking, indel realignment, and base quality recalibration, as previously described49. Single nucleotide variants and small indels were called with GATK HaplotypeCaller and annotated using ANNOVAR50, dbSNP (v138), 1000 Genomes (August 2015), NHLBI Exome Variant Server (EVS), and ExAC (v3)51. The MetaSVM algorithm, annotated using dbNSFP version 2.952, was used to predict deleteriousness of missense variants (annotated as “D-Mis”) using software defaults53. Variant calls were reconciled between Yale and HMS prior to downstream statistical analyses.
Kinship analysis
Relationship between proband and parents was estimated using the pairwise identity-by-descent (IBD) calculation in PLINK54. The IBD sharing between the proband and parents in all trios is between 45% and 55%.
Principal component analysis
To determine the ethnicity of each sample, we used the EIGENSTRAT55 software to analyze tag SNPs in cases, controls, and HapMap subjects as described before56. Because all subjects who carried the p.Met364Thr RGs in GDF1 were self-reported Ashkenazi Jewish (AJ), we utilized an additional software package, LASER57, which can accurately infer worldwide continental ancestry from sequencing data. To validate their reported AJ ancestry and to determine the number of AJ in cases and controls, we first downloaded genome-wide SNP array data for 471 AJ Individuals from the Gene Expression Omnibus database (accession no. GSE23636)58 and then merged this data with 938 unrelated individuals from the Human Genome Diversity Project provided with LASER. We then clustered our cases and controls with these 1,409 samples whose ancestral information was known and determined which individuals in our cohort best cluster with known AJ using LASER.
Variant filtering
We filtered RGs for rare (MAF ≤ 10−3 across all samples in 1000 Genomes, EVS, and ExAC) homozygous and compound heterozygous variants that exhibited high quality sequence reads (pass GATK Variant Score Quality Recalibration [VSQR], have a minimum 8 total reads total for both proband and parents, and have a genotype quality [GQ] ≥ 20). Only LoF variants (nonsense, canonical splice-site, frameshift indels, and start loss), D-Mis, and non-frameshift indels were considered potentially damaging to the disease. For probands whose parents’ WES data were not available, only homozygous variants were analyzed. Synonymous variants were also filtered using the same criteria and analyzed separately to determine whether there is an inflation of background rate.
DNMs were called by Yale using the TrioDenovo59 program and by HMS as previously described49, and filtered using the same criteria, which have been shown to yield a specificity of 96.3% as described previously49. These hard filters include: (1) an in-cohort MAF ≤ 4×10−4; (2) a minimum 10 total reads total, 5 alternate allele reads, and a minimum 20% alternate allele ratio in the proband if alternate allele reads ≥ 10 or, if alternate allele reads is < 10, a minimum 28% alternate ratio; (3) a minimum depth of 10 reference reads and alternate allele ratio < 3.5% in parents; and (4) exonic or canonical splice-site variants.
For the LoF heterozygous variants, we filtered for rarity (MAF ≤ 10−5 across all samples in 1000 Genomes, EVS, and ExAC) and high-quality heterozygotes (pass GATK VQSR, minimum 8 total reads, GQ score ≥ 20, mapping quality [MQ] score ≥ 59, and minimum 20% alternate allele ratio in the proband if alternate allele reads ≥ 10 or, if alternate allele reads is < 10, a minimum 28% alternate ratio). Additionally, variants located in segmental duplication regions (as annotated by ANNOVAR50), RGs, and DNMs were excluded. Of particular note, all LoF heterozygous variants that met aforementioned criteria in 226 singletons were also included in the LoF heterozygous burden analysis even though an unknown proportion of these filtered variants could be de novo or compound heterozygous events. Finally, in silico visualization was performed on: (1) calls in the H-CHD set, (2) calls in the LoF-intolerant gene set (pLI ≥ 0.9), (3) variants that appear at least twice, and (4) variants in the top 50 significant genes from our burden analysis
Estimation of the expected number of recessive and dominant variants
We implemented a polynomial regression model coupled with a one-tailed binomial test to quantify the enrichment of damaging RGs in a specific gene or gene set in cases, independent of controls. Details about the modeling of the distribution of recessive and dominant variant counts are in the Supplementary Note. The expectation of the RG count for each gene was calculated using the fitted values from the polynomial model by the formula below:
where ‘i’ denotes the ‘ith’ gene and ‘N’ denotes the total number of RGs. For a given gene set, the expected RG count was based on the sum of fitted values for the gene set.
