Abstract
Purpose of the review
Increasingly, there is a need for examining exposure disease associations in large, diverse datasets to understand the complex determinants of pediatric disease and disability. Recognizing that children’s health research consortia will be important sources of big data, it is crucial for the pediatric research community to be knowledgeable about the challenges and opportunities that they will face. This review will provide examples of existing children’s health consortia; highlight recent pooled analyses conducted by children’s health research consortia; address common challenges of pooled analyses and provide recommendations to advance collective research efforts in pediatric research.
Recent findings
Formal consortia and other collective-science initiatives are increasingly being created to share individual data from a set of relevant epidemiological studies to address a common research topic. There are practical challenges to the participation of investigators within consortia that need to be addressed, including providing centralized data management, addressing barriers to data sharing and harmonization of data across studies and respecting the data ownership of original investigators.
Summary
Researchers who access consortia with data centers will be able to go far beyond their initial hypotheses and potentially accomplish research that was previously thought infeasible or too costly.
Key phrases: children’s health research consortia, pooled analyses, data sharing, ontology
Introduction
There is a need for examining exposure disease associations in large, diverse datasets in order to understand the complex determinants of pediatric disease and disability. Combining data across studies has great potential to advance pediatric research by increasing the sample size and scope of scientific hypotheses examined. Part of this process involves groups of scientists collaborating to address more complex and ambitious research questions than can be achieved by a single investigator [1]. Collective study approaches are important as more data can improve assessment of weak risk factors in health outcomes that may have large public health implications. Larger datasets also facilitate the investigation of interactions between exposures and the identification of vulnerable sub-groups within the population, which smaller individual studies may not have adequate power to detect. Additionally, decisions in health and public policy are best supported by findings observed in several studies or in large datasets with large, diverse populations.
Most collective study approaches in children’s health have either combined summary statistics using meta-analyses or combined primary data from several studies in pooled analyses. Meta-analyses provide an objective risk estimate of effect of interest but can be subject to publication bias. Pooled analyses combine the actual individual subject level data so that novel and potentially more detailed analyses are possible than what was conducted by each individual study. However, differences in data collection, variable construction and limitations of data sharing can limit researchers’ ability to conduct pooled analyses. To address some of these concerns, formal consortia and other collective-science initiatives are increasingly being created to share individual data from a set of relevant epidemiological studies to address a common research topic under the concept that the joint effort of many individual groups can accomplish far more than working alone.
As consortia grow in size, the quantity and complexity of data collected also rises. The challenges and opportunities presented by the task of combined analysis of diverse data types are now commonly referred to as biomedical big data science [2]. Indeed, big data techniques and aggregated data sets have already given us new power to analyze data sets in novel ways [3, 4]. Recognizing that children’s health research consortia will be important sources of big data, it is crucial for the pediatric research community to be knowledgeable about the challenges and opportunities that they will face.
This review will provide examples of existing children’s health consortia; highlight recent pooled analyses conducted by children’s health research consortia; address common challenges of pooled analyses of disparate data and provide recommendations to advance collective research efforts in pediatric research. Throughout this review, whenever appropriate we will use the experience of the Children’s Health Exposure Analysis Resource (CHEAR) Data Center [5] to provide some insight into these issues.
Current approaches to pediatric consortia and pooled-analyses in children’s health research
There has been a growing interest in establishing pediatric research consortia and other collective science efforts in order to advance children’s health research [6]. Pediatric research consortia can be organized around a specific disease (e.g. Pediatric Diabetes Consortium [7]), a common research field (e.g. Sanford Children’s Genomic Medicine Consortium [8]) or as a methodological resource within a specific field (e.g. FaceBase Consortium [9]). An example of the third type of consortia, CHEAR is a comprehensive set of resources to enable NIH-funded researchers to include or broaden analyses of environmental exposures in their existing studies of children’s health. It is composed of exposure assessment laboratories, a coordinating center and the Data Repository, Analysis, and Science Center (referred to herein as Data Center). In addition to formal consortia constructed in the early stages of research, there have been a number of examples of ongoing or completed epidemiologic studies with common themes that begin to collectively organize in order to conduct pooled analyses. The Environmental Health Risks in European Birth Cohorts [10] is an example of this type of consortia and includes efforts to compile inventories of birth cohorts across Europe and develop methods to conduct analyses across studies. Many of these children’s health research consortia have pooled their individual studies’ data to address a common hypothesis. This approach provides greater statistical power to address not only the main exposure-outcome relationships, but also exposure interactions that could not be addressed using an individual study’s population, when the general association is similar across studies.
