Summary
Nuclear movements are important for multiple cellular functions and are driven by polarized forces generated by motor proteins and cytoskeleton. During skeletal myofiber formation or regeneration, nuclei move from the center to the periphery of the myofiber for proper muscle function. Centrally located nuclei are also found in different muscle disorders. Using theoretical and experimental approaches, we demonstrate that nuclear movement to the periphery of myofibers is mediated by centripetal forces around the nucleus. These forces arise from myofibril contraction and cross-linking that “zip” around the nucleus in combination with tight regulation of nuclear stiffness by lamin A/C. In addition, an Arp2/3 complex containing Arpc5L together with γ-actin is required to organize desmin to cross-link myofibrils for nuclear movement. Our work reveals that centripetal forces exerted by myofibrils squeeze the nucleus to the periphery of myofibers.
Introduction
Nuclear positioning within cells is important for multiple cellular activities during development, immune response, tissue homeostasis and regeneration 1. Moreover, nuclear positioning defects result in multiple human disorders such as lissencephaly, deafness, and muscle disorders 2,3. The mechanisms of nuclear positioning involve cytoskeletal networks and motor proteins. In most cases, the nucleus is connected to the cytoskeleton by nuclear envelope proteins or by the nuclear pore complex 4–8. Microtubules or actin growing towards the nucleus, as well as active diffusion can also drive nuclear movement 9–11.
The most diversified example of nuclear positioning occurs during skeletal muscle differentiation. Muscle fibers (myofibers) are multinucleated cells that are formed by the fusion of mononucleated muscle precursor cells (myoblasts)12. Nuclei are initially in the center of the myofiber and then move towards the myofiber periphery during myogenesis13.
We recently showed that nuclear positioning within myofibers is required for proper muscle function 14. In addition, several monogenic muscle disorders and regenerating muscle exhibit centrally located nuclei 15–18. The architecture of skeletal muscle is designed for its contractile purpose. Sarcomeres, the contractile unit, are connected to one another longitudinally by the z-line and form bundles named myofibrils that span the entire length of the myofiber 19. These myofibril bundles are cross-linked by desmin networks connected at the z-line. Desmin is a cytoplasmic intermediate filament that is expressed in muscle cells in replacement of vimentin and both these intermediate filaments share similar molecular and cellular functions 20,21. Myofibrils are surrounded by transversal triads, specialized junctions composed of t-tubules and sarcoplasmic reticulum. These structures are responsible for excitation-contraction (EC) coupling, allowing the transduction of electric membrane potentials into muscle contractions 22. Myofibrils are formed during gestation and surround the centrally located nuclei. The positioning of nuclei at the periphery occurs near birth whereas transversal triads are only formed at a later stage in the first postnatal weeks 23.
We recently found that BIN1/amphiphysin-2 (mutated in centronuclear myopathies) interacts with N-WASP and both are required for actin-dependent peripheral nuclear movement and transversal triad formation 24. These results imply a role for actin that is nucleated by the Arp2/3 complex25–27, in nuclear movement to the periphery. Here we confirm this hypothesis and provide the mechanism of nuclear movement to the periphery of myofibers.
Results
Nuclei are squeezed by myofibrils when moving to the myofiber periphery
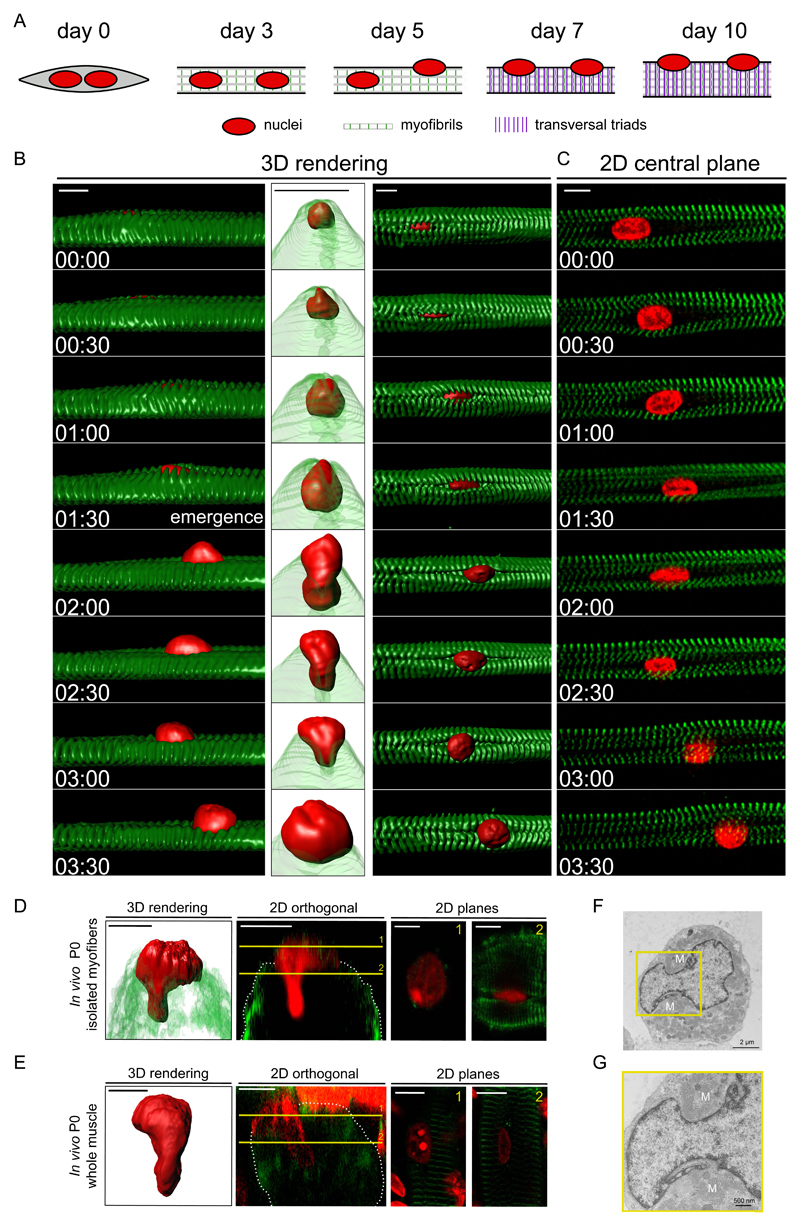
To investigate the mechanism of nuclear movement to the periphery of muscle cells, we used an in vitro myofiber system that recapitulates the embryonic and neonatal development of muscle fibers 23,24,28. After 3 days, myofibers with myofibrils exhibit centrally located nuclei and no transversal triads. Between days 4-6, nuclei move to the periphery followed by transversal triad organization at day 7 (Fig. 1A).
Figure 1. Visualizing nuclear movement to the periphery.
A. Timeline of muscle differentiation in the in vitro system used to study peripheral nuclear positioning and transversal triad formation. Nuclei are in red, myofibrils in white (with z-lines in green) and transversal triads as purple lines. Day 3: myofibril formation. Day 5: initiation of peripheral nuclear positioning. Day 7: transversal triad formation.
B. Kymograph from a time-lapse movie of a 5-day myofiber depicting peripheral movement of a nucleus (H2B-iRFP, red) through myofibrils (YFP-α-actinin, green). Left: view from the side, surface three-dimensional (3D) rendering. Middle: view from the right side, with transparent myofibrils 3D rendering. Right: view from the top, surface three-dimensional rendering. Time, hh:mm. Scale bar, 10 μm.
C. Two dimensional (2D) view of the central plane of a kymograph from a time-lapse movie of a 5-day myofiber depicting peripheral movement of a nucleus (H2B-iRFP, red) through myofibrils (YFP-α-actinin, green). Scale bar, 10 μm.
D. Representative image of a nucleus squeezing to the periphery from an in vivo isolated myofiber of a newborn mouse and stained for myofibrils (α-actinin, green) and nucleus (red). Left: 3D rendering. Middle left: 2D orthogonal view, yellow lines represent slices seen in right side panels. Middle right: 2D plane from yellow slice 1. Right: 2D plane from yellow slice 2. Scale bar, 10 μm. Image shown is representative of 2 experiments.
E. Representative image of a nucleus squeezing to the periphery after performing a clearing protocol of a whole muscle in a newborn mouse and stained for myofibrils (α-actinin, green) and nucleus (red). Left: 3D rendering. Middle left: 2D orthogonal view, yellow lines represent slices seen in right side panels. Middle right: 2D plane from yellow slice 1. Right: 2D plane from yellow slice 2. Scale bar, 10 μm. Image shown is representative of 2 experiments.
F. Transversal electron micrograph of a 4.5 day myofiber showing a nucleus beginning to protrude towards the periphery. Scale bar, 2 µm. “M” annotate myofibrils.
G. Transversal electron micrograph of a 4.5 day myofiber showing a nucleus during budding to the periphery. Scale bar, 500 nm. 4x Magnifications corresponding to the yellow square is shown below the image. “M” annotate myofibrils. Image shown is representative of 4 experiments.
We performed 3D time-lapse spinning disk microscopy of these differentiated in vitro myofibers labeled for myofibrils and nuclei between day 4.5 and 5.5. Before movement to the periphery, centrally located nuclei are surrounded by myofibril bundles (Fig. S1A and Supplementary movie S1). The movement of the nucleus to the periphery begins with the emergence of an elongated nuclear wrinkle through a narrow gap in between myofibrils (Fig. 1B,C). With time the wrinkle increases into a protrusion and the nucleus undergoes a dramatic deformation. This results in the nucleus being expelled from the middle of the myofibril bundle to the myofiber periphery in the direction of the nuclear protrusion over a period of 145 +/- 41 min, with an average speed of 2.4 μm/h (Supplementary movie S2). The deformation and protrusion of nuclei reaching the periphery of myofibers was also observed in vivo (Fig. 1D,E) and by electron microscopy (EM) (Fig. 1F,G).
Desmin cross-links myofibrils for nuclear movement to the myofiber periphery
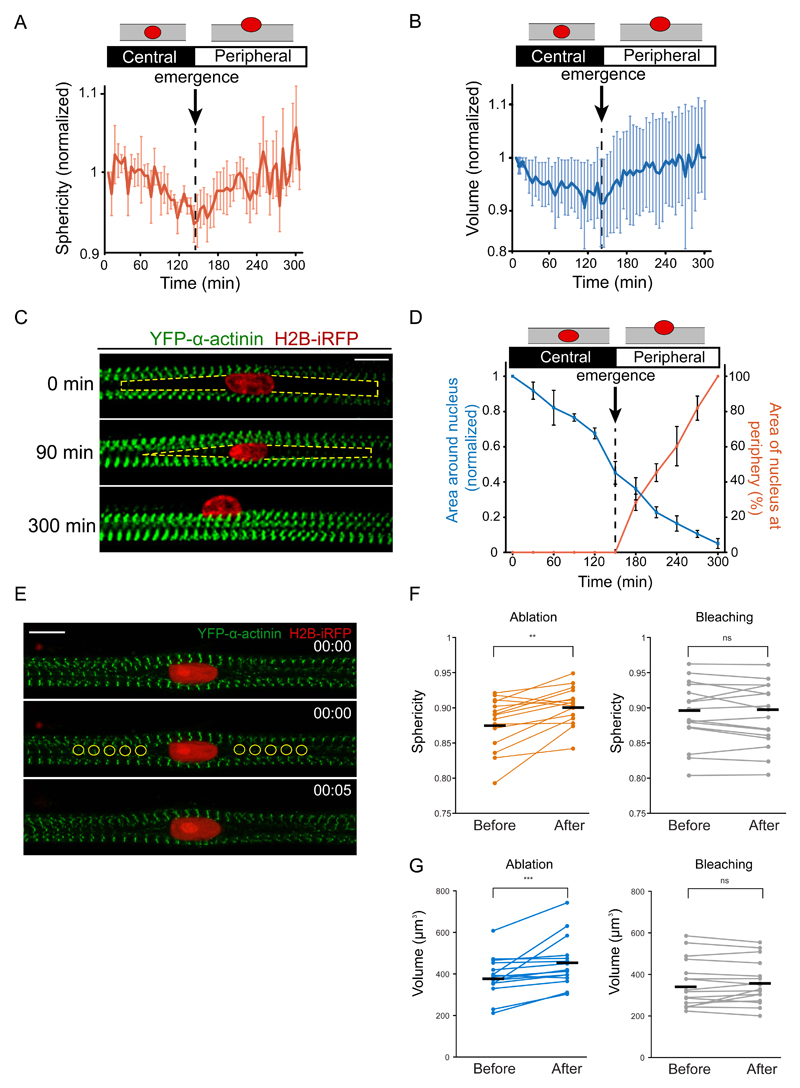
Quantification of 3D nuclear shape reveals that the nucleus sphericity and volume decrease prior to protrusion and emergence before being re-established as the nucleus starts to emerge at the periphery (Fig. 2A,B, S1B,C).
