Abstract
The multidrug resistance 1 gene (MDR1) encodes P-glycoprotein (Pgp), a member of the ATP-binding cassette (ABC) transporter family that confers tumor drug resistance by actively effluxing a number of anti-tumor agents. We have previously shown that MDR1 transcription is regulated by epigenetic events such as histone acetylation, and have identified the histone acetylase P/CAF and the transcription factor NF-Y as the factors mediating the enzymatic and DNA anchoring functions, respectively, at the MDR1 promoter. It has also been shown that MDR1 activation is accompanied by increased methylation on lysine 4 of histone H3 (H3K4). In this study, we have further the investigation of histone methylation in MDR1 regulation and function. We show that the Mixed Lineage Leukemia 1 (MLL1) protein, a histone methyltransferase specific for H3K4, is required for MDR1 promoter methylation, as knockdown of MLL1 resulted in a decrease in MDR1 expression. The regulation of MDR1 by MLL1 has functional consequences in that downregulation of MLL1 led to increased retention of the Pgp-specific substrate DIOC2(3), as well as increased cellular sensitivity to several Pgp substrates. Regulation of MDR1 by MLL1 was dependent on the CCAAT box within the proximal MDR1 promoter, similar to what we had shown for MDR1 promoter acetylation, and also requires NF-Y. Finally, overexpression of the most prevalent MLL fusion protein, MLL-AF4, led to increased MDR1 expression. This is the first identification of a histone methyltransferase and its leukemogenic rearrangement that regulates expression of an ABC drug transporter, suggesting a new target for circumvention of tumor multidrug resistance.
INTRODUCTION
It has been well established that chromatin regulates gene expression through the dynamic modifications of histone residues. The repertoire of histone modifications includes acetylation, phosphorylation, methylation, sumolation and ubiquitination; the complex interplay of these chromatin modifications establishes a “histone code” that serves as a recognition and recruitment signal for other transcription modifying proteins, thereby dictating the structure and function of the chromatin fiber and contributing to the activation or repression of specific genes (1–3).
Our laboratory has had a long-standing interest in the transcriptional regulation of the MDR1 gene in response to both external and internal stimuli. The MDR1 gene product, P-glycoprotein (Pgp), is the best-studied member of the ABC family of transporters, which also includes the MDR-associated proteins (MRPs) and the breast cancer resistance protein (BCRP) (4, 5). Pgp was first identified due to its overexpression in drug resistant tumor cells, where it functions as a broad range drug transporter, thereby conferring resistance to many important chemotherapeutic agents including vinblastine, doxorubicin, and paclitaxel (4, 5). In addition to its well-described role as a mediator of tumor drug resistance, it has been found in normal human tissues and has been implicated in the response of cells to more general apoptotic stimuli (6), in the bio-distribution of drugs (7, 8), and in the transport of natural substances, including peptides, phospholipids, cholesterol and steroids (4, 9, 10). Given its varied and critical roles in both tumor and normal cells, it is not surprising that the expression of Pgp is highly regulated from inception to degradation.
We previously demonstrated that a variety of signaling pathways that lead to MDR1 activation are integrated by an MDR1 enhancesome that is anchored to the promoter via the DNA binding proteins NF-Y and SP1, which interact with an inverted CCAAT box (−82 to −73) and GC box (−56 to −42), respectively (11, 12). More recently, we and others have shown that histones associated with the MDR1 proximal promoter are acetylated in response to multiple inducers (11, 13), and that this acetylation is dependent on the recruitment of the histone acetyltransferase P/CAF to the MDR1 enhancesome through interaction with NF-Y (11). In the present study, we have continued to explore the role of chromatin modifications in MDR1 expression by interrogating a role for histone methyltransferases in the regulation of this gene. Histone methylation, like histone acetylation, occurs on lysine residues within histone tails and also plays a role in the recruitment of multiple transcriptional effectors to specific promoters (2). While there are several potential methylation sites within histone tails, there is a very strong correlation between the degree of trimethylation on lysine 4 of histone H3 (H3K4me3) at the 5’ ends of genes and their transcription rate, RNA polymerase II occupancy and histone acetylation (2, 14, 15).
Herein we show that MDR1 H3K4 methylation is dependent on the methyltransferase MLL1, as knockdown of MLL1 decreased the constitutive expression of MDR1, increased cellular retention of MDR1 substrates, and sensitized cancer cells to chemotherapeutic agents. Notably, an MLL fusion protein expressed in some forms of leukemia also induced MDR1 expression; current studies are directed at querying a correlation between MDR1 expression and the expression of MLL1 fusion protein in leukemias that harbor these translocations. This is the first identification of a histone methyltransferase that regulates a member of the ABC family of drug transporters, and provides a potential new target for modulation of anti-cancer drug sensitivity/resistance.
MATERIALS AND METHODS
Cell Lines
HeLa cells (CCL2, a human cervical adneocarcinoma cell line) were purchased from ATCC, grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with10% fetal bovine serum (FBS) (Atlanta Biologicals). Phoenix amphotropic cells were a gift from Dr. Dan R. Littman (New York University, NY) and were grown in DMEM containing 10% FBS. Cells were frozen down at an early passage, and are checked routinely for mycoplasm contamination. Parental HEK 293 and Jurkat cells, 293- and Jurkat-MLL-AF4 Tet-on cell lines were a gift from Dr. Nancy J. Zeleznik-Le (Loyola University, IL). 293-MLL-AF4 cells were maintained in DMEM plus 10% FBS, 5 µg/ml blasticidin, and 100 µg/ml hygromycin. Jurkat-MLL-AF4 cells were maintained in RPMI 1640 medium with 10% FBS, 10 µg/ml blasticidin, and 200 µg/ml hygromycin. Expression of transgenes was induced using 1 or 2 µg/ml doxycycline (16). Parental 293 and Jurkat cells were maintained in the same media as their sublines without blasticidin and hygromycin.
Plasmids and siRNA
pCXN2 plasmids expressing flag-tagged full-length MLL1 (f-MLL1) or MLL1 with deletion of its SET (catalytic) domain (f-MLL1-ΔSET) were obtained from Dr. Jay Hess (University of Michigan, MI). Empty vector was created by digesting pCXN2-f-MLL1 with XbaI and BglII, followed by blunt end re-ligation of the resulting plasmid lacking MLL1 cDNA. The MDR1 promoter-luciferase constructs containing the full-length MDR1 promoter (−1202/+118), the serial deletions, the CCAAT-box mutant, the GC mutant, as well as the NF-Y dominant negative construct (NF-Y(DN)) have been previously described (11, 12, 17). The siGenome SMARTpool MLL1 siRNA and SiControl non-targeting siRNA #1 were purchased from Dharmacon.
Transfection and Luciferase Assays
Lipofectamine 2000 and oligofectamine (Invitrogen) were used to transfect plasmid and siRNA oligos, respectively, according to the manufacturer’s instructions. Cells were lysed and processed as recommended using the Luciferase Assay System (Promega). Luciferase activity was normalized to protein concentration, which was determined using the Bio-Rad Protein Assay, and to the luciferase activity of pGL2B empty vector. Experiments were repeated in triplicate a minimum of 3 times.