Several recent children’s health pooled analyses, addressing a wide variety of exposure-outcome associations, have been published [11–15]. Each of these pooled analyses have identified strengths and limitations encountered through the use of this approach. Table 1 provides a summary of these selected publications. Overall, these studies have been able to address children’s health issues where the existing body of published literature has not provided conclusive results and achieved greater statistical power to address these issues by combining several individual studies. However, the process of pooling studies that may not have collected their data identically poses several limitations. For example, missing data, a universal problem in epidemiologic data, can pose additional problems when pooling data if a potential confounding variable cannot be included in an analysis because it only available in some, but not all of the studies. Additionally, data harmonization of variables intended to capture the same information using different response levels needs to be carefully considered during pooling efforts. Similar variables have to be reduced to the lowest common set of responses, e.g., a multilevel smoking history variable must be converted to “ever/never smoked” for all studies to have a comparable measure, resulting in lost information. Often, these strengths and weakness were noted by the authors of the pooled analyses (Table 1).
Table 1.
Recently published children’s health study pooled analyses
| First Author (Publication date) | Study description | Primary study aim | Author identified strengths and limitations |
|---|---|---|---|
| Casas (2015) [11] | 9377 mother–child pairs enrolled in 14 study populations from 11 European birth cohorts | Explore exposure–response relationship between PCB-153 and p-p′-DDE and birth outcomes; to evaluate whether any no exposure–effect level and susceptible subgroups exist; and to assess the role of maternal gestational weight gain. | Able to harmonize common potential confounders but with loss of some particular and valuable cohort characteristics. Only one PCB congener 153 was measured in all cohorts. There are 209 PCB congeners that differ in structure and mechanism of action and hence may have different health outcomes. Large sample size allowed better description of the shape of the relationship and identification of effect modifiers. |
| Iszatt (2015) [12] | Up to 2,487 children from 7 European birth cohorts | Using biomarker concentrations of polychlorinated biphenyl 153 (PCB-153) (n = 2,487), and p,p′-dichlorodiphenyldichloroethylene (p,p′-DDE) (n = 1,864), estimate prenatal and postnatal persistent organic pollutants (POPs) exposure using a validated pharmacokinetic model and examine POP exposure association with infant growth from birth to 24 months in singleton term children. | The largest study to date using pooled data across larger samples of individuals with heterogeneous and distinct prenatal/postnatal exposure profiles. Compared with single-cohort studies, the pooled design had:
Although variance inflation factors were < 5, variance doubled when prenatal and postnatal were mutually adjusted, suggesting collinearity. |
| Buckley (2016) [13] | 707 children from three prospective cohort studies enrolled in the US between 1998 and 2006 | Examine associations of prenatal urinary phthalate metabolite concentrations and body mass index (BMI) assessed in children between ages 4 and 7 years and evaluated differences by child’s sex. | Pooling data from three independent cohorts with notable variation in population characteristics strengthened the robustness of the findings. Provided a large sample size to assess heterogeneity of associations by hypothesized modifying factors. Potential bias due to missing covariate data and loss to follow-up may exist. |
| Engel SM. (2016) [14] | pooled analysis of four birth cohorts (children’s centers; n = 936) | Evaluate associations of prenatal exposure to organophosphorus pesticides (OPs) with mental and psychomotor development of children 24 months of age, taking into account both genetic and demographic susceptibility factors. | Both confounder adjustment and examination of heterogeneity were limited to covariates and characteristics that were shared by all centers. Although pooling these cohorts afforded more power to investigate gene–environment interactions, power was still limited in stratified analyses. Pooled analyses improved the ability to determine whether there is an overall effect of OP exposure experienced in diverse settings on child neurodevelopment both for policy and for research purposes. |
| Stratakis (2016) [15] | Multicenter, population-based birth cohort study from 1996 to 2011 in 9 European countries and the US 26,184 pregnant women and their children | To examine whether fish intake in pregnancy is associated with offspring growth and the risk of childhood overweight and obesity. | Strengths include large sample size, centralized data analysis following a consensus protocol, standardized exposure definition, and harmonized information about child outcomes and potential confounders. Potential confounding variables were defined as similarly as possible among the cohorts. |