Figure 2. Myofiber cross-linking drives nuclear movement to the periphery.
A. Quantification of the sphericity of nuclei during nuclear movement to the periphery in 5-day myofibers. Dashed line corresponds to the emergence of the nucleus at the myofiber periphery (n = 8 myofibers Data from 6 independent experiments were combined. Error bars correspond to s.e.m. Source data is available in Supplementary Table 3.
B. Quantification of the volume of nuclei during nuclear movement to the periphery in 5-day myofibers. Dashed line corresponds to the emergence of the nucleus at the myofiber periphery (n = 8 myofibers). Data from 6 independent experiments were combined. Error bars correspond to s.e.m. Source data is available in Supplementary Table 3.
C. Kymograph from a time-lapse movie of a 5-day myofiber depicting area around the nucleus in between myofibrils (yellow dashed line) during nuclear movement to the periphery with the nucleus (H2B-iRFP, red) migrating through myofibrils (YFP-α-actinin, green). Scale bar, 10 μm.
D. Quantification of the area in between myofibrils around the nucleus (blue line) and the percentage of area of the nucleus at the periphery over time (orange line) in 5-day myofibers (n = 8 myofibers). Error bars correspond to s.e.m. Source data is available in Supplementary Table 3.
E. Representative images of a 4.5-day myofiber time-lapse movie before and after laser ablation with myofibrils depicted in green (YFP-α-actinin) and the nucleus in red (H2B-iRFP). Yellow circles represent ablation areas performed at 00:00 (hh:mm). Scale bar, 10 μm. Image shown is representative of 6 experiments.
F. Dot plot showing changes in nuclei sphericity before and after ablation (orange) or bleaching (black). Black lines represent the average. Data from 3 independent experiments were combined (n = 15 nuclei). Wilcoxon matched-pairs signed ranked test was used to determine statistical significances, where ** P < 0.01, NS, not significant. Source data is available in Supplementary Table 3.
G. Dot plot showing changes in nuclei sphericity before and after ablation (orange) or bleaching (black). Black lines represent the average. Data from 3 independent experiments were combined and error bars represent s.e.m from (n = 15 nuclei). Wilcoxon matched-pairs signed ranked test was used to determine statistical significances, where *** P < 0.001, NS, not significant. Source data is available in Supplementary Table 3.
When analyzing the middle plane of the myofiber, we observed an area next to the nucleus where myofibrils were not present due to deflection around the nucleus (Fig. 1C, 2C), which was also observed by EM (Fig. S1D). This area decreases prior to nuclear movement as though myofibrils cross-link and “zip” together towards the nucleus (Fig. 2D). Next we used laser ablation to disrupt cross-linking between myofibrils labeled with YFP-α-actinin. Upon ablation with high powered laser, both nuclear sphericity and volume increased, which was not observed upon bleaching with low powered laser (Fig. 2E-G, S1E). Both adjacent myofibrils distance and myofiber thickness increased upon ablation (Fig. S1F). We fixed the cells after ablation and stained for α-actinin to confirm that high power laser was ablating myofibrils whereas low power laser was only bleaching the YFP signal without disrupting myofibrils (Fig. S1E). Overall, this data suggest an increase in the forces applied to the nucleus by myofibril zipping around the nucleus before the initiation of nuclear movement to the periphery.
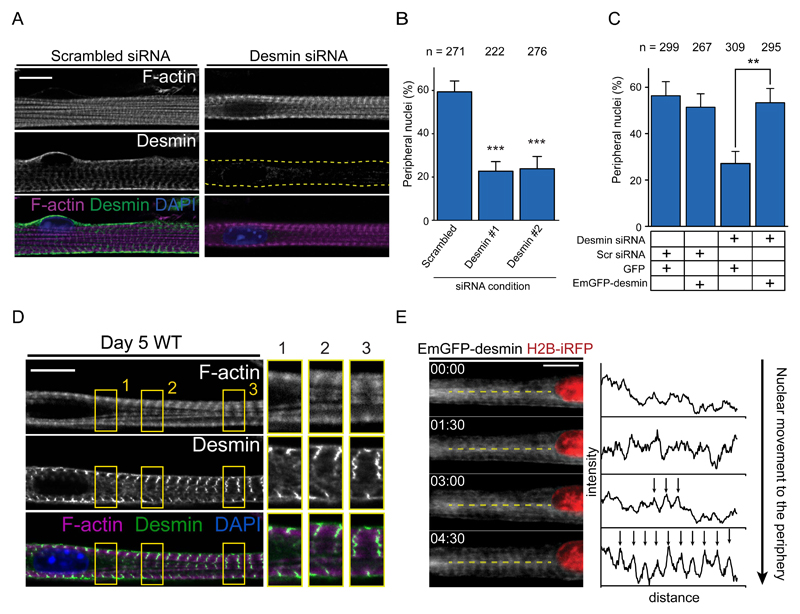
Desmin is an intermediate filament known to cross-link myofibrils at the z-line and was implicated in nuclear positioning in muscle 18,29–31. Depletion of desmin by siRNA-mediated knock down impairs nuclear movement to the periphery (Fig. 3A,B and Fig. S2A,B) without disruption of myofibril structure or contraction frequency (Fig. S2C-E). Expression of human EmGFP-desmin, resistant to mouse siRNA, restored nuclear movement (Fig. 3C). Furthermore, the desmin network is already organized at the z-line before nuclear movement but only in sections of the myofibers away from centrally located nuclei. Desmin is disorganized near centrally located nuclei in the area devoid of myofibrils (Fig. 3D). When we visualized EmGFP-desmin during nuclear movement to the periphery we observed the organization of desmin in striations towards the nucleus, similar to myofibril zipping (Fig. 1C, 3E). These data suggest that myofibril cross-linking and zipping by desmin leads to the squeezing of the nucleus to the cell periphery.
Figure 3. Desmin crosslinks myofibrils to induce nuclear movement to the periphery.
A. Representative immunofluorescence image of a 10-day myofiber knocked down for scrambled or desmin and stained for F-actin (phalloidin, magenta), desmin (green) and DAPI (nucleus, blue). Scale bar, 10 μm.
B. Quantification of peripheral nuclei positioning in 10-day myofibers knocked down for scrambled or desmin. Data from 3 independent experiments were combined and error bars represent s.e.m from indicated n nuclei for each cohort. Unpaired t-test was used to determine statistical significances, where *** P < 0.001. Source data is available in Supplementary Table 3.
C. Quantification of peripheral nuclei positioning and traversal triads in 10-day myofibers knocked down for scrambled or desmin and transfected with either GFP or Em-GFP-desmin. Data from 3 independent experiments were combined and error bars represent s.e.m from indicated n nuclei for each cohort. Unpaired t-test was used to determine statistical significances, where ** P < 0.01.
D. Representative immunofluorescence image of a 4.5-day myofiber knocked stained for F-actin (phalloidin, magenta), desmin (green) and DAPI (nucleus, blue). Arrow indicates organized desmin whereas arrowhead indicates disorganized desmin. 2x Magnifications corresponding to the yellow squares are showed on the right corresponding to areas near the nucleus with disorganized desmin either without (1.) or with myofibrils (2.), or areas away from the nucleus with organized desmin and myofibrils (3.) . Scale bar, 10 μm. Image shown is representative of 3 experiments.
E. Kymograph from a time-lapse movie of a 5-day myofiber depicting desmin organization, Emerald-desmin (EmGFP-desmin, gray) during nuclear movement to the periphery (H2B-iRFP, red). Yellow dashed lines represent the region used to perform line scans plotted on the right. Arrows highlight the transversal organization of desmin. Time, hh:mm. Scale bar, 10 μm. Source data is available in Supplementary Table 3.
γ-actin is required for nuclear movement and desmin organization
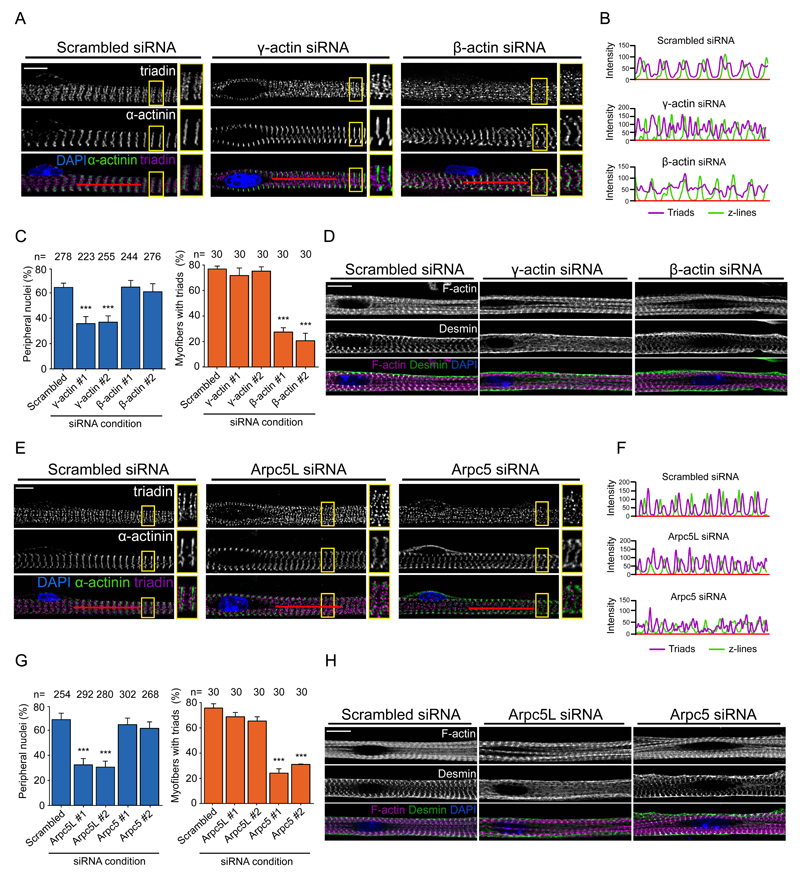
We next explored how nuclear positioning to the periphery is regulated, taking into account our previous study showing that amphiphysin-2/BIN1, which is mutated in centronuclear myopathies, triggers peripheral nuclear positioning via N-Wasp and actin 24. Six actin isoforms are expressed in mammals of which three are found in skeletal muscle: α-, β- and γ-actin. α-actin is the main sarcomeric actin, required for sarcomere formation. We therefore focused our analysis on the cytoplasmic actins namely γ- and β-actin, known to have different functions (Fig. S3A) 32,33. We found that siRNA-mediated depletion of γ-actin, but not β-actin, inhibits nuclear positioning (Fig. S3B-D, 4A,C). In contrast, β-actin depletion, but not γ-actin, inhibits transversal triad formation (Fig. 4A-C). We confirmed these results by performing rescue experiments with siRNA-resistant GFP-tagged γ-actin and β-actin (Fig. S3E-G).
