Abstract
Flaviviruses, such as Dengue, Japanese encephalitis, West Nile, Yellow Fever, and Zika viruses, are serious human pathogens that cause significant morbidity and mortality globally each year. Flaviviruses are single-stranded, positive-sense RNA viruses, and encode two multidomain proteins, NS3 and NS5, that possess all enzymatic activities required for genome replication and capping. NS3 and NS5 interact within virus-induced replication compartments to form the RNA genome replicase complex. Although the individual enzymatic activities of both proteins have been extensively studied and are well characterized, there are still gaps in our understanding of how they interact to efficiently coordinate their respective activities during positive strand RNA synthesis and capping. Here, we discuss what is known about the structures and functions of the NS3 and NS5 proteins and propose a preliminary NS3:NS5:RNA interaction model based on a large body of literature about how the viral enzymes function, physical restraints between NS3 and NS5, as well as critical steps in the replication process.
Graphical abstract
Introduction
The genus Flavivirus is the largest among the family Flaviviridae with 53 different species (1). Flaviviruses can be further divided into non-vector, tick-borne and mosquito-borne clusters (2,3). Mosquito-borne flaviviruses, such as dengue virus (DENV), West Nile virus (WNV), yellow fever virus (YFV) and Japanese encephalitis virus (JEV) have become an increasing public health concern over the last decades since their global incidence has grown dramatically. For example, WNV was not present in the Americas prior to its introduction in New York in 1999. However, it has rapidly spread over the continent and can be found in many Western hemisphere countries today (4–6). Another example is the recent emergence of Zika virus (ZIKV). ZIKV has been known since the 1950s, but until 2007 no significant outbreaks and only few cases had been reported. In 2007, the first large Zika outbreak occurred on Yap Island. In 2013–2014, there were outbreaks in four other groups of Pacific Islands: French Polynesia, Easter Island, Cook Islands, New Caledonia. The current Zika epidemic began in early 2015 in Brazil and is now affecting most countries in Central and South America (7,8) and is intruding into North America.
Today, more than half of the world’s population is exposed to at least one flavivirus. In most cases, flaviviruses cause flu-like symptoms including fever, headache, muscle and joint pain, nausea, vomiting, and rash (9–13). Only a small number of infections result in severe disease like high fever, seizures, encephalitis, paralysis, jaundice (YFV), haemorrhage (DENV and YFV), coma and ultimately death (9–12). DENV is endemic in over 100 countries, putting 3.9 billion people at risk of infection. Each year, 390 million dengue infections occur, and 500,000 patients with dengue haemorrhagic fever require hospitalization, 2.5% of which die from the infection (11). YFV is endemic in 47 countries with a combined population of over 900 million people, resulting in an estimated 200,000 cases and 30,000 deaths each year (12,14). JEV is endemic in 24 countries, exposing more than 3 billion to the risk of infection and causing 68,000 clinical cases each year. Among those with severe symptoms, the fatality rate is especially high with up to 30% (13,600 – 20,400 deaths annually), and 20–30% of survivors suffer permanent neurologic or psychiatric sequelae (10). Currently, there are no specific antiviral drugs available against flaviviruses, and treatment consists of supportive care (9–12). The large numbers of annual cases and fatalities emphasize the need for effective antiviral drugs and/or vaccines. In order to develop novel methods of treatment and prevention, understanding how flavivirus genome replication functions on a molecular level is critical.
Flavivirus genome organization and life cycle
Flaviviruses have a positive-sense, single-stranded RNA genome of approximately 11kb that contains a 5’ cap, 5’ and 3’ untranslated regions (UTRs), and a single open reading frame (ORF). The ORF codes for a large polyprotein that is co- and post-translationally cleaved into three structural (C, prM, E) and eight non-structural (NS1, NS2A, NS2B, NS3, NS4A, 2K, NS4B, NS5) proteins. The structural proteins form the virus particle and play a role in virus entry into the host cell as well as assembly and release of new virions. The capsid protein binds the genomic RNA to form the nucleocapsid core, and the E and prM glycoproteins are viral surface proteins attached to the host-derived lipid envelope. The non-structural proteins form the viral replication complex inside the host cell (15). Only NS3 and NS5 are known to possess enzymatic activities. NS3 encodes serine protease (16–18), RNA helicase (19,20), nucleotide triphosphatase (NTPase) (21–23) and RNA triphosphatase (RTPase) (24,25) enzymatic activities. NS5 encodes methyl- and guanylyltransferase (MTase and GTase) (26–28) enzymatic activities as well as an RNA-dependent RNA polymerase (RdRp) (29,30). NS2A, NS2B, NS4A, 2K and NS4B are transmembrane proteins located within the endoplasmic reticulum (ER) membrane (31–34). Certain regions of NS2B, NS4A and NS4B also interact with NS3 and thus anchor the replication complex formed by NS3 and NS5 to the ER membrane (35–39). In addition, NS2B serves as an essential cofactor for the NS3 protease (40,41) and NS4B blocks interferon α/β signaling (42). NS4A induces ER membrane rearrangements that are thought to be involved in forming the viral replication compartment (33,43) while NS1 has been suggested to participate in genome replication by associating with the luminal side of the replication compartment and immune evasion through modulation of host defense mechanisms (44–46).
After virus entry, the viral genome is translated into the viral polyprotein on the rough ER (47). NS1, prM and E are translated into the ER lumen, the transmembrane domains of NS2A, NS2B, NS4A, 2K, and NS4B are translated into the ER membrane, and C, NS3 and NS5 remain in the cytoplasm (15,48). Localization of prM, E and NS1 to the ER lumen is ensured by signal peptides within the C-terminal hydrophobic domain of the C protein as well as the C-terminal transmembrane domains of prM and E. NS2A, NS2B, NS4A, 2K, and NS4B are inserted into the ER membrane due to their hydrophobic nature and multiple transmembrane domains. Together, NS2A and NS2B possess five transmembrane domains, causing NS3 to be localized on the cytosolic face of the ER membrane. NS5 is also localized at the ER membrane towards its cytosolic face, given that NS4A, 2K and NS4B possess a total of six transmembrane domains (15). The polyprotein is cleaved into individual viral proteins by host signalases inside the ER lumen and the viral protease NS2B-NS3 in the cytoplasm (48). The non-structural proteins form the replicase complex in virus-induced replication compartments on the ER membrane (49,50). The viral genome is first transcribed into a negative sense RNA, thus forming a dsRNA replication intermediate. This intermediate then serves as template for the synthesis of a large number of capped (+) ssRNA viral genomes (reviewed in (51)). The newly generated viral genomes are then used for further translation of viral proteins, generation of sfRNA (52), or association with structural proteins and incorporation into new virions which are transported through the trans-Golgi network and finally released from the host cell by exocytosis (15).
Viral RNA replication is a major process during the viral life cycle. Whereas negative strand synthesis is relatively well understood, the mechanisms of positive strand synthesis and capping by the NS3:NS5 replicase complex remains elusive.
Non-structural protein 3
The flavivirus non-structural protein 3 (NS3) is one of the two viral proteins with enzymatic activity. NS3 is well conserved among the Flaviviridae with a sequence identity of approximately 65% between WNV, DENV, JEV, ZIKV and YFV. The NS3 protein contains two domains, an N-terminal protease domain and a C-terminal helicase domain, which are coupled via a short flexible linker (53).
There have been multiple studies on the effect of the NS3 protease and helicase domains on each other. Most studies suggest a functional coupling of the two domains based on the observation that helicase activity is significantly increased in the full-length protein compared to the helicase domain alone (53–57). However, there has also been evidence that the presence of the protease domain has little influence on the activities of the helicase domain (58,59). These discrepancies might be due to the different nature of the protein constructs used in each study or differing experimental conditions.