Addressing Challenges to Pooled Data Analyses
Creating Centralized Data Management for Construction and Maintenance of Datasets
As data has played a more central role in enabling research [16], often consortia will have a single data center that provides services to the member organizations through a central repository for the data collected by the members as well as statistical and data analytic services for joint research projects. The National Heart, Lung, and Blood Institute (NHLBI) has created a compendium of best practices for data coordinating centers [17]. Among the key elements identified as requirements for a well-functioning data center, emphasis is put on data management including security, data systems, reporting and analysis as well as quality assurance/quality control. The research productivity of the consortium is dependent on the effectiveness of the data center to ingest, organize, analyze and share data. Recognizing this critical role, the CHEAR Data Center [5], for example, has focused on developing a comprehensive data repository geared to facilitate maximum analysis, sharing, and interoperability of exposure data analyzed within the CHEAR network, following the FAIR (Findable, Accessible, Interoperable, and Re-usable) principles [18]. Specifically, the CHEAR Data Center has created standardized templates for lab and epidemiologic data, so data across studies can be automatically ingested and harmonized. Data can be accessed by study investigators from anywhere through the use of web portals and cloud-based technologies for data storage and deidentified publicly-available data.
Promoting Data Sharing while Maintaining Data Security
The sharing of data derived from human subjects—making them both transparent and accessible to others—raises a host of ethical, scientific, and process questions that are not always present in other areas of science, such as physics, geology, or chemistry [19]. Concerns about protection of the privacy of research participants whose data are shared beyond the original investigators are often raised. One must verify that the original informed consent of research participants from whom the data were collected permits data sharing beyond the original purpose. Investigators who submit their data to data repositories should also verify that appropriate data security, confidentiality, and privacy measures are in place for protection of research participants.
The creation of a public data repository requires a formal data sharing document that outlines requirements of both the submitter and the entity that will hold the data. This document should clearly state best practices in computer security standards to ensure the protection of privacy and confidentiality of the participant. Practices must be in place to ensure verification of informed consent, an embargo period and safe computer practices on both the research user the data and repository. Additionally, all data deposited in a public repository should be de-identified using best practices, such as the standards detailed in the HIPAA privacy rule [20]. Recent research suggests that re-identification of data de-identified using those standards was very low [21].
The open access and community approach is similar to that of collaborative software development, where the focus is on availability and communication [22]. The NIH has invested in large-scale “data commons” efforts with the overall goal of making digital objects and tools available to foster research collaboration. Two recent efforts include the Office of Data Science’s data commons platform [23] and National Cancer Institute’s (NCI) newly unveiled Genomic Data Commons [24]. NIH Office of Data Science aims “to enable biomedical research as a digital enterprise through which new discoveries are made and knowledge generated by maximizing community engagement and productivity” [25].
Data Harmonization Across Studies to Allow Pooling
As discussed above, pooling data across different studies depends on variables having similar response levels. This process can be greatly facilitated by utilizing standardized terms within a field and/or using those terms to find similarities between variables across studies [6]. Ontologies can play a key role in this process. Ontologies are often defined as specifications of conceptualizations; they contain explicit, computer-understandable descriptions of term meanings and inter-relationships. As a result, ontologies provide a common vocabulary for variables and concepts across different studies, facilitating the pooling of data across studies. For example, Figure 1 shows the variable constructions for maternal education from three hypothetical studies. Mapping each study’s variable levels to the education terms contained within the ontology illustrates that pooling across the studies will require a variable with the 3 education terms that are common across the studies. This could be managed manually for three studies, but as consortia grow to tens and potentially hundreds of studies, having an ontology enables the final variable construction to be automatically generated, ensuring consistency and reducing time and computer programming for the consortium’s data center.
Figure 1.