Alternatively, RG can also be modeled separately as compound heterozygotes or homozygotes without the need for regression fits. In this method, the expected number of compound heterozygotes for each gene is derived from distributing the observed number of RGs, N, across all genes according to the ratio of the squared de novo probabilities:
The expected number of homozygotes is derived similarly, but using the linear ratio of de novo probabilities:
The total number of expected RG for each gene is the sum of the derived expected compound heterozygous and homozygous values.
For rare LoF heterozygous variants, we found that the number of LoF heterozygous variants in a gene was inversely correlated with the pLI score obtained from the ExAC database. To control for the potential confounding effect due to the pLI score, we stratified genes into 5 subsets by pLI quantiles: (1) those with a pLI score between 0 and the first quantile (pLI = 3.1×10−5); (2) those with a pLI score between the first quantile and the second quantile (pLI = 2.9×10−2); (3) those with a pLI score between the second quantile and the third quantile (pLI = 0.71); (4) those with a pLI score between third quantile and 1; (5) those without a pLI score. For each set, the expected number of LoF heterozygous variants for a gene was estimated by the following formula:
where ‘j’ denotes the ‘jth’ gene, ‘k’ denotes the ‘kth’ set, and ‘L’ denotes the total number of LoF heterozygous variants. The expected number of heterozygous variants closely match the observed number of heterozygous variants in each gene in cases and controls (Supplementary Figure 2).
Statistical analysis
Gene-set enrichment analysis
To test for over-representation of a gene set without controls and correction for consanguinity, a one-tailed binomial test was conducted by comparing the observed number of variants to the expected count estimated using the method detailed above. Assuming that our exome capture reagent captures N genes and the testing gene set contains M genes, then the p-value of finding k variants in this gene set out of a total of x variants in the entire exome is given by
where p= (Σgene set Expected Valuei)/(Σall genes Expected Valuej). Enrichment was calculated as the observed number of genotypes/variants divided by the expected number of genotypes/variants.
Gene-based binomial test
A one-tailed binomial test was used to compare the observed number of damaging variants within each gene was compared to the expected number estimated using the approach detailed above. Enrichment was calculated as the number of observed damaging genotypes/variants divided by the expected number of damaging genotypes/variants.
De novo enrichment analysis
The R package ‘denovolyzeR’ was used for the analysis of DNMs based on a mutation model developed previously60, 61. The probability of observing a DNM in each gene was derived as described previously49, except that the coverage adjustment factor was based on the full set of 2,645 case trios or 1,789 control trios (separate probability tables for each cohort). The overall enrichment was calculated by comparing the observed number of DNMs across each functional class to expected under the null mutation model. The expected number of DNMs was calculated by taking the sum of each functional class specific probability multiplied by the number of probands in the study, multiplied by two (diploid genomes). The Poisson test was then used to test for enrichment of observed DNMs versus expected as implemented in denovolyzeR60. For gene set enrichment, the expected probability was calculated from the probabilities corresponding to the gene set only.
To estimate the number of genes with > 1 DNM, 1 million permutations were performed to derive the empirical distribution of the number of genes with multiple DNMs. For each permutation, the number of DNMs observed in each functional class was randomly distributed across the genome adjusting for gene mutability. The empirical p value is calculated as the proportion of times that the number of recurrent genes from the permutation is greater than or equal to the observed number of recurrent genes.