Western Blot Analysis
Cells were harvested in lysis buffer (50 mM Tris-HCl (pH 7.4), 100 mM NaCl, 0.5% sodium deoxycholate, and 0.5% NP-40) containing complete EDTA-free protease inhibitor cocktail (Roche). Protein lysates were separated by SDS-PAGE and blotted with appropriate antibodies: MLLN (N4.4) and MLLC (9–12) (Millipore), Pgp (C219) (Abcam) and α-tubulin (B-5-1-2) (Sigma). Protein bands were visualized on a Bio-Rad Chemidoc XRS using the ECL Advance Western Blotting Kit (GE Healthcare).
MLL1 Knockdown by shRNA
shRNA against MLL1 (5’-GTGCCAAGCACTGTCGAAA-3’) and control scrambled shRNA (5’-GCGCGCTTTGTAGGATTCG-3’) were cloned into the viral vector pQCXIP gfp super forward, a gift from Dr. Liang Zhou (Northwestern University, IL), as described previously (18, 19). Phoenix amphotropic cells were transfected with the retroviral constructs using calcium phosphate (20). Viral supernatants from transfected phoenix cells were used to infect HeLa cells for 2 d before being subjected to puromycin selection at 0.6 µg/ml.
Chromatin Immunoprecipitation (ChIP)
ChIP was carried out using the EZ CHIP kit (Millipore) following the manufacturer’s instructions. Basically, chromatin crosslinked with 0.5% formaldehyde was sheared to an average length of 500 bp by sonicating 10 times with 10-sec pulse followed by 60-sec recovery period at a setting of 5 in a Fisher Scientific 550 Sonic Dismembrator. Sheared chromatin was immunoprecipitated with anti-H3K4me3, rabbit IgG (Millipore) or no antibody overnight at 4°C with rotation. Purified DNA was subjected to PCR using MDR1 promoter-specific primers P (forward: 5’-ACCTGTTTCGCAGTTTCTCG-3’; reverse: 5’-CCTCTGCTTCTTTGAGCTTG-3’), MDR1 5’ coding region-specific primers T (forward: 5’-AACTCTGCCTTCGTGGAGAT-3’; reverse: 5’-ATCCATTCCGACCTGAAGAG-3’), GAPDH promoter-specific primers (provided in the kit), and HoxA7 promoter-specific primers (forward: 5’-GAGCCTCCAGGTCTTTTTCC-3’; reverse: 5’-ACACCCCCAGATTTACACCA-3’). PCR products were visualized following 2.0% agarose gel electrophoresis.
mRNA Analysis
RNA was isolated using Trizol reagent (Invitrogen). Semi-quantitative RT-PCR was performed using the Superscript One-Step RT-PCR kit (Invitrogen). The sequences of the forward and reverse primers were 5’-AACGGTTTCAGCTGCCTCTA-3’ and 5’-TTTGGGTCACCTGAACTTCC-3’ (MLL1); 5’-CCCATCATTGCAATAGCAGG-3’ and 5’-GTTCAAACTTCTGCTCCTGA-3’ (MDR1); 5’-CCCCTGGCCAAGGTCATCCATGACAA-3’ and 5’-GGCCATGAGGTCCACCACCCTGTTGC-3’ (GAPDH); 5’-CACCTACTACAGGACCGCCAA-3’ and 5’-GGGGTTTGTTCACTGTCACTGTCC (MLL-AF4).
DIOC2(3) Retention Assay
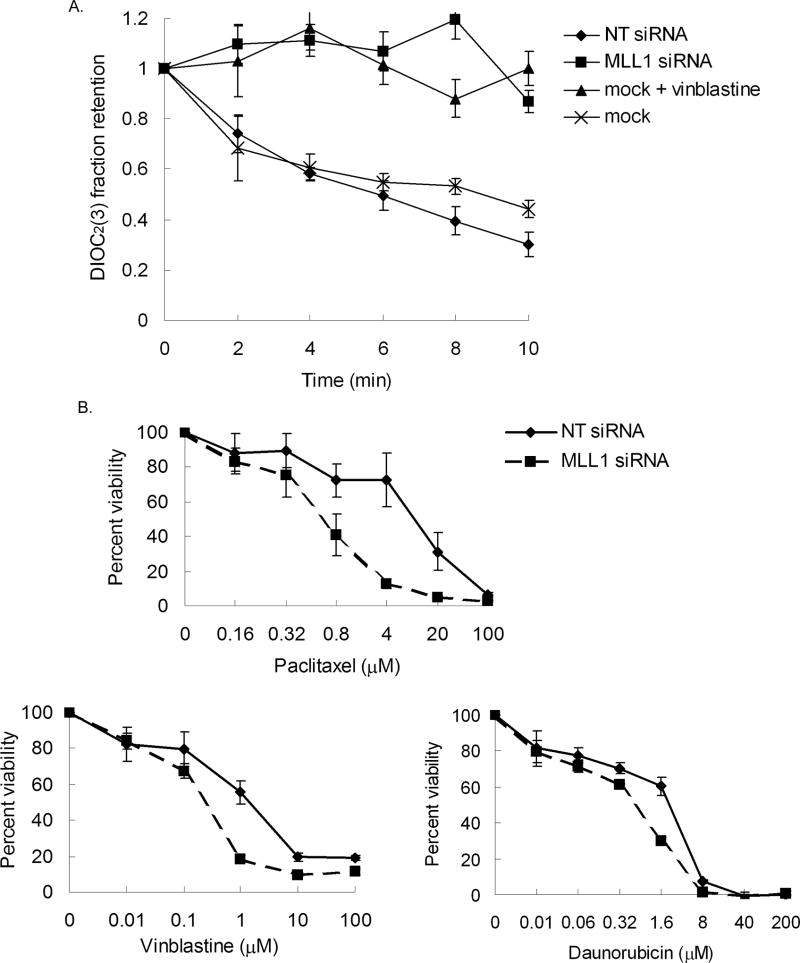
Seventy-two hours post-transfection of siRNA, cells were trypsinized, washed and resuspended at 1 × 106 cells/ml in DMEM medium (without phenol red) containing 1% BSA, then incubated with 2.2 µM DIOC2(3) for 30 minutes on ice. Cells were then washed and reuspended in 1 ml aliquots at 2.5 × 105 cells/ml in cold 1% BSA medium containing either 22 µM vinblastine or 0.1% DMSO, then incubated at 37°C for the times indicated in Figure 5A. Transport of dye was stopped by the addition of ice-cold 1% BSA medium. The cells were immediately centrifuged, washed and resuspended in 250 µl of ice-cold 1% BSA medium. Fluorescence was measured in black-walled 96-well microtiter plates using a fluorescence plate reader at an excitation wavelength of 485 nm and an emission wavelength of 530 nm. Each experiment was conducted in duplicate and repeated three times.
