Abstract
Skeletal muscle atrophy is associated with a disruption in protein turnover involving increased protein degradation and suppressed protein synthesis. Although it has been well studied that the IGF-1/PI3K/Akt pathway plays an essential role in the regulation of the protein turnover, molecule(s) that triggers the change in protein turnover still remains to be elucidated. TRB3 has been shown to inhibit Akt through direct binding. In this study, we hypothesized that TRB3 in mouse skeletal muscle negatively regulates protein turnover via the disruption of Akt and its downstream molecules. Muscle-specific TRB3 transgenic (TRB3TG) mice had decreased muscle mass and fiber size, resulting in impaired muscle function. We also found that protein synthesis rate and signaling molecules, mTOR and S6K1, were significantly reduced in TRB3TG mice, whereas the protein breakdown pathway was significantly activated. In contrast, TRB3 knockout mice showed increased muscle mass and had an increase in protein synthesis rate, but decreases in FoxOs, atrogin-1, and MuRF-1. These findings indicate that TRB3 regulates protein synthesis and breakdown via the Akt/mTOR/FoxO pathways.
Keywords: TRB3, protein degradation, protein synthesis, atrophy
INTRODUCTION
Skeletal muscle plays many important roles in general health, including whole-body metabolism and mobility not only in the elderly but also in young individuals [1]. Moreover, reduced muscle mass resulting from several critical conditions, such as diabetes, cancer, and obesity, has substantially contributed to poor quality of life and increased mortality [2–4]. Therefore, a better understanding of the signaling molecules that regulate muscle mass could enhance future treatments and significantly reduce a healthcare burden.
The maintenance of muscle mass is a direct consequence of muscle protein turnover, involving the homeostatic regulation of protein accretion and breakdown and it directly impacts muscle growth and atrophy. Insulin and insulin-like growth factor-1 (IGF-1) are key anabolic hormones that regulate muscle protein homeostasis through the PI3K/Akt signaling pathway [5–7]. Particularly, Akt controls both anabolic and catabolic signals in skeletal muscle via its downstream proteins, mammalian target of rapamycin (mTOR) and forkhead box O families (FoxOs), respectively [6, 8–10]. mTOR mediates protein synthesis through the phosphorylation of ribosomal S6 kinase 1 (S6K1) and 4EBP1, and it promotes cell proliferation and growth in response to anabolic stimuli, including insulin, IGF-1, and nutrients [6, 11]. On the other hand, FoxOs regulate catabolic processes and their activities are suppressed by the activation of Akt [12]. FoxOs activates protein breakdown in skeletal muscle through the ubiquitin-proteasome system by upregulating the expression of two muscle-specific E3 ubiquitin ligases, atrogin-1 and MuRF-1, also called atrogenes [9, 10]. However, signaling molecules that regulate protein turnover in the IGF-1/PI3K/Akt pathway have not been fully described.
A mammalian Drosophila tribbles homolog 3 (TRB3) is a pseudokinase that lacks an enzymatic domain of protein kinase [13]. Multiple tissues including skeletal muscle have displayed an increase in expression of TRB3 under various stressful conditions, such as fasting, ER stress, and insulin resistance [13–17]. In mouse myoblast C2C12 cells, TRB3 overexpression directly disrupts Akt signaling and delays muscle cell differentiation [14]. In vivo studies have established that TRB3 regulates glucose homeostasis and ER stress-induced insulin resistance in skeletal muscle by inhibiting Akt activity [17]. Although numerous studies have demonstrated that TRB3 negatively regulates the Akt signaling pathway to influence metabolism, TRB3’s effects on downstream pathways of Akt, such as protein synthesis and breakdown, in skeletal muscle have not been elucidated.
In the present study, we hypothesized that overexpression of TRB3 would disrupt protein turnover through the suppression of Akt activation in skeletal muscle. Our findings show that muscle-specific TRB3 transgenic (TRB3TG) mice exhibited impaired muscle function and decreased muscle mass, resulting from suppressed mTOR/S6K1 signaling and increased FoxO1 and FoxO3a activation. Further, the mRNA expression of atrogin-1 and MuRF-1 was elevated in TRB3TG mice. In contrast, the TRB3 knockout mice tended to have increased muscle mass and protein synthesis rate, while the mRNA expression of atrogenes was significantly suppressed. These results suggest that TRB3 plays a prominent role in the regulation of skeletal muscle protein balance and function.
