Abstract
Secreted frizzled-related protein 2 (SFRP2) is a pro-angiogenic factor expressed in the vasculature of a wide variety of human tumors, and modulates angiogenesis via the calcineurin-dependent Nuclear Factor of Activated T-cells cytoplasmic 3 (NFATc3) pathway in endothelial cells. However, until now, SFRP2 receptor for this pathway was unknown. In the present study, we first used amino acid alignments and molecular modeling to demonstrate that SFRP2 interaction with frizzled-5 (FZD5) is typical of Wnt/FZD family members. To confirm this interaction, we performed co-immunofluorescence, co-immunoprecipitation, and ELISA binding assays, which demonstrated SFRP2/FZD5 binding. Functional knock-down studies further revealed that FZD5 is necessary for SFRP2-induced tube formation and intracellular calcium flux in endothelial cells. Using protein analysis on endothelial cell nuclear extracts, we also discovered that FZD5 is required for SFRP2-induced activation of NFATc3. Our novel findings reveal that FZD5 is a receptor for SFRP2, and mediates SFRP2-induced angiogenesis via calcineurin/NFATc3 pathway in endothelial cells.
Keywords: Wnt, calcium signaling, endothelial cells, calcineurin, angiosarcoma
INTRODUCTION
Secreted frizzled-related proteins (SFRPs) are important regulators of a variety of developmental and pathologic processes. The preponderance of in vivo evidence supports that SFRP2 stimulates tumor growth. Gain of function studies have shown that SFRP2 strongly promotes tumor growth of intracranial glioma [1], renal cell carcinoma [2], lung cancer [3], melanoma [4], and osteosarcoma [5]. Overexpression of transfected SFRP2 in MCF7 breast adenocarcinoma cells increased their resistance to apoptotic signals in vitro [6]. Likewise, SFRP2 is a key factor in chemotherapy resistance of damaged tumor microenvironment [7].
One mechanism through which SFRP2 stimulates tumor growth is through induction of angiogenesis. SFRP2 protein is overexpressed in the vasculature of a wide variety of human tumors, including breast cancer, angiosarcoma, prostate cancer, hepatocellular carcinoma, lung cancer, renal cell carcinoma, ovarian cancer, and pancreatic cancer [8,9]. SFRP2 stimulates angiogenesis in vitro by increasing endothelial cell tube formation and migration, and, in vivo, in a mouse Matrigel® angiogenesis assay [9]. Furthermore, a monoclonal SFRP2 antibody inhibits the growth of triple negative breast cancer and angiosarcoma in vivo, coincident with a reduction in tumor angiogenesis [10]. This growing evidence supports the hypothesis that SFRP2 is a therapeutic target for cancer treatment.
In elucidating the mechanism through which SFRP2 stimulates angiogenesis, a novel pathway was identified: the noncanonical Wnt/calcineurin/nuclear factor of activated T-cells c3 (NFATc3) pathway. Silencing of NFATc3 in endothelial cells blocked SFRP2-induced endothelial tube formation [11]. NFAT is a transcription factor that plays an important role in facilitating angiogenic responses, including vascular endothelial growth factor (VEGF)-induced angiogenesis [12,13]. However, the receptor through which SFRP2 mediates NFATc3 activation and stimulates angiogenesis is unknown. We propose that SFRP2 may mediate these events by directly binding to one of the ten frizzled G-protein coupled transmembrane receptors (GPCR). Furthermore, we hypothesize that the specific frizzled GPCR is frizzled 5 (FZD5), which has been reported to be involved with both calcineurin activation [14] and vascular development [15].
Binding of Wnt5a to FZD5 leads to a transient release of Ca2+ from the endoplasmic reticulum which ultimately results in the phosphorylation and nuclear translocation of NFAT [14,16]. FZD5 has been shown to be necessary for angiogenesis of the yolk sac and placenta [15] and regulates retinal vascular development [17,18]. FZD5 null mice are embryonic lethal because embryonic blood vessels do not connect with the maternal vessels [15]. Given that FZD5 is involved in activating Wnt/calcium signaling and vascular development, we hypothesized that FZD5 could also mediate SFRP2-induced NFATc3 activation and angiogenesis. In this study, we report that SFRP2 ligand binds to FZD5 receptor and co-localizes at the endothelial cell membrane. Furthermore, SFRP2/FZD5 interaction is necessary for SFRP2-mediated calcium/calcineurin/NFAT activation and angiogenesis.
MATERIALS AND METHODS
Cell culture
2H11 mouse endothelial cells and SVR (SVEN 1 ras) angiosarcoma cells (ATCC®, Manassas, VA, USA) were cultured in Opti-MEM™ (Life Technologies, Grand Island, NY, USA) with 5% and 8% FBS (SH30071.03; Hyclone Logan, Utah) respectively, and 1% penicillin/streptomycin (v/v) (Corning, Corning, NY, USA). MS1 (MILE SEVEN1) mouse pancreatic endothelial cells (ATCC®) were cultured in DMEM containing high glucose, L-Glutamine (ATCC®), with 10% FBS (ATCC®) and 1% penicillin/streptomycin (v/v) (Corning). Cells were cultured at 37°C in an incubator with 5% CO2 and 95% humidity. HMEC-1 human endothelial cells (ATCC®, Manassas, VA, USA) were cultured in MCDB131 medium (Gibco, Grand Island, NY, USA) with 10ng/ml EGF (Gibco, Grand Island, NY, USA), 1ug/ml hydrocortisone (Tocris, Bristol, UK), 10nM Glutamine, (Gibco, Grand Island, NY, USA), 10% FBS (Omega Scientific, Tarzana, CA), 1% penicillin/streptomycin (v/v) (Corning, Corning, NY, USA). 2H11 endothelial cells were chosen for study due to their expression of known tumor endothelial markers and several well-recognized endothelial cells markers. They respond to known anti-angiogenic agents, and have been used for in vitro angiogenesis assays to evaluate potential angiogenic properties [19]. This cell type also responds to exogenous SFRP2 stimulation [10]. SVR angiosarcoma cells were chosen as a second cell line for their high endogenous levels of SFRP2 [9] and because they form malignant angiosarcomas in vivo [20], while MS1 cells have lower SFRP2 levels [9] and form benign hemangiomas in vivo [20].
Antibodies and proteins
The following antibodies were purchased from Abcam, Cambridge, MA, USA: rabbit anti-Frizzled 5 antibody (ab75234), goat anti-SFRP2 antibody (ab77618), rabbit anti-NFATc3 antibody (ab93628), rabbit anti-TATA binding protein TBP antibody – ChIP Grade (ab63766). Mouse β-catenin antibody (BD1480) was purchased from Santa Cruz Biotechnology Inc., Dallas, TX, USA (sc-59893), and rabbit β-Tubulin antibody from Cell Signaling Technology, Danvers, MA, USA (2146). Rabbit anti-actin (A2103), and rabbit anti-SFRP2 antibody (HPA002652) used for co-immunoprecipitation studies were from Sigma-Aldrich, St Louis, MO, USA. Mouse anti-FRZD5 (H00007855-M01) and rabbit anti-FZD5 (H00007855-D01P) used in co-immunoprecipitation studies were from Abnova, Taipei City, Taiwan. Control goat IgG (AB-108-C) was from R&D Biosystems, Minneapolis, MN, USA, and control mouse IgG2a (400224B) was from Biolegend, San Diego, CA, USA. Secondary antibodies including anti-mouse immunoglobulin G (IgG), horseradish peroxidase (HRP)-linked whole antibody (NA931), ECL anti-rabbit IgG, and HRP-linked whole antibody (NA934) were purchased from GE Healthcare Bio-Sciences Corp. Recombinant mouse and human SFRP2 protein (rmSFRP2 and rhSFRP2, respictively) were provided by the Protein Expression and Purification Core Lab at University of North Carolina at Chapel Hill. Frizzled-5 Fc Fusion, FZD5-Fc (HFZ5FC-050) was purchased from ACRO Biosystems, Newark, DE, USA. Recombinant human Wnt5a (rhWnt5a; MBS692220) and recombinant mouse Wnt5a (rmWnt5a; MBS2011413) were purchased from MyBiosource, San Diego, CA, USA.
