Abstract
Objective:
Alcohol use disorders (AUDs) are associated with decreased gray matter, and neuroinflammation is one mechanism through which alcohol may confer such damage, given that heavy alcohol use may promote neural damage via activation of toll-like receptor 4 (TLR4)-mediated inflammatory signaling cascades. We previously demonstrated that TLR4 is differentially methylated in AUD compared with control subjects, and the present study aims to extend this work by examining whether TLR4 methylation moderates the relationship between alcohol use and gray matter.
Method:
We examined TLR4 methylation and gray matter thickness in a large sample (N =707; 441 males) of adults (ages 18–56) reporting a range of AUD severity (mean Alcohol Use Disorders Identification Test score = 13.18; SD = 8.02). We used a series of ordinary least squares multiple regression equations to regress gray matter in four bilateral brain regions (precuneus, lateral orbitofrontal, inferior parietal, and superior temporal) on alcohol use, TLR4 methylation, and their interaction, controlling for demographic, psychological, and other substance use variables.
Results:
After we corrected for multiple tests, a significant Alcohol × TLR4 Methylation interaction emerged in the equations modeling left precuneus and right inferior parietal gray matter. Follow-up analyses examining the nature of these interactions demonstrated a significant negative association between alcohol and precuneus and inferior parietal gray matter in individuals with low TLR4 methylation, but no relationship between alcohol and gray matter in the high methylation group.
Conclusions:
These findings suggest that TLR4 methylation may be protective against the damage conferred by alcohol on precuneus and inferior parietal gray matter, thereby implicating TLR4 for further investigation as a possible AUD treatment target.
Neuroimaging research supports the negative impact of chronic alcohol exposure on brain structure and function, and considerable evidence indicates an association between alcohol use disorders (AUDs) and gray matter loss (e.g., Paul et al., 2008; Pfefferbaum et al., 1992). Although the molecular mechanism(s) driving the effects of alcohol on the brain are not fully understood, human and animal studies implicate neuroinflammation as a contributing factor (Crews et al., 2015). In brief, inflammation occurs in the brain and periphery to defend against toxins such as ethanol. The immune system relies on pattern-recognition receptors, such as toll-like receptor 4 (TLR4), to identify pathogens and activate immune cells. Ethanol and its metabolites can bind to TLR4 in the brain and initiate neuroinflammatory signaling cascades (Alfonso-Loeches et al., 2010; Lewis et al., 2013).
In the normal immune response, downstream mediators of this cascade serve a protective, regulatory function. However, uncontrolled activation of the TLR4-mediated inflammatory response (e.g., because of chronic ethanol use) is associated with neuroinflammation, reduced neuroprotection and neuronal repair, and increased neurodegeneration (Guerri & Pascual, 2013). Individuals with AUDs suffer pronounced damage in frontal brain regions (Moselhy et al., 2001). Therefore, examining TLR4 activity in these regions has become a topic of recent AUD studies.
Of note, intermittent ethanol was associated with increased frontal cortical TLR4 in mice (Vetreno & Crews, 2012), and binge-like ethanol administration in rats increased TLR4 gene expression in the prefrontal cortex (PFC), which was linked to increased inflammatory cytokines and alterations in myelin protein levels (Pascual et al., 2014). Further, eliminating TLR4 receptors in mice was shown to prevent PFC damage and cognitive deficits following ethanol treatment (Montesinos et al., 2015), thereby strengthening the link between alcohol-related TLR4 activity and frontal brain damage. Evidence supporting this link in humans is preliminary, but one study found an association between age at drinking onset and upregulated TLR4 in rodent and human postmortem orbitofrontal cortex (OFC; Vetreno et al., 2013). Given that preliminary evidence lends support for the link between alcohol and frontal TLR4 activity in humans, further investigation of the role of TLR4 in mediating alcohol-related frontal damage is warranted.
Relatedly, epigenetic mechanisms including DNA methylation may mediate neuroadaptations via impacting gene transcription (Starkman et al., 2012). Methylation of genes that code for molecular mediators of the inflammatory cascade may be particularly relevant in AUDs. Indeed, TLR4 is hypermethylated in AUD subjects compared with controls (Hagerty et al., 2016). Another study found that binge-like ethanol treatment in adolescent mice produced epigenetic changes associated with increased anxiety, altered reward responses, and increased alcohol preference (Montesinos et al., 2016). This study also found that TLR4-deficient mice were protected from molecular and behavioral alterations of ethanol in the brain, indicating a role for TLR4 in the epigenetic changes and behavioral effects of alcohol.
The present study explores whether TLR4 methylation moderates the effect of alcohol on gray matter thickness in four regions of interest (ROIs). The precuneus, superior temporal (ST), and inferior parietal (IP) were selected because research has linked alcohol use to functional impairments and decreased volume in these regions (e.g., Courtney et al., 2014; Fein et al., 2009; Momenan et al., 2012; Pfefferbaum et al., 1998; Telesford et al., 2015; Yang et al., 2016). Lateral OFC was chosen given the association between alcohol and TLR4 expression in the OFC in animals and postmortem human brain tissue (Vetreno et al., 2013). Further, in a study of 436 adults, we previously found negative associations between AUD and gray matter in these four areas (Thayer et al., 2016).
Following our work demonstrating that TLR4 is hypermethylated in AUD (Hagerty et al., 2016), and given that TLR4 methylation may suppress TLR4 expression, thereby downregulating neuroinflammatory signaling, we hypothesize that TLR4 methylation will confer protective benefits on gray matter. Thus, individuals with lower methylation will show a negative association between alcohol dependence severity and gray matter in the precuneus, IP, ST, and OFC compared to individuals with higher methylation.