MATERIALS AND METHODS
Animal Care and Use
Female (10–11-week-old) C57BL/6 wild type (WT), TRB3TG [18], and whole-body TRB3 knockout (TRB3KO) mice [17, 19] were used for all experiments. All animals were maintained on a standard chow diet (no. 8604, Harlan Teklad Diet, Madison, WI, USA) and allowed ad libitum access to food and water. They were housed under 12 h (7AM – 7PM) of light per day in temperature-controlled environment. All protocols were approved by the Institutional Animal Care and Use Committee of the University of South Carolina.
Western Blot Analysis
Gastrocnemius (GAS) muscles were collected and protein lysates were prepared for Western blot analysis as previously described [17, 20]. Primary and secondary antibodies were purchased from commercial sources, including p-Akt Thr308 (9275) and Ser473 (9271), Akt (9272), p-mTOR Ser2448 (2971), mTOR (2972), p-S6K1 Thr389 (9205), S6K1 (9202), p-FoxO1 Ser256 (9461), FoxO1 (2880), p-FoxO3a Ser253 (13129), FoxO3a (12829), GAPDH (2118) from Cell Signaling Technology (Danvers, MA, USA), Atrogin-1 (AP2041), MuRF-1 (MP3401) from ECM (Versailles, KY, USA), TRB3 (ST1032; Calbiochem, San Diego, CA, USA), antipuromycin (MABE343; EDM Millipore Billerica, MA, USA), anti-rabbit (NA934; GE healthcare, Pittsburgh, PA, USA) IgG- and anti-mouse (61-0220; Invitrogen, Carlsbad, CA, USA) IgG2a-horesradish peroxidase conjugated secondary antibodies. The specific bands were visualized using ECL reagents (NEL105001EA; PerkinElmer, Waltham, MA, USA), and the intensity of bands was quantified using Image Studio Lite (LI-COR Biosciences, Lincoln, NE, USA).
Real-Time Polymerase Chain Reaction Analysis
Tibialis anterior (TA) muscles were used for real-time PCR analysis as previously described [17]. Total RNA was extracted using Trizol reagent (15596018; Life Technologies, Carlsbad, CA, USA) and cDNA was synthesized from 4 µg of total RNA by High Capacity cDNA kit (Life Technologies, Carlsbad, CA, USA). SYBR green PCR master mix (4309155; Life Technologies, Carlsbad, CA, USA) carried cDNA and primers. Primer sequences were as follows: TBP forward, 5’-ACCCTTCACCAATGACTCCTATG-3’, and TBP reverse, 5’-TGACTGCAGCAAATCGCTTGG-3’; TRB3 forward, 5’-TCTCCTCCGCAAGGAACCT-3’, and TRB3 reverse, 5’-TCTCAACCAGGGATGCAAGAG-3’; Atrogin-1 forward, 5’-GCAGAGAGTCGGCAAGTC-3’, and Atrogin-1 reverse, 5’-CAGGTCGGTGATCGTGAG-3’; MuRF-1 forward, 5’-GGAACCTGCTGGTGGAAAACATC-3’, and MuRF-1 reverse, 5’-CGTCTTCGTGTTCCTTGCACATC-3’; Cathepsin-L forward, 5’-GTGGACTGTTCTCACGCTCAAGG-3’, and Cathepsin-L reverse, 5’-TCCGTCCTTCGCTTCATAGGT-3’. TBP expression served as an internal control and relative levels of mRNA expression were normalized to its expression.
In vivo Protein Synthesis Measurement
The rate of in vivo protein synthesis was determined by measuring puromycin incorporation into a nascent peptide as previously described [21]. Briefly, mice received an intraperitoneal injection of 0.04 µmol/g body weight with puromycin (540411; EDM Millipore, Billerica, MA, USA) dissolved in 100 µl of PBS 30 minutes before euthanasia. Tissues were collected and stored at −80 °C for future analysis. Western blot was used to determine the amount of puromycin incorporation.
Muscle Cross-Sectional Area Analysis
Frozen TA muscles were sectioned (10-µm) on a cryostat microtome and affixed to slides. Sections were stained with hematoxylin and eosin according to a previous study [22, 23]. Fibers were analyzed by ImageJ 1.48v (National Institutes of Health, Bethesda, MD, USA). Approximately 200 fibers were traced per sample, as determined by a plateau in standard deviation in order to determine the proximate fiber number and cross-sectional area.
Statistical Analysis
Data are expressed as means ± standard error of the mean. Student t-test and two-way ANOVA followed by the Bonferroni Post hoc analysis were used to determine significance. P < 0.05 was considered statistically significant.