Gain and loss of function studies with FZD5
Stable transfection of 2H11 endothelial cells with FZD5 shRNA
To silence FZD5 in endothelial cells, we used HuSH shRNA plasmids containing FZD5 (OriGene, Rockville MD, USA). The constructs were provided in pGFP-V-RS vectors. Four different FZD5 shRNA constructs containing the following sequences were tested: TTCCTTCTGGCAGGCTTCGTGTCACTCTT, GAGGCATCGGCTACAACCTGACGCACATG, ACCGTTGCCACCTTCC TCATTGACATGGA, and GGTCATCCTGTCGCTCACCTGGTTCTTGG. Control shRNA constructs were pGFP-V-RS empty vector (OriGene). 2H11 endothelial cells were seeded in DMEM with 10% FBS at a density of 2×104 cells/well and incubated overnight. For transfection with FZD5 shRNA plasmids or empty vector, 1 μg of shRNA plasmid was diluted in 500 μl of Opti-MEM medium and mixed thoroughly. PLUS™ reagent (Invitrogen, Grand Island, NY, USA) was mixed and 5 μl was added to the diluted shRNA. The solution was mixed gently and incubated at RT for 5 minutes. Lipofectamine® reagent was mixed gently and 15 μl was added to the solution containing PLUS™ reagent and shRNA, mixed, and incubated at RT for 30 minutes. 500 μl of SOC solution (Corning Cellgo, Manassas, VA, USA) was added drop wise to the cells and gently mixed by rocking. The cells were incubated at 37°C for 48 h. After transfection, the media was replaced and cells were selected with 3 μg/ml puromycin (Gibco, Grand Island, NY, USA) in DMEM with 10% FBS. The selective medium was changed every 2 to 3 days. After selection the cells were kept in full growth medium containing 3 μg/ml of puromycin. Western blot analysis on whole cell lysates was performed to confirm the downregulation of FZD5. In these assays, whole cell lysates were extracted with the Mammalian Protein Extract reagent (M-PER; Thermo Fisher Scientific, Waltham, MA, USA) as described in the manufacturer’s manual. The most efficient downregulation of FZD5 was obtained with the following shRNA construct: TTCCTTCTGGCAGGCTTCGTGTCACTCTT. Puromycin was removed from the medium in tube formation and migration assays two days prior to initiating the experiments.
Stable transfection of MS1 endothelial cells with GFP and FZD5/GFP lentivirus
MS1 cells were transfected with pLenti-GIII-CMV-GFP-2A-Puro (Applied Biological Materials Inc., Richmond, BC, Canada) alone (control) or with a FZD5 sequence [21] following the manufacturer’s protocol (Applied Biological Materials Inc.). Briefly, 5×104 cells/well were incubated overnight in complete medium containing DMEM with 10% FBS (ATCC®, Manassas, VA, USA). The following day, the medium was replaced by fresh complete medium mixed with polybrene (8 μg/mL; Millipore, Danvers, MA, USA), ViralPlus Transduction Enhancer G698 (1:100; Applied Biological Materials Inc.), and GFP or FZD5/GFP lentivirus. After an overnight incubation, the medium was removed and the cells were incubated for another night in fresh complete medium. The following day, puromycin (2 μg/ml; Gibco, Grand Island, NY, USA) was added to the medium. The expression of FZD5 was verified by western blot and GFP expression was verified using the fluorescent EVOS microscope (Life Technologies, Carlsbad, CA, USA).
Generation of recombinant human SFRP2 for SFRP2/FZD5 binding studies
Plasmid construction
Human recombinant SFRP2 was produced by the University of North Carolina Chapel Hill Protein Expression and Purification Core in order to measure the binding affinity between SFRP2 and FZD5. rhSFRP2 expression plasmid was prepared by the gene synthesis and cloning services of Genewiz (South Plainfield, NJ, USA). DNA encoding the full length rhSFRP2 nucleotide sequence with a C-terminal hexahistidine tag was synthesized (human codon optimized) and cloned into the pIRES2-AcGFP1 vector (Clontech Laboratories, Inc., Mountain View, CA, USA) via 5′ Nhe1 and 3′ BamH1 restriction sites, creating pIRES2-Ac-GFP1-hSFRP26HIS plasmid. A Kozak sequence (GCCACC) was added after the Nhe1 restriction site and immediately before the starting ATG codon of the rhSFRP2 coding sequence.
Stable transfection of HEK 293f cell line
HEK293f cells were transfected with the pIRES2-Ac-GFP1-hSFRP26HIS plasmid using Lipofectamine (Invitrogen, Grand Island, NY, USA), following the manufacturer’s protocols. Briefly, 0.5×106 HEK293f cells in 2% FBS Freestyle 293 Expression medium (Invitrogen). The following day, the medium was replaced by lipofectamine diluted in Opti-MEM (Invitrogen). pIRES2-Ac-GFP1-hSFRP26HIS plasmid DNA was diluted into Opti-MEM. DNA and Lipofectamine were then mixed and incubated at RT for 20 min before the mixture was added to the plate. The next day, the medium was removed and replaced with fresh 2% FBS Freestyle 293 expression media supplemented with geneticin (Invitrogen) at 2mg/ml. After 10 days, the medium was aspirated and replaced every 3–4 days with fresh 2% FBS Freestyle 293 expression medium + geneticin (1:25). Colonies were apparent after 10 days. Cells were resuspended into fresh Freestyle 293 + geneticin (1:25), and transferred to a shaker flask, shaken at 134 rpm, 8% CO2, 37° C. These cells were subjected to 2 rounds of FACS sorting at the UNC FACS core facility, where the top 30% of the cells with green fluorescent protein (GFP) signal were collected and expanded. The result was a >95% GFP positive cellular pool, designated HEK293f-hSFRP2, that was ready to be used for recombinant human SFRP2 (rhSFRP2) protein production.
Expression and purification of rhSFRP2
The HEK293f-SFRP2 cells were cultured in Freestyle293 expression media supplemented with geneticin (1:100) to a density of 1×106 in a volume of 600mL in a 2.8L fernbach flask (shaken at 134 rpm, 8% CO2, 37°C). Valproic acid was added to a final concentration of 4mM. On days 3, 5, and 7 the culture was supplemented with 100 ml Freestyle expression media which also contained 20 ml 10% BOOST 1 SUPPLEMENT (Thermo Fisher Scientific, Waltham, MA, USA), geneticin (50 mg/ml) and glutamax. Cell viability was monitored via trypan blue staining until viability decreased below 50%, at which point the supernatant was harvested by centrifugation followed by 0.22 μm filtration into a sterile vessel. This media was then concentrated and buffer exchanged into 50mM Na Phosphate pH 7.4, 500mM NaCl, 25mM Imidazole buffer, and subjected to Ni+2 affinity chromatography. The resulting peak from the elution step was concentrated and subjected to gel filtration chromatography. The sizing column was equilibrated with 50mM Na Phosphate pH 7.4, 150mM NaCl, 15% glycerol buffer. The fractions containing rhSFRP2 were pooled and concentrated.
Microplate Solid Phase Protein Binding ELISA Assay
A microplate solid phase protein binding assay [22] was used to determine the Kd and EC50 between rhSFRP2 and FZD5-Fc fusion protein, and compared to the Kd and EC50 between rhWnt5a and FZD5-Fc. Flat-bottom Ni+2 coated 96-well microplates (Thermo Fisher Scientific, Waltham, MA, USA) were blocked with 0.05% bovine serum albumen (BSA) in phosphate buffered saline (PBS) overnight at 4°C. A concentration range of his-tagged rhSFRP2 and rhWnt5a diluted in PBS (pH 7.4) from concentrations of 24 nM to 1500 nM was incubated overnight at 37°C (n=3 per dose). The plates were washed 4 times with 150μl/well of PBS. Then, 100 nM of FZD5-Fc fusion protein in PBS was incubated in the plate with rhSFRP2 at 37°C for 1.5 hours. Plates were washed 4 times, and subsequently incubated with 50 μl/well of secondary antibody (goat anti-rabbit from Southern Biotech, Birmingham, Alabama, cat no 4030-05), diluted 1:20,000 in PBS with 0.05% BSA for 1 hour at RT. After plates were washed 5 times, each well was incubated with 50μl K-Blue TMB substrate (Neogen, Lexington, KY, USA) for 5 minutes in the dark. Absorbance was read at 450 nm. EC50 estimates from data were derived via unconstrained nonlinear regression analysis with variable slope using GraphPad Prism and converted to Kd using the Cheng-Prusoff equation where agonist concentration and EC50 were equal [23]. Statistical differences between SFRP2/FZD5 binding and WNT5a/FZD5 binding were calculated using a two-tailed student’s t-test, with a p ≤ 0.05 being significant. Results are expressed as means ± standard error of the mean (sem).
Molecular modeling of the SFRP2/FZD5 heterodimer interaction
Modeling and visualizations were performed using MOE Version 2014.9 (Chemical Computing Group). Sequence alignments were performed using ClustalW with the BLOSOM62 similarity matrix in Bioedit 7.2.5. Amino acid sequences used were human SFRP2 (NP_003004.1) and human FZD5 (BAB60959.1). After alignment, homology modeling was performed using MOE Homology Model. Ten total models were generated and the best model was selected using medium grain intermediates, GB/VI scoring, and Amber12:ETH force field. The final model was protonated at pH 7.4. The template PDB:4F0A structure resolution was 3.25Å, while the homology model overall root-mean square deviation (RMSD) to 4F0A was 0.6Å between FZD5 and FZD8; 1.14Å between SFRP2 and Wnt8. For detection of inter-molecular interactions, hydrogen bonds used a cutoff of 0.5kcal/mol, ionic bonds used a cutoff of −0.5kcal/mol, and the distance cutoff was 4.5Å.