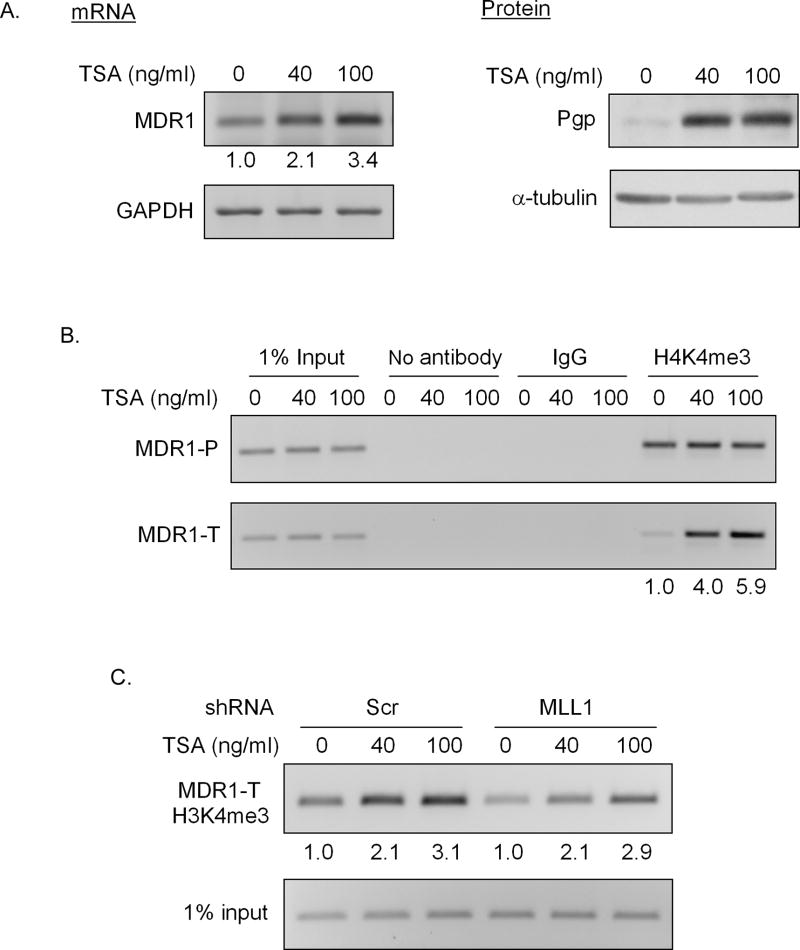
Figure 5. MLL1 is not involved in TSA induced H3K4 trimethylation at the MDR1 locus.
Treatment of HeLa cells with 40 or 100 ng/ml TSA for 24h following with A, RT-PCR analysis of relative MDR1 and GAPDH mRNA levels (left panel), and western blot analysis of Pgp and α-tubulin levels (right panel) and B, ChIP analysis of H3K4me3 levels at the MDR1 locus. Immunoprecipitated chromatin was amplified with PCR using MDR1 promoter-specific primers (MDR1-P) and MDR1 coding region-specific primers (MDR1-T). The induction fold of MDR1 mRNA and coding region-associated H3K4me3 levels with TSA treatment over the mock treatment (0) was shown at the bottom. C, ChIP analysis of H3K4me3 levels at the MDR1 coding region in Scr and MLL1 shRNA transduced HeLa cells following treatment with 40 or 100 ng/ml TSA for 24h. The induction fold of H3K4me3 levels with TSA treatment over mock treatment (0) was shown at the bottom.
Cytotoxicity Assay
Seventy-two hours post-transfection of siRNA, cells were trypsinized and reseeded in 96-well microtiter plates in replicates of six in growth medium without penicillin/streptomycin. After 24 hours, cells were treated with paclitaxel, vinblastine, daunorubicin, or methotrexate at the concentrations shown in Figure 5B for 2 hours, then washed twice and incubated in medium without penicillin/streptomycin for an additional 70 hours. Cell viability was measured using the CellTiter 96 Aqueous Non-Radioactive Cell Proliferation Assay (Promega) according to manufacturer’s instructions. Experiments were repeated a minimum of three times.
RESULTS
MLL1 regulates MDR1 transcription
We and others have shown that histones associated with the MDR1 gene are regulated by epigenetic changes, including acetylation (11, 12) and methylation (13). While the histone acetyltransferase P/CAF has been shown to be responsible for acetylation of MDR1 proximal promoter histones, the methyltransferase responsible for creating the methylation signature at the MDR1 locus is unknown. To investigate this we surveyed the known human enzymes that have been shown to trimethylate H3K4, including members of the MLL and SET1/COMPASS families (15). MLL1, a member of the trithorax group (TrxG) of histone methyltransferases, is required for normal development due to its role as the major regulator of Hox gene transcription (21, 22). It is a 3969 amino acid nuclear protein with a complex domain structure; the full-length protein undergoes site-specific proteolysis to generate a mature MLL1 protein consisting of a non-covalent heterodimer of two associated subunits: N-terminal MLLN (300kDa) and C-terminal MLLC (180kDa) (23, 24). Methylation of H3K4 is mediated through a conserved SET domain within the C terminus (25, 26). Although Hox genes were the first identified and best studied direct targets of MLL1, this enzyme has been found associated with a subset of other RNA polymerase II-occupied promoters (27–30).
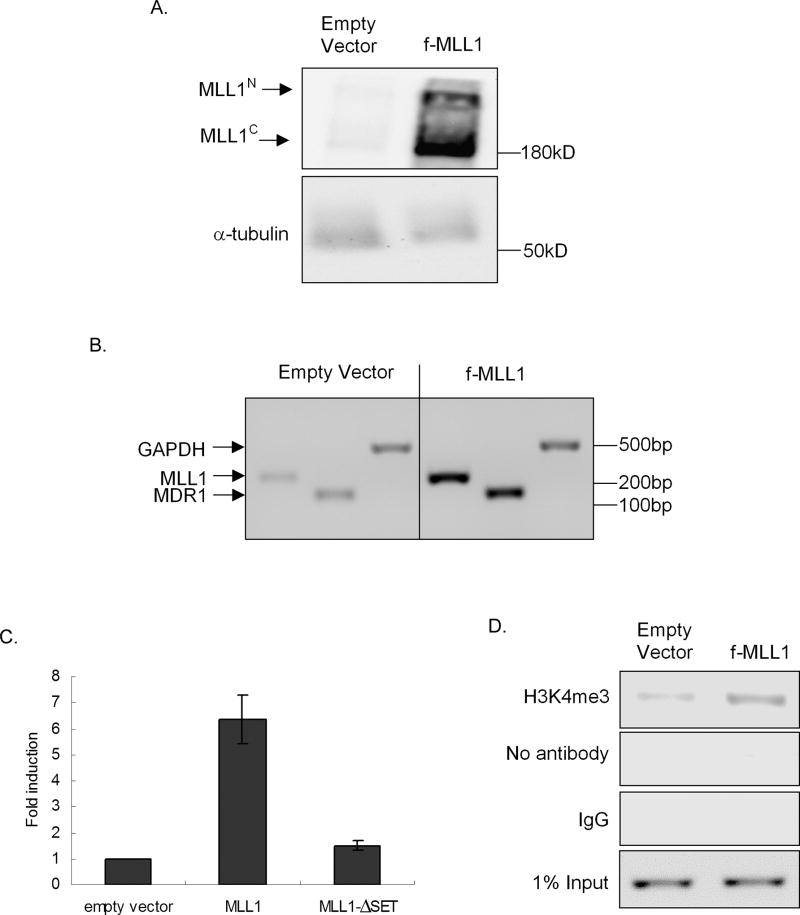
To query a role for MLL1 in endogenous MDR1 transcription, a mammalian expression construct containing flag-tagged full-length MLL1 (f-MLL1) or an MLL1 construct containing a deletion of the SET domain (f-MLL1-ΔSET) was transiently transfected into HeLa cells. Overexpression of f-MLL1, demonstrated at the levels of protein (Fig. 1A) and mRNA (Fig 1B), led to an ~ 5-fold increase in MDR1 mRNA levels relative to cells transfected with empty vector (Fig. 1B, compare left and right panels), with minimal effect on GAPDH mRNA level. Overexpression f-MLL1-ΔSET had no discernible effect on MDR1 gene expression (date not shown), indicating a requirement for MLL1 catalytic activity in the regulation of MDR1 expression.