To examine whether any individual gene contain more DNMs than expected, the expected number of DNMs for each functional class (LoF, D-Mis, and LoF+D-Mis) was calculated from the corresponding probability adjusting for cohort size. The Poisson test was then used to compare the observed DNMs for each gene versus expected. For each gene, we compared the statistical significance across LoF, D-Mis, and LoF+D-Mis and reported the most significance statistical values. The Bonferroni multiple-testing threshold is, therefore, equal to 8.8×10−7 (0.05/(3×18,989)).
Meta-analysis of damaging de novo and LoF heterozygous variants
The Fisher’s method62 with 4 degrees of freedom was performed to combine p-values from damaging DNMs and LoF heterozygous variants. We calculated p-values for damaging DNMs in each gene by comparing the observed number of damaging DNMs to the expected number in a respective gene under the null mutation model. We calculated p-values for LoF heterozygous variants using the one-tailed binomial test to compare the observed number of LoF heterozygous variants to the expected number adjusted for LoF de novo probabilities.
Estimating the number of genes with more than one recessive genotype
One million permutations were performed to derive the empirical distribution of the number of genes with multiple damaging RGs. For each permutation, the number of observed damaging RGs (N = 467) was randomly distributed across the genome using the fitted values from the polynomial model for each gene. The empirical p value is calculated as the proportion of times that the number of recurrent genes from the permutation is greater than or equal to the observed number of recurrent genes (N = 44). Similarly, 1 million permutations were conducted on synonymous RGs as an ancillary analysis.
Estimating the number of overlapping genes with damaging/LoF de novo mutations between CHD and autism cohorts
A permutation test was performed to assess the enrichment of overlapping genes with damaging/LoF DNMs shared between the CHD and autism cohorts. Given that the observed numbers of genes with DNMs in the CHD and autism cohorts are N1 and N2, respectively, and the observed number of overlapping genes is M, we sampled N1 genes from all genes in the CHD cohort and N2 genes from all genes in the autism cohorts without replacement using the probability of observing at least one DNM as weight. The number of overlapping genes, P, was determined in each iteration of the simulation. A total of 1,000,000 iterations were conducted to construct the empirical distribution. The empirical number of overlapping genes was calculated by taking the average of the number of overlapping gens across all iterations. The empirical p-value was calculated as follows:
Gene ontology enrichment analysis
The complete list of genes which harbored LoF/damaging variants were input into GOrilla63 (http://cbl-gorilla.cs.technion.ac.il/) to identify enriched GO terms compared to the background set of genes (M=18,715). A false-discovery rate (FDR; represented as q value) of 0.1 was used as cutoff.
Case vs. control comparison
For FLT4 and SMAD6, we compared the burden of LoF alleles in all European cases to all non-Finnish subjects in the ExAC database. Only LoF variants with a global (i.e. across all individuals) MAF < 10−5 were extracted from ExAC for comparison. The total number of alleles evaluated per gene was taken as the median of the allele numbers reported for all positions in a gene. A two-sided Fisher’s exact test was used to compare the frequency of LoF variants in FLT4 and SMAD6.
URLs
GATK: (https://www.broadinstitute.org/gatk/); TrioDeNovo: (http://genome.sph.umich.edu/wiki/Triodenovo); DenovolyzeR: (http://denovolyzer.org); Plink: (http://pngu.mgh.harvard.edu/~purcell/plink); MetaSVM/ANNOVAR: (http://annovar.openbioinformatics.org); NHLBI ESP: (http://evs.gs.washington.edu/EVS/); ExAC03: (http://exac.broadinstitute.org) Contact the authors for the in-house pipelines
Data availability
Whole-exome sequencing data have been deposited in the database of Genotypes and Phenotypes (dbGaP) under accession number phs000571.v1.p1, phs000571.v2.p1, and phs000571.v3.p2
Supplementary Material
Acknowledgments
The authors are enormously grateful to the patients and families who participated in this research. We thank the following team members for outstanding contributions to patient recruitment: A. Julian, M. Mac Neal, Y. Mendez, T. Mendiz-Ramdeen, C. Mintz (Icahn School of Medicine at Mount Sinai); N. Cross (Yale School of Medicine); J. Ellashek and N. Tran (Children’s Hospital of Los Angeles); B. McDonough, J. Geva, M. Borensztein (Harvard Medical School), K. Flack, L. Panesar, N. Taylor (University College London); E. Taillie (University of Rochester School of Medicine and Dentistry); S. Edman, J. Garbarini, J. Tusi, S. Woyciechowski, (Children’s Hospital of Philadelphia); D. Awad, C. Breton, K. Celia, C. Duarte, D. Etwaru, N. Fishman, M. Kaspakoval, J. Kline, R. Korsin, A. Lanz, E. Marquez, D. Queen, A. Rodriguez, J. Rose, J.K. Sond, D. Warburton, A. Wilpers, and R. Yee (Columbia Medical School). We are grateful to Joseph Ekstein and Dor Yeshorim for provision of anonymized DNA samples. The authors thank Shiuan Wang for critical discussion.