NS3 structures
The three-dimensional structures of individual domains as well as full-length NS3 proteins of various Flaviviruses have been resolved (Figure 1). Different conformations and relative orientations of the two domains have been reported (53,54), suggesting that different conformational states could exist during virus replication. RNA binding has been shown to be one event in the replication cycle that induces a conformational change in the NS3 protein (60,61).
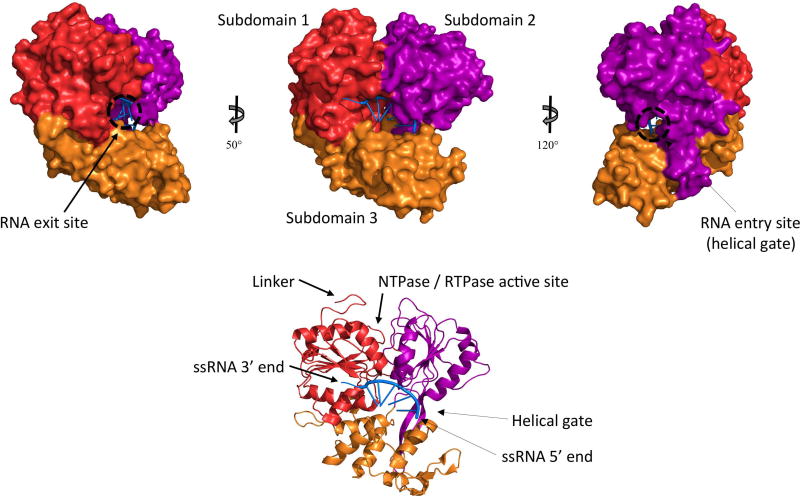
Figure 1. Structure of the NS3 helicase domain.
The crystal structure of the DENV NS3 helicase domain bound to ssRNA (pdb 2jlu) is shown. Subdomains 1, 2 and 3 are colored in red, purple and orange, respectively, and RNA is colored in blue. The 5’ end of the (−) ssRNA is close to the helical gate where dsRNA is separated into individual strands and the negative strand enters the RNA binding tunnel. The 3’ end of the (−) ssRNA is in proximity to the RNA exit site, from where it will be fed into the NS5 RdRp for replication.
NS3 protease domain
The N-terminal third of the NS3 protein constitutes the protease domain, which contains four regions of homology with serine proteases. Three of these regions constitute the catalytic triad and the fourth plays a role in substrate binding. The strictly conserved catalytic triad is formed by residues H51, D75 and S135 (DENV2 numbering) and is located in a cleft situated between two beta-barrels (36). In addition to the catalytic residues, the substrate binding pocket includes residues D129, F130, Y150, N152 and G153 (DENV numbering) (62,63). Substrate specificity has been suggested to be due to an aspartic acid residue at the bottom of the binding pocket (62). NS3 cleavage sites within the viral polyprotein have the consensus sequence of two basic residues followed by a residue with a small side chain, although the exact sequence depends on the cleavage site within the polyprotein as well as on the virus (64).
In order to form an active protease, the NS2B cofactor is essential (65). A β-strand of the central hydrophilic region of NS2B (residues 51–57, DENV numbering) folds into the N-terminal β-barrel of the NS3 protease to stabilize it (36). In addition, the C-terminal region of NS2B (residues 75–85, DENV numbering) undergoes a conformational change upon substrate binding to form a stabilizing β-hairpin that becomes part of the active site (35,36). The active NS2B-NS3 protease is required for cleavage at the NS2A/NS2B, NS2B/NS3, NS3/NS4A, and NS4B/NS5 junctions (40,66). Furthermore, it has been suggested that the cleavage of the C-terminal membrane-spanning segment of the capsid protein that takes place after several basic residues could be mediated by the NS2B-NS3 protease (67).
Since the NS2B protein has multiple transmembrane domains in addition to the central hydrophilic region that interacts with the NS3 protease domain (32), the NS2B:NS3 interaction probably tethers NS3 to the membrane, thus anchoring the replicase complex to the membrane of the replication compartments (68). In fact, both NS2B and NS3 have been found to be associated with virus-induced membrane structures (50,69), and the membrane requirement of the DENV NS3 protease has been shown to be dependent on NS2B, suggesting that the localization of NS3 to membranes is mediated by its interaction with NS2B (70).
NS3 helicase domain
The C-terminal helicase domain of NS3 harbors three enzymatic activities: RNA helicase, nucleoside triphosphatase (NTPase) and RNA triphosphatase (RTPase) (19,23,25). NTPase and RTPase activities have been shown to share the same catalytic residues (24,71,72) whereas dsRNA unwinding, although driven by ATP hydrolysis, takes place at a distinct site known as helical gate (73). The RTPase reaction constitutes the first step in the synthesis of the cap structure at the 5’ end of newly synthesized viral genomes, which also serve as viral mRNAs, by removing the γ-phosphate from the newly synthesized RNA and generating 5’ diphosphorylated RNA (25). The RNA helicase activity unwinds the dsRNA replication intermediate in order to release the newly generated viral genome (positive strand) and to make the negative strand available as template for another round of viral genome synthesis. In addition, the helicase activity removes secondary structures from the viral RNA, especially in the 5’ and 3’ untranslated regions (68,74). An unstructured 3’ end is necessary for replication complex assembly, and a 5’ protruding end is required for an efficient RNA triphosphatase (RTPase) reaction during the synthesis of the cap structure (75). The energy required for the helicase activity is derived from nucleotide hydrolysis at the NTPase active site, but the mechanism which couples these two activities remains elusive (74). NS3 has been shown to hydrolyse the gamma-phosphate of all four NTPs, although preferring purines (76).
The NS3 helicase domain is further divided into three subdomains (Figure 1). Subdomains 1 and 2 contain seven conserved helicase motifs characteristic of DEXH/D box family helicases, including Walker A and Walker B motifs (56,55,77). Residues from these subdomains form one side of the ssRNA binding tunnel with the other side being subdomain 3 (60). The access site to this ssRNA binding tunnel is located between the α-helix 2 of subdomain 2, the α-helix 6 of subdomain 3 and the β-hairpin protruding from subdomain 2 toward subdomain 3 (73). This site is also known as the helical gate since this is where dsRNA is split into two ssRNA strands. The β-hairpin disrupts base stacking between the two strands so that the 3’ end of the negative strand enters the RNA binding tunnel and the 5’ end of the positive strand is forced to move along the protein’s surface (60,74). After going through the RNA binding channel, the 3’ end of the negative strand exits the NS3 protein close to the junction between subdomains 1 and 3, which is at the opposite side of the protein than the helical gate between subdomains 2 and 3 (Figure 1) (60). The dsRNA unwinding site is approximately 30Å away from the ATPase catalytic site (73), and it is still unclear how RNA unwinding and ATP hydrolysis are coupled (68,74). The NTPase/RTPase active site is a basic pocket located close to the interface between the helicase (subdomains 1 and 2) and protease domains (53). The Walker A motif, more precisely residues G198, K199 and T200 (DENV2 numbering), as well as the Walker B motif play essential roles in substrate binding and coordination of the Mg2+ ion, respectively (58,71,55). Residues R457, R458, R460 and R463 (DENV2 numbering) have also been shown to be important for ATPase and RTPase activities (72). The phosphate product generated during NTPase and RTPase reactions has been proposed to exit the active site through a channel lined by residues P195, A316, T317, P326, A455 and Q456 (DENV numbering) (60).
Two non-structural proteins, NS4A and NS4B, serve as allosteric cofactors for the NS3 helicase domain. They enhance its activity by reducing the amount of ATP required and by facilitating the dissociation of ssRNA, respectively (37,38). Since these two proteins also possess transmembrane domains (33,34), their interactions with NS3 likely contribute to anchoring the replicase complex to the ER membrane. Moreover, NTPase activity has been shown to be enhanced by the presence of ssRNA (19,22,78) and both NTPase and RTPase activities have been shown to be stimulated by NS5 (76,78), indicating that allosteric interaction between NS3 and other factors have significant effects on NS3 activity.