RESULTS
Muscle-specific overexpression of TRB3 decreases skeletal muscle mass and impairs muscle function
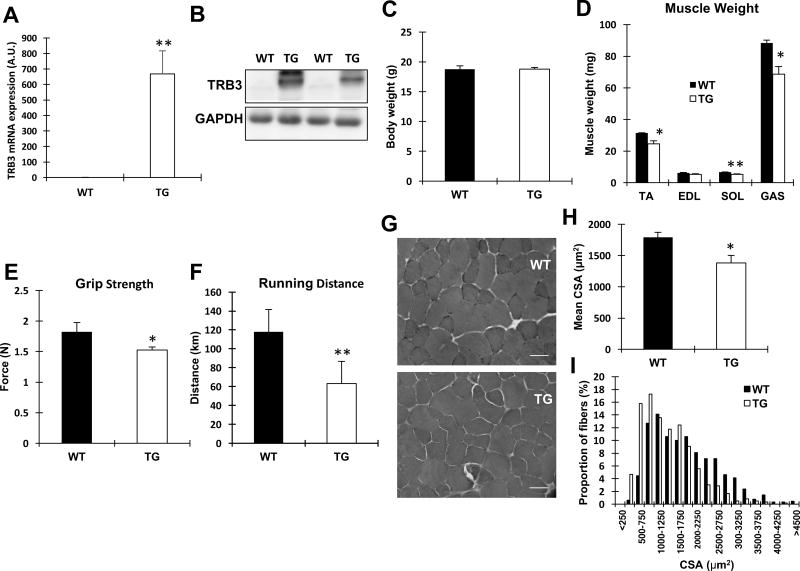
To determine the effects of TRB3 on skeletal muscle mass and function, we studied TRB3TG mice that have previously been examined [18]. The expression of TRB3 mRNA and protein was significantly elevated in TA and GAS, respectively, confirming the overexpression in mouse skeletal muscle (Fig. 1A and B). Body weights were similar between WT and TRB3TG mice (Fig. 1C). However, TRB3TG mice showed a significant decrease in muscle mass (Fig. 1D). TRB3TG mice also displayed a significantly lower (16%) grip strength compared to WT (Fig. 1E). Furthermore, we individually housed WT and TRB3TG mice in wheel cages for 8 weeks to examine the capacity of voluntary wheel exercise. TRB3TG mice performed poorly with a 46% decrease in total running distance compared to WT (Fig. 1F). Taken together, these data suggest that TRB3 plays important roles in skeletal muscle mass regulation and function.
Figure 1. Effects of TRB3 on muscle mass and function in mice.
(A–B) Ten- to eleven-weekold female wild type (WT) and muscle-specific TRB3 transgenic (TG) mice were used to determine TRB3 mRNA and protein expression from tibialis anterior and gastrocnemius, respectively. (C) Body weight was similar between WT and TG. (D) Weights of tibialis anterior (TA), extensor digitorum longus (EDL), soleus (SOL), and gastrocnemius (GAS) muscles were measured. (E–F) Muscle function was evaluated by measuring grip strength and voluntary wheel exercise capacity for 8 weeks. (G) Frozen TA muscles were sectioned in 10-µm-thickness by a cryostat microtome and stained with hematoxylin and eosin. Original magnification: ×400; scale bar: 50 µm. (H) Fibers were traced and analyzed by ImageJ in order to determine the mean CSA. (I) The frequency distribution of the fibers in wild type and TRB3TG mice was determined. Data are the means ± S.E.M (n = 5–6 per group). * P < 0.05, ** P < 0.01 vs. WT
TRB3 decreases muscle cross-sectional area in mouse skeletal muscle
To investigate if the decreased muscle mass in TRB3TG mice is associated with an alteration in fiber size, we measured the cross-sectional area (CSA) in WT and TRB3TG mice. Consistent with decreased muscle weight (Fig. 1D), TRB3TG mice showed a marked decrease (23%) in mean CSA compared to WT (Fig. 1G and H). Furthermore, the percentage of fibers less than 1000 µm2 was increased in TRB3TG mice, while WT littermates were likely to have larger fibers (Fig. 1I). These data indicate that TRB3 adversely affects myofiber size in mouse skeletal muscle.
