Summary
Measurements of dissolved, ascorbate-reducible and total Mn by ICP-OES revealed significantly higher concentrations during estuarine turbidity maxima (ETM) events, compared with non-events in the Columbia River. Most probable number (MPN) counts of Mn-oxidizing or Mn-reducing heterotrophs were not statistically different from that of other heterotrophs (103–104 cells ml−1) when grown in defined media, but counts of Mn oxidizers were significantly lower in nutrient-rich medium (13 cells ml−1). MPN counts of Mn oxidizers were also significantly lower on Mn(III)-pyrophosphate and glycerol (21 cells ml−1). Large numbers of Rhodobacter spp. were cultured from dilutions of 10−2 to 10−5, and many of these were capable of Mn(III) oxidation. Up to c. 30% of the colonies tested LBB positive, and all 77 of the successfully sequenced LBB positive colonies (of varying morphology) yielded sequences related to Rhodobacter spp. qPCR indicated that a cluster of Rhodobacter isolates and closely related strains (95–99% identity) represented approximately 1–3% of the total Bacteria, consistent with clone library results. Copy numbers of SSU rRNA genes for either Rhodobacter spp. or Bacteria were four to eightfold greater during ETM events compared with non-events. Strains of a Shewanella sp. were retrieved from the highest dilutions (10−5) of Mn reducers, and were also capable of Mn oxidation. The SSU rRNA gene sequences from these strains shared a high identity score (98%) with sequences obtained in clone libraries. Our results support previous findings that ETMs are zones with high microbial activity. Results indicated that Shewanella and Rhodobacter species were present in environmentally relevant concentrations, and further demonstrated that a large proportion of culturable bacteria, including Shewanella and Rhodobacter spp., were capable of Mn cycling in vitro.
Introduction
Estuaries are highly productive, dynamic environments that feature steep salinity, temperature, chemical and turbidity gradients, and contribute vital trace minerals and organic matter to food webs in coastal environments. The Columbia River estuary, in particular, is the largest source of freshwater, by volume (60–90%), to the California Current System between the San Francisco Bay and the Strait of Juan de Fuca (Barnes et al., 1972), and it also provides a major source of dissolved manganese and nutrients to the coastal waters (Aguilar-Islas and Bruland, 2006; Bruland et al., 2008). Water circulation and density stratification, controlled by river discharges, tides and shelf winds, enable entrapment of suspended particulate matter in estuarine turbidity maxima (ETM) in the north, south and central channels of the Columbia River estuary (Gelfenbaum, 1983; Jay and Smith, 1990; Jay and Musiak, 1994; Small and Prahl, 2004). ETM are generated because the net flux of deep water upstream into the estuary allows negatively buoyant particulate matter to remain trapped in the estuary in dense clouds of turbidity (Crump et al., 1998). It is in these ETM where much of the enzymatic activity and biogeochemical cycling is thought to occur, for example rapid cycling of trace metals and organic matter by bacteria (Crump and Baross, 1996; Prahl et al., 1998). Suspended particulate matter supports the bulk of microbial decomposition and thus contributes available organic matter to the thriving ecosystem. Crump and colleagues have determined that particle attached bacteria are 10–100 times more active than free living bacteria (Crump and Baross, 1996; Crump et al., 1998; 1999). In addition, Klinkhammer and McManus have proposed that the ETM contain an active community of Mn reducers (Klinkhammer and McManus, 2001). They suggested that a release of organic matter may be catalysed in part by Mn-reducing bacteria in anoxic pockets of suspended particles. We hypothesized that such a release of dissolved Mn in oxic waters should allow for Mn oxidation. Due to emerging evidence indicating the importance of Mn(III) in the environment (Trouwborst et al., 2006), we further hypothesized that Mn(III)-oxidizing bacteria should also be present.