Non-structural protein 5
The Flavivirus non-structural protein 5 (NS5) is the other viral protein with enzymatic activities. It is the largest and most conserved protein among the Flaviviridae with a sequence identity of approximately 68% between WNV, DENV, JEV, ZIKV and YFV. Its N-terminal capping enzyme domain and C-terminal RNA-dependent RNA polymerase (RdRp) domain are coupled via a short linker (79).
NS5 structures
Numerous crystal structures of isolated capping enzyme and RdRp NS5 domains as well as the full-length protein from multiple Flaviviruses have been solved (Figure 2, Figure 3). Some studies reveal a compact structure of the NS5 protein whereas others indicate a more extended structure, which suggests that NS5 possesses a certain flexibility regarding the relative orientation of the two domains, and that it may undergo conformational changes during the RNA replication cycle (79–81). The inter-domain linker seems to play a central role in the adoption of different conformations, since it is well-ordered in some crystal structures but disordered and without a secondary structure in others (79,81). Moreover, the highly conserved GTR sequence of the MTase domain (residues 261–263, DENV numbering) next to the linker region has been proposed to act as a pivot between the two domains (79).
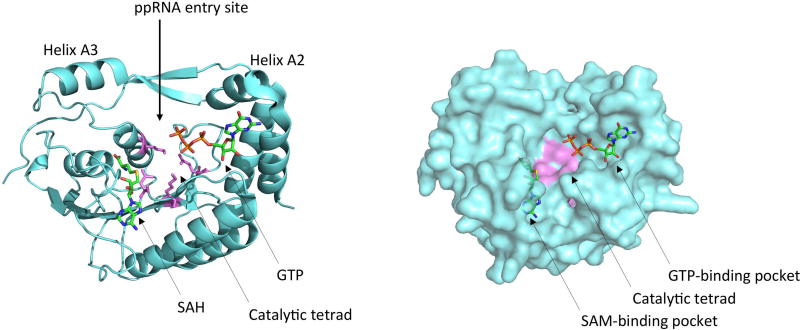
Figure 2. Structure of the NS5 capping enzyme domain.
The crystal structure of the YFV NS5 capping enzyme domain bound to GTP and SAH (pdb 3evd) is shown. The capping enzyme domain is colored in light blue, the catalytic tetrad in violet, and GTP and SAH are colored by element. The RNA entry site and substrate binding pockets are indicated.
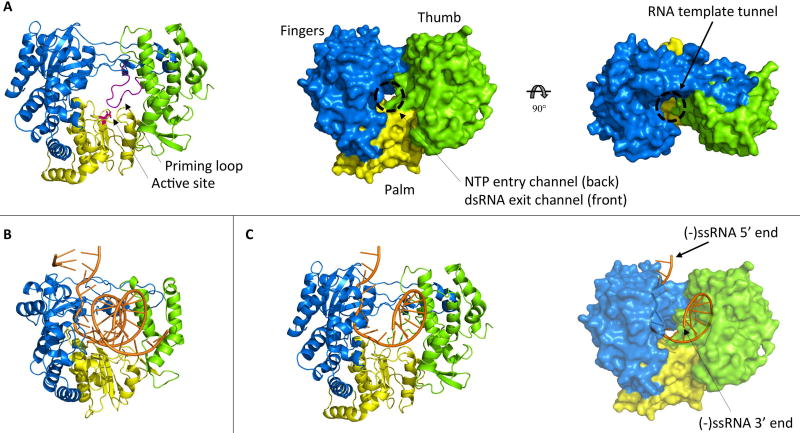
Figure 3. Structure of the NS5 RNA-dependent RNA polymerase domain.
(A) The crystal structure of the JEV NS5 RdRp (pdb 4k6m) is shown. The fingers, palm and thumb subdomains are colored in blue, yellow and green, respectively. In the cartoon representation, the priming loop is colored in purple and the two catalytic aspartic acid residues are shown as magenta colored sticks. In the surface representations, the RNA template tunnel and the NTP entry channel / dsRNA exit channel are indicated. (B) The structure of the poliovirus RdRp elongation complex after multiple nucleotide addition cycles is shown (pdb 3ola). (C) The negative strand from the poliovirus RdRp structure was modelled into the JEV RdRp structure. As expected, the 5’ end of the (−) ssRNA is close to the entry of the RNA template tunnel and the 3’ end is exiting the active site through the dsRNA exit channel.
NS5 capping enzyme domain
The capping enzyme domain of NS5 executes three steps in the synthesis of the 5’ cap structure of the viral genome. First, it transfers a GMP to the 5’ diphosphate end of nascent viral RNA with its GTase activity (28). In the second and third steps, it methylates the cap structure at the N7 position of the guanine as well as at the 2’-O position of the ribose of the first nucleotide with its MTase activity, using S-adenosyl-L-methionine (SAM or AdoMet) as a methyl donor (26,27). There are two different substrate binding pockets within the capping enzyme (Figure 2). These pockets are 12–15Å apart and they are linked by a positively charged surface groove that most likely binds the viral RNA during guanylyltransferase and methyltransferase reactions (26,82–84). The 5’ diphosphorylated RNA (ppRNA) substrate has been shown to be bound by residues F25, K30, R57, K181 and R212 (DENV2 numbering) which are clustered around the base of helix A2, suggesting that the ppRNA might enter into the capping enzyme through the groove between helices A2 and A3 (85). In addition to ppRNA, the guanylyltransferase reaction requires a second substrate: GTP. The GTP binding pocket which includes residues K14, L17, N18, L20, F25, K29 and S150 (DENV2 numbering) has been proposed to be the active site for the GTase activity (26,82). Interestingly, several residues are involved in binding both GTP and ppRNA, and phosphates from GTP and phosphates from ppRNA have been shown to compete for the same binding site on the capping enzyme, indicating that the two substrates for the guanylyltransfer reaction are bound at different times during capping (85). It has been suggested that the enzyme first binds GTP to form a GMP-enzyme intermediate before binding ppRNA and catalyzing the transfer of GMP to the RNA (28,85), although the precise kinetic parameters of both steps have not been fully determined. The resulting GpppRNA is then methylated at the guanine N7 position as well as at the ribose 2’-O position to form completely capped m7GpppAmRNA, with the N7-methylation occurring prior to the 2’-O-methylation (27,83). In both cases, the methyl donor is SAM which binds to a distinct pocket comprising residues S56, G86, W87, T104, L105, D131, V132, I147 and Y219 (DENV2 numbering) (26), and the conserved residues K61-D146-K181-E217 (DENV2 numbering) make up the catalytic tetrad (27). The RNA substrate needs to be repositioned between the two methyltransferase reactions, and the GTP binding pocket has been suggested to accommodate the RNA cap structure during 2’-O-methylation (83).
NS5 RNA-dependent RNA polymerase domain
The RNA-dependent RNA polymerase (RdRp) domain of NS5 replicates the viral RNA genome (30,86). It contains six conserved RdRp motifs and it adopts the canonical right-hand conformation with thumb, palm and fingers subdomains (Figure 3A) (87,88). The 3’ end of the template RNA enters the active site (G662-D663-D664 motif, DENV numbering), which is located within the palm subdomain close to the interface with the two other subdomains, through a tunnel between the fingers and thumb subdomains. A second channel is formed by the fingers and palm subdomain. It also connects to the active site, is approximately perpendicular to the first channel, and goes across the entire protein. NTPs enter the active site through the back of this channel, and the front of this tunnel would be the exit route for the dsRNA product (87–89). However, this exit site is blocked in all observed structures, and a conformational change prior to elongation from a closed state to an open state would be required to accommodate nascent dsRNA in this exit channel (87,89). To date, no NS5 RdRp structures with RNA have been solved, but the RdRp elongation complex from poliovirus has been crystallized (90). This structure can be used as a template to model the negative strand RNA into the NS5 RdRp domain (Figure 3C). The 5’ end of the (−) ssRNA is in proximity to the RNA template tunnel whereas the 3’ end is close to the dsRNA exit channel. This suggests that the 3’ end of the negative strand, which serves as template during positive strand synthesis, is fed into the NS5 RdRp active site through the RNA template tunnel and exits the active site though the dsRNA exit channel after replication has occurred, thus indicating directionality of RNA replication.