Method
Sample and procedures
Participants were pooled from four studies on substance use that included saliva collection (for DNA methylation analysis) and neuroimaging. Alcohol dependence was not an inclusionary criterion for the present analysis, as our goal was to obtain a sample of individuals with a wide range of alcohol dependence symptom severity scores. Exclusionary criteria included brain injury, loss of consciousness, history of bipolar or psychotic disorder, or a positive pregnancy test. Subjects were instructed to abstain from smoking for 2 hours and drinking alcohol for 24 hours before scanning and were required to pass a breath alcohol analysis test before participation. Written informed consent, approved by the University of New Mexico Human Research Committee, was obtained from all participants. In total, 707 participants had complete structural imaging, alcohol use, and methylation data.
Measures
Subjects completed a demographics measure as part of a battery assessing substance use and psychological variables. Information from this measure was used for covariates in the present analyses. Subjects also completed the Alcohol Dependence Scale (ADS; Kivlahan et al., 1989), which assesses alcohol dependence symptom severity, and the Alcohol Use Disorders Identification Test (AUDIT; Saunders et al., 1993), a 10-item measure addressing current and lifetime alcohol problems. Subjects completed the Timeline Followback (TLFB; Sobell & Sobell, 1992), which obtains estimates of daily alcohol, cigarette, and drug use. This instrument requires subjects to recall the number of drinks consumed each day over the prior 60 or 90 days. Marijuana and tobacco use data were also collected from the TLFB (n = 701) for use as covariates. Cigarette use was defined as the average number of cigarettes per day (n = 701).
Because some subjects completed 90-day TLFB measures and some completed 60-day measures, all TLFB variables were adjusted to correspond to a 30-day TLFB. Because days of marijuana use was skewed, marijuana use was recoded following our prior work (Thayer et al., 2016), along a 5-point scale corresponding to none, less than once per week, about once or twice per week, about 3–5 times per week, and about 6–7 times per week. Most participants (n = 671) completed the Fagerström Test of Nicotine Dependence (FTND; Heatherton et al., 1991), which assesses severity of nicotine dependence.
Most subjects (n = 686) completed the Beck Depression Inventory-II (BDI-II; Beck et al., 1996), a 21-item measure of depression severity, and the Beck Anxiety Inventory (n = 683) (BAI; Beck & Steer, 1991), to assess for anxiety over the past week.
Image acquisition
Magnetic resonance imaging (MRI) was performed on a 3T Siemens Trio (Erlangen, Germany) whole-body scanner with a 12-channel radio frequency coil. A high-resolution T1-weighted structural image was acquired with a 5-echo multi-echo MP RAGE (magnetization-prepared rapid gradient-echo) sequence with TE (echo time) = 1.64, 3.50, 5.36, 7.22, and 9.08 milliseconds; TR (repetition time) = 2.53 seconds; TI (time for inversion) = 1.20 seconds; flip angle = 7°; NEX (number of excitations) = 1; slice thickness = 1 mm, 33 slices; FOV (field of view) = 256 × 256 mm; resolution = 256 × 256 × 176; voxel size =1 × 1 × 1 mm; and pixel bandwidth = 650 Hz.
Surface-based morphometry
FreeSurfer v5.1 (https://surfer.nmr.mgh.harvard.edu) was used to perform cortical reconstruction and segmentation. Methods included motion correction (Reuter et al., 2010), removal of non-brain tissue using a hybrid watershed/ surface deformation procedure (Ségonne et al., 2004), automated Talairach transformation, segmentation of cortical and subcortical structures (Fischl et al., 2002, 2004), intensity normalization (Sled et al., 1998), tessellation of the gray/white-matter boundary, automated topology correction (Fischl et al., 2001; Ségonne et al., 2007), surface deformation using intensity gradients, and parcellation into cortical and subcortical structures (Dale et al., 1999; Fischl & Dale, 2000). Visual quality control verified skull stripping and gray/white-matter boundary segmentation following procedures in previous work (Weiland et al., 2014). Average cortical thickness over parcellated cortical structures was extracted from automated outputs.
DNA collection, extraction, and storage
Two milliliters of saliva was collected using DNA Collection Kits (Cat. No. ORG-500) from DNA Genotek (Ottawa, Ontario, Canada; www.dnagenotek.com) and stored according to manufacturer’s guidelines until the time of DNA extraction. The DNA was extracted per manufacturer instructions using DNA Genotek’s prepIT DNA extraction kit (Cat. No. PT-L2P-45). DNA was quantified using Invitrogen’s (Thermo Fisher Scientific, Waltham, MA) Qubit™ dsDNA BR Assay Kit (Cat. No. Q-32853) and cryogenically stored at -80 °C.
Methylation assay
To determine the methylation of CpG (5’-C-phosphate-G-3’) sites near the TLR4 promotor, pyrosequencing was performed at EpigenDx (Worcester, MA). Pyrosequencing quantitatively monitors real-time incorporation of nucleotides through the enzymatic conversion of released pyrophosphate into a proportional light signal (Tost & Gut, 2007). The assay covered 4 CG dinucleotides in the first exon after the 5’ untranslated transcription start site, ranging from +27 to +51 in reference to the translational start site (Figure 1). Site analysis was based on the ability to generate primers located around CpG islands and that meet the requirements for accurate pyrosequencing. All primers are owned by EpigenDx (Hopkinton, MA).
Figure 1.
Assayed CpG sites fell within the first exon of the toll-like receptor 4 (TLR4) gene. Selected CpG sites were chosen on the basis of their proximity to the transcription start site (TSS) and location within an important regulatory region of the gene.
DNA was bisulfite treated using a proprietary bisulfite salt solution. When isolation from cells or tissue is carried out, cells are resuspended in 40 μL 1X proteinase K digestion buffer and digested with proteinase K (40 μg) at 50 °C for 30 minutes. Cell debris is pelleted by centrifugation at 14,000 × g for 10 minutes, and 20 μL is used in the bisulfite conversion reaction. DNA is diluted to 45 μL, and 5 μL of 3 N NaOH is added followed by a 30-minute incubation at 42 °C to denature the DNA. 100 μL of bisulfite salt solution is added to DNA and incubated for 14 hours at 50 °C. Bisulfite-treated DNA is purified using Zymogen DNA columns and eluted 20 μL of T1E0.2 pH 8.0, and 1 μL is used for each polymerase chain reaction (PCR).