Figure 1. MLL1 overexpression induces MDR1 expression and tri-methylation of H3K4 at the MDR1 promoter.
A, Western blot analysis of MLL1N, MLL1C, and α-tubulin levels and B, RT-PCR analysis of relative MLL1, MDR1, and GAPDH mRNA levels in HeLa cells 24 hours following transfection with f-MLL1 plasmid. C, MDR1 transcriptional activity was measured by luciferase activity, 24h post-cotransfection of a luciferase reporter construct driven by the MDR1 promoter with empty vector, f-MLL1, or f-MLL1-ΔSET. Luciferase activities were measured and normalized to protein concentration and luciferase activities of empty vectors (pGL2B and pCXN2). D, ChIP analysis of H3K4me3 levels at the MDR1 promoter in HeLa cells 24 hours following transfection with f-MLL1 plasmid. Chromatin was immunoprecipitated with anti-H3K4me3. Negative controls include chromatin precipitated with no antibody or rabbit IgG. Immunoprecipitated chromatin was amplified with PCR using MDR1 promoter-specific primers. 1% input DNA was amplified to ensure that equal amounts of DNA were subjected to CHIP analysis.
To verify the role of MLL1 in MDR1 transcription, f-MLL1 or f-MLL1-ΔSET was co-transfected with a luciferase reporter driven by the MDR1 promoter (pMDR1-1202-Luc) (11). Promoter activity as measured by luciferase expression 24 hours post transfection indicated that MLL1 overexpression correlated with an approximately 6.5-fold induction of MDR1 transcription (Fig. 1C); this activation was dependent on the presence of the SET domain, supporting a requirement for MLL1 methyltransferase activity. ChIP analysis verified that this f-MLL1-induced increase in MDR1 mRNA expression was accompanied by ~ 4-fold increase of H3K4me3 at the proximal promoter region of the MDR1 gene (Fig. 1D), further supporting a role for MLL1 in MDR1 activation. Again, overexpression of f-MLL1-ΔSET had no effect on the MDR1 promoter associated H3K4me3 levels (date not shown).
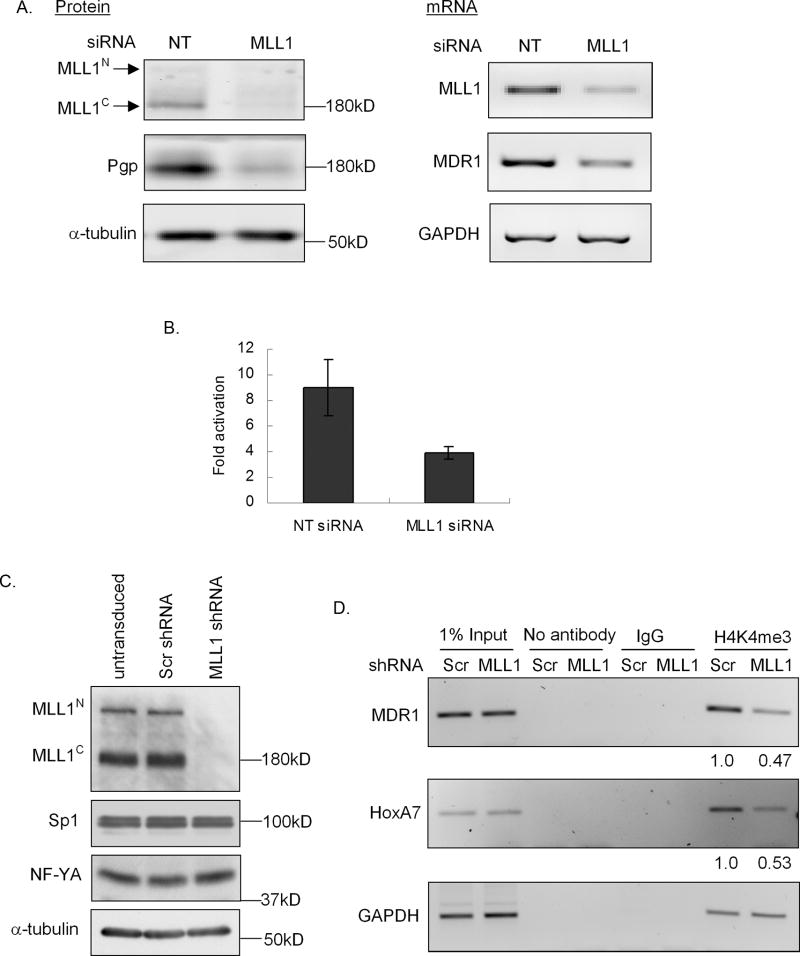
Next, transient knockdown of MLL1 was performed in HeLa cells and effects on the MDR1 promoter were determined. Downregulation of MLL1 protein (> 95%) with MLL1 siRNA was accompanied by an approximately 5-fold decrease in Pgp protein levels (Fig. 2A, left panel) as compared to non-targeting siRNA-transfected cells. RT-PCR analysis also revealed an approximately 5-fold decrease in MDR1 mRNA; GAPDH mRNA levels were unchanged (Fig. 2A, right panel). siRNA-mediated MLL1 knockdown also decreased the expression of the MDR1 promoter-driven luciferase reporter (> 50%) relative to what was observed following treatment with non-targeting siRNA (Fig. 2B).
Figure 2. MLL1 knockdown leads to decreased MDR1 expression and trimethylation of H3K4 at the MDR1 promoter.