This work was supported by the U01 HL098153 and Grant UL1TR000003 from the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, grants to the Pediatric Cardiac Genomics Consortium (U01-HL098188, U01-HL098147, U01-HL098153, U01-HL098163, U01-HL098123 and U01-HL098162), the NIH Centers for Mendelian Genomics (5U54HG006504), the Howard Hughes Medical Institute (RPL and CES) and the Simons Foundation (WKC). SCJ was supported by the James Hudson Brown-Alexander Brown Coxe Postdoctoral Fellowship at the Yale University School of Medicine. JH was supported by the John S. LaDue Fellowship at Harvard Medical School and is a recipient of the Alan Lerner Research Award at the Brigham and Women’s Hospital. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Heart, Lung, and Blood Institute, the National Center for Research Resources or the NIH.
Footnotes
AUTHOR CONTRIBUTIONS
Study design: M.B., W.K.C., M.T.F., B.D.G., E.G., J.R.K., R.P.L., J.G.S., C.E.S.
Cohort ascertainment, phenotypic characterization and recruitment: M.B., W.C., W.K.C., J.D., A.G., B.D.G., E.G., W.G., J.H., R.K., S.M., J.W.N., G.A.P., A.E.R., M.R., C.E.S.
Exome sequencing production and validation: K.B., C.C., R.P.L., S.M.M., I.R.T., J.Z.
Exome sequencing analysis: M.B., R.D.B., S.R.DP., S.C.J., J.H., W.C.H., J.K., R.P.L., S.M., S.M.M., H.Q., C.E.S., J.G.S., M.C.S., S.J.S., Y.S., W.S.W., M.Y., S.Z., X.Z.
Statistical analysis: J.H., S.C.J., R.P.L., Q.L., S.M., C.E.S., S.W., M.Y., H.Z., S.Z.
S.H. performed the biophysical simulation for GDF1.
Writing and review of manuscript: M.B., W.K.C., M.T.F., B.D.G., E.G., J.H., S.C.J., J.R.K., C.W.L., R.P.L., Q.L., C.E.S., D.S., J.G.S., J.Y., S.Z.
Co-senior authors: M.B., R.P.L., C.E.S.
All authors read and approved the manuscript.