Figure 4. Actin and Arp isoforms play different roles in myofiber differentiation.
A. Representative immunofluorescence images of a 10-day myofiber knocked down for scrambled, γ-actin or β-actin and stained for triadin (triad marker, magenta), α-actinin (myofibrils/Z-line marker, green) and DAPI (nucleus, blue). Scale bar, 10 μm. 2x Magnifications corresponding to the yellow squares are showed in the right of each image. Image shown is representative of 3 experiments.
B. Line scans of red line depicted in A to visualize the organization of transversal triad doublets (magenta) and z-line (green) in scramble, γ-actin or β-actin knocked down myofibers.
C. Quantification of peripheral nuclei positioning (left) and transversal triad formation (right) in 10-day myofibers knocked down for scrambled, γ-actin or β-actin. Data from 3 independent experiments were combined and error bars represent s.e.m from indicated n nuclei or myofibers for each cohort. Unpaired t-test was used to determine statistical significances, where *** P < 0.001. Source data is available in Supplementary Table 3.
D. Representative image of 5-day myofibers knocked down for scramble, γ-actin or β-actin and stained for F-actin (phalloidin, magenta), desmin (green) and DAPI (nucleus, blue). Scale bar, 10 μm. Image shown is representative of 3 experiments.
E. Representative immunofluorescence images of 10-day myofibers knocked down for scrambled, Arpc5L or Arpc5 and stained for triadin (triad marker, magenta), α-actinin (myofibrils/Z-line marker, green) and DAPI (nucleus, blue). Scale bar, 10 μm. 2x Magnifications corresponding to the yellow squares are showed in the right of each image. Image shown is representative of 3 experiments.
F. Linescan of red line depicted in A to visualize the organization of transversal triad doublets (magenta) and of z-line (green) in scramble, Arpc5l or Arpc5 knocked down myofibers.
G. Quantification of peripheral nuclei positioning (left) and transversal triad formation (right) in 10-day myofibers knocked down for scrambled, Arpc5L or Arpc5. Data from 3 independent experiments were combined and error bars represent s.e.m from indicated n nuclei or myofibers for each cohort. Unpaired t-test was used to determine statistical significances, where *** P < 0.001. Source data is available in Supplementary Table 3.
H. Representative image of 5-day myofibers knocked down for scramble, Arpc5L or Arpc5 and stained for F-actin (phalloidin, magenta), desmin (green) and DAPI (nucleus, blue). Scale bar, 10 μm. Image shown is representative of 3 experiments.
The actin cytoskeleton associates and organizes vimentin intermediate filaments in different cellular events such as cell migration34,35. As vimentin and desmin share multiple structural, biochemical and physiological properties, actin could be involved in the organization of desmin in skeletal muscle20. Accordingly, γ-actin, but not control or β-actin depleted myofibers lack desmin organization at the z-line (Fig. 4D).
The Arp2/3 complex is required for nuclear movement and desmin organization
We previously showed that N-Wasp, which stimulates the actin nucleating activity of the Arp2/3 complex 25–27 is required for peripheral nuclear positioning 24. We therefore tested the role of the Arp2/3 complex in nuclear movement 25–27(Fig S4A). Depletion of Arpc2, essential for Arp2/3 nucleation activity 36,37, inhibits nuclear movement, but not myofibril formation (Fig. S4B-E). Furthermore, transversal triad formation which occurs later on during myofiber differentiation is also inhibited (Fig. S4B-E). These results were confirmed by Arp2/3 complex inhibition with CK666 38 (Fig. S4E).
Arp2/3 complexes differ in their actin assembly activity depending on the isoforms of Arpc1 (Arpc1A and Arpc1B) and Arpc5 (Arpc5 and Arpc5L) 39. We found that depletion of Arpc1A or Arpc1B prevents both peripheral nuclear positioning and transversal triad formation (Fig. S4F,G). However, depletion of Arpc5L, but not Arpc5, only inhibits nuclear positioning (Fig. 4E-G). In contrast, loss of Arpc5, but not Arpc5L blocks transversal triad formation but not nuclear positioning (Fig. 4E-G and Fig. S4H-J). We additionally found that Arpc5L, but not control or Arpc5 depleted myofibers lack desmin organization at the z-line (Fig. 4H).
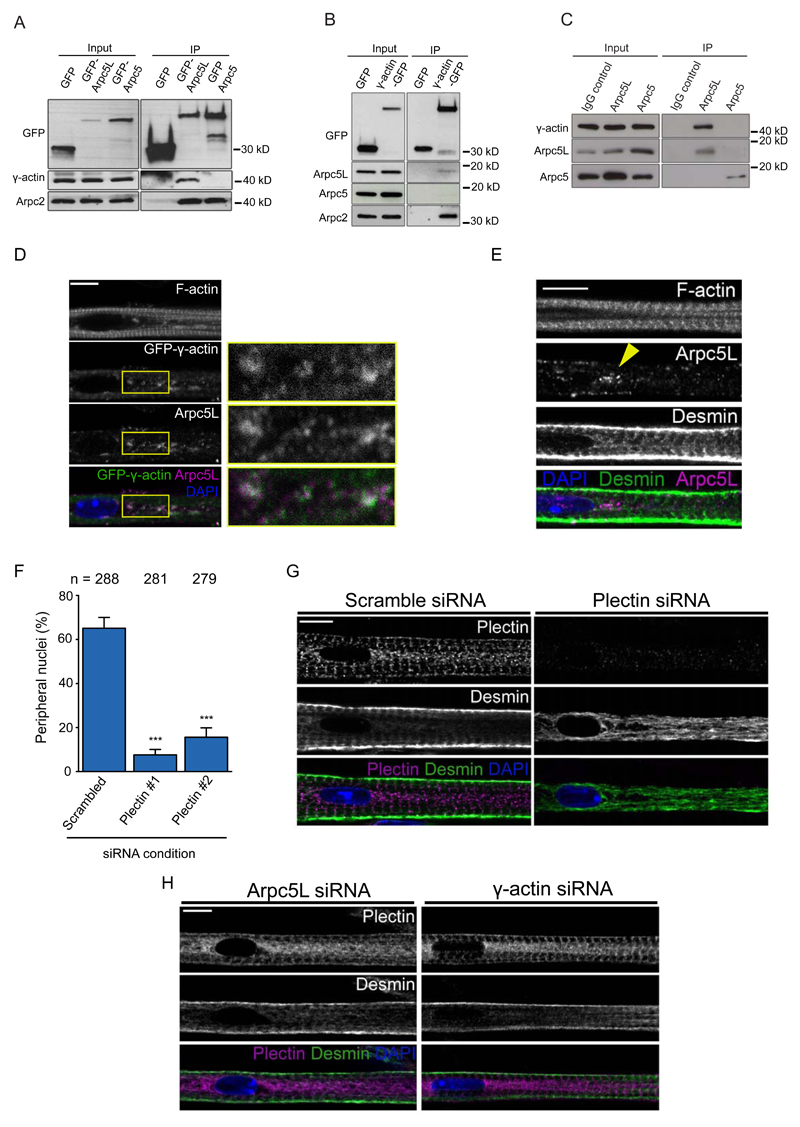
Our finding that γ-actin and Arpc5L are involved in nuclear positioning, prompted us to test if these two proteins interact in myofibers. We found that γ-actin co-immunoprecipitates (co-IP) with GFP-tagged Arpc5L but not Arpc5 (Fig. 5A). Conversely, Arpc5L, but not Arpc5, co-IP with GFP-γ-actin (Fig. 5B). In all situations, Arpc2 was also present in the co- IP. Finally, endogenous γ-actin co-immunoprecipitates with endogenous Arpc5L, but not Arpc5 (Fig. 5C). Consistent with this, Arpc5L and γ-actin co-localize in small patches between myofibrils near centrally located nuclei in 5 day myofibers (Fig. 5D). In contrast, Arpc5 and β-actin are transversally organized (Fig. S5A,B). The number of γ-actin patches was reduced upon depletion of Arpc5L, but not Arpc5 (Fig. S5C,D). The co-localization of Arpc5L and γ-actin occurs in areas depleted of myofibrils where desmin is not organized (Fig. 5E). We also determined that Arpc5L and γ-actin depletion does not affect desmin expression (Fig. S5E,F), however it impairs desmin dynamic as measured by fluorescence recovery after photobleaching (FRAP) of EmGFP-desmin (Fig. S5G,H).
Figure 5. Arpc5L and γ-actin interact to crosslink myofibrils.
A. Western blots with indicated antibodies of GFP, GFP-Arpc5L or GFP-Arpc5 immunoprecipitated from 5-day myofibers. Experiments were performed 3 times.
B. Western blots with indicated antibodies of GFP and GFP-γ-actin immunoprecipitated from 5-day myofibers. Experiments were performed 3 times.
C. Western blots with indicated antibodies of IgG control, Arpc5L and Arpc5 immunoprecipitated from 5-day myofibers. Experiments were performed 3 times.
D. Representative immunofluorescence images of 5-day myofibers expressing GFP-γ-actin (green) and immunostained for Arpc5L (magenta) in 5-day myofibers (left). Scale bar, 10 μm. 3x Magnifications corresponding to the yellow rectangle are shown on the right. Image shown is representative of 3 experiments.
E. Representative immunofluorescence images of 5-day myofibers stained for Arpc5L (magenta), desmin (green) and F-actin. Yellow arrowhead represents area depleted of myofibrils in which desmin is disorganized and Arpc5L is enriched. Scale bar, 10 μm. Image shown is representative of 3 experiments.
F. Quantification of peripheral nuclei positioning in 10-day myofibers knocked down for scrambled or plectin. Data from 3 independent experiments were combined and error bars represent s.e.m from indicated n nuclei for each cohort. Unpaired t-test was used to determine statistical significances, where *** P < 0.001. Source data is available in Supplementary Table 3.
G. Representative immunofluorescence image of a 4.5-day myofiber knocked down for scrambled or plectin and stained for plectin (magenta), desmin (green) and DAPI (nucleus, blue). Scale bar, 10 μm. Image shown is representative of 3 experiments.
H. Representative immunofluorescence image of a 4.5-day myofiber knocked down for Arpc5L or γ-actin and stained for plectin (magenta), desmin (green) and DAPI (nucleus, blue). Scale bar, 10 μm. Image shown is representative of 3 experiments.
To further understand how the Arp2/3 complex and γ-actin organize desmin, we explored the role of plectin, a cytoskeletal linker that connects desmin to the z-line 40. We found that plectin is required for nuclear positioning and desmin organization, but not for myofibril formation (Fig. 5F,G and Fig. S5I-K). Plectin is at the z-line just before nuclear movement, independently of Arpc5L or γ-actin (Fig. 5H). Furthermore, plectin is already at the z-line prior to desmin and peripheral nuclear movement (Fig. S5L). This shows that Arpc5L and γ-actin organize the desmin cytoskeleton downstream or in parallel to plectin for nuclear movement.