A, Western blot analysis of MLL1N, MLL1C, Pgp, and α-tubulin levels (left panel), and RT-PCR analysis of relative MLL1, MDR1, and GAPDH mRNA levels (right panel) in HeLa cells 72 hours following transfection with non-targeting (NT) or MLL1 siRNA. B, MDR1 transcriptional activity was measured as luciferase activity. Cells were transfected with an MDR1 promoter-luciferase construct 72 hours following transfection with 100 nM NT siRNA or MLL1 siRNA. 24 hours post transfection, luciferase activities were measured and normalized to protein concentration and luciferase activity of pGL2B. C, Western blot analysis of MLL1N, MLL1C, NF-YA, Sp1 and α-tubulin levels in untransduced, scrambled shRNA (Scr shRNA) and MLL1 shRNA transduced HeLa cells following puromycin selection for 5 days. D, ChIP analysis of H3K4me3 levels at the MDR1, HoxA7 and GAPDH promoters in Scr and MLL1 shRNA transduced HeLa cells 5 days following puromycin selection. The decreased fold of H3K4me3 levels in MLL1 shRNA transduced cells over scrambled shRNA transduced cells was shown at the bottom.
Finally, ChIP assays were performed to evaluate the effect of MLL1 knockdown on the levels of H3k4me3 associated with the MDR1 promoter. Retrovirus-mediated transduction of an MLL1-specific short hairpin RNA (shRNA) stably reduced the expression of MLL1 as compared to the control, scrambled shRNA (Scr-shRNA) (Fig. 2C). As shown in Fig 2D, knockdown of MLL1 by shRNA resulted in a 2–3-fold decrease of H3K4me3 levels associated with the MDR1 promoter as well as the HoxA7 promoter, which has been previously identified as an MLL1 target gene (31). The GAPDH promoter-associated H3K4me3 levels were unchanged in both MLL1 and scrambled shRNA transduced cells (Fig. 2D). Taken together, these data demonstrate that MDR1 transcriptional activation is accompanied by trimethylation of promoter-associated H3K4, and that this methylation is mediated at least in part by catalytically active MLL1.
MLL1 regulation of MDR1 transcription involves NF-Y
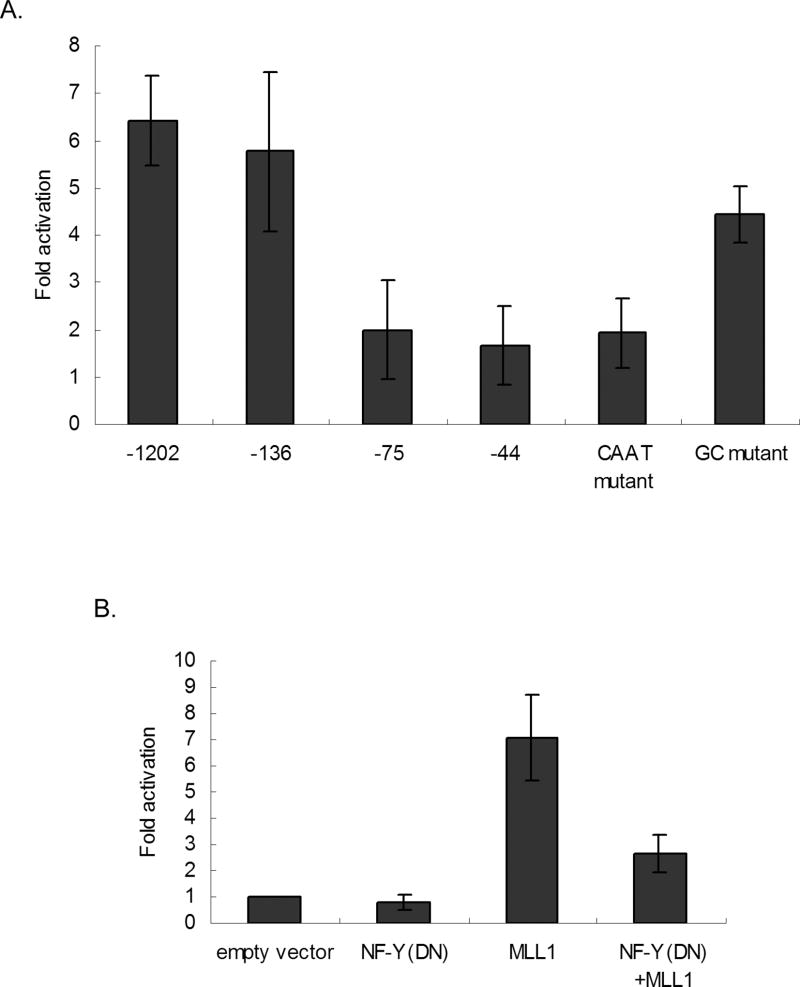
MDR1 gene expression can be activated by a variety of stimuli that converge on a region of the MDR1 proximal promoter that we refer to as the MDR1 “enhancesome” (32, 33). To determine whether MLL1 regulates MDR1 transcription through the MDR1 enhancesome, a series of MDR1 promoter deletion constructs were transiently co-transfected with MLL1 into HeLa cells. Analysis of luciferase activity 24 hours post transfection revealed that deletion of sequences from −1202 to −136 had no significant effect on MDR1 transcriptional activation by MLL1. However, deletion of sequences from −136 to −75 nearly abolished activation of MDR1 by MLL1 (Fig. 3A).
Figure 3. The CCAAT box and the NF-Y transcription factor are required for optimal MLL1 activation of MDR1 transcription.
A, MDR1 transcriptional activity was measured 24h post-co-transfection of f-MLL1 and a luciferase reporter construct driven by different deletions or point mutations of the MDR1 promoter. Luciferase activities were measured and normalized to protein concentration and luciferase activities of empty vectors (pGL2B and pCXN2). B, MDR1 transcriptional activity was measured by luciferase activity 24h post-cotransfection of a luciferase reporter construct driven by the MDR1 promoter with empty vector, f-MLL1, a dominant negative NF-Y (NF-Y(DN)), or both. Luciferase activities were measured and normalized to protein concentration and luciferase activities of empty vectors (pGL2B and pCXN2).
The sequence between −136 and −44 encompasses an inverted CCAAT box and a GC-rich region, both of which have been shown to be responsible for activation of the MDR1 promoter through interactions with NF-Y and Sp1, respectively (11, 12). To determine whether MLL1 activates MDR1 through either of these elements, f-MLL1 was co-transfected with an MDR1 promoter construct mutated in either the inverted CCAAT box or the GC box. Luciferase activity determined 24 h post transfection revealed that the induction of MDR1 transcription by MLL1 was dependent on the inverted CCAAT box (Fig. 3B), while the GC mutant had a relatively minor effect on the induction of MDR1 by MLL1. We and others have shown that the transcription factor NF-Y binds to the inverted CCAAT box and mediates transcriptional activation of MDR1 (11, 12, 34). To investigate a role for NF-Y in MLL1 regulation of MDR1 transcription, a dominant negative NF-Y expression construct, NF-Y(DN) (36), was co-transfected with f-MLL1 and the pMDR1-1202-Luc reporter into HeLa cells. Downregulation of NF-Y activity by the dominant negative protein led to a decrease in MDR1 activation by MLL1, implicating NF-Y in this process (Fig. 3B). Importantly, MLL1 knockdown did not affect the expression of either NF-Y or Sp1 (Fig. 2C).