Co-localization studies
For cellular co-localization of SFRP2 ligand and FZD5 receptor, we performed co-immunofluorescence labeling on SVR angiosarcoma cells, which constitutively make high amounts of SFRP2 [9], and on 2H11 mouse endothelial cells control or stimulated for 1 minute with 10nM rhSFRP2 (UNC Chapel Hill, NC). 5×104 cells/cm2 were plated on chambered coverslips (Thermo Fisher Scientific, Waltham, MA, USA), and, following treatment with SFRP2, were fixed with 4% paraformaldehyde (Alfa-Aesar, Tewksbury, MA, USA), permeabilized in PBS with 0.25% Triton X-100 (Sigma-Aldrich, St Louis, MO, USA). Then, they were washed twice with sterile deionized water, and incubated overnight at 4°C with a combination of rabbit anti-SFRP2 (Sigma-Aldrich; HPA002652) and mouse anti- FZD5 (Sigma-Aldrich; WH0007855M1) antibodies. The next day, the slides were washed 3 times in PBS for 30 minutes each, and incubated with anti-rabbit IgG-FITC (Sigma-Aldrich; F0382) and anti-mouse IgG-Alexa Fluor 546 (Thermo Fisher Scientific; A21133) secondary antibodies for 45 minutes. After, slides were washed twice with PBS, incubated for 5 minutes in Hoechst 33342 (Thermo Fisher Scientific; 62249), washed again with PBS, and stored in PBS overnight prior to imaging. Immunostained cells were visualized using the Olympus FV10i confocal microscope (Olympus, Pittsburg, PA, USA)[24]. Images were analyzed and prepared for publication using Fiji ImageJ 1.51e[25]. Secondary only controls used the same parameters as both primary and secondary antibody conditions. Pearson’s correlation coefficient analysis was performed using Coloc2 in Fiji ImageJ for each condition (n = 9 cells from at least four fields for each samples). In parallel, analysis using Li ICQ and Spearman Rank were performed and showed nearly identical results (data not shown). Differences between samples were estimated using a two-tailed student’s t-test, with a p ≤ 0.05 being significant.
Co-immunoprecipitation for SFRP2/FZD5
2H11 cells transfected with control (sham) or FZD5 shRNA, and MS1 cells transfected with GFP control or FZD5/GFP lentivirus, were treated for 0, 5 or 15 minutes with 10 nM of rhSFRP2. Cell lysates were generated using RIPA buffer (1% Triton-X100; 0.1% Deoxycholate; 150 nM NaCl; 50 mM Tris-HCl; 50 mM NaF; 1 mM Na3VO4; 1 tablet protease inhibitor cocktail from Roche Diagnostics, Indianapolis, IN, USA) and each sample was sonicated for 15s. Protein G magnetic beads (Dynabeads; Thermo Fisher Scientific, Waltham, MA, USA) were washed 3 times for 1h each with RIPA buffer in rotation at 4°C. For 2H11 cells, each protein sample (100μg) was mixed with 5μl of beads, 0.5μg of goat anti-SFRP2 antibody (Abcam, Cambridge, MA, USA) or control goat IgG (R&D Systems, Minneapolis, MN, USA), and placed in rotation overnight at 4°C. For MS1 cells, each protein sample (100μg) was mixed with 5μl of beads, 0.5μg of mouse anti- FZD5 antibody (Abnova, Taipei city, Taiwan) or control mouse IgG2a (R&D Systems), and placed in rotation overnight at 4°C. Beads were then washed 3 times for 1h each with RIPA buffer at 4°C. They were finally isolated and incubated in a 1.5X solution containing LDS sample buffer and sample reducing agent (Thermo Fisher Scientific). Proteins were separated on Bolt 10% Bis-Tris PLus gels (Thermo Fisher Scientific) and transferred on a polyvinylidene difluoride membrane. For 2H11 samples, after blocking for 1h in 5% milk/TBST 0.05%, membranes were incubated with either a mouse anti-FZD5 (1:1000; Sigma-Aldrich, St Louis, MO, USA), or a rabbit anti-SFRP2 (1:1000; Sigma-Aldrich), overnight at 4°C. For MS1 samples, after blocking for 1h in 5% milk/TBST 0.1%, membranes were incubated with either a rabbit anti-FZD5 (1:1000; Abnova, Taipei City, Taiwan), or a rabbit anti-SFRP2 (1:1000; Sigma-Aldrich), overnight at 4 °C. Membranes were then washed in TBST 0.1% and probed with either HRP-conjugated anti-mouse (1:5000; Cell Signaling, Danvers, MA, USA) or anti-rabbit (1:5000; SouthernBiotech, Birmingham, AL, USA) secondary antibodies. Proteins were visualized using westernbright ECL (Advansta, Menlo Park, CA, USA). In parallel, western blot analyses were performed on whole protein lysates using a rabbit anti-SFRP2 (1:1000; Sigma-Aldrich) and a goat anti-FZD5 (1:1000; Santa Cruz Biotechnology Inc., Dallas, TX, USA), as well as a rabbit anti-actin antibody (1:1000; Sigma-Aldrich). Secondary antibodies were: HRP-conjugated anti-rabbit (1:5000; SouthernBiotech, Birmingham) or anti-goat (Thermo Fisher Scientific).
Endothelial tube formation assay
The shFZD5 and sham transfected 2H11 cells or MS1 GFP, MS1 FZD5/GFP cells were plated in Opti-MEM™ with 10% FBS and allowed to settle for 24 hrs. Quiescence was induced by maintaining the cells in Opti-MEM™ with 2% FBS for 18 hrs. 2×104 or 3×104 cells/well, respectively, were added to the Matrigel® matrix according to the In Vitro Angiogenesis Assay protocol (Millipore, Bedford, MA, cat# ECM625). Treated cells were incubated in Opti-MEM™ with 2.5% FBS and SFRP2 10nM. Untreated cells were given fresh Opti-MEM™ with 2.5% FBS. After 4 to 6h of incubation at 37°C, 5% CO2, images were acquired with the 4× objective lens of EVOS FLc Imaging System (ThermoFisher Scientific, Waltham, MA).
To compare effects of blocking FZD5 on SFRP2 induced tube formation between mouse and endothelial cells, we plate 2H11 cells as described above in Matrigel®. HMEC-1 cells were quiesced for 24 hours in MCDB131 medium with 1% FBS, with growth factors as described in the cell culture section. Cells were treated with control medium, or hSFRP2 30nM, or FZD5-FC fusion protein 500 ng/ml, or combination of hSFRP2 30nM plus FZD5-FC 500ng/ml (preincubated for 90 minutes at 37° in CO2 incubator on a rocker). All drugs were added to cells followed with a 30 minute pre-incubation prior to adding to Matrigel®. After 4 to 6h of incubation at 37°C, 5% CO2, images were acquired with the 100X per field of vision using EVOS FLc Imaging System (ThermoFisher Scientific, Waltham, MA).
Branch points were counted using field of view per image with Image J Angiogenesis Analyzer software (National Institutes of Health, Bethesda, MD, USA). Results are expressed as means ± sem. Data were analyzed using one-way repeated measure ANOVA with Bonferroni posttest with significance being when p < 0.05.
Intracellular free calcium assay
shFZD5 and sham transfected 2H11 cells were plated the night before at 8,000 cells/well. Two hours prior to the experiment, culture medium was removed and replaced with serum free Opti-MEM™. The wells were loaded with the calcium-sensitive probe, Calcium-5, for 1 hour at 37°C. Fluorescence responses to agonists (rmSFRP2 (1–30 nM), or recombinant mouse Wnt5a (30–1000 ng/ml)) were measured with a fluorescence imaging plate reader (FLIPRtetra Molecular Devices, Sunnyvale, CA, USA). Responses were expressed as peak heights of the fluorescence signal in relative RFU. The calcium ionophore A23187 (3 μM) was used to quantify maximum attainable fluorescence. RFU kinetic reduction experiments were conducted using four replicates per condition (genotype and concentration), for each of the two plates per experiment. Experiments were replicated on a second occasion resulting in a total of 16 RFU measures for each condition, with the exception of Wnt5a 1000ng/ml which was interrogated on a single occasion yielding 8 RFU measures. Data were evaluated for evidence of batch (plate and experiment) effects using boxplots. Conformity to approximate normality was examined graphically using histograms and normal quantile-quantile plots. Data analysis was performed using analysis of covariance (ANACOVA) using the combination of RFU measures from both experiments as the dependent variable in a single model. Independent factors included genotype (sham vs. shFZD5), concentration (1, 3, 10 and 30 nM for rmSFRP2; 30, 100, 300 and 1000 ng/ml for rmWnt5A), and their two-way interaction. We adjusted for the effect of plate (nested within experiment) as supported by our evaluation of batch effects (Supplemental Fig. 1 A, B). Estimates of average (SE) RFU for sham and shFZD5 conditions were obtained based on appropriately defined linear functions of estimated model parameters. Statistical significance of differences comparing sham to shFZD5 conditions was evaluated using model-based linear contrasts. Graphical analyses were performed using R version 3.2.3. Data analyses were performed using SAS version 13.2. No adjustment was made for multiple comparisons. Statistical significance was defined as p ≤ 0.05.