The NS5 RdRp initiates RNA synthesis by a de novo mechanism (91). Residues 782–809 (DENV numbering) form the priming loop which protrudes from the thumb subdomain toward the active site, thus partially blocking the template RNA tunnel and allowing only ssRNA to enter the catalytic site (88,89). Residues W795 and H798, which are conserved among Flaviviruses, form stacking interactions with nucleotides and hence provide an initiation platform (88,92). H798 forms stacking interactions with the initiating ATP nucleotide (92), and W795 binds a GTP molecule at the de novo GTP-binding site (i-1 position) which is located at a distance of 6–7Å from the catalytic site (89,93). This GTP has been shown to be crucial for initiation, since the need of a high GTP concentration during de novo initiation has been observed, regardless of the nucleotide sequence of the template RNA (94). More precisely, this GTP nucleotide mimics a nascent RNA strand and interacts with the initiating ATP to stabilize it at the i position and thus to contribute to the proper positioning of its 3’ hydroxyl group which is involved in the formation of a phosphodiester bond with the second nucleotide at the i+1 positon (89). Two catalytic magnesium ions, which are coordinated by aspartic acid residues at the active site (D533 and D664, DENV numbering), hold the phosphates of the incoming nucleotide in place during the nucleophilic attack of the 3’-O of the first nucleotide on the α-phosphate of the second nucleotide (87–89). Once the first phosphodiester bond is formed, the RdRp switches to an open conformation and the GTP nucleotide is released from the i-1 position, thus vacating the dsRNA exit tunnel and allowing the elongation of the nascent dsRNA. Motif G of the fingertip region has been proposed to be involved in translocation of the RdRp along the template RNA, but the exact mechanism of translocation remains elusive (89,95).
Protein-protein interactions within the replicase complex
Given that positive strand RNA synthesis and capping requires both NS3 and NS5, and that the functions of these two enzymes are supported by the other non-structural proteins, multiple interactions between non-structural proteins have been identified. As discussed above, NS2B, NS4A and NS4B are cofactors for the NS3 protease and helicase domains, and therefore interact with NS3 (35–39). Furthermore, these three proteins anchor NS3 to the ER membrane, since they possess transmembrane regions in addition to hydrophilic regions that bind NS3 (32–34). None of the small transmembrane non-structural proteins interacts with NS5, and NS5 is therefore recruited to the membrane-associated replicase complex only via its interaction with NS3 (96).
NS3 and NS5 have been shown to interact in vitro by various binding experiments such as pull-down (97–99), ELISA (100,101), surface plasmon resonance (102) and AlphaScreen assay (103). Their interaction in vivo has also been confirmed with multiple techniques such as co-immunoprecipitation using extracts from infected cells (76,97,100,103,104), yeast-two hybrid (98,105), co-localization and bimolecular fluorescence complementation (106). The results from these studies demonstrate that there is a direct interaction between NS3 and NS5 that does not require any other viral protein (97). Several studies have suggested that the NS3:NS5 interaction is mediated by the NS5 RdRp domain (98–101,105) and the NS3 helicase domain (98,100), although a contribution of the protease domain has also been observed (102,103). A schematic NS3:NS5 interaction model in which the interaction is mediated by the NS3 helicase subdomain 3 and the NS5 RdRp thumb subdomain has been proposed (101). However, the SAXS data on which this model is based is severely biased by the artificial covalent fusion of the NS5 320–341 peptide to the C-terminus of the helicase subdomain 3 via a short linker, and data and conclusions from this study might therefore not be correct. Furthermore, the proposed model does not address how the two proteins would actually work together at the molecular level, and fails to consider RNA as an important component of the replicase complex. Further studies with full-length NS3 and NS5 proteins as well as the replicative form of viral RNA are needed to elucidate the NS3:NS5 interaction within the replicase complex during positive strand synthesis.
Development of a detailed NS3:NS5 replication complex model
Understanding how the components of the replicase complex are associated will help define the mechanism of positive strand synthesis and potentially uncover new points for therapeutic intervention. However, it has not been explicitly shown that NS3 and NS5 directly interact during virus replication, primarily due to limitations of tagging the proteins within replication compartments while maintaining replication. In addition, it has not been clearly demonstrated that the NS3:NS5 interaction is essential for virus replication. Nevertheless, the enzymatic activities of both NS3 and NS5 are required for replication, and they most likely interact within the replicase complex in order to coordinate their enzymatic activities for a more efficient replication.
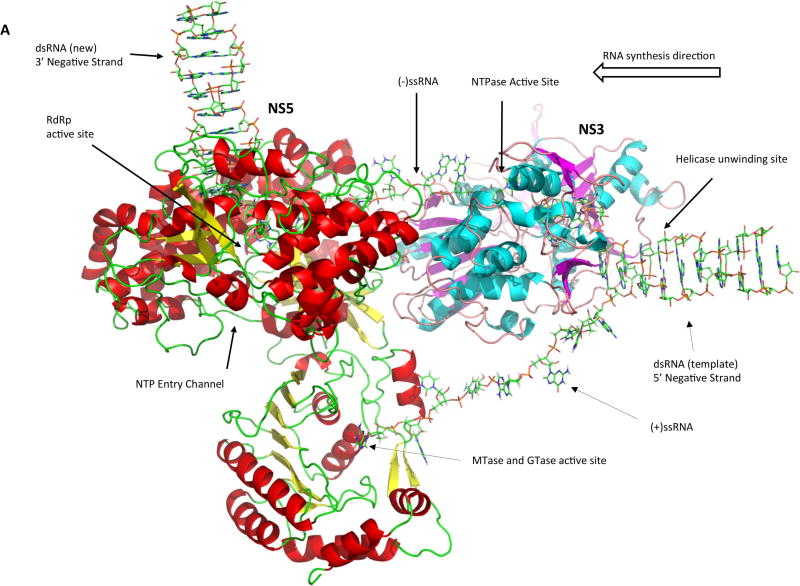
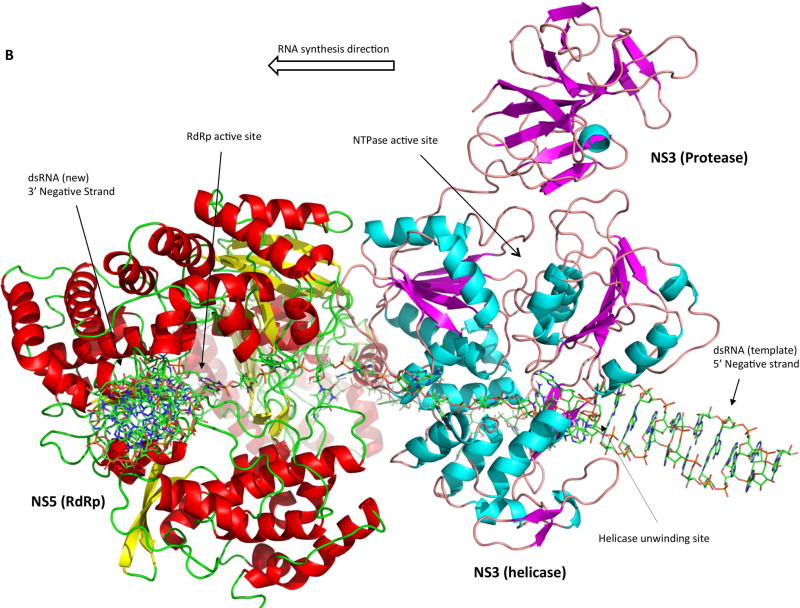
We developed a preliminary NS3:NS5:RNA interaction model based on the known crystal structures as well as enzymatic activities of the two proteins, and we used this information to infer how they might interact in order to efficiently coordinate their activities. Homology models of full-length NS3 and NS5 were generated on the Swissmodel Server (107) using sequences from the African Lineage II West Nile virus strain WN 956 D117 3D (108) and pdb files 2VBC of DENV NS3 (53) and 4K6M of JEV NS5 (79) were used as structure templates. Homology structures were then imported into PyMOL and hand-manipulated such that the 3’ end of the ssRNA in the NS3 protein would feed into the palm domain of the NS5 RdRp. The resulting structure is a model of the viral replication complex during positive sense RNA synthesis (Figure 4A). The dsRNA replication intermediate is separated into two individual strands by the dsRNA passing through the NS3 helicase helical gate, the 5’ diphosphorylated end of the positive strand being guided into the NS5 capping enzyme to undergo GTase and MTase reactions for the formation of a complete cap structure, and the 3’ end of the negative strand being guided into the NS5 polymerase domain to serve as template for the synthesis of a new positive strand of viral RNA (Figure 4B). NS3 and NS5 interact via their helicase and RdRp domain, respectively. The RNA-binding tunnels of both proteins are aligned, and the RNA would act as a rotational axis on the NS3:NS5 interaction. Moreover, pulling of RNA through the RdRp domain may pull NS3 into closer proximity to the RdRp.