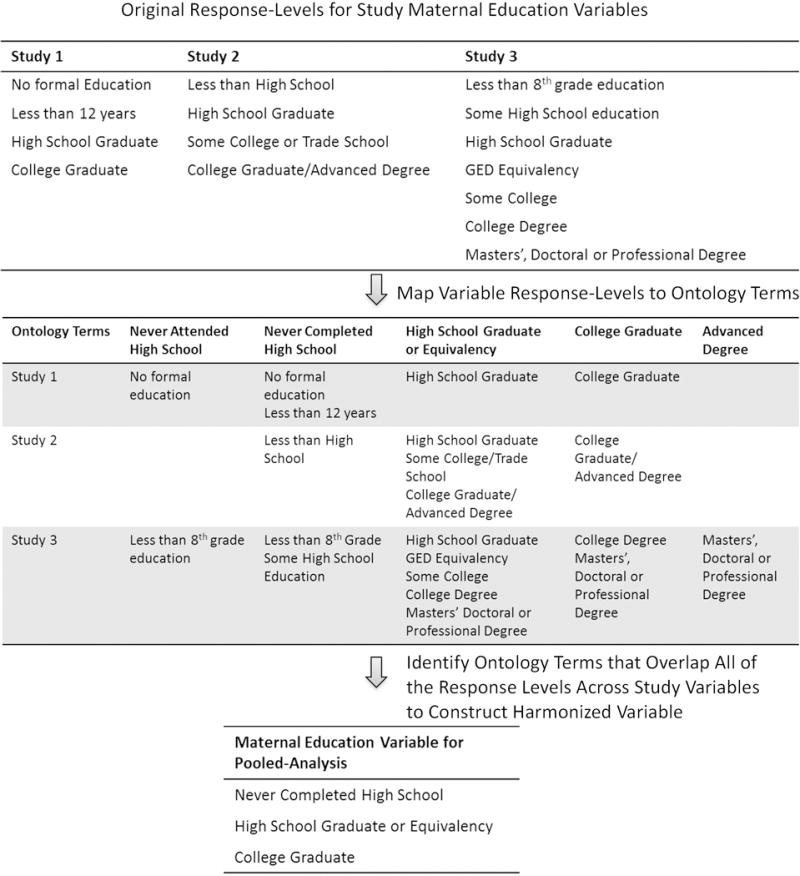
Construction of Harmonized Variables Across Disparate Studies using Standardized Terms within an Ontology. Using maternal education as an illustrative example, variable harmonization starts by detailing the response levels of the variable across disparate studies and then mapping to common terms that encompass all of the response levels. These terms then form the response-levels of the new, harmonized variable that can be used in pooled analyses.
Ontologies not only promote harmonization of data across studies, they also enable technologies that can query the entire data warehouse of consortia using standard terms. This enables researchers to construct customized datasets that pool data across studies. In the CHEAR effort, using the terms within the ontology, end-users can browse the types of data available within CHEAR studies using a dynamically-generated study browser. Through this portal, users can identify the specific studies which contain their variables of interest and download a customized, and harmonized dataset across all studies within the CHEAR network.
Respecting Data Ownership by Original Investigators
Harmonization of data across studies is greatly facilitated when all data are shared, including covariates, and are in raw form, before individual study decisions on categorization or aggregation create inconsistencies across studies. Despite numerous commentaries and standing NIH policies promoting data-sharing, many investigators remain reluctant to share raw data [26–29]. Data “ownership” is often cited as a primary concern of many investigators [27, 28]. There is general agreement that researchers who originally collect and create the data have a legitimate expectation to publish before the data are shared. Investigators need to be reassured that the sizable investments in time and resources used to collect data will be recognized. This can be achieved through the use of embargo periods that prevent data-release before main study results publication thus allowing the investigator the opportunity to publish and for timely data-sharing.
Recognizing investigator concerns, CHEAR will allow an embargo period that ends after publication of the first analysis that used data generated by the CHEAR resources. Furthermore, CHEAR has implemented a data submission agreement and data sharing agreement, based on existing documents from NIH funded projects, e.g., National Database of Autism Research [30] and the NIH Genetics Data Sharing Plan [31], so that investigators are fully informed about the project policies that they are agreeing to through their use of CHEAR. To enable any digital object to be located, the California Digital Library has a service called EZID, which creates unique Digital Object Identifiers (DOI) [32]. We have employed this service on the CHEAR data repository, so that all datasets are automatically given this identifier, which enables the original researcher to receive “credit” for the work required to collect, clean and upload the data.