Western blot for NFATc3 in shFZD5 transfected endothelial cells
The shFZD5 and sham transfected 2H11 mouse endothelial cells were grown to 80–90% confluency in Opti-MEM™ supplemented with 5% FBS (ATCC®, Manassas, VA, USA). Cells were serum starved in Opti-MEM™ with 2% FBS overnight. The following day the media was changed to Opti-MEM™ with 2% FBS and supplements. Control cells received media alone; SFRP2-treated cells were incubated with rmSFRP2 10nM for 1h. Nuclear extracts were prepared using NE-PER nuclear and cytoplasmic extraction reagent as described in the manufacturer’s manual (Pierce Biotechnology, Rockford, IL). Nuclear fractions were confirmed on western blot using the loading control, TATA binding protein TBP antibodies (Abcam, Cambridge, UK), which is a nuclear marker. Protein concentration was measured using Bio-Rad Protein Assay at OD595 (Bio-Rad Laboratories, Hercules, CA, USA). Equal amounts of protein (20μg) were loaded onto SDS-PAGE gels. Proteins were transferred to polyvinylidene difluoride membrane, and western blotting was carried out using a primary anti-rabbit NFATc3 antibody (Abcam), with HRP-conjugated rabbit IgG as the secondary antibody (Southern Biotech, Birmingham, AL, USA). The ECL Advance substrate was used for visualization (GE Healthcare Bio-Sciences, Piscataway, NJ, USA).
Migration assay
Human microvascular endothelial cells (HMEC-1), human triple negative breast cancer cells (MDA-MB-231), and 2H11 mouse endothelial cells were tested for migration using a trans-well assay, following the manufacturer’s protocol (Corning, Corning, NY, USA). For each cell line the lower well was filed with 750 μL of medium containing 0.5X FBS, used as an attractant for the cells. Cells were detached with trypsin, suspended in serum-free medium and counted, before being diluted to the following concentrations: 5 × 104 cells/mL (HMEC-1), 3 × 104 cells/mL (2H11 and MDA-MB-231). From this suspension, 500 μL were added to the upper well. Cells were then treated either with rhSFRP2 (30 nM), rhFZD5-Fc 500ng/ml or a combination of both. Reagents were pre-incubated for 1.5 hour on shaker at 37°C. Each condition, including a control, was tested in triplicate in each assay and each assay for each cell line was repeated 3 times. Cells were incubated at 37°C, 5% CO2 for 24h. To stop the migration process, trans-wells were immersed in PFA 4% for 10 min, RT. Cells were then stained using a solution of crystal violet in trans-wells that were incubated for15 minutes at room temperature. Trans-wells were finally rinsed three times with de-ionized water to remove the excess of crystal violet. Cells that had not migrated were removed from the upper side of the filter using a Q-tip. Analysis was performed as follow: 5 fields from each well were photographed using an EVOS FLc microscope (Thermo Fisher Scientific, Waltham, MA, USA). Cells in each field were counted using Fiji-Image J software. For each well, the number of cells from the five fields was added. In each experiment and for each treatment condition, the total number of cells from each of the three wells was averaged and converted into a percentage of cells for a given treatment, compared to control. Statistics between conditions, for each cell line, were performed on the average percentages of cells (for each treatment and compared to control) from three separate experiments. Data were analyzed using one-way repeated measure ANOVA with Bonferroni posttest with significance being when p < 0.05, n=9, combined date from 3 experiments).
RESULTS
Molecular homology model of SFRP2/FZD5
Wnt and SFRP proteins belong to the same superfamily of secreted protein ligands that activate FZD GPCRS and are structural and functional homologues. This allowed us to create a homology model of interaction between SFRP2 and FZD5 (Fig. 1). The alignment and models indicate that SFRP2 and FZD5 interact with the same topology as Wnt8 and FZD8. Using the heterodimeric X-ray of Wnt8 and FZD8 (PDB: 4f0A) [26], a 3D molecular homology model was built in order to better understand the critical binding domains for these two proteins. The alignment of xWnt8 to hSFRP2 indicates that there are 57 similarities and 63 identities for SFRP2, compared to Wnt8, for a total similarity of 41% (Supplemental Fig. 2). For the FZD GPCRs, the similarity is even closer. The difference between human and mouse FZD8 is only 3% and the difference between human and mouse FZD5 is also 3% (Supplemental Fig. 3), while the difference between human FZD8 and FZD5 is 20%. For mouse FZD8 versus human FZD5, the overall similarity is 67%. The major differences between human or mouse FZD8 and FZD5 is that FZD5 is missing three stretches of amino acids that would be just N-terminal of the first transmembrane segment, the first extracellular loop, and the C-terminal tail (data not shown). For these two protein pairs, the similarities and identities were distributed homogenously throughout the sequence, which provided confidence and accuracy of the homology model.
Figure 1. The theoretical topology of interaction between SFRP2 and FZD5 is similar to other Wnt/FZD family members.
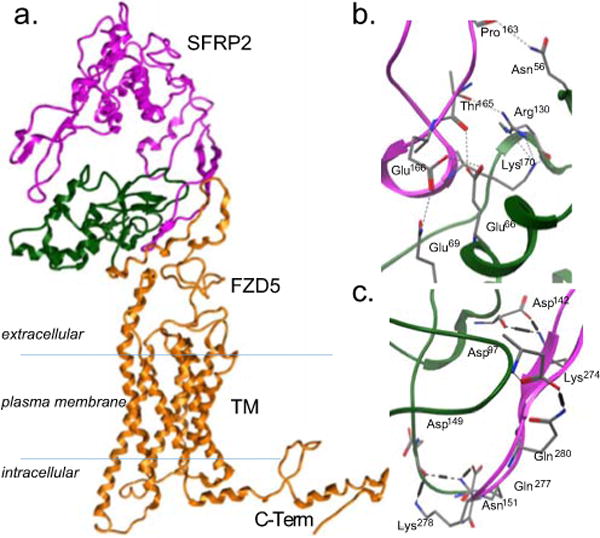
A) Full length homology model of SFRP2 and FZD5. CRD, the cysteine rich domain of FZD5, is colored green. TM, the transmembrane domain of FZD5, is colored orange along with the intracellular domains. B, C) Detail of twelve intermolecular interactions between SFRP2 and the CRD of FZD5.
The interaction of the FZD GPCR cysteine rich domain (CRD) forms a helical globule that is gripped by the ligand as it forms a C-shaped clamp with two extended loops (SFRP2 161–176 and 270–285), interacting with the receptor on opposite faces of the CRD (Fig. 1a). The specific interactions in the two sets of hydrogen bonds include (SFRP2- FZD5) Pro163-Asn56, Thr165-Glu66/Arg130, Glu166-Glu66/Gln69 and Lys170-Arg130 for the first set (Fig. 1b) and Lys274-Asp142, Gln277-Asp140/Asn151, Lys278-Asp149, Gln280-Asp97 for the second set, and a potential ionic bond between Lys274-Asp142 (Fig. 1c).
SFRP2 and FZD5 co-localize at the surface of endothelial cells
To assess the ability of SFRP2 and FZD5 to interact, we used immunocytochemistry and high resolution confocal imaging on 2H11 cells untreated or treated with rhSFRP2 (Fig. 2a–h). Modest co-localization was visible in untreated 2H11 cells (Fig. 2a–c). However, after a one minute treatment with rhSFRP2, both proteins strongly co-localized, as depicted by the presence of a yellow staining at the membrane of the cells (Fig. 2e–g). Co-localization analysis indicated a statistically significant difference (p = 0.0003, n = 9 cells in 4 fields) between the Pearson’s coefficients of control (0.47 +/− 0.003) and SFRP2-treated (0.81 +/− 0.03) 2H11 cells (Fig. 3). Importantly, the interaction between SFRP2 and FZD5 was associated with rapid morphological changes such as increased cell-cell adhesion, which is a typical characteristic of tube formation. The co-localization was confirmed in SVR angiosarcoma cells (Fig. 2i–k, Fig. 3; Pearson’s coefficient 0.79 +/− 0.02), which endogenously express high levels of SFRP2 [9].
Figure 2. SFRP2 and FZD5 co-localize at the surface of 2H11 and SVR cells.