MLL1 regulates MDR1-mediated drug export and drug resistance
Overexpression of MDR1 has been shown to confer resistance to a vast array of chemotherapeutic agents due to its ability to increase drug efflux, thereby decreasing intracellular accumulation (4, 5). Our data showing that MLL1 can activate MDR1 expression suggests that MLL1 can modulate drug transport and, hence, drug resistance. To examine the functional consequences of MDR1 regulation by MLL1, the effect of MLL1 knockdown on drug transport and cellular drug sensitivity was examined. As shown in Fig. 4A, MLL1 knockdown by siRNA, and subsequent MDR1 downregulation, led to a marked increase in retention of DIOC2(3), a highly specific Pgp substrate (35), as compared to cells transfected with non-targeting siRNA. The retention of DIOC2(3) in MLL1 knockdown cells was comparable to that in mock-transfected cells treated with 22 µM vinblastine, a competitive inhibitor of Pgp-mediated DIOC2(3) efflux, confirming the specificity of this assay.
Figure 4. MLL1 knockdown increases MDR1-dependent dye retention and sensitivity of cells to MDR1 substrates.
A, The retention was measured by the fraction of DIOC2(3) remaining in cells, 72 hours post-transfection of mock, non-targeting siRNA (NT siRNA), or MLL1 siRNA. As a control, the retention of DIOC2(3) in the mock transfected cells was performed in the presence of 22 µM vinblastine, a competitive inhibitor of dye efflux. Bars represent +/− standard deviation of 3 independent experiments. B, Cells were incubated with different concentrations of paclitaxel, vinblastine, or daunorubicin for 2 hours, following transfection with either NT siRNA or MLL1 siRNA. Percent of cell viability was measured by MTS assay 70 hours following drug incubation. Bars represent +/− standard deviation of 3 independent experiments.
The increase in dye retention following knockdown of MLL1 was also realized at the level of cellular sensitivity to Pgp substrates. As shown in Fig. 4B, knockdown of MLL1 led to a 5–15-fold increase in cytotoxicity to the Pgp drug substrates paclitaxel, vinblastine, and daunorubicin Notably, there was no significant change in sensitivity to the non-Pgp substrate, methotrexate (IC50 = 16 +/− 5.1 µM for cells transfected with non-targeting siRNA versus IC50 = 11 +/− 3.6 µM for cells transfected with MLL1 specific siRNA, data not shown). This minimizes the possibility that the effect of MLL1 on drug sensitivity is due to a more general effect on cell death pathways. Taken together, these data confirm that differential expression of MLL1 can affect MDR1 expression and in turn affect the sensitivity of cells to Pgp substrates, effectively reversing drug resistance.
MLL1 is not involved in TSA-induced H3K4me3 of the MDR1 gene
Trichostatin A (TSA), a histone deacetylase inhibitor and MDR1 transcriptional activator (11, 13) has been shown to induce H3K4me3 within discrete regions of the MDR1 locus (13). We have repeated these experiments in Hela cells, with interesting results (Fig. 5). TSA induction of MDR1 expression in HeLa cells (Fig 5A) was accompanied by increased H3K4me3 level in the coding region only (PCR primers were targeted 493–687 nt downstream of the transcription starting site) (Fig. 5B). Knockdown of MLL1 resulted in a 2-fold decrease of H3K4me3 level at the MDR1 coding region (Fig. 5C, compare lanes 1 and 4). However, treatment of MLL1 shRNA-transduced cells with TSA induced similar increases in H3K4me3 levels at the MDR1 coding region (Fig. 5C), suggesting that MLL1 is not involved in TSA-induced H3K4me3 of the MDR1 locus.
The MLL-AF4 fusion protein regulates MDR1 transcription
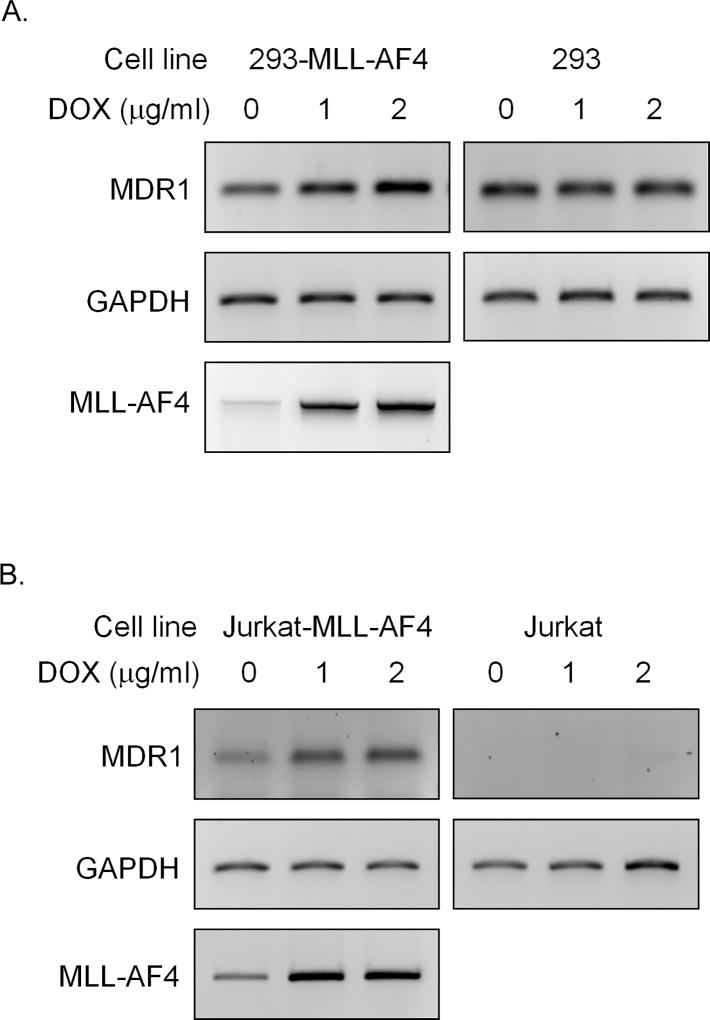
The MLL1 gene is a frequent target for recurrent chromosomal translocations found in human acute leukemias (36, 37). Patients with MLL-rearranged acute lymphoblastic leukemia (ALL) often have a particularly poor prognosis (38). Remarkably, more than 50 translocation partners have been identified and MLL-AF4 is one of the most frequent MLL rearrangements, accounting for 34% and 90% of pediatric and adult MLL-translocation-bearing ALL, respectively (36). To determine whether MLL fusion proteins could regulate MDR1 expression during leukemogenesis, the effect of MLL-AF4 on MDR1 expression was examined in 293 and Jurkat stable cell lines that conditionally express MLL-AF4 following doxycycline (DOX) induction (16). MLL-AF4 mRNA was expressed from 2 to 48 h after DOX induction as determined by RT-PCR, and peaked by 12 h (data not shown); therefore, 12 h of DOX exposure was used in subsequent studies. As shown in Fig. 6A, MDR1 mRNA levels were increased up to 3-fold following DOX treatment of 293-MLL-AF4 cells. Exposure of parental 293 cells to DOX did not affect MDR1 expression, supporting a role for MLL-AF4 in this event. Similar results were seen in Jurkat-MLL-AF4 cells and their parental counterparts (Fig. 6B). These data suggest a possible role for MLL fusion proteins in the regulation of MDR1 transcription during leukemogenesis.