Creating Incentives for Participation in Collective Science
Investigators also have expressed concerns on how participation in collective efforts can affect career progression and receiving appropriate credit for resulting work [29, 33]. Metrics used to evaluate promotion and tenure are often focused on the individual’s contribution through first-authored papers and submission of grants as principal investigators [33]. Some of these concerns can be addressed through the adoption of thoughtful authorship guidelines within collective efforts, such as the CHEAR publication policy [34]. However, there is a real need for academic institutions to begin recognizing the data contributions individuals make to collective science. The NIH Office of Data Science recognizes this need, and is working to change the culture so recognition is possible [35]. Metrics that quantify the participation in collective efforts should be added to the standard promotion metrics. For example, in an effort to promote interdisciplinary research, Benson et al recommended up-weighting publications in journals representative of other academic fields [36]. The same strategy could be used for non-first author publications that resulted from participation in consortia or other collective-science activities. As the amount of consortia-based research continues to grow in order to address the complex research questions of pediatric health and disease, there is a need for creative strategies to ensure that investigators’ efforts are appropriately recognized and rewarded.
Discussion
The CHEAR Data Center and other pediatric consortia and collective-science efforts are addressing many of the challenges of combining data across studies. We encourage all children’s health researchers to consider what research questions could be addressed with the greater power and increased diversity facilitated by combining data across studies. These efforts could be encouraged by strengthening the collective resources in children’s health. For example, a centralized repository of children’s cohort descriptions would allow for the identification of studies with similar approaches and related data thereby facilitating pooled analyses. Similar efforts have been conducted in Europe and should be implemented within the US. Such a repository could be constructed from the published detailed summaries of epidemiologic cohorts. A PubMed search using the terms “cohort profile” and “children” identified 116 publications that contained this type of information. Table 2 provides a summary of the six cohort profiles published in 2016 [37–42]. While inclusion in such a repository would require further investigation of many of the issues discussed above, including data-sharing permissions and investigator interest, we encourage pediatric societies and individual researchers to begin to think about how shared resources can be created to serve the larger pediatric research community.
Table 2.
Recently published children’s health cohort study descriptions
| Cohort Study Name | Cohort Profile First Author | Study Design | Recruitment and follow-up | Geographic location | Study Aims |
|---|---|---|---|---|---|
| The VicGeneration (VicGen) study: An Australian oral health birth cohort. [37] | Johnson S | Birth cohort, 465 mother-child and 1 father-child dyad | Seven waves of data collection have been conducted over 8 years (2008–15) | State of Victoria, Australia | Explore three significant areas of research in child oral health: describing the natural history of caries (decay) from infancy to 5 years; characterization of the salivary microbiota in infants, from 1 month of age through to early childhood, and that of their primary caregiver; and examine the relative contributions of biological, environmental and socio-behavioural factors. |
| The Norwegian Mother and Child Cohort Study (MoBa). [38] | Magnus P | Pregnancy cohort with a family design; as of Sept 2015, the cohort includes more than 114 000 children, 95 000 mothers and 75 000 fathers | Recruited pregnant mothers and fathers-to-be from 1999 through 2008; Follow-up include questionnaire completion during pregnancy, and when the child was 6 months, 18 months, 3, 5, 7, 8 and 13 years old as well as linkages to health registries. | Norway, includes 50 of Norway’s 52 hospitals with maternity units. | The main aim of MoBa is to detect causes of serious diseases through estimation of specific exposure-outcome associations especially among the children but also among parents. |
| Hamamatsu Birth Cohort for Mothers and Children (HBC Study). [39] | Takagai S | Birth cohort; 1258 neonates (including 38 twins) from 1138 mothers. | Recruited between November 2007 and March 2011; Follow-up started during pregnancy and continued during infancy at 1, 4, 6, 10, 14, 18, 24, 32, and 40 months, 4.5 years, 6 years and 8 years. Further follow-up visits are also planned. | All but five of the neonates were born at the University Hospital of Hamamatsu University School of Medicine, Japan. | Designed to elucidate the early developmental trajectories of children living in the community in Japan. |