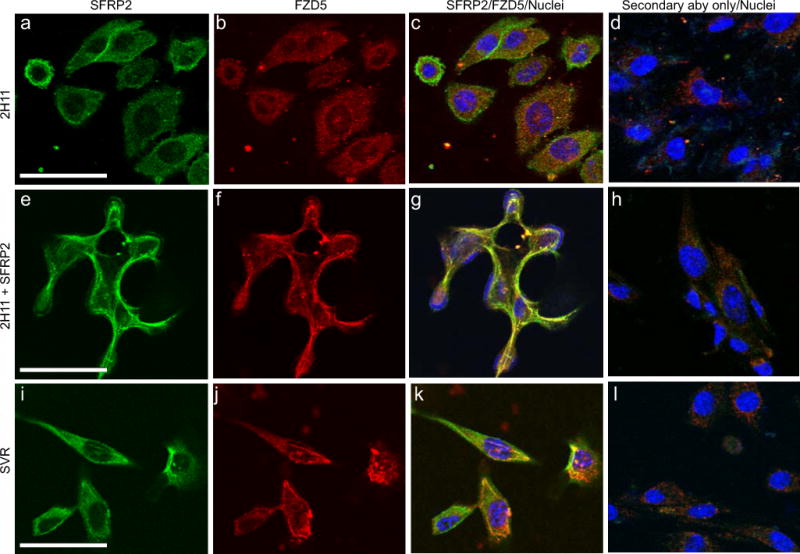
Fluorescence confocal microscopy was used to detect localization of SFRP2 (A, E, I) and FZD5 (B, F, J) in 2H11 (A–H) and SVR (I–L) cells. 2H11 cells were either untreated (A–D) or treated with 10 nM SFRP2 (E–H) for 1 minute. SFRP2 (green) FZD5 (red) were visualized using an FITC- or AlexaFluor546-labeled secondary antibody, respectively. Nuclei (blue) were identified using Hoeschst 33342. Merged images (C, G, K) showing the co-localization of SFRP2 and FZD5 (yellow) at the surface of SFRP2-treated 2H11 cells (G), at the surface SVR cells which highly express endogenous SFP2 (K), but not at the surface of untreated 2H11 cells (C). Controls were treated with secondary antibody only, with the primary antibody omitted (D, H, I). Scale bar in A, E and I represents 30μm.
Figure 3. Intracellular colocalization analysis of SFRP2 and FZD5 in 2H11 and SVR cells.
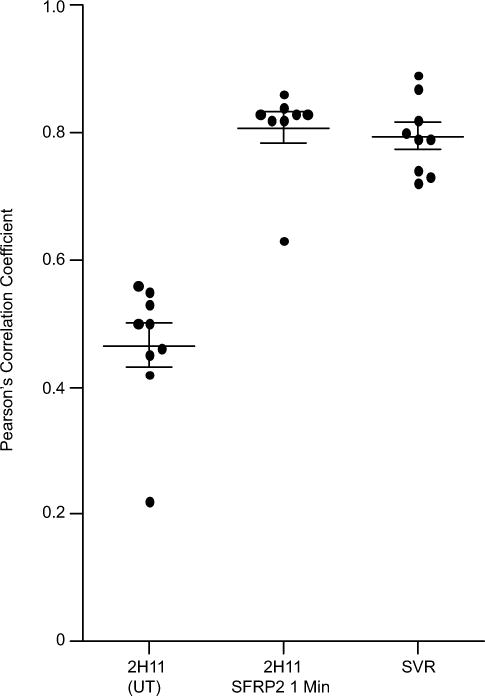
The vertical scatter plot shows individual Pearson’s correlation coefficients with the mean of the coefficients and standard error of the mean (n = 9 cells from at least four fields for all samples). Analysis indicates that in 2H11 cells, a pool of SFRP2 and FZD5 colocalize. This phenomenon is rapidly amplified by the addition of exogenous SFRP2 and reaches the levels of colocalization observed in SVR cells, which express sustained endogenous levels of SFRP2. Difference between 2H11 control and 2H11 exposed for one minute to exogenous rhSFPR2 was significant (two tailed t-test; p = 0.0003).
SFRP2 interacts with FZD5 in endothelial cells
To further confirm the interaction between SFRP2 and FZD5, we performed co-immunoprecipitation experiments on lysates from 2H11 (Fig. 4a, b) and MS1 endothelial cells (Fig. 4c, d) treated with rhSFRP2. The endogenous levels of SFRP2 and FZD5 were first measured in 2H11 control or transfected with shFZD5 (Fig. 4a). The levels of FZD5 were lower in shFZD5-transfected cells, compared to control. SFRP2 was then pulled down from lysates of control or shFZD5-transfected 2H11 cells (Fig. 4b; lane 2, 3, 5, 6). Comparatively, FZD5 was co-immunoprecipitated only after a 15 minute rhSFRP2 treatment of control 2H11 cells (Fig. 4b, lane 3). However, it was not pulled down from equivalently treated shFZD5 cell lysates (Fig. 4b, lane 4–6). To confirm the specificity of the SFRP2 antibody, an IgG was used as a negative control in this assay (Fig 4b, lane 7–12). The reverse experiment was conducted on MS1 cells GFP control or FZD5/GFP. The increased levels of FZD5 were confirmed in FZD5/GFP whole cell lysates, compared to control (Fig. 4c). FZD5 was specifically pulled down from FZD5/GFP lysates and, to a much lower extent, from GFP control lysates (Fig. 4d, lane 1–6). However, SFRP2 co-immunoprecipitated with FZD5 only in lysates from FZD5/GFP MS1 cells treated with rhSFRP2 for 5 and 15 minutes (Fig. 4d, lane 2, 3). Again, an IgG negative control was used to confirm the specificity of the FZD5 antibody (Fig. 4d, lane 7–12).
Figure 4. SFRP2 and FZD5 co-immunoprecipitate in 2H11 and MS1 cells.
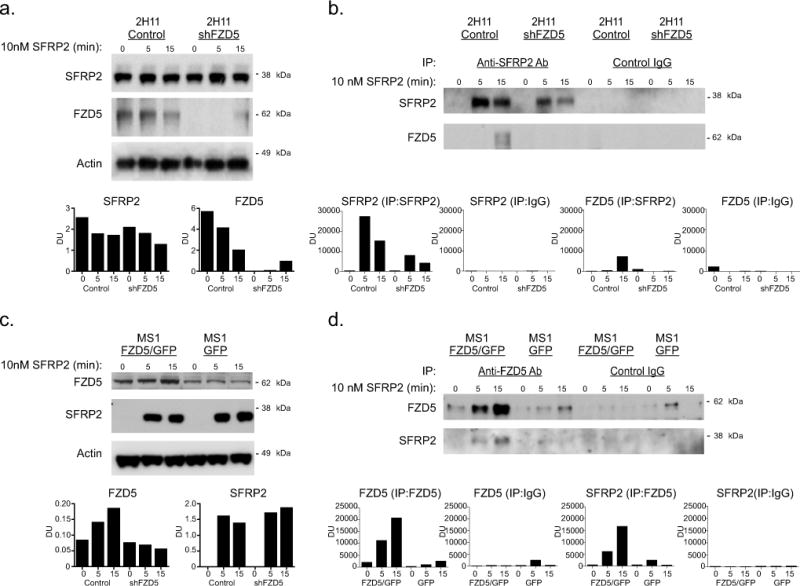
A) Western blot analysis on samples from sham (control) and shFZD5-transfected 2H11 cells treated for 0, 5 and 15 min with 10nM SFRP2, showing the levels of SFRP2 and FZD5. Actin was used as a loading control. Dosimetry units (DU) normalized to actin. B) Sham and shFZD5 samples from 2H11 cells treated for 0, 5 and 15 with 10nM SFRP2 were immunoprecipitated with an anti-SFRP2 antibody (lanes 1 to 6) or a control IgG (lanes 7 to 12) and the levels of SFRP2 and FZD5 were then measured by western blot. C) Western blot analysis on samples from GFP (control) and FZD5/GFP expressing MS1 cells treated for 0, 5 and 15 min with 10nM SFRP2, showing the levels of SFRP2 and FZD5. Actin was used as a loading control, and DU are normalized to actin. D) GFP (control) and FZD5/GFP samples from MS1 cells treated for 0, 5 and 15 with 10nM SFRP2 were immunoprecipitated with an anti-SFRP2 antibody (lanes 1 to 6) or a control IgG (lanes 7 to 12) and the levels of SFRP2 and FZD5 were then measured by western blot.
SFRP2/FZD5 ELISA Assay
In order to confirm a direct interaction between SFRP2 and FZD5, we measured Kd, EC50 and Hill coefficient of rhSFRP2 binding to rhFZD5 in an ELISA-based assay using Ni+2 coated 96-well microplates [22]. Since Wnt5a direct binding to FZD5 has been verified by bio-layer interferometry[27], we used Wnt5a binding to FZD5 as an internal control in our assay. rhSFRP2 and rhFZD5-Fc fusion protein were found to bind with a Kd value of 103 +/− 6 nM, an EC50 of 205 +/− 12 nM, and a Hill coefficient of 1.9, indicating that FZD5 is a high affinity receptor for SFRP2 (Fig. 5). In comparison, the Kd of rhWNT5a for rhFZD5-Fc was 108 +/− 4 nM, an EC50 of 216 +/− 8 nM, and a Hill coefficient of 6.3. Overall, these results validate the ability of SFRP2 to bind directly to FZD5 receptor. Importantly, SFRP2 and Wnt5a bind to FZD5 with similar affinities.