PCR was performed with 0.2 μM of each primer, and one PCR primer was biotinylated to purify the final PCR product using Sepharose beads. The PCR product was bound to Streptavidin Sepharose HP (GE Healthcare Life Sciences, Pittsburgh, PA), and the Sepharose beads containing the immobilized PCR product were purified, washed, and denatured using a 0.2 M NaOH solution and rewashed using the Pyrosequencing Vacuum Prep Tool (Pyrosequencing, Qiagen). 0.2 μM pyrosequencing primer was annealed to the purified single-stranded PCR product. 10 μL of the PCR products were sequenced by Pyrosequencing PSQ96 HS System (Pyrosequencing, Qiagen) following the manufacturer’s instructions (Pyrosequencing, Qiagen, Germantown, MD). The methylation status of each locus was analyzed individually as a T/C SNP using QCpG software (Pyrosequencing, Qiagen) (Brakensiek et al., 2007; England & Pettersson, 2005; Liu et al., 2007).
Analyzed DNA was presented as percent methylation at each of the four TLR4 CpGs. These four CpGs were all highly correlated (all rs ≥ .98). Thus, the percent methylation at each CpG was averaged to form the average methylation score used in regressions. Using the average of these highly correlated variables was preferable to examining each methylation site independently, thus reducing measurement error and decreasing the likelihood of type I error.
Gray matter regions of interest
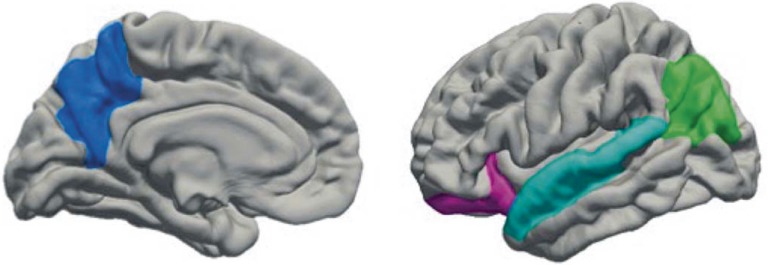
Our aim was to extend prior findings (Thayer et al., 2016) linking alcohol to gray matter by examining TLR4 methylation as a moderator. Because the 436 subjects in the previous study comprise a subset of the 707 subjects included in the present analysis, we have chosen to examine gray matter in brain regions that were associated with alcohol use in our prior study, to focus on the involvement of methylation. ROI selection was also informed by prior research. The gray matter variables tested in Thayer et al. (2016) were calculated by multiplying surface area and cortical thickness. Follow-up analyses indicated that cortical thickness was driving the effect. Thus, we examined cortical thickness across these four regions (Figure 2).
Figure 2.
Gray matter regions of interest. Freesurfer regions of interest (bilateral, but illustrated here in the left hemisphere) were precuneus (blue), lateral orbitofrontal (magenta), superior temporal (cyan), and inferior parietal (green).
Data analyses
Regression models.
Using ordinary least squares multiple regression, we regressed brain variables on age, gender, intracranial volume, ADS total, mean-centered TLR4 methylation (averaged across the 4 CpGs), and ADS × TLR4 interaction. We regressed each region independently on the same predictors, examining right and left hemispheres separately, modeling a total of eight equations. To examine whether results are consistent across multiple measures of alcohol dependence, we ran the same eight models replacing ADS with AUDIT total. To examine whether results were consistent with a measure of alcohol consumption, we ran the same models using TLFB drinks per drinking day as the alcohol variable.
Follow-up analyses.
For all significant interactions, post hoc analyses were planned to clarify the direction of effects. High and low methylation groups were created based on the median methylation score, and high and low drinking groups were created based on the median ADS score. This procedure was repeated using the AUDIT total.
Results
Sample characteristics
Participants were predominantly male (62.4%), with a mean age of 31.13 years (SD = 9.49; range: 18–56; Table 1), and were representative of the racial background in the southwestern United States. Participants spanned a wide range of alcohol dependence severity, with ADS total scores from 0 to 43 (maximum = 47). AUDIT scores ranged from 0 to 39 (maximum = 40), and 72.7% of subjects scored 8 or more, which is the standard cutoff indicating hazardous consumption. A total of 13.6% of subjects endorsed anxiety symptoms of moderate or greater severity, and 13.4% endorsed depression symptoms of moderate or greater severity. Sixty-four percent reported smoking cigarettes at least once in the period covered by the TLFB, and 41% reported using marijuana at least once. Age and ADS total score were not significantly correlated controlling for sex, marijuana use, cigarette use, depression, and anxiety. However, age and AUDIT score were correlated (r = .144, p < .001) controlling for these covariates.
Table 1.
Sample demographics
| Characteristic | M (SD) or % |
| Age | 31.1 (9.5) |
| Male gender | 62.40% |
| Intracranial volume, mm3 | 1,551,550.6 (226,949.2) |
| Race | |
| White | 56.30% |
| Latino | 15.70% |
| Biracial/mixed | 13.20% |
| Native American | 7.60% |
| Black | 2.70% |
| Asian/Pacific Islander | 1% |
| Unknown/declined to state | 3.40% |
| ADS total score | 9.2 (7.4) |
| AUDIT total score | 13.2 (8.0) |
| TLFB | |
| Drinks per drinking day | 5.8 (3.8) |
| Total drinks | 81.37 (85.18) |
| Total drinking days | 13.08 (8.73) |
| Marijuana use frequency | 1.9 (1.3) |
| Cigarettes per day | 6.2 (8.2) |
| BDI total score | 9.5 (8.4) |
| BAI total score | 7.9 (8.2) |
| FTND total score | 2.27 (2.69) |
Notes: ADS = Alcohol Dependence Score; AUDIT=Alcohol Use Disorders Identification Test; TLFB = Timeline Followback; BDI = Beck Depression Inventory; BAI = Beck Anxiety Inventory; FTND = Fagerstrom Test for Nicotine Dependence.