| The EDEN mother-child cohort on the prenatal and early postnatal determinants of child health and development. [40] | Heude B | Birth cohort; 2002 pregnant women were enrolled in the study (1034 women from Nancy and 968 from Poitiers) | Recruitment extended from 2003 to 2006; followed-up for up to 8 years. | Two university maternity clinics, in Nancy and Poitiers, France | The overall objective was to examine the relations and potential interactions between maternal exposures and health status during pregnancy, fetal development, health status of the infant at birth and the child’s health and development |
| The Saguenay Youth Study (SYS). [41] | Pausova Z | Two-generational study of adolescents and their parents (n= 1029 adolescents and 962 parents) | Recruitment and assessment took place over a 10-year period (2003–12) and a second follow-up period took place from 2012 and 2015. | Genetic founder population of the Saguenay Lac St Jean region of Quebec, Canada | Investigating the etiology, early stages and trans-generational trajectories of common cardiometabolic and brain diseases. The ultimate goal of this study is to identify effective means for increasing healthy life expectancy. |
| The Health Outcomes and Measures of the Environment (HOME) study. [42] | Braun JM | Prospective pregnancy and birth cohort; 401 pregnancy women were enrolled. | Recruitment between March 2003 and January 2006; Follow-up during pregnancy and when children were 5 weeks and 1, 2, 3, 4, 5 and 8 years old. | Greater Cincinnati OH metropolitan area | To determine whether early life environmental chemical exposures influence children’s health |
Conclusions
We have discussed the need for collective approaches in studying children’s health and briefly reviewed the current methodology of combining studies and presented many issues that arise when collaborative research takes place. There are practical challenges to the participation of investigators within these frameworks that need to be addressed for them to work. Pooling of multiple datasets in a cohesive and integrated manner can be made more effective through a data center and we have used the CHEAR Data Center as an example that has the infrastructure to enable the measurement and integration of environmental exposures from multiple children’s health studies. Researchers who access consortia with data centers will be able to go far beyond their initial hypotheses and potentially accomplish research that was previously thought infeasible or too costly.
Key points.
Children’s health research consortia are sources of large, diverse datasets which are important for examining exposure disease associations however, it is crucial for the pediatric research community to be knowledgeable about the challenges and opportunities of consortia.
Pooling of multiple datasets in a cohesive and integrated manner can be made more effective through a data center and we have used the CHEAR Data Center as an example that has the infrastructure to enable integration of environmental exposures from multiple children’s health studies.
Researchers who access consortia with data centers will be able to go far beyond their initial hypotheses and potentially accomplish research that was previously thought infeasible or too costly.
Acknowledgments
None.
Financial support and sponsorship: This work was supported by: National Institute of Environmental Health Sciences: 1U2CES026555 and 5P30ES023515
Footnotes
Conflicts of interest: None declared.
References
- 1.Thompson A. Thinking big: large-scale collaborative research in observational epidemiology. Eur J Epidemiol. 2009;24(12):727–31. doi: 10.1007/s10654-009-9412-1. [DOI] [PubMed] [Google Scholar]
- 2.National Institutes of Health. What is Big Data? Data Science at NIH. 2016 [Available from: https://datascience.nih.gov/bd2k/about/what]
- 3.Yoo C, Ramirez L, Liuzzi J. Big data analysis using modern statistical and machine learning methods in medicine. Int NeurourolJ. 2014;18(2):50–7. doi: 10.5213/inj.2014.18.2.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan J, Han F, Liu H. Challenges of Big Data Analysis. NatlSci Rev. 2014;1(2):293–314. doi: 10.1093/nsr/nwt032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.National Institute of Environmental Health Sciences. Data Repository, Analysis, and Science Center. CHEAR Program. 2016 [Available from: https://chearprogram.org/about/datarep.] This comprehensive environmental exposure resource is a groundbreaking effort to increase the use of environmental biomarkers in children’s health research.