Figure 4. Model of the replicase complex formed by NS3, NS5 and viral RNA during positive strand synthesis.
(A) Front view. (B) Top view. The incoming dsRNA replication intermediate is denatured by the NS3 helicase. The 5’ end of the positive strand is guided into the NS5 capping enzyme domain active site for the synthesis of the 5’ cap structure. The 3’ end of the negative strand flows through the NS3 RNA binding tunnel and then into the NS5 RdRp active site to serve as template for the synthesis of a new positive strand.
There are multiple positive aspects to our model. Since crystal structures for both NS3 and NS5 are known, potential interactions can be easily visualized. The model is based on a large body of literature about how the viral enzymes function, and physical restraints (ssRNA) as well as critical steps in the replication process were used to guide the development of the model. The substrate entry channels for NS3 (ATP) and NS5 (nucleotides) are on approximately the same side, which would probably be important for substrate access to the enzymes if the replicase complex is anchored within the neck of the membranous replication compartment. Finally, the model follows Occam’s Razor and is a testable model even in the absence of a co-crystal structure.
There are a few potential caveats to our model. The current model assumes a 1:1 stoichiometry of NS3 and NS5, but it is possible that the complex is 2:2, 2:1 or some other permutation, although it is not currently known due to limitations in the availability to resolve the atomic-level structure of the membranous viral replication compartments. In addition, the model does not address membrane association of the replication complex, although anchoring of NS3 to the ER membrane via NS2B, NS4A and NS4B would be the most likely scenario. Finally, both proteins may undergo conformational changes during the virus life cycle, and the crystal structures that served as template for our homology structures may not be in the right conformation for positive strand RNA synthesis.
Nevertheless, the model we present is a good foundation for the precise characterization of the NS3:NS5 interaction. Future studies can use this model as a starting point for long time-scale all-atom molecular dynamics simulations that can precisely define the dynamic interactions between NS3, NS5, and RNA and how the interactions affect the structures of the replicase complex components. One of the major limitations of just using crystallography is that these structures are static snapshots in time, and may not represent physiologically relevant structures or conformations. Additionally, using models as we have presented allow for examination using molecular dynamics simulations. These simulations can uncover dynamical allosteric interactions between the replicase components using rigorous biophysical parameters, interactions which are difficult to trap in crystallography experiments. These computational data can then be used to direct mutagenesis analyses of recombinant NS3, NS5, and RNA interactions using traditional techniques such as pull downs and SPR, and mutations that show effects on the stability of the replicase complex in vitro can be introduced into infectious viruses to generate empirically validated models of the replicase complex. Data from these experimental studies can be iteratively fed back into the interaction model until it most likely represents the actual protein-protein interaction. Ultimately, development of an atomic level model of the flavivirus replicase complex may lead to the identification of compounds that might interrupt NS3:NS5 interaction or allosteric interactions and thus be interesting new candidates for antiviral drug development.
Acknowledgments
This work was supported by a graduate training award from the Faculté de médecine et des sciences de la santé de l’Université de Sherbrooke to C.B., a grant from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2016-03916) to M.B. and a grant from the National Institutes of Health (5 R01 AI114675) to B.J.G.
References
- 1.International Committee on Taxonomy of Viruses. Virus Taxonomy: 2015 Release [Internet] 2015 Available from: http://www.ictvonline.org/virusTaxonomy.asp.
- 2.Kuno G, Chang G-J, Tsuchiya KR, Karabatsos N, Cropp CB. Phylogeny of the Genus Flavivirus. J Virol. 1998;72(1):73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasilakis N, Weaver SC. Flavivirus transmission focusing on Zika. Curr Opin Virol. 2016;22:30–5. doi: 10.1016/j.coviro.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chancey C, Grinev A, Volkova E, Rios M. The Global Ecology and Epidemiology of West Nile Virus. Biomed Res Int. 2015;2015:376230. doi: 10.1155/2015/376230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciota AT, Kramer LD. Vector-Virus Interactions and Transmission Dynamics of West Nile Virus. Viruses. 2013;5(12):3021–47. doi: 10.3390/v5123021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elizondo-Quiroga D, Elizondo-Quiroga A. West Nile Virus and its Theories, a Big Puzzle in Mexico and Latin America. J Glob Infect Dis. 2013;5(4):168–75. doi: 10.4103/0974-777X.122014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO. Situation report: Zika virus, Microcephaly, Guillain-Barré Syndrome. 24 November 2016 [Internet] 2016 Available from: http://www.who.int/emergencies/zika-virus/situation-report/24-november-2016/en/
- 8.WHO. The History of Zika Virus [Internet] 2017 Available from: http://www.who.int/emergencies/zika-virus/history/en/
- 9.WHO. West Nile virus: Fact sheet No 354 (July 2011) [Internet] 2011 Available from: http://www.who.int/mediacentre/factsheets/fs354/en/#.
- 10.WHO. Japanese encephalitis: Fact sheet No 386 (December 2015) [Internet] 2015 Available from: http://www.who.int/mediacentre/factsheets/fs386/en/
- 11.WHO. Dengue: Fact sheet (July 2016) [Internet] 2016 Available from: http://www.who.int/mediacentre/factsheets/fs117/en.