A theoretical model for nuclear movement to the myofiber periphery
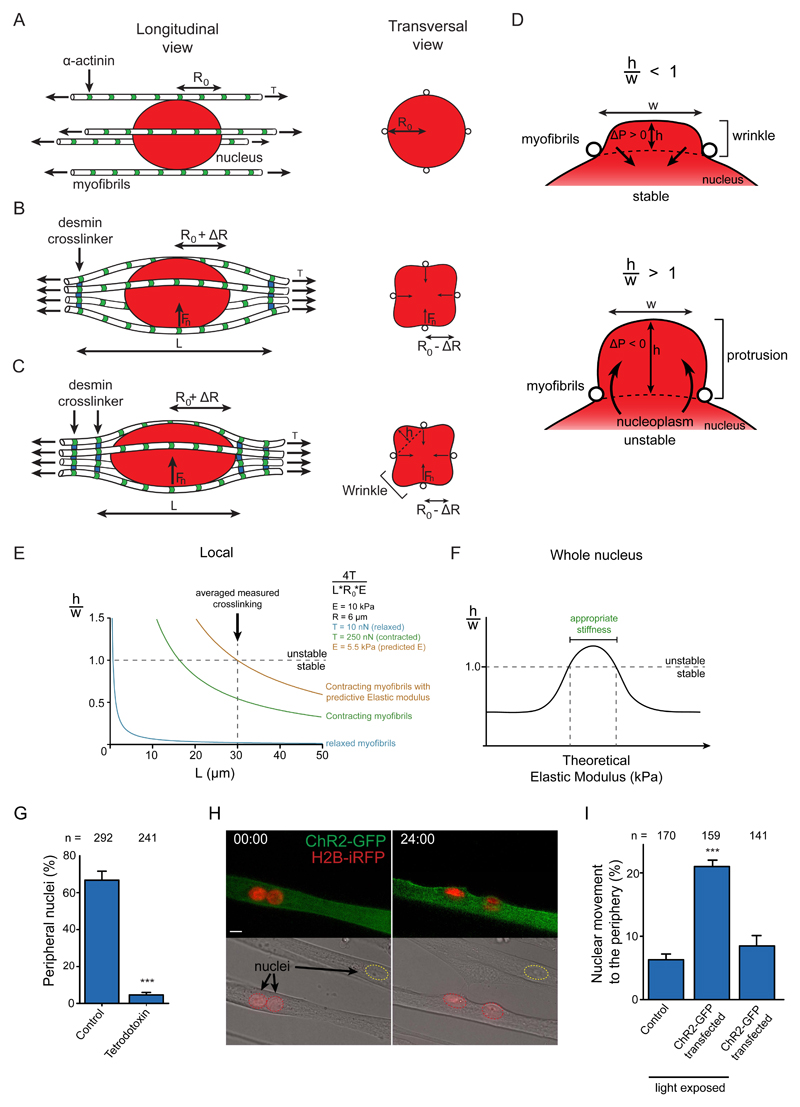
We developed a theoretical model of nuclear movement to the periphery. Before peripheral movement, the nucleus (of radius R0 ≈ 6 μm) is wrapped by a bundle of N ≈ 30 myofibrils 41 and centered along the symmetry axis of the bundle. Each myofibril is modeled as an active spring, whose relaxed tension (T ≈ 1 to 10 nN) is induced by myofibrils at the measured sarcomere length of 2.7 μm (Fig. 6A) 42. Myofibril cross-linking (L) induces centripetal forces Fn on the nucleus towards the myofiber axis of the order of Fn ≈ 4TR0/L for each myofibril (Fig. 6B,C). Denoting γ the surface tension of the nucleus (assumed viscous at the time scale of the experiments), the amplitude of nuclear deformation can be estimated as ΔR ≈ 4NTR0/(γL) ≈ 0.1 to 1 μm, where we made use of the effective Young’s modulus E ≈ γ/R0 at low deformation. This results in the increase of the wrinkle size, which is between myofibrils, as was experimentally confirmed by 3D time-lapse confocal microscopy and EM (Fig. S6A,B and Supplementary movie S3). As soon as the height h of one of these wrinkles is of the order of its width 2πR/N, the wrinkle is destabilized and forms a protrusion into which all the nucleus content flows, so that the nucleus finally leaks out of the myofibril bundle (Fig. 6D and Fig. S6C). The threshold is given equivalently by h ≈ w or ER0 = 4T/L. Given the above orders of magnitude, this threshold is not reached with cross-linking alone (Fig.6E). In order to reach the instability threshold, other parameters such as myofibril tension T and elastic modulus (E) must vary 43 (Fig. 6E). The model also predicts that 2 adjacent nuclei cannot move to the periphery (Fig. S6D) which explains the requirement of nuclei to spread longitudinally before peripheral movement 24.
Figure 6. Theoretical model of peripheral nuclear movement.
A-C. Schematic of a nucleus (red) surrounded by myofibrils (white) before (A) and during (B and C) peripheral migration. Longitudinal view (left) and transversal view (right). R0, radius of the undeformed nucleus. ΔR, amplitude of radial deformation. Fn= force applied by myofibrils on the nucleus. L, length between crosslinkers (blue). h, height of a nuclear wrinkle.
D. Transversal schematic of a stable nuclear wrinkle (top) and unstable nuclear protrusion (bottom). w, width of a nuclear wrinkle/protrusion. ΔP, difference in hydrostatic pressure between nucleus and wrinkle upon an increase of h.
E. Model of the stability of wrinkles relative to force on the nucleus. The scaled wrinkle size h/w is plotted as a function of L based on the function 4T/L*R0*E. Above the threshold h/w ≈ 1, the nucleus is moved to the periphery. Different situations are ploted: relaxed myofibrils (blue), contractile myofibrils (green), contractile myofibrils with predicted nucleus Elastic modulus (E) (Orange).
F. Model for the stability of wrinkles relative to global nuclear stiffness, i.e Elastic modulus. The scaled wrinkle size h/w is plotted as a function of global E. The model analyses both large and small E regimes; the intermediate regime is here extrapolated.
G. Quantification of peripheral nuclei positioning in 10-day myofibers treated with tetrodotoxin from day 4.5. Data from 3 independent experiments were combined and error bars represent s.e.m from indicated n nuclei for each cohort. Unpaired t-test was used to determine statistical significances, where *** P < 0.001.
H. Representative images of the first (left) and last (right) frame of a 24-hour time-lapse movie of untransfected as well as ChR2-GFP (green) and H2B-iRFP (red) transfected myofibers exposed to blue light Nuclei from transfected (red dashed line) and untransfected (yellow dashed line) myofibers are indicated. . 00:00 (hh:mm) corresponds to 3.5-day myofibers. Scale bar, 10 μm. Image shown is representative of 4 experiments.
I. Quantification of nuclei migrating to the periphery as in B., and expressing ChR2-GFP not exposed to light. Data from 6 independent experiments were combined and error bars represent s.e.m from indicated n nuclei for each cohort. Unpaired t-test was used to determine statistical significances, where *** P< 0.001. Source data is available in Supplementary Table 3.
Myofibril tension T can increase up to 25 fold upon contraction reaching predicted values of T = 250 N (Fig. S6D)43. On the other hand, for the entirety of the nucleus, decreases in elastic modulus E as predicted by the model would enforce large deformations needing an adaptation of the analysis. The nucleus would adopt a cylindrical shape, squeezed between almost undeformed myofibrils. The instability will therefore only be reached for γ < 4NTrf3 /(R03L), where rf is the typical radius of the myofibrils bundle in the absence of the nucleus. In a situation with large and small E regimes, the instability is not reached with the biophysical parameters of the model. Therefore, the decrease of nuclear stiffness predicted by the theoretical model to be required to reach the instability threshold probably only occurs locally. Thus, peripheral nuclear movement only occurs within a window of nuclear stiffness for the entire nucleus (Fig. 6F).
This theoretical model predicts that alongside myofibril crosslinking, nuclear spreading, myofiber contraction and changes in nuclear stiffness (globally and locally) play a part in nuclear movement to the periphery.
Nuclear movement requires myofiber contraction
To test the involvement of contraction in nuclear movement, we blocked myofibril contraction with tetradotoxin and found that peripheral nuclear position is inhibited (Fig. 6G). We also found that optogenetically-inducing muscle contraction using ChR2-GFP in premature myofibers (3.5-days) promotes peripheral nuclear movement (Fig. 6H,I, Fig. S6E and Supplementary movie S4,5) 44. Optogenetically-induced contractions did not promote nuclear movement in the absence of Arpc5L and γ-actin, although contractions (optogenetically-induced and spontaneous) were not impaired (Fig. S6F-I). Myofibril contraction is therefore required for peripheral nuclear positioning as predicted by the theoretical model and it is not dependent on Arpc5L or γ-actin.
Nuclear stiffness is important for nuclear movement
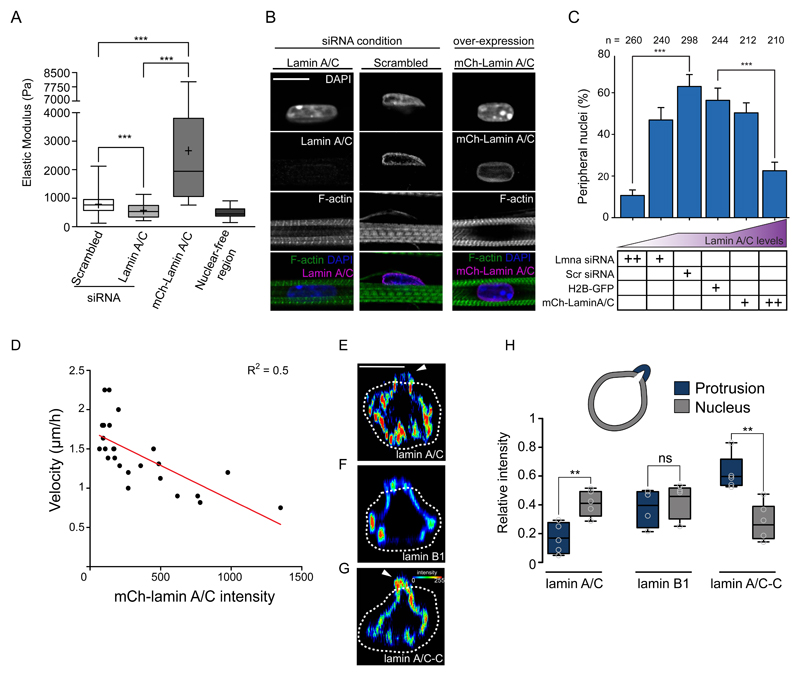
The observed nuclear deformation during movement to the periphery and the theoretical model suggest a role for nuclear stiffness in nuclear movement. We manipulated the stiffness of myonuclei either by siRNA mediated knockdown of lamin A/C (to decrease stiffness)(Fig. S7A,B) or by mCherry-lamin A/C (mCh-lamin A/C) overexpression (to increase stiffness), as previously showed 45,46. Changes in nuclear stiffness upon lamin A/C manipulations were directly confirmed by atomic force microscopy (AFM) (Fig. 7A) and by measuring nuclear deformation during optogenetically-induced contractions (Fig. S7C-E). We found that either the loss or overexpression of lamin A/C decreases the number of peripheral nuclei (Fig. 7B,C and Fig. S7F). In contrast the downregulation of lamin B1, which is not involved in nuclear stiffness 45, did not decrease peripheral nuclei (Fig. S7G-J). Moreover, re-expression of human mCh-lamin A/C in lamin A/C siRNA treated myofibers rescued peripheral nuclear positioning (Fig. S8A,B). In lamin A/C siRNA treated myofibers with only one nucleus expressing mCh-lamin A/C, a greater proportion of mCh-lamin A/C nuclei were at the periphery (59%, n=22) compared to the lamin A/C negative nuclei (19.97%, n=169)(Fig.S8A). Furthermore, the localization of nuclear envelope proteins Nesprin-1 and SUN2 were not affected by knock down or overexpression of lamin A/C (Fig. S8C,D) demonstrating that the LINC complex nuclear localization is independent of lamin A/C in myofibers. Myofibril integrity was also unaffected by lamin A/C knockdown (Fig. S8E). Finally, the levels of overexpressed mCherry-lamin A/C were inversely correlated with the velocity of nuclear movement to the periphery (Fig. 7D). Overall, these data support that variations in nuclear stiffness (decrease or increase) impair nuclear movement as predicted by our theoretical model.