- 6**.Kahn MG, Bailey LC, Forrest CB, Padula MA, et al. Building a Common Pediatric Research Terminology for Accelerating Child Health Research. Pediatrics. 2014;133(3):516–25. doi: 10.1542/peds.2013-1504. This article presents the importance of ontologies and controlled terminiologies within pediatric health resarch and includes helpful examples to illustrate these concepts to researchers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pediatric Diabetes Consortium. :2016. Available from: http://pdc.jaeb.org/
- 8.Children’s Health Clinics of Minnesota. Children’s Minnesota collaborates with five children’s hospitals to improve pediatric health. Children’s Hospitals and Clinics of Minnesota. 2016 Available from: https://www.childrensmn.org/2016/09/14/press-release-sandord-genomic-consortium/
- 9.Welcome. FaceBase. 2016 Available from: https://www.facebase.org/
- 10*.Environmental Health Risks in European Birth Cohorts. 2016 [Available from: http://www.enrieco.org/.] The website for this coordinated initiative by the European Union provides an inventory of the multiple birth cohorts conducted within Europe, including links to the websites of the individual cohorts participating in ENRIECO.
- 11.Casas M, Nieuwenhuijsen M, Martinez D, Ballester F, et al. Prenatal exposure to PCB-153, p,p′-DDE and birth outcomes in 9000 mother-child pairs: exposure-response relationship and effect modifiers. Environ Int. 2015;74:23–31. doi: 10.1016/j.envint.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 12.Iszatt N, Stigum H, Verner MA, White RA, et al. Prenatal and Postnatal Exposure to Persistent Organic Pollutants and Infant Growth: A Pooled Analysis of Seven European Birth Cohorts. Environ Health Perspect. 2015;123(7):730–6. doi: 10.1289/ehp.1308005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buckley JP, Engel SM, Braun JM, Whyatt RM, et al. Prenatal Phthalate Exposures and Body Mass Index Among 4- to 7-Year-old Children: A Pooled Analysis. Epidemiology. 2016;27(3):449–58. doi: 10.1097/EDE.0000000000000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engel SM, Bradman A, Wolff MS, Rauh VA, et al. Prenatal Organophosphorus Pesticide Exposure and Child Neurodevelopment at 24 Months: An Analysis of Four Birth Cohorts. Environ Health Perspect. 2016;124(6):822–30. doi: 10.1289/ehp.1409474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stratakis N, Roumeliotaki T, Oken E, Barros H, et al. Fish Intake in Pregnancy and Child Growth: A Pooled Analysis of 15 European and US Birth Cohorts. JAMA Pediatr. 2016;170(4):381–90. doi: 10.1001/jamapediatrics.2015.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharya S, Andorf S, Gomes L, Dunn P, et al. ImmPort: disseminating data to the public for the future of immunology. ImmunolRes. 2014;58(2–3):234–9. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- 17.National Heart Lung and Blood Institute. Compendium of Best Practices for Data Coordinating Centers - NHLBI. NIH. 2016 Available from: https://www.nhlbi.nih.gov/research/resources/compendium.
- 18**.Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. This manuscript formally presents the FAIR (findable, accessible, interoperable and reusable) principles that were created to advance scientific discovery by improving the infrastructure of data management. The principles are clearly defined and examples of their implementation are provided as examples for other researchers who aim to implement these principles relative to the data produced by their own research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Pool R, Rusch E. Principles and obstacles for sharing data from environmental health research: Workshop summary. The National Academies Press; Washington, DC: 2016. A summary of the workshop that explored the key concerns, principles and obstascles to sharing of data used in support of environmental health research and policy making. [PubMed] [Google Scholar]
- 20.OCR. Guidance regarding methods for de-identification of protected health information in accordance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule. 2012 Available from: http://www.hhs.gov/sites/default/files/ocr/privacy/hipaa/understanding/coveredentities/De-identification/hhs_deid_guidance.pdf.
- 21.El Emam K, Jonker E, Arbuckle L, Malin B. A Systematic Review of Re-Identification Attacks on Health Data. PLoS ONE. 2011;6(12):e28071. doi: 10.1371/journal.pone.0028071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wikipedia. Collaborative software development model. 2016 Available from: https://en.wikipedia.org/wiki/Collaborative_software_development_model.
- 23.National Institutes of Health. Commons Home Page. Data Science at NIH. 2016 Available from: https://datascience.nih.gov/commons.