- 12.WHO. Yellow fever: Fact sheet No 100 (May 2016) [Internet] 2016 Available from: http://www.who.int/entity/mediacentre/factsheets/fs100/en/
- 13.WHO. Zika virus: Fact sheet (June 2016) [Internet] 2016 Available from: http://www.who.int/entity/mediacentre/factsheets/zika/en/
- 14.WHO. Yellow fever: Fact sheet No 100 (March 2014) [Internet] 2014 Available from: http://www.who.int/mediacentre/factsheets/fs100/en/
- 15.Lindenbach BD, Thiel H-J, Rice CM. Flaviviridae: The Viruses and Their Replication. Fields Virology. (5) 2007:1101–52. [Google Scholar]
- 16.Chambers TJ, Weir RC, Grakoui A, McCourt DW, Bazan JF, Fletterick RJ, et al. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci U S A. 1990;87(22):8898–902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preugschat F, Yao C-W, Strauss JH. In vitro Processing of Dengue Virus Type 2 Nonstructural Proteins NS2A, NS2B, and NS3. J Virol. 1990;64(9):4364–74. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wengler G, Czaya G, Färber PM, Hegemann JH. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J Gen Virol. 1991;72(4):851–8. doi: 10.1099/0022-1317-72-4-851. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Clum S, You S, Ebner KE, Padmanabhan R. The Serine Protease and RNA-Stimulated Nucleoside Triphosphatase and RNA Helicase Functional Domains of Dengue Virus Type 2 NS3 Converge within a Region of 20 Amino Acids. J Virol. 1999;73(4):3108–16. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Utama A, Shimizu H, Morikawa S, Hasebe F, Morita K, Igarashi A, et al. Identification and characterization of the RNA helicase activity of Japanese encephalitis virus NS3 protein. FEBS Lett. 2000;465(1):74–8. doi: 10.1016/s0014-5793(99)01705-6. [DOI] [PubMed] [Google Scholar]
- 21.Takegami T, Sakamuro D, Furukawa T. Japanese Encephalitis Virus Nonstructural Protein NS3 Has RNA Binding and ATPase Activities. Virus Genes. 1995;9(2):105–12. doi: 10.1007/BF01702653. [DOI] [PubMed] [Google Scholar]
- 22.Warrener P, Tamura JK, Collett MS. RNA-Stimulated NTPase Activity Associated with Yellow Fever Virus NS3 Protein Expressed in Bacteria. J Virol. 1993;67(2):989–96. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wengler G, Wengler G. The Carboxy-Terminal Part of the NS 3 Protein of the West Nile Flavivirus Can Be Isolated as a Soluble Protein after Proteolytic Cleavage and Represents an RNA-stimulated NTPase. Virology. 1991;184(2):707–15. doi: 10.1016/0042-6822(91)90440-m. [DOI] [PubMed] [Google Scholar]
- 24.Bartelma G, Padmanabhan R. Expression, Purification, and Characterization of the RNA 5’-triphosphatase activity of Dengue Virus Type 2 Nonstructural Protein 3. Virology. 2002;299(1):122–32. doi: 10.1006/viro.2002.1504. [DOI] [PubMed] [Google Scholar]
- 25.Wengler G, Wengler G. The NS 3 Nonstructural Protein of Flaviviruses Contains an RNA Triphosphatase Activity. Vol. 197. Virology. 1993:265–73. doi: 10.1006/viro.1993.1587. [DOI] [PubMed] [Google Scholar]
- 26.Egloff M-P, Benarroch D, Selisko B, Romette J-L, Canard B. An RNA cap (nucleoside-2’-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21(11):2757–68. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, et al. West Nile Virus 5’-Cap Structure Is Formed by Sequential Guanine N-7 and Ribose 2’-O Methylations by Nonstructural Protein 5. J Virol. 2006;80(17):8362–70. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Issur M, Geiss BJ, Bougie I, Picard-Jean F, Despins S, Mayette J, et al. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA. 2009;15(12):2340–50. doi: 10.1261/rna.1609709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guyatt KJ, Westaway EG, Khromykh AA. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J Virol Methods. 2001;92(1):37–44. doi: 10.1016/s0166-0934(00)00270-6. [DOI] [PubMed] [Google Scholar]
- 30.Tan B-H, Fu J, Sugrue RJ, Yap E-H, Chan Y-C, Tan YH. Recombinant Dengue Type 1 Virus NS5 Protein Expressed in Escherichia coli Exhibits RNA-Dependent RNA Polymerase Activity. Virology. 1996;216(2):317–25. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- 31.Xie X, Gayen S, Kang C, Yuan Z, Shi P-Y. Membrane Topology and Function of Dengue virus NS2A protein. J Virol. 2013;87(8):4609–22. doi: 10.1128/JVI.02424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Li Q, Wong YL, Liew LSY, Kang C. Membrane topology of NS2B of dengue virus revealed by NMR spectroscopy. Biochim Biophys Acta. 2015;1848(10):2244–52. doi: 10.1016/j.bbamem.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Miller S, Kastner S, Krijnse-Locker J, Bühler S, Bartenschlager R. The Non-structural Protein 4A of Dengue Virus Is an Integral Membrane Protein Inducing Membrane Alterations in a 2K-regulated Manner. J Biol Chem. 2007;282(12):8873–82. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 34.Miller S, Sparacio S, Bartenschlager R. Subcellular Localization and Membrane Topology of the Dengue Virus Type 2 Non-structural Protein 4B. J Biol Chem. 2006;281(13):8854–63. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- 35.Aleshin AE, Shiryaev SA, Strongin AY, Liddington RC. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007;16(5):795–806. doi: 10.1110/ps.072753207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, et al. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13(4):372–3. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 37.Shiryaev SA, Chernov A V, Aleshin AE, Shiryaeva TN, Strongin AY. NS4A regulates the ATPase activity of the NS3 helicase: a novel cofactor role of the non-structural protein NS4A from West Nile virus. J Gen Virol. 2009;90(9):2081–5. doi: 10.1099/vir.0.012864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umareddy I, Chao A, Sampath A, Gu F, Vasudevan SG. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J Gen Virol. 2006;87(9):2605–14. doi: 10.1099/vir.0.81844-0. [DOI] [PubMed] [Google Scholar]
- 39.Zou J, Lee LT, Wang QY, Xie X, Lu S, Yau YH, et al. Mapping the Interactions between the NS4B and NS3 Proteins of Dengue Virus. J Virol. 2015;89(7):3471–83. doi: 10.1128/JVI.03454-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chambers TJ, Grakoui A, Rice CM. Processing of the Yellow Fever Virus Nonstructural Polyprotein: a Catalytically Active NS3 Proteinase Domain and NS2B Are Required for Cleavages at Dibasic Sites. J Virol. 1991;65(11):6042–50. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falgout B, Pethel M, Zhang Y-M, Lai C-J. Both Nonstructural Proteins NS2B and NS3 Are Required for the Proteolytic Processing of Dengue Virus Nonstructural Proteins. J Virol. 1991;65(5):2467–75. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muñoz-Jordán JL, Sánchez-Burgos GG, Laurent-Rolle M, García-Sastre A. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A. 2003;100(24):14333–8. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. Regulated Cleavages at the West Nile Virus NS4A-2K-NS4B Junctions Play a Major Role in Rearranging Cytoplasmic Membranes and Golgi Trafficking of the NS4A Protein. J Virol. 2006;80(9):4623–32. doi: 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mackenzie JM, Jones MK, Young PR. Immunolocalization of the Dengue Virus Nonstructural Glycoprotein NS1 Suggests a Role in Viral RNA Replication. Virology. 1996;220(1):232–40. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 45.Muylaert IR, Chambers TJ, Galler R, Rice CM. Mutagenesis of the N-Linked Glycosylation Sites of the Yellow Fever Virus NS1 Protein: Effects on Virus Replication and Mouse Neurovirulence. Virology. 1996;222(1):159–68. doi: 10.1006/viro.1996.0406. [DOI] [PubMed] [Google Scholar]