Figure 5. SFRP2 and Wnt5a bind FZD5.
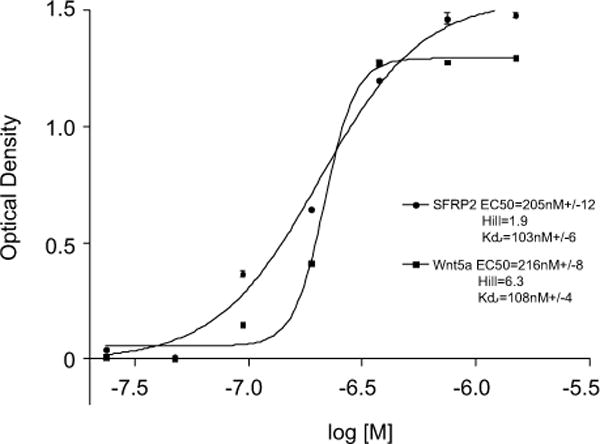
Graphic representation of rhSFRP2 and rhWnt5a binding to FZD5-Fc showing changes in 450 nm optical density (OD450) as a function of the log of ligand concentrations. To do this, A microplate solid phase protein binding assay was performed using a concentration range of his-tagged recombinant human SFRP2 and Wnt5a from 24nM – 1500nM (x axis). The experiment was repeated twice for a total of n = 6. Kd, EC50 and Hill coefficient were calculated and compared for SFRP2 and Wnt5a binding to FZD5.
FZD5 mediates SFRP2-induced endothelial tube branching
Our earlier studies revealed that SFRP2 stimulates endothelial tube formation [9]. For loss of function studies, we used 2H11 cells that have higher protein levels of FZD5 (Fig. 4a). For gain of function studies, we used MS1 cells that have lower FZD5 protein levels (Fig. 4c). To determine whether FZD5 is the receptor involved in this function, we first used 2H11 sham and shFZD5 endothelial cells in a 4 hour tube formation assay (Fig. 6a). rhSFRP2 treatment of sham transfected cells resulted in an increase in the number of branch points compared to untreated sham transfected cells (665 ± 23 vs. 476 ± 6; p<0.05, n=3), and the ability of shFZD5 2H11 cells to form tubes was reduced. Furthermore, as opposed to sham-transfected cells, rhSFRP2 treatment did not result in an increase of branch points by shFZD5 transfected cells (98 ± 6 vs. 101 ± 14; p=NS, n=3). To confirm the role of FZD5 on SFRP2 function, we tested the effects of SFRP2-induced tube formation on MS1 GFP and FZD5/GFP cells (Fig. 6b). rhSFRP2 treatment on GFP control cells didn’t promote tube formation. Likewise, in the absence of rhSFRP2, the overexpression of FZD5 didn’t increase the ability of MS1 cells to form tubes, compared to control cells. However, when FZD5/GFP transfected cells were treated with rhSFRP2, the number of branch points increased significantly (untreated FZD5/GFP cells: 76 ±20; FZD5/GFP cells + SFRP2: 427 ±74, p<0.05). Therefore, FZD5 is necessary to mediate SFRP2-dependent endothelial tube formation.
Figure 6. FZD5 is necessary for SFRP2-induced cellular and molecular function.
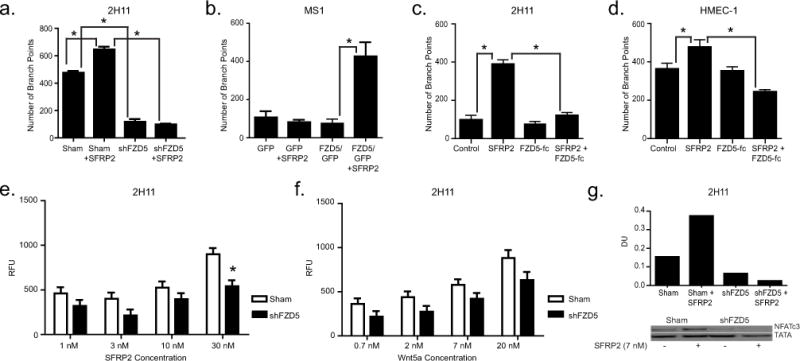
A, B) Bar graphs showing the number of branch points (± SEM) in tube formation assays using 2H11 (A) and MS1 cells (B). Sham (control) and shFZD5-transfected 2H11 cells (A), or GFP (control) and FZD5/GFP-expressing MS1 cells (B) were plated in a Matrigel® tube formation assay and treated with either control media or SFRP2 10nM for 4 hours. Experiments were repeated 3 times (n = 12 total). C, D) Bar graphs showing the number of branch points (± SEM) in tube formation assays using 2H11 (C) and HMEC-1 endothelial cells (D). Cells were plated in a tube formation assay and treated with either control media, SFRP2 30nM, FZD5-FC 500 ng/ml, or SFRP2 30nM + FD5-FC 500 ng/ml for 4 hours. Experiments were repeated 3 times (n=12 for HMEC-1 and n=9 for 2H11). E,F) Bar graphs showing calcium levels measured (RFU ± SEM) in sham and shFZD5-transfected 2H11 cells depending on the concentrations of SFRP2 (E; 1–30nM) or Wnt5a (F; 0.7–20nM). Each experiment was repeated 4 times (n = 16). G) Western blot showing the levels of nuclear NFATc3 in sham (control) and shFZD5-transfected 2H11 cells treated with recombinant media or mouse recombinant SFRP2 (10nM) for 1 hour. TATA was used as an internal control, and DU (dosimetry units) normalized to TATA. *: p-value<0.05.
To compare effects of blocking FZD5 on SFRP2 induced tube formation between mouse and human endothelial cells, we used a FZD5-FC fusion protein, which is an IgG protein that contains the FZD5 binding site. SFRP2 statistically significantly induced tube formation in both mouse 2H11endothelial cells (p<0.05, Fig 6c) and human HMEC-1 cells (p<0.05, Fig. 6d). The FZD5-FC fusion protein blocked SFRP2 induced tube formation in HMEC-1 cells (p<0.05, Fig 6d), and in 2H11 cells (p<0.05, Fig. 6c). For 2H11 cells the experiment was performed in triplicate 3 times (n=9), and for HMEC-1 cells the experiment was performed in quadruplicate and repeated 4 times (n=12). This demonstrates that FZD5 mediates SFRP2 induced tube formation in both mouse and human endothelial cells.
FZD5 mediates SFRP2-induced NFATc3 nuclear accumulation in endothelial cells
When calcineurin is activated by ligand/receptor binding, NFAT translocates to the nucleus, where it activates genes involved in angiogenesis [12,13]. Our previous studies showed that SFRP2 stimulated angiogenesis by activating the calcineurin/NFATc3 pathway [9–11]. To determine whether FZD5 was involved in this pathway, we compared nuclear NFATc3 protein levels in sham transfected versus shFZD5-transfected 2H11 cells treated or untreated with rmSFRP2. After a one hour treatment of sham-transfected 2H11 endothelial cells with rmSFRP2, nuclear NFATc3 levels increased substantially (Fig. 6g). However, in shFZD5-transfected cells, rmSFRP2 had no effect on nuclear NFATc3 levels. We conclude that FZD5 is required for SFRP2-mediated effects on NFATc3 signaling.
FZD5 mediates SFRP2-induced intracellular calcium release in endothelial cells
To confirm the effects on calcineurin/NFATc3 pathway, changes in intracellular calcium levels induced by SFRP2 or WNT5a were measured in sham and shFZD5-transfected 2H11 cells loaded with the calcium-sensitive fluorescent probe, Calcium-5. Fluorescence levels were measured following cell exposure to rmSFRP2 1–30 nM (Fig. 6e), or rmWNT5a 0.7–20 nM (Fig. 6f). Differences in the distribution of RFU by plate and date of experiment are shown in boxplots for rmSFRP2 and rmWnt5a (Supplemental figure 1). rmSFRP2 exposure induced a concentration-dependent increase in intracellular free calcium in sham 2H11 cells (Fig. 6e). In shFZD5 2H11 cells, rmSFRP2-induced calcium release was reduced with a maximum effect at 30 nM (p=0.0002). A concentration-dependent increase in intracellular free calcium was also observed upon exposure to rmWNT5a (Fig. 6f). However, while this calcium release was reduced in FZD5 silenced cells, the difference was not statistically significant. Therefore, in endothelial cells, SFRP2 modulates intracellular calcium levels through a FZD5-dependent mechanism.