- 24.National Cancer Institute. Home. NCI Genomic Data Commons. 2016 Available from: https://gdc.cancer.gov/
- 25.Bonazzi V. BD2K and the Commons: ELIXR All Hands. 2016 Available from: http://www.slideshare.net/VivienBonazzi/bd2k-and-the-commons-elixr-all-hands.
- 26.Coady SA, Wagner E. Sharing individual level data from observational studies and clinical trials: a perspective from NHLBI. Trials. 2013;14:201. doi: 10.1186/1745-6215-14-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27*.Budin-Ljøsne I, Isaeva J, Maria Knoppers B, Marie Tassé A, et al. Data sharing in large research consortia: experiences and recommendations from ENGAGE. European Journal of Human Genetics. 2014;22(3):317–21. doi: 10.1038/ejhg.2013.131. Reporting on the results of a survey of participants in a large international consortia, this article highlights potential barriers on data-sharing. Lessons learned from the ENGAGE consortia can help other consortia formulate their own data-sharing policies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearce N, Smith AH. Data sharing: not as simple as it seems. Environ Health. 2011;10:107. doi: 10.1186/1476-069X-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samet JM. Data: To Share or Not to Share? Epidemiology. 2009;20(2):172–4. doi: 10.1097/EDE.0b013e3181930df3. [DOI] [PubMed] [Google Scholar]
- 30.National Database for Autism Research - Home. 2016 Available from: https://ndar.nih.gov/index.html.
- 31.National Institutes of Health. Genomic Data Sharing. 2016 Available from: https://gds.nih.gov/index.html.
- 32.The Regents of the University of California. EZID Home. 2016 Available from: http://ezid.cdlib.org/
- 33.Ness RB. “Big” Science and the Little Guy. Epidemiology. 2007;18(1):9–12. doi: 10.1097/01.ede.0000249529.86885.5a. [DOI] [PubMed] [Google Scholar]
- 34.National Institute of Environmental Health Sciences. Children’s Health Exposure Analysis Resource (CHEAR) 2016 Available from: https://www.niehs.nih.gov/research/supported/exposure/chear/
- 35.Bourne PE. pebourne: Professional Developments Worth Sharing. 2016 Available from: https://pebourne.wordpress.com/
- 36.Benson MH, Lippitt CD, Morrison R, Cosens B, et al. Five ways to support interdisciplinary work before tenure. Journal of Environmental Studies and Sciences. 2016;6(2):260–7. [Google Scholar]
- 37.Johnson S, Carpenter L, Amezdroz E, Dashper S, et al. Cohort Profile: The VicGeneration (VicGen) study: An Australian oral health birth cohort. Int J Epidemiol. 2016 doi: 10.1093/ije/dyw024. [DOI] [PubMed] [Google Scholar]
- 38.Magnus P, Birke C, Vejrup K, Haugan A, et al. Cohort Profile Update: The Norwegian Mother and Child Cohort Study (MoBa) Int J Epidemiol. 2016;45(2):382–8. doi: 10.1093/ije/dyw029. [DOI] [PubMed] [Google Scholar]
- 39.Takagai S, Tsuchiya KJ, Itoh H, Kanayama N, et al. Cohort Profile: Hamamatsu Birth Cohort for Mothers and Children (HBC Study) Int J Epidemiol. 2016;45(2):333–42. doi: 10.1093/ije/dyv290. [DOI] [PubMed] [Google Scholar]
- 40.Heude B, Forhan A, Slama R, Douhaud L, et al. Cohort Profile: The EDEN mother-child cohort on the prenatal and early postnatal determinants of child health and development. Int J Epidemiol. 2016;45(2):353–63. doi: 10.1093/ije/dyv151. [DOI] [PubMed] [Google Scholar]
- 41.Pausova Z, Paus T, Abrahamowicz M, Bernard M, et al. Cohort Profile: The Saguenay Youth Study (SYS) Int J Epidemiol. 2016 doi: 10.1093/ije/dyw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braun JM, Kalloo G, Chen A, Dietrich KN, et al. Cohort Profile: The Health Outcomes and Measures of the Environment (HOME) study. Int J Epidemiol. 2016 doi: 10.1093/ije/dyw006. [DOI] [PMC free article] [PubMed] [Google Scholar]