Members of the Shewanellaceae are well known for dissimilatory metal reduction (Nealson and Saffarini, 1994; Gralnick et al., 2006), and are often found in redox interfaces where Mn cycling is important, such as those occurring in the Black Sea, Amazon River, Halifax Harbor, Oneida Lake, and in deep-sea sediments (Nealson and Scott, 2006; Hau and Gralnick, 2007). Both Mn reduction (Nealson and Scott, 2006) and Mn oxidation (Staudigel et al., 2006; this work; C.R. Anderson, R.E. Davis, N.S. Bandolin, A.M. Baptista and B.M. Tebo, in preparation) have been detected among Shewanella species. However, Mn(II) oxidation by Shewanella has received little attention, and it may or may not be an environmentally relevant process. DiChristina and DeLong mention involvement in ferromanganese deposits as a possibility for Shewanella putrefaciens, based on circumstantial evidence (DiChristina and DeLong, 1993), and Shewanella oneidensis MR-1 has been shown to have a chemotactic response towards Mn(II) (Bencharit and Ward, 2005). Further, it has been hypothesized that bacteria may oxidize Mn in order to store the oxides for use as alternative electron acceptors when suboxic or anoxic conditions arise (Tebo, 1983; de Vrind et al., 1986, Brouwers et al., 2000a). Shewanella may be one of a few genera (along with Bacillus and Pseudomonas) that would be capable of this, based on our current knowledge of microbial physiology, i.e. the ability of some of these organisms to grow with electron acceptors other than oxygen. Both of these reactions, Mn oxidation and Mn reduction, play important roles in carbon cycling in the environment (Nealson and Myers, 1992; Sunda and Kieber, 1994; Klinkhammer and McManus, 2001). Mn is also an important electron shuttle or redox mediator, due to the fact that reduced Mn(II) is readily soluble and can diffuse into the upper oxic zone, whereas oxidized Mn(IV) is a solid precipitate and can sink back down into the sediment (Nealson and Myers, 1992; Nealson and Saffarini, 1994).
Manganese(II)-oxidizing bacteria can accelerate the rate of Mn oxidation by several orders of magnitude over abiotic oxidation, and the Mn oxides produced are biogeochemically active (Tebo et al., 2005). Mn oxides are highly reactive and can oxidize humics or other organics (Sunda and Kieber, 1994), scavenge reactive oxygen species or heavy metals, and serve as electron acceptors in anaerobic respiration (Tebo et al., 2004). Mn(II) oxidizers are phylogenetically diverse and representatives from the alpha (Anderson et al., 2009a), beta (Emerson and Ghiorse, 1992) and gamma (Kepkay and Nealson, 1987) classes of the Proteobacteria, as well as from the Gram-positives (van Waasbergen et al., 1996) have been known for some time. Prior to this study, members of the Roseobacter clade among the Alphaproteobacteria, but not species of Rhodobacter were known to oxidize Mn(II). For example, culture-based studies in the deep-sea identified several new species from among the Roseobacter clade that are capable of oxidizing Mn(II), including strains SPB1, SPB6 and LOB8 (Templeton et al., 2005). Further, studies in Elkhorn Slough near Monterey Bay, California noted several related strains including UAzPsJAC-6, UAzPsK-5, AzwK-3b and UAzPsJAC-1b (Hansel and Francis, 2006).
Members of the species Rhodobacter form a monophyletic clade within the larger Rhodobacteraceae family of which Roseobacter spp. are also members. Roseobacteraceae are among the most easily cultivated and are known to be important in dimethylsulfoniopropionate degradation, carbon monoxide oxidation, aerobic anoxygenic photosynthesis, Fe(II) oxidation, and production of bioactive compounds and quorum sensing molecules (Giovannoni and Rappé, 2000; Buchan et al., 2005; Wagner-Döbler and Biebl, 2006; Poulain and Newman, 2009). They occupy a diversity of ecological niches including ocean surface layers, coastal regions, submarine basalts, sea ice and estuaries, but become less abundant or absent in freshwater and terrestrial habitats (Buchan et al., 2005, and references therein). Based on molecular techniques, members of the Roseobacter lineage often dominate or at least represent a large abundance, from 10% to 40% of the population, of the SSU rRNA profiles in offshore Mediterranean (Acinas et al., 1999), oceanic (Gonzalez et al., 2000) and coastal waters (Cottrell and Kirchman, 2000; Dang and Lovell, 2002; Suzuki et al., 2004).