Figure 6. MLL-AF4 induces MDR1 expression.
RT-PCR analysis of relative MDR1, MLL-AF4 and GAPDH mRNA levels without (0) or with (1, 2 µg/ml) DOX treatment for 12 h in A, 293-MLL-AF4 cells (left panel) and parental 293 cells (right panel), and in B, Jurkat-MLL-AF4 cells (left panel) and parental Jurkat cells (right panel).
Discussion
We have shown that histone methylation regulates MDR1 expression, and that the histone methyltransferase, MLL1, is involved in this regulation. Induction of MDR1 transcriptional activity by wild type MLL1 is dependent on the SET domain, demonstrating a requirement for the methyltransferase activity of MLL1 for this effect. This is consistent with what has already been shown for other MLL1 targets, such as Hox genes (25, 26) that require H3K4 methylation for recruitment of the basal transcription machinery and transcriptional initiation. It has been suggested that this requirement could be more important for a subset of genes, including those lacking TATA or other canonical core promoter elements (29, 39). Interestingly, all of the human drug-related transporters examined to date, including MDR1, lack an appropriately positioned TATA box (32, 33), suggesting that recruitment of the basal machinery to these genes could be enhanced by prior methylation of H3K4 by MLL1.
The CCAAT box that we have previously identified as part of the MDR1 enhancesome is required for MLL1 regulation of MDR1 transcription (11, 12). We and others have shown that the transcription factor NF-Y interacts with this element upon MDR1 gene activation (11, 12, 34). Here we show that NF-Y is also involved in the transcriptional regulation of MDR1 by MLL1, although how this is accomplished is not yet clear. In one scenario MLL1 may be recruited to the MDR1 enhancesome by NF-Y, which has been shown to be involved in the positioning of chromatin histone methyl marks, and required for MLL complex recruitment and H3K4 methylation on other CCAAT element containing promoters (40). While we have not yet been able to demonstrate a direct interaction between NF-Y and MLL1, we recognize the technical difficulties with these studies and additional experiments will be required to rule this out. Alternatively, MLL1 could be recruited to the MDR1 promoter by other factors and subsequent H3K4 methylation may provide a mark for recruitment of multiple transcriptional activators that recognize and bind to this histone modification to facilitate gene transcription (2, 15). Either scenario could explain our data demonstrating a requirement for NF-Y in MLL1-mediated MDR1 activation; the interplay between these critical factors is currently under investigation. Moreover, it should be noted that > 95% knockdown of MLL1 only partially decreased MDR1 H3K4me3 as detected by ChIP analysis. Therefore, the possibility remains that, while MLL1 is clearly involved in MDR1 transcriptional regulation, other histone methylases may also be required to achieve the full complement of methylated residues.
Indeed, we have also shown that TSA induces H3K4me3 levels of the MDR1 gene through its coding region. This is consistent with other reports showing that HDAC inhibitors can induce specific increases in H3K4 methylation in many cell types (41, 42). What is interesting is our observation that MLL1 is not required for TSA-induced methylation of the coding region, suggesting a role for another methyltransferase in creating this methyl mark. MLL4, a member of the MLL methyltransferase family, has been identified as the major activity responsible for the addition of H3K4me3 with HDAC inhibitors treatment (42). Whether MLL4, or another methyltransferase, is involved in TSA-induced H3K4me3 of the MDR1 locus in under investigation.
A particularly relevant finding in this study was that the transcriptional regulation of MDR1 by MLL1 has functional consequences. Knockdown of MLL1 leads to increased retention of a Pgp-specific dye as well as increased cellular sensitivity to the Pgp substrates paclitaxel, vinblastine, and daunorubicin. This has greater significance considering that many leukemias have a high prevalence of MLL dysregulation. In some cases, this is achieved through gene amplification (43), but in the majority of cases, MLL1 is found as part of a chromosomal translocation (36, 37). An initially perplexing observation was that the MLL found in translocations lacked the catalytic SET domain, yet was still capable of activating MLL1 target genes with no effect on H3K4 methylation (25). The current thinking is that at least a subset of MLL fusion partners, including AF4, can recruit DOT1L, a histone H3K79 methyltransferase to activate target genes and provoke leukemogenesis with enhanced H3K79me2 (36, 44–47). In this study, we showed that MDR1 expression is upregulated by MLL-AF4 with almost the same efficiency as wild type MLL1. While the mechanism by which this occurs is still under investigation, this is the first evidence that MLL fusions may contribute to the regulation of drug resistance during leukemogenesis. While there is some contradictory data, the expression and activity of MDR1 has been identified as predictors of the treatment outcome in childhood acute lymphoblastic leukemia in many clinical studies (48–50). To date, there have been no efforts to correlate MDR1 expression with the expression of MLL1 or MLL fusion proteins. Our data suggest that the analysis of the impact of MLL1 fusion proteins on expression of MDR1 and other drug transporters is warranted.
In conclusion, this is the first report of a histone methyltransferase involved in the regulation of an ABC drug transporter, and the first indication that translocations involved in leukemogenesis may also impact drug resistance in this tumor type. Thus, MLL1 provides a novel target for epigenetic therapy of cancer to circumvent multidrug resistance.
Acknowledgments
We thank members of the Scotto laboratory as well as our CINJ colleagues for insightful discussions. This research was supported by the Laboratory Support Service Shared Resource of The Cancer Institute of New Jersey (P30-CA072720).