FZD5 mediates endothelia and breast cancer cell migration
Migration is an important process during tumor growth as it allows tumor cells and cells from the micro-environment to constantly reorganize the structure of the primary tumor, in order to facilitate its expansion in local tissues. In later stages, migration is a required mechanism for metastatic spread towards local and distant organs. SFRP2 promotes migration of both endothelial and tumor cells[10,11]. To test whether this function of SFRP2 involves FZD5, we used a trans-well migration assay. We first asked whether rhSFRP2 would promote the migration of a mouse 2H11 endothelial cells (Fig. 7a). hSFRP2 significantly increased their migration compared to control by 27 +/− 2.5 %; p < 0.05. While rhFZD5-Fc didn’t have any substantial anti-migratory effect on its own, rhSFRP2 with rhFZD5-Fc completely abrogated rhSFRP2-induced migration (p<0.05). To determine if SFRP2 effect on murine endothelial cell migration was a general function of SFRP2 in human cellular models, we performed the same assay on the HMEC-1 human endothelial cell line (Fig. 7b). Again, rhSFRP2 treatment increased cell migration (27 +/− 2.1 %; p < 0.05) and this effect was blocked by rhFZD5-Fc (p<0.05). Finally, to determine whether SFRP2 function is dependent on FZD5 in tumor cells, we used the MDA-MB-231 breast cancer cell line (Fig. 7c). hSFRP2 promoted migration in the same proportions as it did for murine and human endothelial cells (29 +/− 2.7%; p < 0.05), and this effect was inhibited again by a pre-incubation with rhFZD5-Fc (p<0.05). Each experiment was performed in triplicate and repeated 3 times (n=9). Together, our results support a general pro-migratory function for SFRP2 and suggest that this function is FZD5-dependent.
Figure 7. FZD5 is necessary for SFRP2 induced cell migration.
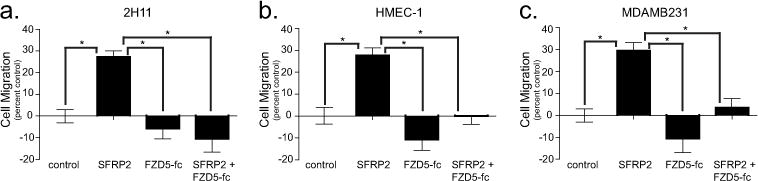
(A) 2H11 (1.5 × 104 cells), (B) HMEC-1 (2.5 × 104 cells), and (C) MDA-MB-231 cells (1.5 × 104 cells) control, treated with rhSFRP2 (30 nM), rhFZD5-Fc (500 ng/ml), or a combination of both were allowed to migrate for 24h in a trans-well assay. As depicted in the bar graphs (A–C), rhSFRP2 significantly promoted the migration of endothelial (A, B) and tumor cells (C) in similar proportions (~25%). rhFZD5-Fc alone had no significant effect on cell migration. However, when pre-incubated with rhSFRP2, rhFZD5-fc blunted the pro-migratory effects of rhSFRP2. Results represent the percent of migrating cells (+/− SEM), compared to control, and are the average of 3 independent experiments (n = 9). *: p<0.05.
DISCUSSION
Previously, the known functions of SFRPs were ascribed to their ability to antagonize Wnt signaling by directly interacting with Wnt ligands and acting as a trap. This led to the assumption that SFRPs could have tumor suppressive functions [28]. Conversely, other groups reported that SFRP2 is an agonist of β-catenin [29–32], suggesting a pro-tumoral effect of this ligand. To clarify whether SFRP2 increases or decreases as tumors progress, we previously found that SFRP2 signal intensity increased as a function of tumor volume using SFRP2-targeted molecular imaging of a mouse tumor in vivo [33]. Other studies confirmed our observation showing that overexpression of SFRP2 increases angiogenesis and tumor growth in vitro and in vivo, [8,9,4,6,34,1,7,2,3]. Furthermore, antagonizing SFRP2 with a monoclonal antibody inhibits tumor growth in vivo [10], which establishes the preponderance of evidence that SFRP2 stimulates, rather than suppresses, tumor growth. However, until recently, the signaling pathways used by SFRP2 to promote angiogenesis were unknown.
Recent studies provide evidence that SFRPs also bind proteins distinct from Wnt, and can exert other biological functions (2). For example, SFRP2 functions as an enhancer of collagen processing and cardiac fibrosis by binding to bone morphogenetic protein 1 and regulating the procollagen-C proteinase [35]. SFRP1 functions as an axon guidance cue by interacting with Frizzled-2 in a Wnt-independent manner [36]. In elucidating the SFRP2 signaling pathway for stimulation of angiogenesis, we found that β-catenin was not required [9]. Rather, we identified a novel function of SFRP2: induction of angiogenesis via the calcineurin/NFAT pathway [9–11], and in this study we identified the receptor that mediates this signaling pathway.
We have introduced several lines of evidence in support of our hypothesis that FZD5 is the receptor for SFRP2. First, SFRP2 bound in an ELISA competition assay with a Kd of 103 +/− 6 nM, compared to a Kd of 108 +/− 4 nM between FZD5 and its ligand WNT5a. Second, we used co-immunofluorescence to show SFRP2 co-localized with FZD5 in endothelial and angiosarcoma cells at the plasma membrane. Third, SFRP2 and FZD5 co-immunoprecipated in 2H11 and MS1 endothelial cells. Functionally, when FZD5 is silenced in endothelial cells, SFRP2 fails to induce endothelial cell tube formation, while SFRP2 stimulates endothelial tube formation in control endothelial cells. Silencing SFRP2 in endothelial cells blocks SFRP2-induced NFATc3 activation and intracellular calcium influx. Finally, FZD5 is required for SFRP2 migration in both endothelial and breast cancer cells. Identification of this novel SFRP2/FZD5 signaling pathway and their influence on calcineurin/NFATc3 activation and angiogenesis may increase our understanding of the complex mechanisms involved in tumor growth and may facilitate development of novel therapies to combat cancer.
Supplementary Material
Acknowledgments
We thank the Protein Expression and Purification Core Lab at University of North Carolina at Chapel Hill for production of human SFRP2 recombinant protein. This work was supported by National Institute of Health (1R01CA142657-A1 and 1U01CA189281-01A1 to NK-D), and the Kristin Ann Carr Foundation. Microscopy was supported in part by the Cell and Molecular Imaging Shared Resource, Hollings Cancer Center, Medical university of South Carolina (P30 CA138313).
Footnotes
The authors declare no conflict of interest.
References
- 1.Roth W, Wild-Bode C, Platten M, Grimmel C, Melkonyan HS, Dichgans J, Weller M. Secreted Frizzled-related proteins inhibit motility and promote growth of human malignant glioma cells. Oncogene. 2000;19(37):4210–4220. doi: 10.1038/sj.onc.1203783. [DOI] [PubMed] [Google Scholar]
- 2.Yamamura S, Kawakami K, Hirata H, Ueno K, Saini S, Majid S, Dahiya R. Oncogenic functions of secreted Frizzled-related protein 2 in human renal cancer. MolCancer Ther. 2010;9(6):1680–1687. doi: 10.1158/1535-7163.MCT-10-0012. [DOI] [PubMed] [Google Scholar]
- 3.Xiao X, Xiao Y, Wen R, Zhang Y, Li X, Wang H, Huang J, Liu J, Long T, Tang J. Promoting roles of the secreted frizzled-related protein 2 as a Wnt agonist in lung cancer cells. Oncology reports. 2015;34(5):2259–2266. doi: 10.3892/or.2015.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaur A, Webster MR, Marchbank K, Behera R, Ndoye A, Kugel CH, 3rd, Dang VM, Appleton J, O’Connell MP, Cheng P, Valiga AA, Morissette R, McDonnell NB, Ferrucci L, Kossenkov AV, Meeth K, Tang HY, Yin X, Wood WH, 3rd, Lehrmann E, Becker KG, Flaherty KT, Frederick DT, Wargo JA, Cooper ZA, Tetzlaff MT, Hudgens C, Aird KM, Zhang R, Xu X, Liu Q, Bartlett E, Karakousis G, Eroglu Z, Lo RS, Chan M, Menzies AM, Long GV, Johnson DB, Sosman J, Schilling B, Schadendorf D, Speicher DW, Bosenberg M, Ribas A, Weeraratna AT. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature. 2016;532(7598):250–254. doi: 10.1038/nature17392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Techavichit P, Gao Y, Kurenbekova L, Shuck R, Donehower LA, Yustein JT. Secreted Frizzled-Related Protein 2 (sFRP2) promotes osteosarcoma invasion and metastatic potential. BMC cancer. 2016;16(1):869. doi: 10.1186/s12885-016-2909-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JL, Chang CJ, Chueh LL, Lin CT. Secreted Frizzled Related Protein 2 (sFRP2) Decreases Susceptibility to UV-Induced Apoptosis in Primary Culture of Canine Mammary Gland Tumors by NF-kappaB Activation or JNK Suppression. Breast Cancer Res Treat. 2006;100(1):49–58. doi: 10.1007/s10549-006-9233-9. [DOI] [PubMed] [Google Scholar]