COMPETING FINANCIAL INTERESTS
None
References
- 1.van der Linde D, et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58:2241–7. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 2.Egbe A, Lee S, Ho D, Uppu S, Srivastava S. Prevalence of congenital anomalies in newborns with congenital heart disease diagnosis. Ann Pediatr Cardiol. 2014;7:86–91. doi: 10.4103/0974-2069.132474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marino BS, et al. Neurodevelopmental outcomes in children with congenital heart disease: evaluation and management: a scientific statement from the American Heart Association. Circulation. 2012;126:1143–72. doi: 10.1161/CIR.0b013e318265ee8a. [DOI] [PubMed] [Google Scholar]
- 4.Soemedi R, et al. Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. Am J Hum Genet. 2012;91:489–501. doi: 10.1016/j.ajhg.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glessner JT, et al. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ Res. 2014;115:884–96. doi: 10.1161/CIRCRESAHA.115.304458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaidi S, Brueckner M. Genetics and Genomics of Congenital Heart Disease. Circ Res. 2017;120:923–940. doi: 10.1161/CIRCRESAHA.116.309140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pediatric Cardiac Genomics, C et al. The Congenital Heart Disease Genetic Network Study: rationale, design, and early results. Circ Res. 2013;112:698–706. doi: 10.1161/CIRCRESAHA.111.300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaidi S, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–3. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homsy J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–6. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oyen N, et al. Recurrence of congenital heart defects in families. Circulation. 2009;120:295–301. doi: 10.1161/CIRCULATIONAHA.109.857987. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015 doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prendiville T, Jay PY, Pu WT. Insights into the genetic structure of congenital heart disease from human and murine studies on monogenic disorders. Cold Spring Harb Perspect Med. 2014;4 doi: 10.1101/cshperspect.a013946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sifrim A, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016;48:1060–5. doi: 10.1038/ng.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krumm N, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47:582–8. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu H, et al. VAAST 2.0: improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet Epidemiol. 2013;37:622–34. doi: 10.1002/gepi.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yandell M, et al. A probabilistic disease-gene finder for personal genomes. Genome Res. 2011;21:1529–42. doi: 10.1101/gr.123158.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singleton MV, et al. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am J Hum Genet. 2014;94:599–610. doi: 10.1016/j.ajhg.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reeve JP, Rannala B. DMLE+: Bayesian linkage disequilibrium gene mapping. Bioinformatics. 2002;18:894–5. doi: 10.1093/bioinformatics/18.6.894. [DOI] [PubMed] [Google Scholar]
- 19.Kaasinen E, et al. Recessively inherited right atrial isomerism caused by mutations in growth/differentiation factor 1 (GDF1) Hum Mol Genet. 2010;19:2747–53. doi: 10.1093/hmg/ddq164. [DOI] [PubMed] [Google Scholar]
- 20.Lee SJ. Expression of growth/differentiation factor 1 in the nervous system: conservation of a bicistronic structure. Proc Natl Acad Sci U S A. 1991;88:4250–4. doi: 10.1073/pnas.88.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rankin CT, Bunton T, Lawler AM, Lee SJ. Regulation of left-right patterning in mice by growth/differentiation factor-1. Nat Genet. 2000;24:262–5. doi: 10.1038/73472. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka C, Sakuma R, Nakamura T, Hamada H, Saijoh Y. Long-range action of Nodal requires interaction with GDF1. Genes Dev. 2007;21:3272–82. doi: 10.1101/gad.1623907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ching YH, et al. Mutation in myosin heavy chain 6 causes atrial septal defect. Nat Genet. 2005;37:423–8. doi: 10.1038/ng1526. [DOI] [PubMed] [Google Scholar]
- 24.Hershberger RE, et al. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3:155–61. doi: 10.1161/CIRCGENETICS.109.912345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niimura H, et al. Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly. Circulation. 2002;105:446–51. doi: 10.1161/hc0402.102990. [DOI] [PubMed] [Google Scholar]
- 26.Ikemba CM, et al. Mitral valve morphology and morbidity/mortality in Shone's complex. Am J Cardiol. 2005;95:541–3. doi: 10.1016/j.amjcard.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 27.Theis JL, et al. Recessive MYH6 Mutations in Hypoplastic Left Heart With Reduced Ejection Fraction. Circ Cardiovasc Genet. 2015;8:564–71. doi: 10.1161/CIRCGENETICS.115.001070. [DOI] [PubMed] [Google Scholar]