Figure 7. Contraction and nuclear stiffness are involved in nuclear movement to the periphery.
A. Box plot of nuclear stiffness in 5-day myofibers knocked down for scramble or lamin A/C or transfected with mCh-lamin A/C measured by AFM using nuclear-free region as a control. Data from 4 independent experiments were combined and error bars represent s.e.m from at least 90 measurements for each cohort. Unpaired t-test was used to determine statistical significances, where *** P < 0.001. Source data is available in Supplementary Table 3.
B. Representative immunofluorescence images of 10-day myofibers knocked down for lamin A/C, scrambled or over-expressing mCherry-lamin A/C (mCh-lamin A/C) and stained for lamin A/C (magenta), F-actin (green) and DAPI (nucleus, blue). Scale bar, 10 μm. Image shown is representative of 3 experiments.
C. Quantification of peripheral nuclei positioning in 10-day myofibers knocked down for lamin A/C or scrambled, or over-expressing H2B-mCherry or mCherry-lamin A/C. Data from 3 independent experiments were combined and error bars represent s.e.m from indicated n nuclei for each cohort. Unpaired t-test was used to determine statistical significances, where *** P < 0.001. Source data is available in Supplementary Table 3.
D. Plot correlating velocity of nuclear movement to the periphery with the intensity of the mCherry-lamin A/C signal in time-lapse movies of myofibers expressing mCherry-lamin A/C between 4.5 and 5.5 days. Red line corresponds to the significant linear regression, P = 0.0001 of R2 = 0.5004 for n = 24 nuclei. Source data is available in Supplementary Table 3.
E-G. Orthogonal view of nuclei from 5 day myofibers with protrusion initiation, stained with lamin A/C (E), lamin B1 (F) or lamin A/C-C (G) the intensity signal represented as a heat map. White dashed line represents the outline of myofibrils. White arrowheads represent asymmetry nuclear stiffness. Scale bar, 10 μm.
H. Box plot comparing the intensity of lamin A/C, lamin B1 or lamin A/C-C in the part of the nucleus still inside the myofibril bundle (grey: nucleus) versus the forming protrusion (blue: protrusion). Data from 5 independent experiments were combined and error bars represent s.e.m from n = 5 nuclei for each cohort. Unpaired t-test was used to determine statistical significances, where ** P < 0.01, NS, not significant. Box represents the upper and lower quartile, bar represents median and error bars represent maxima and minima. Source data is available in Supplementary Table 3.
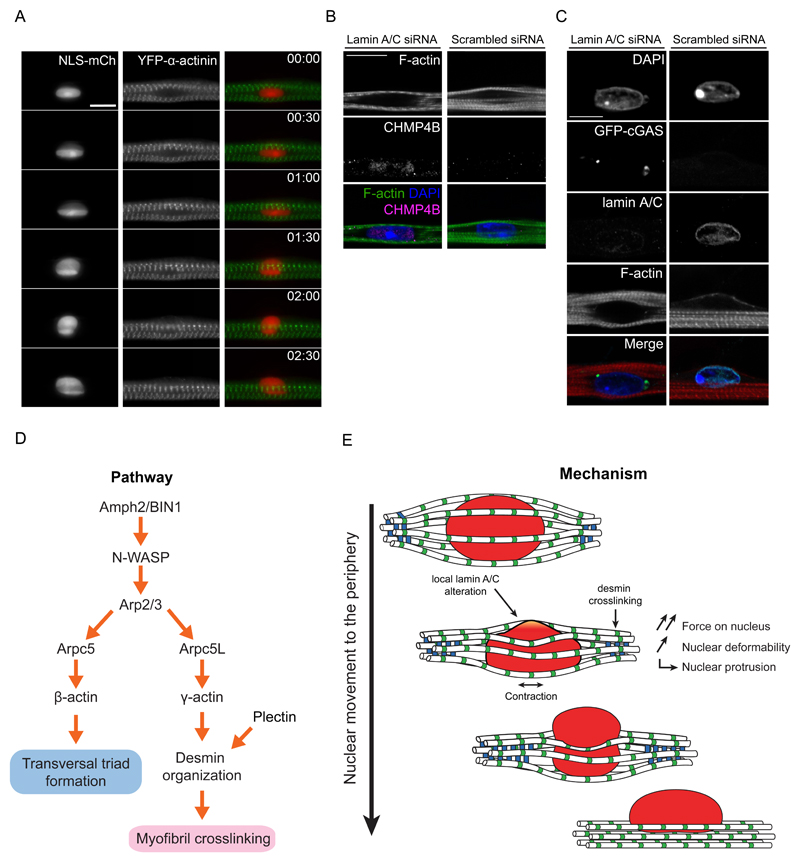
We next investigated if local alterations of nuclear lamins could be associated with nuclear protrusions as predicted by the model. By observing endogenous lamin A/C and lamin B1 during nuclear movement, we found that lamin A/C, but not lamin B1, is asymmetrically distributed at the nuclear protrusion (Fig. 7E,F,H). Furthermore, a reduction of lamin A/C multimerization, detected with an epitope specific antibody (lamin A/C-C), also occurs at the nuclear protrusion (Fig. 7G,H)47. Despite the lamin A/C asymmetry and the extent of nuclear squeezing, we did not observe nuclear envelope rupture evaluated by leakage of NLS-mCh from the nucleus to the cytoplasm (Fig. 8A), recruitment of the ESCRT complex to the nuclear envelope (Fig. 8B), or recruitment of cGAS from the cytoplasm to the nucleus (Fig. 8C) 48,49. Our observations thus advocate that nuclear stiffness, dependent on lamin A/C, is locally involved in peripheral nuclear positioning.
Figure 8. Model of nuclear movement to the periphery in myofibers.
A. Kymograph from a time-lapse movie of a 4-day myofiber depicting movement to the periphery of a nucleus (NLS-mCh, red) surrounded by myofibrils bundles (YFP-α-actinin, green). Note that NLS-mCh is not released from the nucleus to the cytoplasm suggesting that nuclear disruption does not occur during nuclear movement to the periphery. Time, hh:mm. Scale bar, 10 μm.
B. Representative image of 4.5-day myofibers knocked down for lamin A/C or scrambled and stained for myofibrils (F-actin, green), ESCRT III (CHMP4B, magenta) and nucleus (DAPI, blue). Note that the ESCRT complex is not recruited to the nuclear envelope in scramble situation. Lamin A/C siRNA was used as a control. Scale bar, 10 µm. Image shown is representative of 3 experiments.
C. Representative image of 10-day myofibers knocked down for lamin A/C or scrambled and transfected with GFP-cGAS (green) as well as being stained for myofibrils (F-actin, red), lamin A/C (cyan) and nucleus (DAPI, blue). Note that GFP-cGAS is not recruited to the nucleus in scramble situation suggesting that nuclear disruption does not occur. Lamin A/C siRNA was used as a control. Scale bar, 10 µm. Image shown is representative of 3 experiments.
D. Pathway of the molecular players involved in nuclear movement to the periphery and transversal triad formation.
E. Mechanism of nuclear movement to the periphery.
Discussion
Nuclear positioning at the periphery of the cell is a hallmark of functional myofibers. Movement of nuclei from a central position to the periphery is a key step during muscle development and is affected in many muscle disorders, as well as muscle regeneration. We elucidate here the mechanism by which myofibers expel their nuclei to the periphery. We demonstrate that this movement is driven by myofibril cross-linking and contraction, and requires a tight regulation of nuclear stiffness by lamin A/C (Fig. 8D). Furthermore, we found that myofibril crosslinking involves the organization of desmin by an Arpc5L-containing Arp2/3 complex together with γ-actin, (Fig. 8E).
A mechanism of nuclear positioning at the periphery of myofibers
Nuclear movement within cells is driven by intracellular forces. It has always been reported that the machinery generating these forces is initially polarized in order to give a direction to the movement 1. In contrast, the nuclear movement to the periphery of myofibers here described does not involve initial polarized machinery. The forces induced by myofibrils around the nucleus are centripetal and symmetrically distributed around the nucleus. According to our theoretical model and experimental data, the polarization of the movement stems from the formation of a nuclear protrusion that depends on local changes of nuclear stiffness or stochastic alterations of forces exerted by the myofibrils around the nucleus. Thus, the direction of nuclear movement is not only dependent on the cytoplasmic machinery but also on the intrinsic properties of the nucleus. Future work should address if these local changes in nuclear stiffness are stochastic or locally triggered.
A connection between the nucleus and the cytoskeleton is usually required to drive nuclear movements 8. This connection is often mediated by the nuclear envelope proteins Nesprins and SUNs (the LINC complex) that link the cytoskeleton to the nuclear lamina 3,5,6. Our results suggest that a nucleus-cytoskeleton connection is not required for peripheral nuclear movement. Since the LINC complex was implicated in longitudinal spreading of nuclei earlier during myofiber development and anchoring at the plasma membrane, future work should address how the LINC complex is involved in peripheral nuclear positioning 50–55.
Distinct Arp2/3 complexes and actin isoforms have unique roles in skeletal muscle
We also found that distinct populations of Arp2/3 complexes together with specific actin isoforms have segregated functions in skeletal myofibers, nuclear movement and transversal triad formation. Despite having very similar structure, β- and γ-actin isoforms have unique functions in cells, and particularly in skeletal muscle 56–59. Our results strongly suggest that different physiological functions of β- and γ-actin isoforms can be mediated by specific Arp2/3 complex isoforms 39. This specificity can be achieved by compartmentalization of the Arp2/3 and actin complexes, different nucleation activities of Arpc5 isoforms or by different polymerization rates of actin isoforms dependent on Ca2+ concentration 33,39,58.
Perinuclear Arp2/3–dependent actin polymerization around the nucleus was found to disrupt nuclear lamina to allow the nuclei of dendritic cells to pass through constrictions during cell migration60. Our data suggest that nuclear movement to the periphery occurs by a different mechanism. The Arp2/3 complex and γ-actin are important for the organization of desmin to crosslink myofibrils. Such organization is analogous to the mechanisms found in migrating cells where actin retrograde flow organizes the vimentin intermediate filament cytoskeleton via plectin 34,35. Desmin replaces vimentin as the main structural intermediate filament during skeletal muscle differentiation. Its organization transitions from a meshwork early on during myofiber development to being organized at the z-line. We show here that the transition of desmin from a meshwork to being organized at the z-line coincides and is required for nuclear movement 18,29–31.
Nuclear positioning in muscle disorders
Muscle disorders such as centronuclear myopathies, myotonic dystrophy, desminopathies and laminopathies exhibit centrally located nuclei 15–17,24,61,62. Some of the genes encoding the proteins involved in peripheral nuclear positioning are mutated in these disorders. Thus the mechanism of nuclear positioning described here explains why mutations in apparently diverse proteins leads to a common phenotype of centrally located nuclei in different muscle disorders 16,59,63.
Materials and Methods
Cell Culture, animals, transfections and reagents
All procedures using animals were approved by the Institutional ethics committee and followed the guidelines of the National Research Council Guide for the care and use of laboratory animals. In vitro myofibers were generated with primary myoblasts isolated from 5 - 7 day old pups of C57BL/6 mice, using the protocol previously described 1. Cells were grouped regardless of gender. In vivo clearing and myofiber isolation was performed on newborn mice.