TRB3TG mice display a decrease in protein synthesis
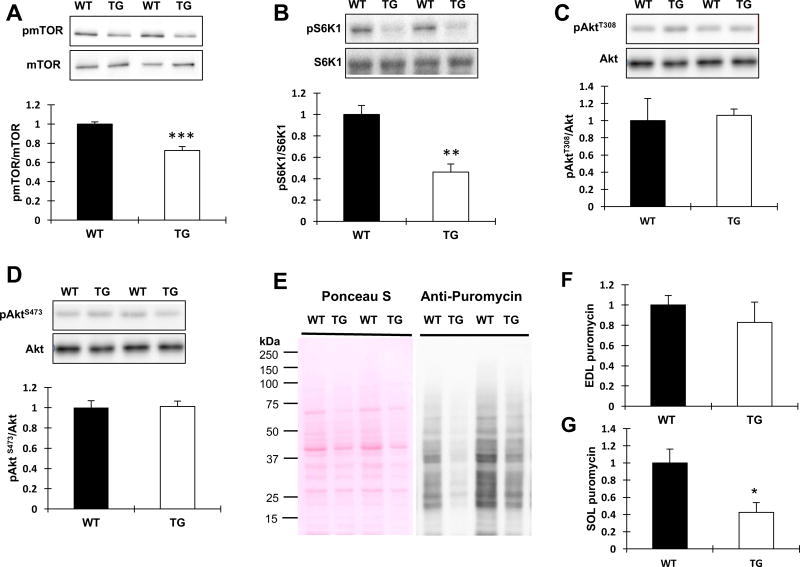
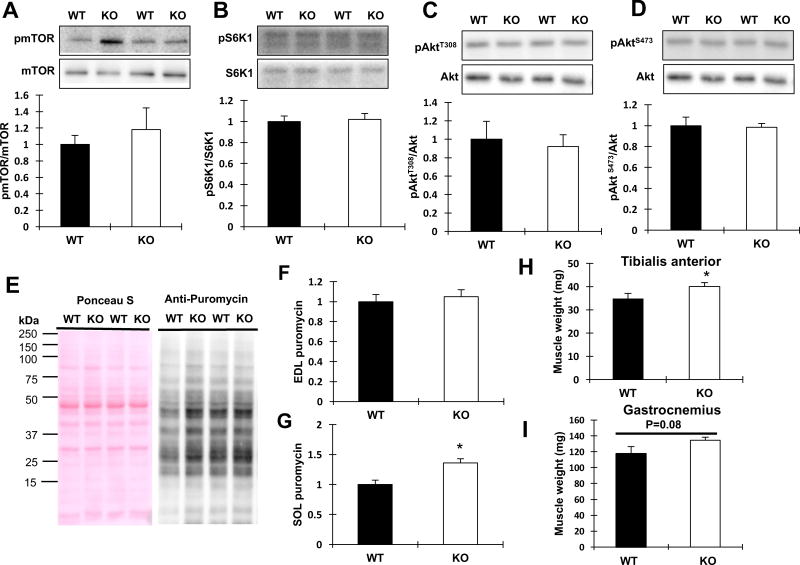
Given that TRB3TG mice had a decrease in muscle mass compared to WT, we next determined if TRB3 overexpression affects signaling molecules involved in protein synthesis. We have previously shown that overexpression of TRB3 in C2C12 myotubes decreases insulin-stimulated Akt phosphorylation through direct binding to Akt [17, 24]. To determine the effect of TRB3 on protein synthesis in skeletal muscle, we studied Akt-regulated downstream proteins, mTOR/S6K1. Overexpression of TRB3 significantly decreased mTOR phosphorylation at Ser2448 compared to WT (Fig. 2A). Consistent with reduced mTOR phosphorylation, the phosphorylation of S6K1 was substantially suppressed (60%) in TRB3TG mice (Fig. 2B). Basal Akt phosphorylation at Thr308 and Ser473 was not different between groups (Fig. 2C and D). To determine how decreased mTOR/S6K1 signaling affects the rate of protein synthesis, WT and TRB3TG mice were given an intraperitoneal injection of puromycin, a structural analog of tyrosyl-tRNA, to measure the rate of protein synthesis as previously described [21]. The protein synthesis rate was reduced in EDL (20%) and SOL (60%) muscles from TRB3TG mice compared to the muscles from WT (Fig. 2 E–G). We further tested the signaling molecules involved in protein synthesis and measured the rate of protein synthesis in TRB3KO mice. Although the level of phosphorylated mTOR was slightly higher in the TRB3KO group (Fig. 3A), we did not observe a significant difference in the Akt/mTOR/S6K1 signaling in TRB3KO mice compared to WT (Fig. 3 A–D). The rate of protein synthesis in SOL muscle in TRB3KO mice was higher than that of WT (Fig. 3G), although there was no difference in protein synthesis rate in EDL muscle (Fig. 3F). Moreover, the muscle weights of TRB3KO mice were increased (Fig. 3H and I) compared to WT, without a change in body weight (unpublished observations by RH Choi and HJ Koh). These results demonstrate that TRB3 regulates the anabolic signaling pathway and the rate of protein synthesis in mouse skeletal muscle.