- 46.Muller DA, Young PR. The flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antiviral Res. 2013;98(2):192–208. doi: 10.1016/j.antiviral.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 47.Stohlman SA, Wisseman CLJ, Eylar OR, Silverman DJ. Dengue Virus-Induced Modifications of Host Cell Membranes. J Virol. 1975;16(4):1017–26. doi: 10.1128/jvi.16.4.1017-1026.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perera R, Kuhn RJ. Structural proteomics of dengue virus. Curr Opin Microbiol. 2008;11(4):369–77. doi: 10.1016/j.mib.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uchil PD, Satchidanandam V. Architecture of the Flaviviral Replication Complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J Biol Chem. 2003;278(27):24388–98. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- 50.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CKE, Walther P, et al. Composition and Three-Dimensional Architecture of the Dengue Virus Replication and Assembly Sites. Cell Host Microbe. 2009;5(4):365–75. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saeedi BJ, Geiss BJ. Regulation of flavivirus RNA synthesis and capping. Wiley Interdiscip Rev RNA. 2013;4(6):723–35. doi: 10.1002/wrna.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, et al. A Highly Structured, Nuclease-Resistant, Noncoding RNA Produced by Flaviviruses Is Required for Pathogenicity. Cell Host Microbe. 2008;4(6):579–91. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 53.Luo D, Xu T, Hunke C, Grüber G, Vasudevan SG, Lescar J. Crystal Structure of the NS3 Protease-Helicase from Dengue Virus. J Virol. 2008;82(1):173–83. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo D, Wei N, Doan DN, Paradkar PN, Chong Y, Davidson AD, et al. Flexibility between the Protease and Helicase Domains of the Dengue Virus NS3 Protein Conferred by the Linker Region and Its Functional Implications. J Biol Chem. 2010;285(24):18817–27. doi: 10.1074/jbc.M109.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mastrangelo E, Milani M, Bollati M, Selisko B, Peyrane F, Pandini V, et al. Crystal Structure and Activity of Kunjin Virus NS3 Helicase; Protease and Helicase Domain Assembly in the Full Length NS3 Protein. J Mol Biol. 2007;372(2):444–55. doi: 10.1016/j.jmb.2007.06.055. [DOI] [PubMed] [Google Scholar]
- 56.Xu T, Sampath A, Chao A, Wen D, Nanao M, Chene P, et al. Structure of the Dengue Virus Helicase/Nucleoside Triphosphatase Catalytic Domain at a Resolution of 2.4 Å. J Virol. 2005;79(16):10278–88. doi: 10.1128/JVI.79.16.10278-10288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu T, Sampath A, Chao A, Wen D, Nanao M, Luo D, et al. Towards the design of flavivirus helicase/NTPase inhibitors: crystallographic and mutagenesis studies of the dengue virus NS3 helicase catalytic domain. Novartis Found Symp. 2006;277:87–97. doi: 10.1002/0470058005.ch7. [DOI] [PubMed] [Google Scholar]
- 58.Assenberg R, Mastrangelo E, Walter TS, Verma A, Milani M, Owens RJ, et al. Crystal Structure of a Novel Conformational State of the Flavivirus NS3 protein: Implications for Polyprotein Processing and Viral Replication. J Virol. 2009;83(24):12895–906. doi: 10.1128/JVI.00942-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gebhard LG, Kaufman SB, Gamarnik AV. Novel ATP-Independent RNA Annealing Activity of the Dengue Virus NS3 Helicase. PLoS One. 2012;7(4):e36244. doi: 10.1371/journal.pone.0036244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27(23):3209–19. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benzaghou I, Bougie I, Picard-Jean F, Bisaillon M. Energetics of RNA binding by the West Nile virus RNA triphosphatase. FEBS Lett. 2006;580(3):867–77. doi: 10.1016/j.febslet.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 62.Bazan JF, Fletterick RJ. Detection of a Trypsin-like Serine Protease Domain in Flaviviruses and Pestiviruses. Virology. 1989;171(2):637–9. doi: 10.1016/0042-6822(89)90639-9. [DOI] [PubMed] [Google Scholar]
- 63.Valle RPC, Falgout B. Mutagenesis of the NS3 Protease of Dengue Virus Type 2. J Virol. 1998;72(1):624–32. doi: 10.1128/jvi.72.1.624-632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yotmanee P, Rungrotmongkol T, Wichapong K, Choi SB, Wahab HA, Kungwan N, et al. Binding specificity of polypeptide substrates in NS2B/NS3pro serine protease of dengue virus type 2: A molecular dynamics Study. J Mol Graph Model. 2015;60:24–33. doi: 10.1016/j.jmgm.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 65.Chambers TJ, Nestorowicz A, Amberg SM, Rice CM. Mutagenesis of the Yellow Fever Virus NS2B Protein: Effects on Proteolytic Processing, NS2B-NS3 Complex Formation, and Viral Replication. J Virol. 1993;67(11):6797–807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin C, Amberg SM, Chambers TJ, Rice CM. Cleavage at a Novel Site in the NS4A Region by the Yellow Fever Virus NS2B-3 Proteinase Is a Prerequisite for Processing at the Downstream 4A/4B Signalase Site. J Virol. 1993;67(4):2327–35. doi: 10.1128/jvi.67.4.2327-2335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lobigs M. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proc Natl Acad Sci U S A. 1993;90(13):6218–22. doi: 10.1073/pnas.90.13.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li K, Phoo WW, Luo D. Functional interplay among the flavivirus NS3 protease, helicase, and cofactors. Virol Sin. 2014;29(2):74–85. doi: 10.1007/s12250-014-3438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh aa. Ultrastructure of Kunjin Virus-Infected cells: Colocalization of NS1 and NS3 with Double-Stranded RNA, and of NS2B with NS3, in Virus-Induced Membrane Structures. J Virol. 1997;71(9):6650–61. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clum S, Ebner KE, Padmanabhan R. Cotranslational Membrane Insertion of the Serine Proteinase Precursor NS2B-NS3(Pro) of Dengue Virus Type 2 Is Required for Efficient in Vitro Processing and Is Mediated through the Hydrophobic Regions of NS2B. J Biol Chem. 1997;272(49):30715–23. doi: 10.1074/jbc.272.49.30715. [DOI] [PubMed] [Google Scholar]
- 71.Benarroch D, Selisko B, Locatelli GA, Maga G, Romette J-L, Canard B. The RNA helicase, nucleotide 5’-triphosphatase, and RNA 5’-triphosphatase activities of Dengue virus protein NS3 are Mg2+dependent and require a functional Walker B motif in the helicase catalytic core. Virology. 2004;328(2):208–18. doi: 10.1016/j.virol.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 72.Sampath A, Xu T, Chao A, Luo D, Lescar J, Vasudevan SG. Structure-Based Mutational Analysis of the NS3 Helicase from Dengue Virus. J Virol. 2006;80(13):6686–90. doi: 10.1128/JVI.02215-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mastrangelo E, Bolognesi M, Milani M. Flaviviral helicase: Insights into the mechanism of action of a motor protein. Biochem Biophys Res Commun. 2012;417(1):84–7. doi: 10.1016/j.bbrc.2011.11.060. [DOI] [PubMed] [Google Scholar]
- 74.Luo D, Vasudevan SG, Lescar J. The flavivirus NS2B-NS3 protease-helicase as a target for antiviral drug development. Antiviral Res. 2015;118:148–58. doi: 10.1016/j.antiviral.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 75.Wang C-C, Huang Z-S, Chiang P-L, Chen C-T, Wu H-N. Analysis of the nucleoside triphosphatase, RNA triphosphatase, and unwinding activities of the helicase domain of dengue virus NS3 protein. FEBS Lett. 2009;583(4):691–6. doi: 10.1016/j.febslet.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 76.Cui T, Sugrue RJ, Xu Q, Lee AKW, Chan Y-C, Fu J. Recombinant Dengue Virus Type 1 NS3 Protein Exhibits Specific Viral RNA Binding and NTPase Activity Regulated by the NS5 Protein. Virology. 1998;246(2):409–17. doi: 10.1006/viro.1998.9213. [DOI] [PubMed] [Google Scholar]
- 77.Wu J, Bera AK, Kuhn RJ, Smith JL. Structure of the Flavivirus Helicase : Implications for Catalytic Activity, Protein Interactions, and Proteolytic Processing. J Virol. 2005;79(16):10268–77. doi: 10.1128/JVI.79.16.10268-10277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yon C, Teramoto T, Mueller N, Phelan J, Ganesh VK, Murthy KHM, et al. Modulation of the Nucleoside Triphosphatase/RNA Helicase and 5’-RNA Triphosphatase Activities of Dengue Virus Type 2 Nonstructural Protein 3 (NS3) by Interaction with NS5, the RNA-dependent RNA Polymerase. J Biol Chem. 2005;280(29):27412–9. doi: 10.1074/jbc.M501393200. [DOI] [PubMed] [Google Scholar]