Cells were transfected with siRNA (20nM) using RNAiMAX (cat # 15338-100), cDNA 1µg/µl using Lipofectamine-LTX Plus reagent (cat# 13778-150) or using Lipofectamine 2000 (cat# 11668-019) following manufacturer instructions (Life Technologies, see individual catalogue numbers). For primary myoblasts, cells were transfected 6 h prior to differentiation in order to promote protein silencing or overexpression effectiveness from the beginning of differentiation.
CK666 (Arp2/3 inhibitor) was obtained from Sigma (cat# SML0006-5MG) and was added to 4.5-day myofibers at a concentration of 50 μM. Tetrodotoxin (contraction inhibitor) was obtained from Sigma (cat# T8024) and was added to 4.5-day myofibers at a concentration of 10 μM.
Plasmids
pcDNA GFP was a gift from Alexis Gautreau (Ecole Polytechnique de Palaiseau). GFP-Arpc5 and GFP-Arpc5L were previously described2. The γ-actin mouse cDNA, synthesized from Life Technologies with silent point mutations to make it resistant against siRNA, was cloned into a pDONR221 Gateway entry vector (Life Technologies) and recombined into a pEGFP_C1-GW destination vector and a pRFP-GW destination vector to generate pEGFP_C1- γ-actin and pRFP-γ-actin plasmids respectively. The β-actin cDNA, synthesized from Life Technologies with silent point mutations to make it resistant against siRNA, was cloned into a pDONR221 Gateway entry vector (Life Technologies) and recombined into a pEGFP-C1-GW destination vector and a pRFP-GW destination vector to generate a pEGFP_C1-β-actin and pRFP-β-actin plasmids respectively. The ChR2-GFP construct was obtained through addgene (Plasmid #20939). YFP-α-actinin plasmid was a gift from Pekka Lappalainen (university of Helsinki). iRFP-H2B was a gift from Mathieu Coppey (Institut Curie). EmGFP-Desmin was obtained through addgene (plasmid #54059). NLS-tdtomato, H2B-mCherry was a gift from the Lammerding lab (Weill Institute). GFP-CGas was a gift from the Piel lab (Institut Curie).
Antibodies
Antibody information is provided in Supplementary table 1.
siRNA
The sequences of siRNAs used in the study are provided in Supplementary table 2.
Western Blotting
Cells were lysed in PBS + 1%SDS and passed through a Qiashredder column (Qiagen) to disrupt DNA. Protein concentration was measured with a BCA kit according to manufacturer instructions (Pierce). Equal amount of sample were boiled in 30ul sample buffer and were loaded on 4-12% pre-cast Bis-Tris gel (Invitrogen) and transferred into nitrocellulose membrane using the iBlot apparatus (Invitrogen). Membranes were blocked with blocking buffer (5% Non Fat Dry Milk, 0.1% Tween in TBS). Primary antibodies were incubated overnight in blocking buffer at 4°C. After three washes with TBS-Tween 0.1%, membranes were incubated with HRP-conjugated secondary antibodies (1 hour at room temperature). Proteins were visualized using ECL reagent (Pierce). All western blots were performed 3 times.
Immunoprecipitation
For immunoprecipitation of GFP-tagged proteins, a GFP-Trap system (Chromotek cat# gtm-20) was used following manufacturer instructions. After thorough washing with the lysis buffer, bound proteins were eluted in sample buffer boiled and analyzed by western blot.
Immunofluorescence and immunohistochemistry
Fluorodishes were fixed in 4% paraformaldehyde for 10 min, permeabilized with tritonx100 (0.5% in PBS) and aspecific sites were blocked with BSA 1% and Goat serum 10% for 30 min. Primary antibodies were added O.N at 4°C in saponin 0.1% and BSA 1% in PBS. Fluorodishes or fibers were washes three times and then incubated with secondary antibodies together with DAPI for 60 minutes. Each experiments were performed at least 3 times.
Lamin quantifications of nuclear positioning
Variability in the strength of the lamin A/C knock down could be observed by immunofluorescence. As such, myofibers were quantified with either complete knocked down (referred as “++”) or partial knock down (referred as “+”). This allowed a quantification of varying levels of lamin A/C and therefore nuclear stiffness. From the Lmna siRNA experiments, when lamin A/C intensity was below 10% of the control intensity, it was categorized as “++” knockdown. When lamin A/C intensity was between 10% and 50% of the control intensity, it was categorized as “+” knockdown. For the overexpression experiments, we normalized mCh-lamin A/C expression relative to the higher expressing mCh-Lamin A/C nucleus (which was 100%). When intensities were between 100% and 50%, it was categorized as “++” overexpression. When intensities were below 50%, it was categorized as “+”.
Isolation of myofibers
TA single fibers were isolated as described 1. TA muscle was explanted from newborn male or female CD1 mice and then digested in DMEM containing 0.2% typeI collagenase (Sigma) for 2 h at 37°C. Mechanical dissociation of fibers was performed using a thin pasteur pipette and followed under a transilluminating-fluorescent stereomicroscope.
Atomic Force Microscopy (AFM)
The elasticity of the cells was measured using an atomic force microscope NanoWizard II (JPK Instruments, Berlin, Germany) mounted on the top of an Axiovert 200 inverted microscope (Carl Zeiss, Jena, Germany). Nanoindentation experiments were carried out on live cells, at 25°C, in Iscove’s Modified Dulbecco’s Media (IMDM). For these measurements non-functionalized OMCL TR-400-type (Olympus, Japan) with a long triangular cantilever and a silicon nitride tips (nominal force constant of 20 pN/nm) (Olympus, Japan) were used. Differential interference contrast (DIC) microscopy and fluorescence microscopy were used to position the tip on the top of the cell nucleus of each cell previously transfected. For every contact between cell and cantilever, the distance between the cantilever and the cell was adjusted to maintain a maximum applied force of 500 pN before retraction. Cell elasticity was measured on one point of each cell adhered to the tissue culture dish (8 force-distance curves per cell), and on approximately 8 cells of 3 different culture dishes. A total of 24 cells were tested for each experimental condition. Data collection for each force-distance cycle was performed at 2 Hz and with a z-displacement range of 8 μm. Nuclear-free regions were used as a comparative control to account for membrane and organelle stiffness. Data acquired on the nanoindentation experiments (force indentation curves) were analyzed to obtain the cells Young’s modulus (E), using JPK Image Processing v. 5.1.8, by the application of the Hertzian model. The probe was modeled as a quadratic pyramid, with a tip angle of 35° (half-angle to face) and a Poisson ratio of 0.50.
Fluorescence recovery after photobleaching (FRAP)
FRAP experiments were performed on a Zeiss LSM 880 confocal microscope from Carl Zeiss equipped with the ZEN gray edition software, using a Plan-Apochromat 63x Oil DIC objective (Numerical aperture: 1.40). Each fiber was imaged during 5 minute periods at 5 second intervals, with two frames being acquired before bleaching. Fluorescence bleaching was conducted using a 488 nm wavelength laser at 100% laser power (maximum laser power) for 20 iterations. The bleached area consisted of a circle with the average width of myofibers. Fluorescence intensities of the bleached regions were obtained using FIJI software and a nonlinear one-phase association curved available in Graphpad Prism was used for data fitting. Data analysis was performed in Microsoft Excel and Graphpad Prism, the data was averaged for at least 8 cells for each of the conditions studied.
Transmission electron microscopy (TEM)
Myofibers were fixed for 1h at 4ºC in 0.1 M sodium cacodylate buffer, pH 7.3, containing 2.5% (v/v) gluteraldehyde and 0.1% (v/v) formaldehyde. Following 1 hour (on ice) post-fixation in 1% (aq.) osmium tetroxide and 30 minutes contrast in block in 1% (aq.) uranyl acetate. Dehydration was made using ethanol gradient (50-70-95-100%). Samples were flat embedded in Durcupan resin and hardened at 60ºC for 72h. For cells in transversal plane (top to bottom of the cell), sections were directly obtained, and for coronal plane of orientation (anterior to posterior) 2 blocks of the same condition were glued together and sectioned in a 90º plane in relation to the flat surface of the blocks, both orientations were sectioned using an ultramicrotome Reichert Supernova (Leica microsystems©), semi-thin sections (500 nm) were stained with toluidine blue for light microscope evaluation. Ultra-thin sections (70 nm), were obtained and collected in formvar coated copper slot grids (AGAR scientific©), and counter-stained with uranyl acetate and lead citrate (Reynold recipe), regions of interest in both orientation planes were sequentially screened in a Hitachi H-7650 transmission electron microscope at 100kV acceleration.
Whole muscle clearing
Tissue clearing is a method to clarify the tissue to replace the dehydrant with a miscible agent 3. The tissue will therefore become translucent for greater 3D imaging. Legs of newborn mice were isolated and fixed overnight in PFA 4%. Legs were pre-treated overnight with (1% triton, 0.5% tween, 0.25%NP-40, 0.25% Na-deoxycholate, 3% BSA, 0.002% NA-azide, 1% urea in PBS). The following day, legs were incubated in CBB before being incubated with primary antibody for 2 days. After primary antibody incubation, legs were washed several time with PBT and left rotated to wash over night. The following day, legs were incubated with secondary antibody for 2 days and then washed with PBT several times and overnight. DAPI was subsequently added for 3 hours and washed several times before starting the clearing treatment. Clearing began by incubating legs with 25% formamide and 10% PEG for an hour, followed by 50% formamide and 20% PEG for an hour and finishing with 50% formamide and 20% PEG overnight. Legs were then mounted with fluoromount and imaged.
Peripheral nuclei position quantification
Quantification was performed as previously described 1. Briefly, myofibers were stained for DAPI and traids and images in Z-stacks with 0.5 mm interval were acquired with a Leica SPE confocal microscope with a 63x 1.3 NA Apo objective. Nuclei extruding the myofiber periphery were scored as peripheral.
Transversal triad quantification
Quantification was performed as previously described 1. Briefly, myofibers were stained for DHPR1, triadin and DAPI and images in Z-stacks with 0.5 mm interval were acquired with a Leica SPE confocal microscope with a 63x 1.3 NA Apo objective. Myofibers having more than 50% of triads organized, where DHPR and Triadin were transversally overlapping,were scored as positive.
Protrusion intensity quantification
Orthogonal images of nuclear buds were generated and line scans were drawn on the nuclear envelop either on part of the nucleus still inside the myofibril bundle or part of the nucleus outside the bundle (bud). For figure S2, line scans were drawn on the bud in contact with the plasma membrane (cap) and the bud which was not (side). Line scan width was 1 µm.
Line scans
Line scan was drawn at the same spot in the kymograph depicting desmin and a plot was generated. Line scan width was 2 µm.
Microscopy
Live imaging was performed using an incubator to maintain cultures at 37°C and 5% CO2 (Okolab) and × 20 0.3 NA PL Fluo dry objective. Epi-fluorescence images were acquired using a Nikon Ti microscope equipped with a CoolSNAP HQ2 camera (Roper Scientific), an XY-motorized stage (Nikon), driven by Metamorph (Molecular Devices). Confocal images were acquired using Leica SPE confocal microscope with a 63x 1.3 NA Apo objective or Zeiss LSM 710 and Zeiss LSM 880 with a 63x 1.4 NA Plan-Apochromat objective. 3D-time-lapse spinning disk microscopy was performed using a Zeiss Cell Observer Spinning Disk system equipped with Z-piezo (Prior), Spinning Disk CSU-X1M 5000 (Yokogawa), 488nm 561nm and 638nm excitation laser, an incubator to maintain cultures at 37°C and 5% CO2 (Pekon), EM-CCD camera Evolve 512 (Photometrics) and a 63x 1.4 NA Plan-Apochromat objective. Fiji was used as an imaging processing software and Adobe illustrator was used to raise figures.