Figure 2. Effect of TRB3 overexpression on protein synthesis in mouse skeletal muscle.
GAS muscles were collected from WT and TRB3TG mice. (A–D) The activations of signaling molecules related to protein synthesis, including mTOR (A), S6K1 (B), and Akt (C and D), were analyzed by Western blot. (E–G) After 5 hours of fasting, mice received an intraperitoneal injection of 0.04 µmol/g body weight with puromycin dissolved in 100 µl of PBS 30 min before euthanasia to measure the rate of protein synthesis. Western blot was used to measure the amount of puromycin incorporation into nascent peptides. (E) Ponceau S staining of puromycin analysis from SOL muscle showed that proteins were equally loaded. EDL (F) and SOL (G) muscles were analyzed. Data are the means ± S.E.M. (n = 5–6 per group). * P < 0.05, ** P < 0.01, *** P < 0.001 vs. WT
Figure 3. Effects of TRB3 knockout on protein synthesis and muscle mass.
Ten- to eleven-week-old female wild type (WT) and whole-body TRB3 knockout (TRB3KO) mice were euthanized and GAS muscles were collected for analyses. (A–D) The levels of phosphorylation of signaling molecules, including mTOR (A), S6K (B), and Akt (C and D) were determined by Western blot. (E) Equally amount of protein lysates was analyzed for puromycin incorporation from SOL muscle. (F–G) The amount of puromycin incursion was determined in EDL (F) and SOL (G) muscles. (H–I) Muscle weights in TRB3KO mice were determined for TA (H) and GAS (I) muscles. Data are the means ± S.E.M. (n = 5–6 per group). * P < 0.05 vs. WT
TRB3TG mice have activated protein degradation and increased atrogin-1 and MuRF-1 mRNA expression
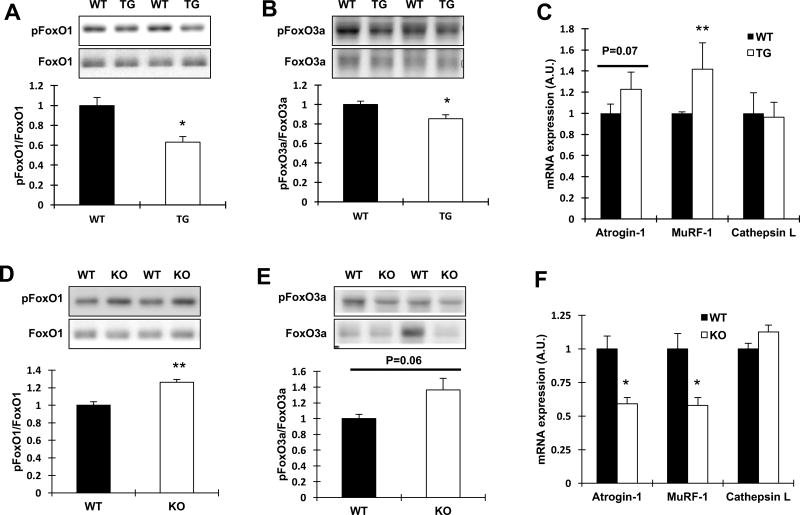
We next determined if TRB3 affects the protein degradation pathway, including the FoxOs and the muscle-specific E3 ubiquitin ligases, atrogin-1 and MuRF-1. TRB3TG mice showed a decrease in phosphorylation of FoxO1 and FoxO3a compared to WT, indicating activated FoxO signaling (Fig. 4A and B). Interestingly, the mRNA expression of MuRF-1 was significantly elevated, and there was a tendency (P=0.07) to increase the atrogin-1 expression in TRB3TG mice without a change in cathepsin L mRNA expression, a marker for lysosomal protease (Fig. 4C). FoxO1 and FoxO3a activities were inhibited in TRB3KO mice compared with WT (Fig. 4D and E). These results also coincided with a significant reduction in atrogin-1 and MuRF-1 mRNA expression, whereas cathepsin L was not altered (Fig. 4F). Taken together, these data indicate that TRB3 regulates FoxOs and protein degradation pathway.
Figure 4. Effects of TRB3 in skeletal muscle on protein degradation signaling and E3 ubiquitin ligases.