Alcohol and TLR4 methylation
In contrast to prior results demonstrating increased TLR4 methylation in AUD versus control individuals (Hagerty et al., 2016), we did not observe a significant correlation between alcohol dependence symptom severity and methylation. However, given the wide range of alcohol use in the present sample, we created “extreme groups” based on ADS score. Using an independent samples t test, we compared TLR4 methylation in the 55 individuals who obtained a score of 0 on the ADS to the 50 individuals with the highest ADS score (21 or above). We observed a difference in methylation between the high ADS group (M = 24.56, SD = 14.95) and the low ADS group (M = 19.72, SD = 9.75) that was significant at the trend level, t(83)= -1.944, p = .055.
Regression results
In the models reported below, slope values are reported as standardized coefficients. Significance was set at p < .05 before correcting for multiple tests. In the model in which right IP was the criterion, the predictors accounted for 27.0% of the variance in gray matter, and a significant ADS × TLR4 interaction emerged (b = .224), t(700) = 2.796, p = .005. In the left precuneus model, the predictors accounted for 28.5% of the variance in gray matter, and there was a significant ADS × TLR4 interaction (b = .238), t(700) = 3.001, p = .003. In the right precuneus model, predictors accounted for 25.1% of the variance in gray matter, and there was a significant ADS × TLR4 interaction (b = .192), t(700) = 2.372, p = .018.
The same significant interactions emerged when each TLR4 CpG was included in the model separately instead of the TLR4 average. Trend-level interactions emerged in models predicting left ST (b = .140), t(700) = 1.701, p = .089; left IP (b = .158), t(700) = 1.906, p = .057; and right OFC (b = .140), t(700) = 1.877, p = .061. Bonferroni-corrected p value threshold was set at .00625 (.05/8). Using this threshold, interactions predicting right IP and left precuneus remained significant, even controlling for BAI total, BDI total, marijuana smoking days, and cigarettes per day and FTND total.
Using AUDIT as the predictor, significant AUDIT × TLR4 interactions emerged for the left precuneus region (b = .219), t(700) = 2.644, p = .008; left IP region (b = .183), t(700) = 2.117, p = .035; and right IP region (b = .252), t(700) = 3.016, p = .003. Only the interaction in the right IP model passed correction. When covariates were included, the interaction remained significant. When covariates were included in the left precuneus model, the interaction term also passed correction (b = .257), t(642) = 2.994, p = .003.
Alcohol consumption regression results
We observed a significant Methylation × TLFB interaction passing Bonferroni correction in the right IP model. The predictors accounted for 27.2% of the variance in gray matter, and there was a significant TLFB × TLR4 interaction (b = .106), t(670) = 3.119, p = .002. When covariates were included, the interaction passed correction.
TLR4 × ADS interaction
To explore the nature of the significant interactions, we created high and low methylation groups based on the median methylation score of 19.92, and high and low ADS groups based on the median ADS score of 8. The mean gray matter values for right IP and left precuneus for individuals in each of four groups are plotted in Figure 3: high drinking/ high methylation (n = 164), high drinking/low methylation (n = 163), low drinking/high methylation (n = 190), and low drinking/low methylation (n = 190). We also examined partial correlations between gray matter and ADS (controlling for age, sex, depression, anxiety, marijuana use, and cigarette use) within high and low methylation groups. No significant association was observed in the high methylation group, but there was a negative correlation between ADS and left precuneus (r = -.208, p < .001) and right IP (r = -.205, p < .001) in the low methylation group.
Figure 3.
Gray matter thickness across alcohol and toll-like receptor 4 (TLR4) methylation groups. (A) Left precuneus thickness in high and low TLR4 methylation groups across individuals with high and low Alcohol Dependence Scale (ADS) scores. (B) Right inferior parietal thickness in high and low TLR4 methylation groups across individuals with high and low ADS scores. (C) Left precuneus thickness in high and low TLR4 methylation groups across individuals with high and low Alcohol Use Disorders Identification Test (AUDIT) scores. (D) Right inferior parietal thickness in high and low TLR4 methylation groups across individuals with high and low AUDIT scores.
TLR4 × AUDIT interaction
To explore the consistency of our results across measures, we created high and low drinking groups based on the median AUDIT score of 12. The mean gray matter values for right IP and left precuneus for individuals in each of four groups are plotted in Figure 3: high drinking/high methylation (n = 167), high drinking/low methylation (n = 171), low drinking/high methylation (n = 187), and low drinking/low methylation (n = 182). We examined the partial correlation between gray matter and AUDIT (controlling for age, sex, depression, anxiety, marijuana, and cigarette use) within high and low methylation groups. No significant association was observed in the high methylation group, but there was a negative correlation between AUDIT and gray matter in left precuneus (r = -.249, p < .001) and right IP (r = -.280, p < .001) in the low methylation group.
Discussion
The present study examined whether TLR4 methylation moderates the relationship between alcohol use severity and gray matter. We hypothesized that greater methylation would be associated with decreased alcohol-related gray matter damage. We observed significant interactions between methylation and alcohol dependence symptom severity predicting gray matter in precuneus and IP ROIs in the direction consistent with our hypothesis. Further, the significant interaction observed between TLFB alcohol consumption and TLR4 methylation suggests that methylation may influence alcohol-related damage in the IP region, perhaps in a dose-dependent fashion. Contrary to our hypotheses, we did not observe significant interactions predicting gray matter in the lateral OFC or ST regions. Given that the OFC was chosen for the present analysis based on preliminary associations between alcohol and TLR4 expression in animal and human postmortem samples (Vetreno et al., 2013), it is possible that this region is not significantly structurally affected by TLR4 activity. In addition, it should be noted that we did observe a trend-level interaction in the ADS model for which left ST was the criterion. Thus, it is possible that the effect of methylation in this region is small and should be investigated further, perhaps using a more severe clinical sample.