- 28.Harrison MJ, Shapiro AJ, Kennedy MP. Congenital Heart Disease and Primary Ciliary Dyskinesia. Paediatr Respir Rev. 2016;18:25–32. doi: 10.1016/j.prrv.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Karkkainen MJ, et al. Missense mutations interfere with VEGFR-3 signalling in primary lymphoedema. Nat Genet. 2000;25:153–9. doi: 10.1038/75997. [DOI] [PubMed] [Google Scholar]
- 30.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan HL, et al. Nonsynonymous variants in the SMAD6 gene predispose to congenital cardiovascular malformation. Hum Mutat. 2012;33:720–7. doi: 10.1002/humu.22030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timberlake AT, et al. Two locus inheritance of non-syndromic midline craniosynostosis via rare SMAD6 and common BMP2 alleles. Elife. 2016;5 doi: 10.7554/eLife.20125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shieh JT, Bittles AH, Hudgins L. Consanguinity and the risk of congenital heart disease. Am J Med Genet A. 2012;158A:1236–41. doi: 10.1002/ajmg.a.35272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaipainen A, et al. Expression of the fms-like tyrosine kinase 4 gene becomes restricted to lymphatic endothelium during development. Proc Natl Acad Sci U S A. 1995;92:3566–70. doi: 10.1073/pnas.92.8.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wamstad JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–20. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paige SL, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–32. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ang SY, et al. KMT2D regulates specific programs in heart development via histone H3 lysine 4 di-methylation. Development. 2016;143:810–21. doi: 10.1242/dev.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Razzaghi H, Oster M, Reefhuis J. Long-term outcomes in children with congenital heart disease: National Health Interview Survey. J Pediatr. 2015;166:119–24. doi: 10.1016/j.jpeds.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oyen N, et al. Prepregnancy Diabetes and Offspring Risk of Congenital Heart Disease: A Nationwide Cohort Study. Circulation. 2016;133:2243–53. doi: 10.1161/CIRCULATIONAHA.115.017465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morishima M, Yasui H, Ando M, Nakazawa M, Takao A. Influence of genetic and maternal diabetes in the pathogenesis of visceroatrial heterotaxy in mice. Teratology. 1996;54:183–90. doi: 10.1002/(SICI)1096-9926(199610)54:4<183::AID-TERA2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 42.Zhu JY, Fu Y, Nettleton M, Richman A, Han Z. High throughput in vivo functional validation of candidate congenital heart disease genes in Drosophila. Elife. 2017;6 doi: 10.7554/eLife.22617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohye RG, et al. Comparison of shunt types in the Norwood procedure for single-ventricle lesions. N Engl J Med. 2010;362:1980–92. doi: 10.1056/NEJMoa0912461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldberg CS, et al. Factors associated with neurodevelopment for children with single ventricle lesions. J Pediatr. 2014;165:490–496 e8. doi: 10.1016/j.jpeds.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–5. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 46.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11 10 1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Genomes Project C, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Homsy J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–6. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong C, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24:2125–37. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 56.Lemaire M, et al. Recessive mutations in DGKE cause atypical hemolytic-uremic syndrome. Nat Genet. 2013;45:531–6. doi: 10.1038/ng.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C, et al. Ancestry estimation and control of population stratification for sequence-based association studies. Nat Genet. 2014;46:409–15. doi: 10.1038/ng.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bray SM, et al. Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. Proc Natl Acad Sci U S A. 2010;107:16222–7. doi: 10.1073/pnas.1004381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei Q, et al. A Bayesian framework for de novo mutation calling in parents-offspring trios. Bioinformatics. 2015;31:1375–81. doi: 10.1093/bioinformatics/btu839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ware JS, Samocha KE, Homsy J, Daly MJ. Interpreting de novo Variation in Human Disease Using denovolyzeR. Curr Protoc Hum Genet. 2015;87:7 25 1–15. doi: 10.1002/0471142905.hg0725s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–50. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fisher RA. Statistical methods for research workers. ix. Oliver and Boyd; Edinburgh, London: 1925. p. 1l. [Google Scholar]
- 63.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-exome sequencing data have been deposited in the database of Genotypes and Phenotypes (dbGaP) under accession number phs000571.v1.p1, phs000571.v2.p1, and phs000571.v3.p2