References
- 1.Lennartsson A, Ekwall K. Histone Modification patterns and epigenetic codes. Biochim Biophys Acta. 2009;1790:863–8. doi: 10.1016/j.bbagen.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 3.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Eckford PD, Sharom FJ. ABC efflux pump-based resistance to chemotherapy drugs. Chem Rev. 2009;109:2989–3011. doi: 10.1021/cr9000226. [DOI] [PubMed] [Google Scholar]
- 5.Stavrovskaya AA, Stromskaya TP. Transport proteins of the ABC family and multidrug resistance of tumor cells. Biochemistry (Moscow) 2008;73:592–604. doi: 10.1134/s0006297908050118. [DOI] [PubMed] [Google Scholar]
- 6.Sakaeda T, Nakamura T, Hirai M, et al. MDR1 up-regulated by apoptotic stimuli suppresses apoptotic signaling. Pharm Res. 2002;19:1323–9. doi: 10.1023/a:1020302825511. [DOI] [PubMed] [Google Scholar]
- 7.Tanigawara Y. Role of P-glycoprotein in drug disposition. Ther Drug Monit. 2000;22:137–40. doi: 10.1097/00007691-200002000-00029. [DOI] [PubMed] [Google Scholar]
- 8.Zhou SF. Structure, function and regulation of P-glycoprotein and its clinical relevance in drug disposition. Xenobiotica. 2008;38:802–32. doi: 10.1080/00498250701867889. [DOI] [PubMed] [Google Scholar]
- 9.Mizutani T, Masuda M, Nakai E, et al. Genuine functions of P-glycoprotein (ABCB1) Curr Drug Metab. 2008;9:167–74. doi: 10.2174/138920008783571756. [DOI] [PubMed] [Google Scholar]
- 10.Aye IL, Singh AT, Keelan JA. Transport of lipids by ABC proteins: interactions and implications for cellular toxicity, viability and function. Chem Biol Interact. 2009;180:327–39. doi: 10.1016/j.cbi.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 11.Jin S, Scotto KW. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol Cell Biol. 1998;18:4377–84. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Z, Jin S, Scotto KW. Transcriptional activation of the MDR1 gene by UV irradiation. Role of NF-Y and Sp1. J Biol Chem. 2000;275:2979–85. doi: 10.1074/jbc.275.4.2979. [DOI] [PubMed] [Google Scholar]
- 13.Baker EK, Johnstone RW, Zalcberg JR, El-Osta A. Epigenetic changes to the MDR1 locus in response to chemotherapeutic drugs. Oncogene. 2005;24:8061–75. doi: 10.1038/sj.onc.1208955. [DOI] [PubMed] [Google Scholar]
- 14.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–8. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia ZB, Popovic R, Chen J, et al. The MLL fusion gene, MLL-AF4, regulates cyclin-dependent kinase inhibitor CDKN1B (p27kip1) expression. Proc Natl Acad Sci USA. 2005;102:14028–33. doi: 10.1073/pnas.0506464102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mantovani R, Li XY, Pessara U, Hooft van Huisjduijnen R, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J Biol Chem. 1994;269:20340–6. [PubMed] [Google Scholar]
- 18.Ramirez-Carrozzi VR, Nazarian AA, Li CC, et al. Selective and antagonistic functions of SWI/SNF and Mi-2 nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–96. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, Cheng EH, Hsieh JJ. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev. 2007;21:2385–98. doi: 10.1101/gad.1574507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swift S, Lorens J, Achacoso P, Nolan GP. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.im1017cs31. Chapter 10:Unit 10.17C. [DOI] [PubMed] [Google Scholar]
- 21.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–43. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 22.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–8. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 23.Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood. 2002;100:3710–8. doi: 10.1182/blood-2002-04-1015. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol. 2003;23:186–94. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 26.Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci USA. 2006;103:6629–34. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guenther MG, Jenner RG, Chevalier B, et al. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci USA. 2005;102:8603–8. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci USA. 2005;102:14765–70. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P, Lin C, Smith ER, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29:6074–85. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim DA, Huang YC, Swigut T, et al. Chromatin remodeling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–33. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra BP, Ansari KI, Mandal SS. Dynamic association of MLL1, H3K4 trimethylation with chromatin and Hox gene expression during cell cycle. FEBS J. 2009;276:1629–40. doi: 10.1111/j.1742-4658.2009.06895.x. [DOI] [PubMed] [Google Scholar]
- 32.Scotto KW, Johnson RA. Transcription of the multidrug resistance gene MDR1: a therapeutic target. Mol Interv. 2001;1:117–25. [PubMed] [Google Scholar]
- 33.Scotto KW. Transcriptional regulation of ABC drug transporters. Oncogene. 2003;22:7496–511. doi: 10.1038/sj.onc.1206950. [DOI] [PubMed] [Google Scholar]
- 34.Okamura H, Yoshida K, Sasaki E, Morimoto H, Haneji T. Transcription factor NF-Y regulates mdr1 expression through binding to inverted CCAAT sequence in drug-resistant human squamous carcinoma cells. Int J Oncol. 2004;25:1031–7. [PubMed] [Google Scholar]
- 35.Minderman H, Vanhoefer U, Toth K, et al. DiOC2(3) is not a substrate for multidrug resistance protein (MRP)-mediated drug efflux. Cytometry. 1996;25:14–20. doi: 10.1002/(SICI)1097-0320(19960901)25:1<14::AID-CYTO2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 36.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications, and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 37.Liedtke M, Cleary ML. Therapeutic targeting of MLL. Blood. 2009;113:6061–8. doi: 10.1182/blood-2008-12-197061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen CS, Sorensen PH, Domer PH, et al. Molecular rearrangements on chromosome 11q23 predominate in infant acute lymphoblastic leukemia and are associated with specific biologic variables and poor outcome. Blood. 1993;81:2386–93. [PubMed] [Google Scholar]
- 39.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–72. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donati G, Gatta R, Dolfini D, Fossati A, Ceribelli M, Mantovani R. An NF-Y-dependent switch of positive and negative histone methyl marks on CCAAT promoters. PloS One. 2008;3:e2066. doi: 10.1371/journal.pone.0002066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Lin Q, Yoon HG, et al. Involvement of histone methylation in regulation of transcription by thyroid hormone receptor. Mol Cell Biol. 2002;22:5688–97. doi: 10.1128/MCB.22.16.5688-5697.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nightingale KP, Gendreizig S, White DA, Bradbury C, Hollfelder F, Turner BM. Cross-talk between histion modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetykation and histone H3K4 methylation. J Biol Chem. 2007;282:4408–16. doi: 10.1074/jbc.M606773200. [DOI] [PubMed] [Google Scholar]
- 43.Reddy KS, Parsons L, Mark L, et al. Segmental amplification of 11q23 region identified by fluorescence in situ hybridization in four patients with myeloid disorders: a review. Cancer Genet Cytogenet. 2001;126:139–46. doi: 10.1016/s0165-4608(00)00406-4. [DOI] [PubMed] [Google Scholar]
- 44.Okada Y, Feng Q, Lin Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–78. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 45.Zeisig BB, Cheung N, Yeung J, So CW. Reconstructing the disease model an epigenetic networks for MLL-AF4 leukemia. Cancer Cell. 2008;14:345–7. doi: 10.1016/j.ccr.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 46.Krivtsov AV, Feng Z, Lemieux ME, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–68. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guenther MG, Lawton LN, Rozovskaia T, et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22:3403–8. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kourti M, Vavatsi N, Gombakis N, et al. Expression of multidrug resistance1 (MDR1), multidrug resistance-related protein 1 (MRP1), lung resistance protein (LRP), and breast cancer resistance protein (BCRP) genes and clinical outcome in childhood acute lymphoblastic leukemia. Int J Hematol. 2007;86:166–73. doi: 10.1532/IJH97.E0624. [DOI] [PubMed] [Google Scholar]
- 49.Dhooge C, De Moerloose B, Laureys G, et al. Expression of the multidrug transporter P-glycoprotein is highly correlated with clinical outcome in childhood acute lymphoblastic leukemia: results of a long-term prospective study. Leuk Lymphoma. 2002;43:309–14. doi: 10.1080/10428190290006080. [DOI] [PubMed] [Google Scholar]
- 50.Marie JP, Legrand O. MDR1/P-GP expression as a prognostic factor in acute leukemias. Adv Exp Med Biol. 1999;457:1–9. doi: 10.1007/978-1-4615-4811-9_1. [DOI] [PubMed] [Google Scholar]