Of note, in contrast to previous findings demonstrating TLR4 hypermethylation in AUD cases versus controls (Hagerty et al., 2016), we did not observe an association between ADS or AUDIT and methylation. However, we hypothesize that the discrepancy may be because individuals in the Hagerty et al. (2016) study met the clinical criteria for alcohol dependence (which was not an inclusionary criterion for the present study), and alcohol-related TLR4 methylation changes may be subtle and more easily observable when comparing severe drinkers to healthy controls. This hypothesis is supported by our near-significant findings from a t test comparing individuals with the lowest versus highest ADS scores. Given that the “low ADS” group comprised individuals recruited for studies on substance use, these subjects likely do not represent true “healthy controls.” Thus, methylation differences may become more apparent when comparing heavy drinkers with healthy individuals. Future work is needed to investigate this question. Last, although greater symptom severity did not predict methylation, methylation did affect the association between alcohol severity and gray matter, suggesting that among drinkers, TLR4 methylation may be a pre-existing, individual difference variable that influences the amount of alcohol-related damage incurred, but is not necessarily influenced by alcohol in a dose-response fashion.
Gaining a deeper understanding of this relationship could have important treatment implications. If TLR4 methylation is a pre-existing individual difference variable, then lower methylation could be considered a risk factor for more severe AUD presentation, perhaps via promoting alcohol-induced cortical damage and subsequent inhibitory control deficits, which in turn promote relapse. TLR4 methylation status could also be examined in a prevention context to identify adolescents or adults at risk for AUD. Further, TLR4 could be targeted pharmacologically among heavy drinkers to mitigate the degree of alcohol-induced cortical damage and perhaps attenuate the cognitive deficits and behavioral sequelae associated with problem drinking and relapse. Further research is needed to inform these potential prevention and treatment options and better characterize the relationship between TLR4 methylation and alcohol-related neural damage across different populations such as adolescents, light drinkers, heavy drinkers, and individuals with chronic alcohol dependence.
Regarding our hypothesis that TLR4 methylation moderates damage conferred by alcohol on gray matter, we observed no significant association between ADS/AUDIT and gray matter in the high methylation group, but we did observe a negative correlation between ADS/AUDIT and gray matter in the low methylation group, suggesting that methylation may protect against gray matter damage in AUD (given that those with greater methylation did not show gray matter damage associated with the ADS/AUDIT score, whereas those with lower methylation show decreased gray matter associated with alcohol use severity). Assuming that TLR4 methylation affects gene expression, results support that TLR4 methylation may downregulate TLR4-mediated inflammatory signaling, thereby mitigating gray matter loss. Although we did not measure TLR4 expression, animal evidence shows that TLR4 methylation is associated with altered TLR4 expression. For example, enhanced TLR4 expression was accompanied by TLR4 promotor demethylation in the context of a systemic inflammatory response (Chang et al., 2015). These findings provide preliminary evidence that TLR4 methylation may decrease TLR4 expression and thereby protect the brain from the negative effects of TLR4-mediated inflammatory signaling in AUDs.
An important next step is to examine functional consequences of TLR4 methylation, particularly regarding IP and precuneus activation during executive functioning tasks. Prior work has demonstrated differences between AUD and controls in these regions across various cognitive tasks. For example, alcohol-dependent individuals show greater IP activation during anticipation of a stop signal (Hu et al., 2015), and AUD severity was negatively associated with IP activation during response inhibition (Claus et al., 2013). In addition, alcohol use was associated with altered precuneus activation during the Stroop task (Hatchard et al., 2015), and AUD severity was positively correlated with precuneus responses during alcohol-taste tasks (Claus et al., 2011; Courtney et al., 2014). Thus, alcohol may be associated with disrupted neural activation in regions important for cognitive control and cue reactivity. Following our results suggesting that TLR4 methylation moderates the relationship between alcohol and gray matter in these ROIs, future functional neuroimaging studies should investigate TLR4 methylation as a moderating factor in the context of alcohol-related functional impairments.
Several methodological issues limit the interpretation of our results. First, we assume that TLR4 methylation is associated with decreased TLR4 expression in the brain, although expression in human brain tissue cannot be quantified in vivo. Animal studies have shown epigenetic downregulation of TLR4 gene expression in peripheral tissues (Takahashi et al., 2009; Zampetaki et al., 2006), but future work is needed to determine whether methylation decreases TLR4 expression in the human brain. Further, bisulfite sequencing cannot differentiate between methylcytosine variants (e.g., 5-hydroxylmethylcytosine) (Huang et al., 2010), which may have distinct regulatory effects (e.g., Wen et al., 2014; Wu et al., 2011). This limitation further contributes to the uncertainty of the relationship between TLR4 methylation and expression and should be addressed in future work.
In addition, although methylation sites were selected based on proximity to the transcription start site and location within a TLR4 regulatory region, we measured methylation in a small region of the gene, and it is possible that methylation within another region would confer stronger effect sizes. Further, it is unclear how methylation at each individual TLR4 CpG site may influence gene expression. In addition, methylation assays were conducted on buccal cells. However, studies have demonstrated an association between buccal and brain tissue methylation (Smith et al., 2015), and we previously found methylation of CpGs (including in TLR4) that differed significantly between AUD and control subjects was highly consistent across brain and buccal tissue (Hagerty et al., 2016).