(A–B) Phosphorylated FoxO1 (A) and 3a (B) were measured in GAS muscles from wild type (WT) and TRB3TG (TG) mice. (C) The mRNA expression of atrogin-1, MuRF-1, and cathepsin L was determined in TA muscle. (D–E) The levels of phosphorylation of FoxO1 (D) and 3a (E) were measured in GAS muscles from WT and TRB3KO (KO) mice. (F) Atrogin-1, MuRF-1, and cathepsin L mRNA expressions were determined in TA muscle. Data are the means ± S.E.M. (n = 5–6 per group). * P < 0.05, ** P < 0.01 vs. WT
DISCUSSION
The balance between anabolic and catabolic signal is critical to maintain muscle mass. Akt, a key protein kinase in insulin signaling, regulates both protein synthesis and degradation pathways. However, it has not yet been clearly elucidated what molecule(s) may regulate Akt signaling. In the current study, we found that TRB3TG mice had reduced muscle mass, decreased fiber size, and impaired muscle function. Protein synthesis rate and signaling molecules, mTOR and S6K1, were significantly decreased in TRB3TG mice. Furthermore, atrogin-1 and MuRF-1 were highly increased in TRB3TG mice compared to WT, indicating an activation of protein degradation. In addition, TRB3KO mice displayed increased muscle mass and also elevated protein synthesis rate, while the protein degradation pathway was significantly suppressed. Taken together, these findings suggest that TRB3 plays a prominent role in the regulation of protein turnover.
Given the proposed role of TRB3 in the inhibition of Akt signaling [13, 24, 25], our findings demonstrate that TRB3 expression in skeletal muscle affects Akt downstream proteins, including mTOR and FoxOs, in order to regulate protein balance. The decreased mTOR/S6K1 phosphorylation and protein synthesis rate in TRB3TG mice were associated with increased FoxOs activation and atrogenes expression, which may influence the reduced muscle mass. We believe that the decreased muscle mass and function in TRB3TG mice is due, at least in part, to the negative regulation of the Akt/mTOR/FoxOs signaling. Earlier studies have reported that the effect of TRB3 on the inhibition of Akt is related to metabolic dysfunctions, such as insulin resistance and glucose intolerance [13, 17]. However, the regulation of downstream pathways of Akt by TRB3 has not been directly tested. Our findings support the hypothesis that TRB3 negatively regulates Akt downstream signaling in mouse skeletal muscle at basal state to disrupt protein balance, leading to a suppression of protein synthesis and an activation of protein breakdown. In the present study, we examined the basal Akt phosphorylation and did not detect a decreased Akt phosphorylation in TRB3TG (Fig 2C and D). Given that Akt is activated by anabolic stimulants, such as insulin and IGF-1 [26], it is possible that the marginal difference of basal Akt phosphorylation in TRB3TG mice was undetectable by the Western blot analysis. Our previous study and others have used insulin treatment to demonstrate that Akt activation is impaired by TRB3 expression while the level of Akt phosphorylation was not different at basal level [17, 27]. For future studies, it will be necessary to determine Akt activation and its downstream signaling molecules in TRB3TG mice at both basal and insulin-stimulated conditions to find a link between Akt activity and protein turnover pathways.
In contrast to our findings, An et al. have reported that overexpression of TRB3 in skeletal muscle results in greater muscle mass in TA, SOL, and GAS and contains more oxidative muscle fibers [18]. The reasons for the discrepancy are not clear. It is possible that different genetic backgrounds resulted in different phenotypes. The TRB3TG mice were generated in the embryos with mixed background [16]. Mice carrying the TRB3 transgene on the mixed background were backcrossed three to four times to C57BL/6 WT mice before used for the previous experiments [18]. We continued to backcross the TRB3TG mice with C57BL/6 WT mice additional three times (six to seven times in total) for the present experiments. In addition, the inconsistent phenotypes might also result from different diet between the previous study (Harlan Teklad Diet #8604) and current study (LabDiet Mouse Diet 9F) as different diet may affect study outcomes, including phenotypes of animal models [28]. Another contradict to previous report is exercise capacity in TRB3TG mice. An et al. has revealed TRB3 transgenic mice increased maximal running capacity compared to WT whereas ours showed significantly decreased total running distance on 8 weeks of voluntary wheel exercise [18]. In addition to the different genetic backgrounds and diets, different exercise capacity results may be due to the different exercise testing protocol. We utilized voluntary wheel cage running for 8 weeks and measured total running distance, but maximal time-to-exhaustion treadmill protocol was applied in the previous experiment and they analyzed maximal exercise capacity in total work. Therefore, depending on the purpose of testing, these two different measurements would produce different results, and it is possible that the same animal model could respond differently as previously described [29, 30].