- 7.Sun Y, Zhu D, Chen F, Qian M, Wei H, Chen W, Xu J. SFRP2 augments WNT16B signaling to promote therapeutic resistance in the damaged tumor microenvironment. Oncogene. 2016;35(33):4321–4334. doi: 10.1038/onc.2015.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhati R, Patterson C, Livasy CA, Fan C, Ketelsen D, Hu Z, Reynolds E, Tanner C, Moore DT, Gabrielli F, Perou CM, Klauber-DeMore N. Molecular characterization of human breast tumor vascular cells. Am J Pathol. 2008;172(5):1381–1390. doi: 10.2353/ajpath.2008.070988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courtwright A, Siamakpour-Reihani S, Arbiser JL, Banet N, Hilliard E, Fried L, Livasy C, Ketelsen D, Nepal DB, Perou CM, Patterson C, Klauber-DeMore N. Secreted Frizzle-Related Protein 2 Stimulates Angiogenesis via a Calcineurin/NFAT Signaling Pathway. Cancer Res. 2009;69(11):4621–4628. doi: 10.1158/0008-5472.CAN-08-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontenot E, Rossi E, Mumper R, Snyder S, Siamakpour-Reihani S, Ma P, Hilliard E, Bone B, Ketelsen D, Santos C, Patterson C, Klauber-DeMore N. A novel monoclonal antibody to secreted frizzled-related protein 2 inhibits tumor growth. MolCancer Ther. 2013;12(5):685–695. doi: 10.1158/1535-7163.MCT-12-1066. doi:1535-7163.MCT-12-1066 [pii];10.1158/1535-7163.MCT-12-1066 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siamakpour-Reihani S, Caster J, Bandhu ND, Courtwright A, Hilliard E, Usary J, Ketelsen D, Darr D, Shen XJ, Patterson C, Klauber-DeMore N. The Role of Calcineurin/NFAT in SFRP2 Induced Angiogenesis-A Rationale for Breast Cancer Treatment with the Calcineurin Inhibitor Tacrolimus. PLoSOne. 2011;6(6):e20412. doi: 10.1371/journal.pone.0020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minami T, Horiuchi K, Miura M, Abid MR, Takabe W, Noguchi N, Kohro T, Ge X, Aburatani H, Hamakubo T, Kodama T, Aird WC. Vascular endothelial growth factor- and thrombin-induced termination factor, Down syndrome critical region-1, attenuates endothelial cell proliferation and angiogenesis. J BiolChem. 2004;279(48):50537–50554. doi: 10.1074/jbc.M406454200. [DOI] [PubMed] [Google Scholar]
- 13.Zaichuk TA, Shroff EH, Emmanuel R, Filleur S, Nelius T, Volpert OV. Nuclear factor of activated T cells balances angiogenesis activation and inhibition1. J ExpMed. 2004;199(11):1513–1522. doi: 10.1084/jem.20040474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De A. Wnt/Ca2+ signaling pathway: a brief overview. Acta BiochimBiophysSin(Shanghai) 2011;43(10):745–756. doi: 10.1093/abbs/gmr079. doi:gmr079 [pii];10.1093/abbs/gmr079 [doi] [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa T, Tamai Y, Zorn AM, Yoshida H, Seldin MF, Nishikawa S, Taketo MM. Mouse Wnt receptor gene FZD5 is essential for yolk sac and placental angiogenesis. Development. 2001;128(1):25–33. doi: 10.1242/dev.128.1.25. [DOI] [PubMed] [Google Scholar]
- 16.Kumawat K, Menzen MH, Bos IS, Baarsma HA, Borger P, Roth M, Tamm M, Halayko AJ, Simoons M, Prins A, Postma DS, Schmidt M, Gosens R. Noncanonical WNT-5A signaling regulates TGF-beta-induced extracellular matrix production by airway smooth muscle cells. FASEB J. 2013;27(4):1631–1643. doi: 10.1096/fj.12-217539. [DOI] [PubMed] [Google Scholar]
- 17.Liu C, Nathans J. An essential role for frizzled 5 in mammalian ocular development. Development. 2008;135(21):3567–3576. doi: 10.1242/dev.028076. doi:dev.028076 [pii];10.1242/dev.028076 [doi] [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Fuhrmann S, Vetter ML. A nonautonomous role for retinal frizzled-5 in regulating hyaloid vitreous vasculature development. Invest OphthalmolVisSci. 2008;49(12):5561–5567. doi: 10.1167/iovs.08-2226. doi:iovs.08-2226 [pii];10.1167/iovs.08-2226 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walter-Yohrling J, Morgenbesser S, Rouleau C, Bagley R, Callahan M, Weber W, Teicher BA. Murine endothelial cell lines as models of tumor endothelial cells. Clin Cancer Res. 2004;10(6):2179–2189. doi: 10.1158/1078-0432.ccr-03-1013. [DOI] [PubMed] [Google Scholar]
- 20.Arbiser JL, Bonner MY, Berrios RL. Hemangiomas, angiosarcomas, and vascular malformations represent the signaling abnormalities of pathogenic angiogenesis. CurrMolMed. 2009;9(8):929–934. doi: 10.2174/156652409789712828. [DOI] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Biesiadecki BJ, Jin JP. A high-throughput solid-phase microplate protein-binding assay to investigate interactions between myofilament proteins. JBiomedBiotechnol. 2011;2011:421701. doi: 10.1155/2011/421701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochemical pharmacology. 1973;22(23):3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 24.Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300(4):C723–742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM. Toward a Shared Vision for Cancer Genomic Data. The New England journal of medicine. 2016;375(12):1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dijksterhuis JP, Baljinnyam B, Stanger K, Sercan HO, Ji Y, Andres O, Rubin JS, Hannoush RN, Schulte G. Systematic mapping of WNT-FZD protein interactions reveals functional selectivity by distinct WNT-FZD pairs. The Journal of biological chemistry. 2015;290(11):6789–6798. doi: 10.1074/jbc.M114.612648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J Cell Sci. 2008;121(Pt 6):737–746. doi: 10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- 29.Esteve P, Sandonis A, Ibanez C, Shimono A, Guerrero I, Bovolenta P. Secreted frizzled-related proteins are required for Wnt/beta-catenin signalling activation in the vertebrate optic cup. Development. 2011;138(19):4179–4184. doi: 10.1242/dev.065839. [DOI] [PubMed] [Google Scholar]
- 30.Gehmert S, Sadat S, Song YH, Yan Y, Alt E. The anti-apoptotic effect of IGF-1 on tissue resident stem cells is mediated via PI3-kinase dependent secreted frizzled related protein 2 (Sfrp2) release. BiochemBiophysRes Commun. 2008;371(4):752–755. doi: 10.1016/j.bbrc.2008.04.151. [DOI] [PubMed] [Google Scholar]
- 31.Lee JL, Chang CJ, Wu SY, Sargan DR, Lin CT. Secreted frizzled-related protein 2 (SFRP2) is highly expressed in canine mammary gland tumors but not in normal mammary glands. Breast Cancer Res Treat. 2004;84(2):139–149. doi: 10.1023/B:BREA.0000018412.83348.ff. [DOI] [PubMed] [Google Scholar]
- 32.Melkonyan HS, Chang WC, Shapiro JP, Mahadevappa M, Fitzpatrick PA, Kiefer MC, Tomei LD, Umansky SR. SARPs: a family of secreted apoptosis-related proteins. Proc NatlAcadSciUSA. 1997;94(25):13636–13641. doi: 10.1073/pnas.94.25.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuruta JK, Klauber-DeMore N, Streeter J, Samples J, Patterson C, Mumper RJ, Ketelsen D, Dayton P. Ultrasound molecular imaging of secreted frizzled related protein-2 expression in murine angiosarcoma. PLoSOne. 2014;9(1):e86642. doi: 10.1371/journal.pone.0086642. doi:10.1371/journal.pone.0086642 [doi];PONE-D-13-36396 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma X, Wang X, Gao X, Wang L, Lu Y, Gao P, Deng W, Yu P, Ma J, Guo J, Cheng H, Zhang C, Shi T, Ma D. Identification of five human novel genes associated with cell proliferation by cell-based screening from an expressed cDNA ORF library. Life sciences. 2007;81(14):1141–1151. doi: 10.1016/j.lfs.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi K, Luo M, Zhang Y, Wilkes DC, Ge G, Grieskamp T, Yamada C, Liu TC, Huang G, Basson CT, Kispert A, Greenspan DS, Sato TN. Secreted Frizzled-related protein 2 is a procollagen C proteinase enhancer with a role in fibrosis associated with myocardial infarction. NatCell Biol. 2009;11(1):46–55. doi: 10.1038/ncb1811. doi:ncb1811 [pii];10.1038/ncb1811 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez J, Esteve P, Weinl C, Ruiz JM, Fermin Y, Trousse F, Dwivedy A, Holt C, Bovolenta P. SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. NatNeurosci. 2005;8(10):1301–1309. doi: 10.1038/nn1547. doi:nn1547 [pii];10.1038/nn1547 [doi] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