Finally, our sample included individuals reporting a wide range of alcohol consumption and dependence severity scores. Although our inclusion of individuals who do not meet alcohol dependence criteria may limit clinical interpretations, sampling a large number of individuals encompassing the full range of possible alcohol use behaviors increases our power to detect effects (Cohen et al., 2003) and increases the generalizability of results to lighter and heavier drinkers. Given that a large percentage of our sample reported hazardous drinking on the AUDIT, it seems likely that these results are relevant for alcohol-dependent individuals. Further research focusing specifically on clinical populations is an important next step.
In summary, we provide initial evidence that TLR4 methylation moderates the relationship between alcohol and brain structure, perhaps via downregulating alcohol-related inflammatory signaling. Future work examining how TLR4 methylation and gene expression in neural tissue affects brain structure and/or function could shed further light on the role of TLR4 methylation in AUD. If TLR4 methylation is a plausible mechanism for decreasing the neuroinflammatory effects of alcohol on gray matter, TLR4 may represent a key target for future pharmacological AUD treatments.
Footnotes
This work is supported by National Science Foundation Graduate Research Fellowships (US; Division of Graduate Education No. 1144083) to Hollis C. Karoly and Sarah L. Hagerty. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Science Foundation. This work is also supported by National Institute on DrugAbuse Grant R36DA040020 to Rachel E. Thayer and R01DA025074 to Kent E. Hutchison, and National Institute on Alcohol Abuse and Alcoholism Grants R01AA012238 and 5R01AA014886-06 to Kent E. Hutchison.
References
- Alfonso-Loeches S., Pascual-Lucas M., Blanco A. M., Sanchez-Vera I., Guerri C. Pivotal role of TLR4 receptors in alcohol-induced neuroinflammation and brain damage. Journal of Neuroscience. 2010;30:8285–8295. doi: 10.1523/JNEUROSCI.0976-10.2010. doi:10.1523/JNEUROSCI.0976-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck A. T., Steer R. A. Relationship between the Beck anxiety inventory and the Hamilton anxiety rating scale with anxious outpatients. Journal of Anxiety Disorders. 1991;5:213–223. doi:10.1016/0887-6185(91)90002-B. [Google Scholar]
- Beck A., Steer R., Brown G. San Antonio, TX: Psychological Corporation; 1996. Manual for the BDI-II. [Google Scholar]
- Brakensiek K., Wingen L. U., Länger F., Kreipe H., Lehmann U.2007Quantitative high-resolution CpG island mapping with Pyrose-quencing reveals disease-specific methylation patterns of the CDKN2B gene in myelodysplastic syndrome and myeloid leukemia Clinical Chemistry 5317–23.. doi:10.1373/clinchem.2007.072629 [DOI] [PubMed] [Google Scholar]
- Chang G., Zhuang S., Seyfert H.-M., Zhang K., Xu T., Jin D., Shen X. Hepatic TLR4 signaling is activated by LPS from digestive tract during SARA, and epigenetic mechanisms contribute to enforced TLR4 expression. Oncotarget. 2015;6:38578–38590. doi: 10.18632/oncotarget.6161. doi:10.18632/oncotarget.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus E. D., Ewing S. W., Filbey F. M., Sabbineni A., Hutchison K. E. Identifying neurobiological phenotypes associated with alcohol use disorder severity. Neuropsychopharmacology. 2011;36:2086–2096. doi: 10.1038/npp.2011.99. doi:10.1038/npp.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus E. D., Feldstein Ewing S. W., Filbey F. M., Hutchison K. E. Behavioral control in alcohol use disorders: Relationships with severity. Journal of Studies on Alcohol and Drugs. 2013;74:141–151. doi: 10.15288/jsad.2013.74.141. doi:10.15288/jsad.2013.74.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J., Cohen P., West S. G., Aiken L. S. 3rd ed. Mahwah, NJ: Lawrence Erlbaum Associates; 2003. Applied multiple regression/correlation analysis for the behavioral sciences. [Google Scholar]
- Courtney K. E., Ghahremani D. G., London E. D., Ray L. A. The association between cue-reactivity in the precuneus and level of dependence on nicotine and alcohol. Drug and Alcohol Dependence. 2014;141:21–26. doi: 10.1016/j.drugalcdep.2014.04.026. doi:10.1016/j.drugalcdep.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews F. T., Sarkar D. K., Qin L., Zou J., Boyadjieva N., Vetreno R. P. Neuroimmune function and the consequences of alcohol exposure. Alcohol Research: Current Reviews. 2015;37:331–351. [PMC free article] [PubMed] [Google Scholar]
- Dale A. M., Fischl B., Sereno M. I. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. doi:10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- England R., Pettersson M.2005Pyro Q-CpG™: Quantitative analysis of methylation in multiple CpG sites by Pyrosequencing® [Application Note] Nature Methods 2. doi:10.1038/nmeth800 [Google Scholar]
- Fein G., Shimotsu R., Chu R., Barakos J. Parietal gray matter volume loss is related to spatial processing deficits in long-term abstinent alcoholic men. Alcoholism: Clinical and Experimental Research. 2009;33:1806–1814. doi: 10.1111/j.1530-0277.2009.01019.x. doi:10.1111/j.1530-0277.2009.01019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Dale A. M. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. doi:10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Liu A., Dale A. M. Automated manifold surgery: Constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Transactions on Medical Imaging. 2001;20:70–80. doi: 10.1109/42.906426. doi:10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- Fischl B., Salat D. H., Busa E., Albert M., Dieterich M., Haselgrove C., Dale A. M. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. doi:10.1016/S0896-6273(02)00569-X. [DOI] [PubMed] [Google Scholar]