In summary, our results demonstrate that TRB3 overexpression in skeletal muscle decreased muscle weight and impaired muscle function that were associated with decreased protein synthesis and increased protein breakdown. Our results suggest that TRB3 plays a key role in muscle mass balance through regulating protein synthesis and breakdown pathways.
Supplementary Material
Highlights.
Muscle-specific TRB3 transgenic (TRB3TG) mice display smaller muscle mass and fiber size.
TRB3TG mice show decreased protein synthesis rates and signaling compared to WT.
Protein degradation pathway is highly activated in TRB3TG mice.
TRB3 knockout mice have higher muscle mass with increased protein synthesis rates.
Acknowledgments
We thank M. Montminy (Salk Institute) for providing the muscle-specific TRB3 transgenic mice, and Regeneron for TRB3 knockout mice. This work was supported by National Institutes of Health grants to H.J.K. (R03AR066825 and P20GM10909), L.J.G. (R01DK099511) and a departmental start-up grant to H.J.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goodpaster BH, Park SW, Harris TB, Kritchevsky SB, Nevitt M, Schwartz AV, Simonsick EM, Tylavsky FA, Visser M, Newman AB. The loss of skeletal muscle strength, mass, and quality in older adults: the health, aging and body composition study, The journals of gerontology. Series A. Biological sciences and medical sciences. 2006;61:1059–1064. doi: 10.1093/gerona/61.10.1059. [DOI] [PubMed] [Google Scholar]
- 2.Wannamethee SG, Shaper AG, Lennon L, Whincup PH. Decreased muscle mass and increased central adiposity are independently related to mortality in older men. The American journal of clinical nutrition. 2007;86:1339–1346. doi: 10.1093/ajcn/86.5.1339. [DOI] [PubMed] [Google Scholar]
- 3.Rantanen T, Harris T, Leveille SG, Visser M, Foley D, Masaki K, Guralnik JM. Muscle strength and body mass index as long-term predictors of mortality in initially healthy men, The journals of gerontology. Series A. Biological sciences and medical sciences. 2000;55:M168–173. doi: 10.1093/gerona/55.3.m168. [DOI] [PubMed] [Google Scholar]
- 4.Martin L, Birdsell L, Macdonald N, Reiman T, Clandinin MT, McCargar LJ, Murphy R, Ghosh S, Sawyer MB, Baracos VE. Cancer cachexia in the age of obesity: skeletal muscle depletion is a powerful prognostic factor, independent of body mass index. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:1539–1547. doi: 10.1200/JCO.2012.45.2722. [DOI] [PubMed] [Google Scholar]
- 5.Latres E, Amini AR, Amini AA, Griffiths J, Martin FJ, Wei Y, Lin HC, Yancopoulos GD, Glass DJ. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J Biol Chem. 2005;280:2737–2744. doi: 10.1074/jbc.M407517200. [DOI] [PubMed] [Google Scholar]
- 6.Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 7.Velloso CP. Regulation of muscle mass by growth hormone and IGF-I. British journal of pharmacology. 2008;154:557–568. doi: 10.1038/bjp.2008.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 9.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 11.Proud CG. Role of mTOR signalling in the control of translation initiation and elongation by nutrients. Curr Top. Microbiol. Immunol. 2004;279:215–244. doi: 10.1007/978-3-642-18930-2_13. 215-44. [DOI] [PubMed] [Google Scholar]
- 12.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 13.Du K, Herzig S, Kulkarni RN, Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 2003;300:1574–1577. doi: 10.1126/science.1079817. [DOI] [PubMed] [Google Scholar]
- 14.Kato S, Du K. TRB3 modulates C2C12 differentiation by interfering with Akt activation. Biochem Biophys. Res Commun. 2007;353:933–938. doi: 10.1016/j.bbrc.2006.12.161. [DOI] [PubMed] [Google Scholar]
- 15.Liew CW, Bochenski J, Kawamori D, Hu J, Leech CA, Wanic K, Malecki M, Warram JH, Qi L, Krolewski AS, Kulkarni RN. The pseudokinase tribbles homolog 3 interacts with ATF4 to negatively regulate insulin exocytosis in human and mouse beta cells. J Clin Invest. 2010;120:2876–2888. doi: 10.1172/JCI36849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi L, Heredia JE, Altarejos JY, Screaton R, Goebel N, Niessen S, Macleod IX, Liew CW, Kulkarni RN, Bain J, Newgard C, Nelson M, Evans RM, Yates J, Montminy M. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science. 2006;312:1763–1766. doi: 10.1126/science.1123374. [DOI] [PubMed] [Google Scholar]