- 79.Lu G, Gong P. Crystal Structure of the Full-Length Japanese Encephalitis Virus NS5 Reveals a Conserved Methyltransferase-Polymerase Interface. PLoS Pathog. 2013;9(8):e1003549. doi: 10.1371/journal.ppat.1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bussetta C, Choi KH. Dengue Virus Nonstructural Protein 5 Adopts Multiple Conformations in Solution. Biochemistry. 2012;51(30):5921–31. doi: 10.1021/bi300406n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao Y, Soh TS, Zheng J, Chan KWK, Phoo WW, Lee CC, et al. A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Inter-domain Interface Essential for Protein Flexibility and Virus Replication. PLoS Pathog. 2015;11(3):e1004682. doi: 10.1371/journal.ppat.1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Egloff M-P, Decroly E, Malet H, Selisko B, Benarroch D, Ferron F, et al. Structural and Functional Analysis of Methylation and 5′-RNA Sequence Requirements of Short Capped RNAs by the Methyltransferase Domain of Dengue Virus NS5. J Mol Biol. 2007;372(3):723–36. doi: 10.1016/j.jmb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 83.Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, et al. Structure and Function of Flavivirus NS5 Methyltransferase. J Virol. 2007;81(8):3891–903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Geiss BJ, Thompson AA, Andrews AJ, Sons RL, Gari HH, Keenan SM, et al. Analysis of Flavivirus NS5 Methyltransferase Cap Binding. J Mol Biol. 2009;385(5):1643–54. doi: 10.1016/j.jmb.2008.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Henderson BR, Saeedi BJ, Campagnola G, Geiss BJ. Analysis of RNA Binding by the Dengue Virus NS5 RNA Capping Enzyme. PLoS One. 2011;6(10):e25795. doi: 10.1371/journal.pone.0025795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Selisko B, Dutartre H, Guillemot J-C, Debarnot C, Benarroch D, Khromykh A, et al. Comparative mechanistic studies of de novo RNA synthesis by flavivirus RNA-dependent RNA polymerases. Virology. 2006;351(1):145–58. doi: 10.1016/j.virol.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 87.Malet H, Egloff M-P, Selisko B, Butcher RE, Wright PJ, Roberts M, et al. Crystal Structure of the RNA Polymerase Domain of the West Nile Virus Non-structural Protein 5. J Biol Chem. 2007;282(14):10678–89. doi: 10.1074/jbc.M607273200. [DOI] [PubMed] [Google Scholar]
- 88.Yap TL, Xu T, Chen Y-L, Malet H, Egloff M-P, Canard B, et al. Crystal Structure of the Dengue Virus RNA-Dependent RNA Polymerase Catalytic Domain at 1.85-Angstrom Resolution. J Virol. 2007;81(9):4753–65. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choi KH, Rossmann MG. RNA-dependent RNA polymerases from Flaviviridae. Curr Opin Struct Biol. 2009;19(6):746–51. doi: 10.1016/j.sbi.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 90.Gong P, Peersen OB. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 2010;107(52):22505–10. doi: 10.1073/pnas.1007626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ackermann M, Padmanabhan R. De Novo Synthesis of RNA by the Dengue Virus RNA-dependent RNA polymerase Exhibits Temperature Dependence at the Initiation but Not Elongation Phase. J Biol Chem. 2001;276(43):39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- 92.Selisko B, Potisopon S, Agred R, Priet S, Varlet I, Thillier Y, et al. Molecular Basis for Nucleotide Conservation at the Ends of the Dengue Virus Genome. PLoS Pathog. 2012;8(9):e1002912. doi: 10.1371/journal.ppat.1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Noble CG, Shi P-Y. Structural biology of dengue virus enzymes: Towards rational design of therapeutics. Antiviral Res. 2012;96(2):115–26. doi: 10.1016/j.antiviral.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 94.Nomaguchi M, Ackermann M, Yon C, You S, Padmanbhan R. De Novo Synthesis of Negative-Strand RNA by Dengue Virus RNA-Dependent RNA Polymerase In Vitro: Nucleotide, Primer, and Template Parameters. J Virol. 2003;77(16):8831–42. doi: 10.1128/JVI.77.16.8831-8842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu J, Liu W, Gong P. A Structural Overview of RNA-Dependent RNA Polymerases from the Flaviviridae Family. Int J Mol Sci. 2015;16(6):12943–57. doi: 10.3390/ijms160612943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klema VJ, Padmanabhan R, Choi KH. Flaviviral Replication Complex: Coordination between RNA Synthesis and 5’-RNA Capping. Viruses. 2015;7(8):4640–56. doi: 10.3390/v7082837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R. Association between NS3 and NS5 Proteins of Dengue Virus Type 2 in the Putative RNA Replicase Is Linked to Differential Phosphorylation of NS5. J Biol Chem. 1995;270(32):19100–6. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 98.Johansson M, Brooks AJ, Jans DA, Vasudevan SG. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-β and the viral helicase, NS3. J Gen Virol. 2001;82(4):735–45. doi: 10.1099/0022-1317-82-4-735. [DOI] [PubMed] [Google Scholar]
- 99.Brooks AJ, Johansson M, John AV, Xu Y, Jans DA, Vasudevan SG. The Interdomain Region of Dengue NS5 Protein That Binds to the Viral Helicase NS3 Contains Independently Functional Importin β1 and Importin α/β-Recognized Nuclear Localization Signals. J Biol Chem. 2002;277(39):36399–407. doi: 10.1074/jbc.M204977200. [DOI] [PubMed] [Google Scholar]
- 100.Moreland NJ, Tay MYF, Lim E, Rathore APS, Lim APC, Hanson BJ, et al. Monoclonal antibodies against dengue NS2B and NS3 proteins for the study of protein interactions in the flaviviral replication complex. J Virol Methods. 2012;179(1):97–103. doi: 10.1016/j.jviromet.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 101.Tay MYF, Saw WG, Zhao Y, Chan KWK, Singh D, Chong Y, et al. The C-terminal 50 Amino Acid Residues of Dengue NS3 Protein Are Important for NS3-NS5 Interaction and Viral Replication. J Biol Chem. 2015;290(4):2379–94. doi: 10.1074/jbc.M114.607341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zou G, Chen Y-L, Dong H, Lim CC, Yap LJ, Yau YH, et al. Functional Analysis of Two Cavities in Flavivirus NS5 Polymerase. J Biol Chem. 2011;286(16):14362–72. doi: 10.1074/jbc.M110.214189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Takahashi H, Takahashi C, Moreland NJ, Chang Y-T, Sawasaki T, Ryo A, et al. Establishment of a robust dengue virus NS3-NS5 binding assay for identification of protein-protein interaction inhibitors. Antiviral Res. 2012;96(3):305–14. doi: 10.1016/j.antiviral.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 104.Chen CJ, Kuo MD, Chien LJ, Hsu SL, Wang YM, Lin JH. RNA-Protein Interactions: Involvement of NS3, NS5, and 3’ Noncoding Regions of Japanese Encephalitis Virus Genomic RNA. J Virol. 1997;71(5):3466–73. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vasudevan SG, Johansson M, Brooks AJ, Llewellyn LE, Jans DA. Characterisation of inter- and intra-molecular interactions of the dengue virus RNA dependent RNA polymerase as potential drug targets. Farmaco. 2001;56(1–2):33–6. doi: 10.1016/s0014-827x(01)01014-x. [DOI] [PubMed] [Google Scholar]
- 106.Yu L, Takeda K, Markoff L. Protein-protein interactions among West Nile non-structural proteins and transmembrane complex formation in mammalian cells. Virology. 2013;446(1–2):365–77. doi: 10.1016/j.virol.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 107.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 108.Pierson TC, Sánchez MD, Puffer BA, Ahmed AA, Geiss BJ, Valentine LE, et al. A rapid and quantitative assay for measuring antibody-mediated neutralization of West Nile virus infection. Virology. 2006;346(1):53–65. doi: 10.1016/j.virol.2005.10.030. [DOI] [PubMed] [Google Scholar]