Area around the nucleus quantification
Images from the middle plane of a z-stack expressing YFP- α-actinin and H2B-iRFP, obtained with spinning disk microscopy were used. One transversal line was drawn at a distance of 30 μm on each side of the nucleus. The dark area on each side of the nucleus between this line, the myofibrils and the nucleus was measured using Fiji. These measurements were performed in time-lapse images. Values were normalized relative to the first time point.
Percentage of nucleus at the periphery
During nuclear movement to the periphery, area of nuclei emerging from the myofiber and total area of the nucleus were assessed. A ratio was obtained and a percentage was calculated over time.
Spontaneous contraction quantifications
5-day myofibers were monitored under a light-microscope with phase contrast. Contracting myofibers were spotted and number of contraction was monitored over a one-minute duration to determine contraction frequency.
Spherical and volumetric assessment
Time-lapse images were reconstructed in 3D and over time using the software Imaris© and surface 3-dimensional rendering was performed. Spherical and volumetric values were extracted from the 3D-render of nuclei at each time point. The rendering was adapted for each time point to the background to retain fluorescence intensity. Sphericity was calculated as defined by Wadell in 1932. Values were normalized relative to the first time point. Sphericity and volume were measured every 5 minutes and normalized to the first point for Fig. 2A and B.
Laser ablation and bleaching
4.5-day myofibers were incubated in a Zeiss LSM 880 microscope at 37°C and 5% CO2. 5 circles were drawn on both sides of nuclei as ROI to be ablated or bleached. A Diode 405-30 laser was used at 100% (max power 30 mW) with a scan speed of and 3 iterations for ablation and an Argon laser was used at 50% (max power 25 mW) with a scan speed of and 3 iterations for bleaching. 5 minutes after ablation or bleaching, cells were acquired in z for 3D reconstruction to analyze volume and sphericity.
Nuclear deformations during contractions
4.5-day myofibers were incubated at 37°C and 5% CO2 in the Zeiss Cell Observer Spinning Disk system. Myofibers were transfected with the optogenetic channel ChR2 and NLS-mCherry or mCherry-lamin A/C as well as knocked down for scramble or lamin A/C. Movies were acquired in stream mode in the 555 channel and blue light was shone on cells at different intervals during acquisition to induce contraction. Nuclei length were then measured between relaxed and contracted state and normalized on myofiber thickness.
Pausing duration quantifications
Nuclear movement velocity along myofibers was calculated using software ICY. Nuclei were considered pausing when they moved less than 0.0015 μm/s for a minimum time of an hour. The cut-off values of pausing considerations (one hour at less than 0.0015 μm/s) was chosen based on visual correlation between nuclear movement and pausing from live imaging movies.
Model
The nucleus of radius R0 ≈ 6 μm (at rest) has a complex rheology. It is often described as a linear elastomer (Young’s modulus E ≈ 1 to 10 kPa), whose elastic component is largely due to the lamina envelope. However for large and slow deformations a viscous behavior is expected, as suggested in other contexts (Racine and Piel). The effective surface tension of the nucleus in skeletal muscle cell can therefore be estimated as γ ≈ R0E. Before peripheral movement, the nucleus is wrapped by a bundle of N ≈ 30 myofibrils and centered along the symmetry axis of the bundle. We assume that each myofibril can be modeled as an active spring, whose passive tension (T ≈ 1 to 10nN) was assessed using 2.7 μm sarcomere length (empirical measurement).
During peripheral migration of the nucleus to the periphery, we assume that myofibrils are crosslinked together through Arpc5L-containing Arp2/3 complexes and γ-actin (with a typical distance between myofibrils of the order of 1 μm) and with a distance L ≈ 30 μm between cross-linking points along the myofiber. This induces a normal force Fn on the nucleus towards the myofiber axis of the order of Fn ≈ 4TR0/L for each myofibril. This force first induces a global deformation of the nucleus, which elongates along the myofiber axis. The deformation can be estimated by balancing the surface energy cost of a deformation of amplitude ΔR of the nucleus, which is dEs ≈ γ ΔR2, and the work of the total normal force exerted by myofibrils given by dW ≈ NFnΔR. This yields ΔR ≈ 4NT/(LE) ≈ 0.1 to 1 μm, which is consistent with observations.
Such conformation of the nucleus squeezed by myofibrils is unstable. Between each myofibril squeezing a wrinkle can form. We now argue that as soon as the height h of one of these wrinkles is of the order of its width 2πR/N, the wrinkle is destabilized forming a bleb and all the nucleus content flows into the bleb, so that the nucleus is expelled out of the bundle. Let us denote by ΔP the difference of hydrostatic pressure between the inside and outside of the nucleus. Laplace law gives ΔP=γC, where C is the mean curvature of a wrinkle. For h < 2πR/N, an increase of the wrinkle height h locally increases the curvature C, leading to a local increase in the hydrostatic pressure in the wrinkle, which subsequently relaxes. Conversely, for h > 2πR/N, an increase of h reduces the curvature C, thereby leading to a decrease in the local hydrostatic pressure in the wrinkle. The nucleoplasm subsequently flows into the wrinkle, which is henceforth destabilized and leads to bleb formation. The threshold of this instability is reached when γR0 ≈ 4TR0/L, or T > ER0L/4; this condition is obtained by balancing normal forces on a myofibril for h ≈ 2πR/N: the normal force induced by the myofibrils is 4TR0/L, while the restoring force induced by the deformed nucleus is of order γR0 when h = 2πR/N. Note that here we assumed a constant surface tension γ upon deformation, which yields a threshold independent of the number of myofibrils. Given the above orders of magnitude, this threshold can be reached in principle, provided that tension takes large values and Young’s modulus E is locally small enough values. This shows that such mechanism of nucleus off centering can indeed be at work. In particular this mechanism shows that the motion of the nucleus to the periphery is favored by an increased tension of myofibrils and a reduced distance between crosslinkers.
When the surface tension is low (i.e at low level of lamin A/C), the nucleus undergoes large deformations and this analysis needs to be adapted. One finds that a cylindrical shape of the nucleus, squeezed between almost undeformed myofibrils, is stable for γ< 4NTrf3 /(R03L), where rf is the typical radius of the myofibrils bundle in absence of nucleus. Altogether, this shows that the mechanism of positioning of the nucleus to the periphery is efficient only within a window of values of nucleus rigidity (here modeled effectively as a surface tension), which is controlled by lamin A/C levels.
Optogenetically-induced contraction
Myofibers were transfected with the ChR2-GFP and iRFP-H2B constructs. Upon blue light exposure, the ChR2 channel opens to become permeable to Na+ ions. This induces a depolymerization of the membrane and a subsequent contraction of the myofiber. To monitor the effect of contraction on peripheral nuclear localization, 3.5-day myofibers were exposed to 100 milliseconds of blue light using a bandpass filter 470/40 every 10 minutes for 24 hours. Brighfield and epifluorescent images were acquired every 10 min. The number of peripheral nuclear positioned over a period of 24 hours was assessed both in transfected and untransfected myofibers. Peripheral nuclei were scored when more than a quarter of the total size of nuclei protruded the membrane. A ratio of nuclei having reached the periphery over total nuclei assessed was obtained and a percentage calculated.
Correlative velocity of nuclei and lamin A/C expression
Time-lapse movies of myofibers overexpressing mCherry-lamin A/C were analyzed by quantifying the overall intensity of nuclei expressing the construct and the time required to reach the periphery (20 minutes per frame). The intensity of lamin A/C was obtained by measuring the integrated fluorescence density which is the sum of all the pixels which equals to the product of Area and Mean Gray Value, subtracted by the background integrated density.
Statistics and Reproducibility
Statistical analysis was performed with Graphpad Prism (version 5.0 of GraphPad Software Inc.). Pair wise comparisons were made with Student’s t-test. In peripheral nuclei positioning analysis and in fiber thickness analysis in myofibers, Student’s t-tests were performed between scramble siRNA and experimental condition. For biochemical experiments using human samples, statistical analysis was performed using the Mann-Whitney U test, Wilcoxon matched-pairs signed ranked test or the unpaired Student’s test and multiple statistical comparisons between samples were performed by two-way analysis of variance followed by a Bonferroni’s t-test posthoc correction to obtain a better evaluation of the variability between samples from the same group and samples from each compared group and statistical significance was set at *P < 0.05. The distribution of data points is expressed as mean ± SEM from three or more independent experiments. For statistics with an n lower than 10, a statistical power test was performed using GPower 3.1 based on the statistical test used. Outliers were detected using Gribbs’ test at a P value < 0.05. Variability arises in the maturity obtained from the in vitro muscle system obtained from primary myoblasts. As such, a minimum of 55% of peripheral nuclei in the control cohorts were required to consider the experiment. Representative images are from at least 3 experiments except for in vivo clearing and myofiber isolation which are from 2 experiments. All blots were produced at least 3 times.
Code availability
Image processing was performed using Fiji. Sphericity and volume of nuclei during movement to the periphery was done using Imaris. Algorithms can be obtained in the following link: http://www.bitplane.com/resources.aspx.
Data Availability
Source data for figure panels Fig. 2A-B, 2D, 2F-G, 3B. 3E, 4C, 4G, 5F, 6I, 7A, 7C, 7D and 7H as well as supplementary figure panels S1B-C, S2E, S3F-G, S4E, S4G, S5D, S5H, S6F, S7D-E, S7F, S7J, S8B are available in Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgements
We thank M-H. Verlhac, M. Dias, J. Pinto, G. Gundersen and S. Tapscott for comments on the manuscript. We thank the Gomes Laboratory for discussions. This work was supported by the European Research Council (E.R.G.), EMBO installation (E.R.G.), the myograd PhD program (W.R.), AIM France (W.R., B.C., E.R.G.), LISBOA-01-0145-FEDER-007391 co-funded by FEDER through POR Lisboa 2020 - Programa Operacional Regional de Lisboa, do PORTUGAL 2020 (E.R.G.), and Fundação para a Ciência e a Tecnologia (E.R.G). M.W. and J.V.G.A. were supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001209), the UK Medical Research Council (FC001209), and the Wellcome Trust (FC001209) as well as by postdoctoral fellowships from FRQS (Fonds de recherche du Québec - Santé), EMBO and the Canadian Institutes of Health Research (CIHR) to J.V.G.A.
Footnotes
Author Contributions:
W.R. carried out experiments and analyzed data; J.M. performed lamin and desmin-related experiments; F.C and N.S carried out AFM experiments. W.R., B.C. and E.R.G. conceived and designed experiments; J.V.G.A. and M.W. provided Arp2/3-related unpublished tools; W.R., R.V. and B.C. designed and executed the physical model; W.R. and E.G. wrote the manuscript with assistance from other authors; All authors participated in the critical review and revision of the manuscript.