- Fischl B., van der Kouwe A., Destrieux C., Halgren E., Ségonne F., Salat D. H., Dale A. M. Automatically parcellating the human cerebral cortex. Cerebral Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. doi:10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Guerri C., Pascual M. Role of toll-like receptor 4 in alcohol-induced neuroinflammation and behavioral dysfunctions. In: Cui C., Grandison L., Noronha A., editors. Neural-immune interactions in brain function and alcohol related disorders. New York, NY: Springer; 2013. pp. 279–306. [Google Scholar]
- Hatchard T., Smith A. M., Halchuk R. E., Longo C. A., Fried P. A., Hogan M. J., Cameron I. Effects of low-level alcohol use on cognitive interference: An fMRI study in young adults. Alcohol. 2015;49:7–13. doi: 10.1016/j.alcohol.2014.07.020. doi:10.1016/j.alcohol.2014.07.020. [DOI] [PubMed] [Google Scholar]
- Hagerty S. L., Bidwell L., Harlaar N., Hutchison K. E. An exploratory association study of alcohol use disorder and DNA methylation. Alcoholism: Clinical and Experimental Research. 2016;40:1633–1640. doi: 10.1111/acer.13138. doi:10.1111/acer.13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heatherton T. F., Kozlowski L. T., Frecker R. C., Fagerström K. O. The Fagerstrom Test for Nicotine Dependence: A revision of the Fagerstrom Tolerance Questionnaire. British Journal of Addiction. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. doi:10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- Hu S., Ide J. S., Zhang S., Sinha R., Li C.-S. R. Conflict anticipation in alcohol dependence—A model-based fMRI study of stop signal task. Neuroimage: Clinical. 2015;8:39–50. doi: 10.1016/j.nicl.2015.03.008. doi:10.1016/j.nicl.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Pastor W. A., Shen Y., Tahiliani M., Liu D. R., Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE. 2010;5(1):e8888. doi: 10.1371/journal.pone.0008888. doi:10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivlahan D. R., Sher K. J., Donovan D. M. The Alcohol Dependence Scale: A validation study among inpatient alcoholics. Journal of Studies on Alcohol. 1989;50:170–175. doi: 10.15288/jsa.1989.50.170. doi:10.15288/jsa.1989.50.170. [DOI] [PubMed] [Google Scholar]
- Lewis S. S., Hutchinson M. R., Zhang Y., Hund D. K., Maier S. F., Rice K. C., Watkins L. R. Glucuronic acid and the ethanol metabolite ethyl-glucuronide cause toll-like receptor 4 activation and enhanced pain. Brain, Behavior, and Immunity. 2013;30:24–32. doi: 10.1016/j.bbi.2013.01.005. doi:10.1016/j.bbi.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Zhang X., So C.-K., Wang S., Wang P, Yan L., Chen X. Regulation of Cdx2 expression by promoter methylation, and effects of Cdx2 transfection on morphology and gene expression of human esophageal epithelial cells. Carcinogenesis. 2007;28:488–496. doi: 10.1093/carcin/bgl176. doi:10.1093/arcin/bgl176. [DOI] [PubMed] [Google Scholar]
- Momenan R., Steckler L. E., Saad Z. S., van Rafelghem S., Kerich M. J., Hommer D. W. Effects of alcohol dependence on cortical thickness as determined by magnetic resonance imaging. Psychiatry Research: Neuroimaging. 2012;204:101–111. doi: 10.1016/j.pscychresns.2012.05.003. doi:10.1016/j.pscychresns.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Montesinos J., Pascual M., Pla A., Maldonado C., Rodríguez-Arias M., Miñarro J., Guerri C. TLR4 elimination prevents synaptic and myelin alterations and long-term cognitive dysfunctions in adolescent mice with intermittent ethanol treatment. Brain, Behavior, and Immunity. 2015;45:233–244. doi: 10.1016/j.bbi.2014.11.015. doi:10.1016/j.bbi.2014.11.015. [DOI] [PubMed] [Google Scholar]
- Montesinos J., Pascual M., Rodríguez-Arias M., Miñarro J., Guerri C. Involvement of TLR4 in the long-term epigenetic changes, rewarding and anxiety effects induced by intermittent ethanol treatment in adolescence. Brain, Behavior, and Immunity. 2016;53:159–171. doi: 10.1016/j.bbi.2015.12.006. doi:10.1016/j.bbi.2015.12.006. [DOI] [PubMed] [Google Scholar]
- Moselhy H. F., Georgiou G., Kahn A. Frontal lobe changes in alcoholism: A review of the literature. Alcohol and Alcoholism. 2001;36:357–368. doi: 10.1093/alcalc/36.5.357. doi:10.1093/alcalc/36.5.357. [DOI] [PubMed] [Google Scholar]
- Pascual M., Pla A., Miñarro J., Guerri C. Neuroimmune activation and myelin changes in adolescent rats exposed to high-dose alcohol and associated cognitive dysfunction: A review with reference to human adolescent drinking. Alcohol and Alcoholism. 2014;49:187–192. doi: 10.1093/alcalc/agt164. doi:10.1093/alcalc/agt164. [DOI] [PubMed] [Google Scholar]
- Paul C. A., Au R., Fredman L., Massaro J. M., Seshadri S., Decarli C., Wolf P. A. Association of alcohol consumption with brain volume in the Framingham study. Archives of Neurology. 2008;65:1363–1367. doi: 10.1001/archneur.65.10.1363. doi:10.1001/archneur.65.10.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum A., Lim K. O., Zipursky R. B., Mathalon D. H., Rosenbloom M. J., Lane B., Sullivan E. V. Brain gray and white matter volume loss accelerates with aging in chronic alcoholics: A quantitative MRI study. Alcoholism: Clinical and Experimental Research. 1992;16:1078–1089. doi: 10.1111/j.1530-0277.1992.tb00702.x. doi:10.1111/j.1530-0277.1992.tb00702.x. [DOI] [PubMed] [Google Scholar]