- 17.Koh HJ, Toyoda T, Didesch MM, Lee MY, Sleeman MW, Kulkarni RN, Musi N, Hirshman MF, Goodyear LJ. Tribbles 3 mediates endoplasmic reticulum stress-induced insulin resistance in skeletal muscle. Nature communications. 2013;4:1871. doi: 10.1038/ncomms2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.An D, Lessard SJ, Toyoda T, Lee MY, Koh HJ, Qi L, Hirshman MF, Goodyear LJ. Overexpression of TRB3 in muscle alters muscle fiber type and improves exercise capacity in mice. Am J Physiol Regul Integr Comp Physiol. 2014;306:R925–933. doi: 10.1152/ajpregu.00027.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamoto H, Latres E, Liu R, Thabet K, Murphy A, Valenzeula D, Yancopoulos GD, Stitt TN, Glass DJ, Sleeman MW. Genetic deletion of Trb3, the mammalian Drosophila tribbles homolog, displays normal hepatic insulin signaling and glucose homeostasis. Diabetes. 2007;56:1350–1356. doi: 10.2337/db06-1448. [DOI] [PubMed] [Google Scholar]
- 20.Fujii N, Boppart MD, Dufresne SD, Crowley PF, Jozsi AC, Sakamoto K, Yu H, Aschenbach WG, Kim S, Miyazaki H, Rui L, White MF, Hirshman MF, Goodyear LJ. Overexpression or ablation of JNK in skeletal muscle has no effect on glycogen synthase activity. Am J Physiol Cell Physiol. 2004;287:C200–C208. doi: 10.1152/ajpcell.00415.2003. [DOI] [PubMed] [Google Scholar]
- 21.Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P, Hornberger TA. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. Faseb j. 2011;25:1028–1039. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardee JP, Mangum JE, Gao S, Sato S, Hetzler KL, Puppa MJ, Fix DK, Carson JA. Eccentric contraction-induced myofiber growth in tumor-bearing mice. Journal of applied physiology (Bethesda, Md. : 1985) 2016;120:29–37. doi: 10.1152/japplphysiol.00416.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClung JM, Davis JM, Wilson MA, Goldsmith EC, Carson JA. Estrogen status and skeletal muscle recovery from disuse atrophy. Journal of applied physiology (Bethesda, Md. : 1985) 2006;100:2012–2023. doi: 10.1152/japplphysiol.01583.2005. [DOI] [PubMed] [Google Scholar]
- 24.Koh HJ, Arnolds DE, Fujii N, Tran TT, Rogers MJ, Jessen N, Li Y, Liew CW, Ho RC, Hirshman MF, Kulkarni RN, Kahn CR, Goodyear LJ. Skeletal muscle-selective knockout of LKB1 increases insulin sensitivity, improves glucose homeostasis, and decreases TRB3. Mol Cell Biol. 2006;26:8217–8227. doi: 10.1128/MCB.00979-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koo SH, Satoh H, Herzig S, Lee CH, Hedrick S, Kulkarni R, Evans RM, Olefsky J, Montminy M. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat. Med. 2004;10:530–534. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- 26.Downward J. Mechanisms and consequences of activation of protein kinase B/Akt. Curr. Opin. Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, Liu J, Tian L, Liu Q, Fu Y, Garvey WT. TRIB3 mediates glucose-induced insulin resistance via a mechanism that requires the hexosamine biosynthetic pathway. Diabetes. 2013;62:4192–4200. doi: 10.2337/db13-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reliene R, Schiestl RH. Differences in animal housing facilities and diet may affect study outcomes-a plea for inclusion of such information in publications. DNA repair. 2006;5:651–653. doi: 10.1016/j.dnarep.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Noble EG, Moraska A, Mazzeo RS, Roth DA, Olsson MC, Moore RL, Fleshner M. Differential expression of stress proteins in rat myocardium after free wheel or treadmill run training. Journal of applied physiology (Bethesda, Md. : 1985) 1999;86:1696–1701. doi: 10.1152/jappl.1999.86.5.1696. [DOI] [PubMed] [Google Scholar]
- 30.Liu YF, Chen HI, Wu CL, Kuo YM, Yu L, Huang AM, Wu FS, Chuang JI, Jen CJ. Differential effects of treadmill running and wheel running on spatial or aversive learning and memory: roles of amygdalar brain-derived neurotrophic factor and synaptotagmin I. J Physiol. 2009;587:3221–3231. doi: 10.1113/jphysiol.2009.173088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.