Competing Financial Interests:
The authors declare no competing financial interests
References
- 1.Gundersen GG, Worman HJ. Nuclear Positioning. Cell. 2013;152:1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horn HF, et al. The LINC complex is essential for hearing. J Clin Invest. 2013;123:740–750. doi: 10.1172/JCI66911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Starr DA, Fridolfsson HN. Interactions Between Nuclei and the Cytoskeleton Are Mediated by SUN-KASH Nuclear-Envelope Bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolhy S, et al. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol. 2011;192:855–871. doi: 10.1083/jcb.201007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–9. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 6.Crisp M. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Splinter D, et al. Bicaudal D2, Dynein, and Kinesin-1 Associate with Nuclear Pore Complexes and Regulate Centrosome and Nuclear Positioning during Mitotic Entry. PLoS Biol. 2010;8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janota CS, Calero-Cuenca FJ, Costa J, Gomes ER. SnapShot: Nucleo-cytoskeletal Interactions. Cell. 2017;169:970–970.e1. doi: 10.1016/j.cell.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Almonacid M, et al. Active diffusion positions the nucleus in mouse oocytes. Nat Cell Biol. 2015;17:470–479. doi: 10.1038/ncb3131. [DOI] [PubMed] [Google Scholar]
- 10.Zhao T, Graham OS, Raposo A, St. Johnston D. Growing Microtubules Push the Oocyte Nucleus to Polarize the Drosophila Dorsal-Ventral Axis. Science. 2012 doi: 10.1126/science.1219147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huelsmann S, Ylänne J, Brown NH. Filopodia-like Actin Cables Position Nuclei in Association with Perinuclear Actin in Drosophila Nurse Cells. Dev Cell. 2013;26:604–615. doi: 10.1016/j.devcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cadot B, et al. Nuclear movement during myotube formation is microtubule and dynein dependent and is regulated by Cdc42, Par6 and Par3. EMBO Rep. 2012;13:741–749. doi: 10.1038/embor.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cadot B, Gache V, Gomes ER. Moving and positioning the nucleus in skeletal muscle – one step at a time. Nucleus. 2015;6:01–09. doi: 10.1080/19491034.2015.1090073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metzger T, et al. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 2012;484:120–124. doi: 10.1038/nature10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azibani F, Muchir A, Vignier N, Bonne G, Bertrand AT. Striated muscle laminopathies. Semin Cell Dev Biol. 2014;29:107–115. doi: 10.1016/j.semcdb.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Romero NB. Centronuclear myopathies: A widening concept. Neuromuscul Disord. 2010;20:223–228. doi: 10.1016/j.nmd.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Fugier C, et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat Med. 2011;17:720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- 18.Clemen CS, Herrmann H, Strelkov SV, Schröder R. Desminopathies: pathology and mechanisms. Acta Neuropathol (Berl) 2013;125:47–75. doi: 10.1007/s00401-012-1057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Striated muscle cytoarchitecture: an intricate web of form and function. Annu Rev Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- 20.Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function and disease. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 21.Lowery J, Kuczmarski ER, Herrmann H, Goldman RD. Intermediate Filaments Play a Pivotal Role in Regulating Cell Architecture and Function. J Biol Chem. 2015;290:17145–17153. doi: 10.1074/jbc.R115.640359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Qusairi L, Laporte J. T-tubule biogenesis and triad formation in skeletal muscle and implication in human diseases. Skelet Muscle. 2011;1:26. doi: 10.1186/2044-5040-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flucher BE, Takekura H, Franzini-Armstrong C. Development of the Excitation-Contraction Coupling Apparatus in Skeletal Muscle: Association of Sarcoplasmic Reticulum and Transverse Tubules with Myofibrils. Dev Biol. 1993;160:135–147. doi: 10.1006/dbio.1993.1292. [DOI] [PubMed] [Google Scholar]
- 24.Falcone S, et al. N-WASP is required for Amphiphysin-2/BIN1-dependent nuclear positioning and triad organization in skeletal muscle and is involved in the pathophysiology of centronuclear myopathy. EMBO Mol Med. 2014;6:1455–1475. doi: 10.15252/emmm.201404436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egile C, et al. Activation of the Cdc42 Effector N-Wasp by the Shigella flexneri Icsa Protein Promotes Actin Nucleation by Arp2/3 Complex and Bacterial Actin-Based Motility. J Cell Biol. 1999;146:1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machesky LM, et al. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarar D, To W, Abo A, Welch MD. The Wiskott–Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr Biol. 1999;9:555–S1. doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]
- 28.Pimentel MR, Falcone S, Cadot B, Gomes ER. In Vitro Differentiation of Mature Myofibers for Live Imaging. J Vis Exp JoVE. 2017 doi: 10.3791/55141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ralston E, et al. Blood vessels and desmin control the positioning of nuclei in skeletal muscle fibers. J Cell Physiol. 2006;209:874–882. doi: 10.1002/jcp.20780. [DOI] [PubMed] [Google Scholar]
- 30.Li Z, et al. Desmin Is Essential for the Tensile Strength and Integrity of Myofibrils but Not for Myogenic Commitment, Differentiation, and Fusion of Skeletal Muscle. J Cell Biol. 1997;139:129–144. doi: 10.1083/jcb.139.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milner DJ, Weitzer G, Tran D, Bradley A, Capetanaki Y. Disruption of muscle architecture and myocardial degeneration in mice lacking desmin. J Cell Biol. 1996;134:1255–1270. doi: 10.1083/jcb.134.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dugina V, Zwaenepoel I, Gabbiani G, Clément S, Chaponnier C. β- and γ-cytoplasmic actins display distinct distribution and functional diversity. J Cell Sci. 2009;122:2980–2988. doi: 10.1242/jcs.041970. [DOI] [PubMed] [Google Scholar]
- 33.Bergeron SE, Zhu M, Thiem SM, Friderici KH, Rubenstein PA. Ion-dependent Polymerization Differences between Mammalian - and -Nonmuscle Actin Isoforms. J Biol Chem. 2010;285:16087–16095. doi: 10.1074/jbc.M110.110130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dupin I, Sakamoto Y, Etienne-Manneville S. Cytoplasmic Intermediate Filaments Mediate Actin-Driven Positioning of the Nucleus. J Cell Sci. 2011;124:865–872. doi: 10.1242/jcs.076356. [DOI] [PubMed] [Google Scholar]
- 35.Jiu Y, et al. Bidirectional Interplay between Vimentin Intermediate Filaments and Contractile Actin Stress Fibers. Cell Rep. 2015;11:1511–1518. doi: 10.1016/j.celrep.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Korobova F, Svitkina T. Arp2/3 Complex Is Important for Filopodia Formation, Growth Cone Motility, and Neuritogenesis in Neuronal Cells. Mol Biol Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gournier H, Goley ED, Niederstrasser H, Trinh T, Welch MD. Reconstitution of Human Arp2/3 Complex Reveals Critical Roles of Individual Subunits in Complex Structure and Activity. Mol Cell. 2001;8:1041–1052. doi: 10.1016/s1097-2765(01)00393-8. [DOI] [PubMed] [Google Scholar]
- 38.Nolen BJ, et al. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abella JVG, et al. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat Cell Biol. 2016;18:76–86. doi: 10.1038/ncb3286. [DOI] [PubMed] [Google Scholar]
- 40.Konieczny P, et al. Myofiber integrity depends on desmin network targeting to Z-disks and costameres via distinct plectin isoforms. J Cell Biol. 2008;181:667–681. doi: 10.1083/jcb.200711058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldspink G. The Proliferation of Myofibrils During Muscle Fibre Growth. J Cell Sci. 1970;6:593–603. doi: 10.1242/jcs.6.2.593. [DOI] [PubMed] [Google Scholar]
- 42.Yoshikawa Y, Yasuike T, Yagi A, Yamada T. Transverse elasticity of myofibrils of rabbit skeletal muscle studied by atomic force microscopy. Biochem Biophys Res Commun. 1999;256:13–19. doi: 10.1006/bbrc.1999.0279. [DOI] [PubMed] [Google Scholar]
- 43.Colomo F, Piroddi N, Poggesi C, Te Kronnie G, Tesi C. Active and passive forces of isolated myofibrils from cardiac and fast skeletal muscle of the frog. J Physiol. 1997;500:535. doi: 10.1113/jphysiol.1997.sp022039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakar MS, et al. Formation and optogenetic control of engineered 3D skeletal muscle bioactuators. Lab Chip. 2012;12:4976–4985. doi: 10.1039/c2lc40338b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lammerding J, et al. Lamins A and C but Not Lamin B1 Regulate Nuclear Mechanics. J Biol Chem. 2006;281:25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 46.Swift J, et al. Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ihalainen TO, et al. Differential basal-to-apical accessibility of lamin A/C epitopes in the nuclear lamina regulated by changes in cytoskeletal tension. Nat Mater. 2015;14:1252–1261. doi: 10.1038/nmat4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denais CM, et al. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raab M, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:359–362. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- 50.Apel ED, Lewis RM, Grady RM, Sanes JR. Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J Biol Chem. 2000;275:31986–95. doi: 10.1074/jbc.M004775200. [DOI] [PubMed] [Google Scholar]
- 51.Espigat-Georger A, Dyachuk V, Chemin C, Emorine L, Merdes A. Nuclear alignment in myotubes requires centrosome proteins recruited by nesprin-1. J Cell Sci. 2016;129:4227–4237. doi: 10.1242/jcs.191767. [DOI] [PubMed] [Google Scholar]
- 52.Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc Natl Acad Sci U A. 2005;102:4359–64. doi: 10.1073/pnas.0500711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson MH, Holzbaur ELF. Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development. 2015;142:218–228. doi: 10.1242/dev.114769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, et al. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 2007;134:901–908. doi: 10.1242/dev.02783. [DOI] [PubMed] [Google Scholar]
- 55.Lei K, et al. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Natl Acad Sci U S A. 2009;106:10207–10212. doi: 10.1073/pnas.0812037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belyantseva IA, et al. Gamma-actin is required for cytoskeletal maintenance but not development. Proc Natl Acad Sci U S A. 2009;106:9703–9708. doi: 10.1073/pnas.0900221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prins KW, Call JA, Lowe DA, Ervasti JM. Quadriceps myopathy caused by skeletal muscle-specific ablation of βcyto-actin. J Cell Sci. 2011;124:951–957. doi: 10.1242/jcs.079848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gokhin DS, Fowler VM. Cytoplasmic gamma-actin and tropomodulin isoforms link to the sarcoplasmic reticulum in skeletal muscle fibers. J Cell Biol. 2011;194:105–120. doi: 10.1083/jcb.201011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sonnemann KJ, et al. Cytoplasmic [gamma]-Actin Is Not Required for Skeletal Muscle Development but Its Absence Leads to a Progressive Myopathy. Dev Cell. 2006;11:387–397. doi: 10.1016/j.devcel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Thiam H-R, et al. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat Commun. 2016;7:10997. doi: 10.1038/ncomms10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zwerger M, et al. Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling. Hum Mol Genet. 2013;22:2335–2349. doi: 10.1093/hmg/ddt079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hnia K, et al. Myotubularin controls desmin intermediate filament architecture and mitochondrial dynamics in human and mouse skeletal muscle. J Clin Invest. 2011;121:70–85. doi: 10.1172/JCI44021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldfarb LG, Dalakas MC. Tragedy in a heartbeat: malfunctioning desmin causes skeletal and cardiac muscle disease. J Clin Invest. 2009;119:1806–1813. doi: 10.1172/JCI38027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Falcone S, et al. N-WASP is required for Amphiphysin-2/BIN1-dependent nuclear positioning and triad organization in skeletal muscle and is involved in the pathophysiology of centronuclear myopathy. EMBO Mol Med. 2014;6:1455–1475. doi: 10.15252/emmm.201404436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abella JVG, et al. Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat Cell Biol. 2016;18:76–86. doi: 10.1038/ncb3286. [DOI] [PubMed] [Google Scholar]
- 66.Richardson DS, Lichtman JW. Clarifying Tissue Clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for figure panels Fig. 2A-B, 2D, 2F-G, 3B. 3E, 4C, 4G, 5F, 6I, 7A, 7C, 7D and 7H as well as supplementary figure panels S1B-C, S2E, S3F-G, S4E, S4G, S5D, S5H, S6F, S7D-E, S7F, S7J, S8B are available in Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding author on reasonable request.