- Pfefferbaum A., Sullivan E. V., Rosenbloom M. J., Mathalon D. H., Lim K. O. A controlled study of cortical gray matter and ventricular changes in alcoholic men over a 5-year interval. Archives of General Psychiatry. 1998;55:905–912. doi: 10.1001/archpsyc.55.10.905. doi:10.1001/archpsyc.55.10.905. [DOI] [PubMed] [Google Scholar]
- Reuter M., Rosas H. D., Fischl B. Highly accurate inverse consistent registration: A robust approach. NeuroImage. 2010;53:1181–1196. doi: 10.1016/j.neuroimage.2010.07.020. doi:10.1016/j.neuroimage.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders J. B., Aasland O. G., Babor T. F., de la Fuente J. R., Grant M. Development of the Alcohol Use Disorders Identification Test (AUDIT): WHO Collaborative Project on early detection of persons with harmful alcohol consumption-II. Addiction. 1993;88:791–804. doi: 10.1111/j.1360-0443.1993.tb02093.x. doi:10.1111/j.1360-0443.1993.tb02093.x. [DOI] [PubMed] [Google Scholar]
- Ségonne F., Dale A. M., Busa E., Glessner M., Salat D., Hahn H. K., Fischl B. A hybrid approach to the skull stripping problem in MRI. NeuroImage. 2004;22:1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. doi:10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Ségonne F., Pacheco J., Fischl B. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Transactions on Medical Imaging. 2007;26:518–529. doi: 10.1109/TMI.2006.887364. doi:10.1109/TMI.2006.887364. [DOI] [PubMed] [Google Scholar]
- Sled J. G., Zijdenbos A. P., Evans A. C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Transactions on Medical Imaging. 1998;17:87–97. doi: 10.1109/42.668698. doi:10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Smith A. K., Kilaru V., Klengel T., Mercer K. B., Bradley B., Conneely K. N., Binder E. B. DNA extracted from saliva for methylation studies of psychiatric traits: Evidence tissue specificity and relatedness to brain. American Journal of Medical Genetics: Part B, Neuropsychiatric Genetics. 2015;168:36–44. doi: 10.1002/ajmg.b.32278. doi:10.1002/ajmg.b.32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell L. C., Sobell M. B. Timeline follow-back: A technique for assessing self-reported alcohol consumption. In: Litten R. Z., Allen J. P., editors. Measuring alcohol consumption. New York, NY: Humana Press; 1992. pp. 41–72. doi:10.1007/978-1-4612-0357-5_3. [Google Scholar]
- Starkman B. G., Sakharkar A. J., Pandey S. C. Epigenetics— beyond the genome in alcoholism. Alcohol Research: Current Reviews. 2012;34:293–305. [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Sugi Y., Hosono A., Kaminogawa S. Epigenetic regulation of TLR4 gene expression in intestinal epithelial cells for the maintenance of intestinal homeostasis. Journal of Immunology. 2009;183:6522–6529. doi: 10.4049/jimmunol.0901271. doi:10.4049/jimmunol.0901271. [DOI] [PubMed] [Google Scholar]
- Thayer R. E., Hagerty S. L., Sabbineni A., Claus E. D., Hutchison K. E., Weiland B. J. Negative and interactive effects of sex, aging, and alcohol abuse on gray matter morphometry. Human Brain Mapping. 2016 doi: 10.1002/hbm.23172. doi:10.1002/hbm.23172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesford Q. K., Laurienti P. J., Davenport A. T., Friedman D. P., Kraft R. A., Daunais J. B. The effects of chronic alcohol self-administration in nonhuman primate brain networks. Alcoholism: Clinical and Experimental Research. 2015;39:659–671. doi: 10.1111/acer.12688. doi:10.1111/acer.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tost J., Gut I. G. DNA methylation analysis by pyrosequencing. Nature Protocols. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. doi:10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- Vetreno R. P., Crews F. T. Adolescent binge drinking increases expression of the danger signal receptor agonist HMGB1 and toll-like receptors in the adult prefrontal cortex. Neuroscience. 2012;226:475–488. doi: 10.1016/j.neuroscience.2012.08.046. doi:10.1016/j.neuroscience.2012.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetreno R. P., Qin L., Crews F. T. Increased receptor for advanced glycation end product expression in the human alcoholic prefrontal cortex is linked to adolescent drinking. Neurobiology of Disease. 2013;59:52–62. doi: 10.1016/j.nbd.2013.07.002. doi:10.1016/j.nbd.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland B. J., Korycinski S. T., Soules M., Zubieta J.-K., Zucker R. A., Heitzeg M. M. Substance abuse risk in emerging adults associated with smaller frontal gray matter volumes and higher externalizing behaviors. Drug and Alcohol Dependence. 2014;137:68–75. doi: 10.1016/j.drugalcdep.2014.01.005. doi:10.1016/j.drugalcdep.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Li X., Yan L., Tan Y., Li R., Zhao Y., Qiao J. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biology. 2014;15:R49. doi: 10.1186/gb-2014-15-3-r49. doi:10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., D’Alessio A. C., Ito S., Wang Z., Cui K., Zhao K., Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes & Development. 2011;25:679–684. doi: 10.1101/gad.2036011. doi:10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Tian F., Zhang H., Zeng J., Chen T., Wang S., Gong Q. Cortical and subcortical gray matter shrinkage in alcohol-use disorders: A voxel-based meta-analysis. Neuroscience & Biobehavioral Reviews. 2016;66:92–103. doi: 10.1016/j.neubiorev.2016.03.034. doi:10.1016/j.neubiorev.2016.03.034. [DOI] [PubMed] [Google Scholar]
- Zampetaki A., Xiao Q., Zeng L., Hu Y., Xu Q. TLR4 expression in mouse embryonic stem cells and in stem cell-derived vascular cells is regulated by epigenetic modifications. Biochemical and Biophysical Research Communications. 2006;347:89–99. doi: 10.1016/j.bbrc.2006.06.055. doi:10.1016/j.bbrc.2006.06.055. [DOI] [PubMed] [Google Scholar]