Multicopper oxidases or haem-containing peroxidases (Anderson et al., 2009b) have been implicated in Mn(II) oxidation among many bacterial genera (Brouwers et al., 2000b) including Pedomicrobium (Ridge et al., 2007), Leptothrix (Corstjens et al., 1997), Bacillus (van Waasbergen et al., 1996, Dick et al., 2008a) and Aurantimonas (Dick et al., 2008b, Anderson et al., 2009b). A multicopper oxidase, CumA, was originally thought to be involved in Mn oxidation in Pseudomonas putida (Brouwers et al., 1999), although perhaps another enzyme is secreted via the two-component regulation system instead (Geszvain and Tebo, 2010). Detecting multicopper oxidase and/or haem peroxidase-type genes in new strains of Mn(II) oxidizers can lend credence to the involvement of these enzyme classes in Mn(II) oxidation, and help elucidate the evolutionary phylogeny of these genes. In this study we evaluated Mn-oxidizing Rhodobacter isolates for the presence of Mox-type genes. Overall, the goals of this study were to determine which bacteria are responsible for Mn(II) and Mn(III) oxidation as well as Mn oxide reduction, and to determine the relative abundance of microbial groups implicated in Mn cycling in the Columbia River estuary and ETM.
Results and discussion
Biogeochemical analyses of dissolved and ascorbate-reducible Mn
Resources of the SATURN collaboratory (Baptista et al., 2008) were used to anticipate ETM timing and locations in the North and South Channel of the estuary (Fig. S1). Prior to sampling, we utilized data-informed daily forecasts of baroclinic circulation (http://www.stccmop.org/datamart/virtualcolumbiariver/forecasts), to predict salinity intrusion length and variability in the North and South channels of the estuary. Details of the models (Zhang et al., 2004; Zhang and Baptista, 2008) and their simulation skill (Baptista et al., 2005; Burla, 2009; Frolov et al., 2009) are described elsewhere. The location of the salt wedge was empirically correlated, via observations of sediment concentrations (http://www.stccmop.org/datamart/observation_network), to the timing of the ETMs, which were expected to occur near the front of the salt wedge (B. Crump and L. Herfort, pers. comm.). Samples were collected in the South Channel on 14 June 2007, on 17 July 2007, and in both the North and South Channels from 14 August through 31 August 2007. Samples were deemed to be either ETM or non-ETM samples by the chief scientist at the time of sampling and in general were defined according to a significant change (greater than 1.5- to 2-fold change) in nephelometric turbidity units (NTU), or a change in the transmitted or reflected light due to changes in suspended particulate matter in conjunction with the time and location of the predicted occurrence (see Figs S2 and S3 for example ETM events). The maximum turbidity measurements observed roughly correlated with the salt wedge and could be found at the front edge (low salinity values c. PSU 5) or rear edge (intermediate salinity values c. PSU 15) of the salt wedge. Turbidity generally subsided 30 min to 2 h after the salt wedge had passed (see Fig. S3A as an example). The change in NTU above baseline during an ETM event was also somewhat variable and ranged from as little as ~1.5-fold (Fig. S2A), to as high as ~15-fold (Fig. S3A). This is not too surprising since concentrations of suspended particulate matter in the Columbia River and other estuarine/river systems have been shown to vary with season, river flow, and tidal cycle and amplitude, and these conditions can affect manganese(II) and particulate manganese concentrations as well (Jay and Smith, 1990; Grabemann et al., 1997; Shiller, 1997; Shiller and Stephens, 2005; Aguilar-Islas and Bruland, 2006).
The range in concentration of dissolved manganese (as defined by passing through a 0.2 μm filter) was as expected, c. 1–100 nM (Fig. 1B), according to previous measurements (Klinkhammer et al., 1997; Klinkhammer and McManus, 2001; Aguilar-Islas and Bruland, 2006). Further, as expected, all but one of the samples showed a significant increase in dissolved manganese concentrations during an ETM event relative to samples taken approximately 2 h before or after the corresponding ETM event. These results support research by Klinkhammer and McManus (2001), who also found increases in dissolved Mn concentrations in correlation with turbidity increases. Reductive dissolution of Mn (Aguilar-Islas and Bruland, 2006) as well as high concentrations of organic matter and detrital material have also been noted in suspended particulate matter in the ETM (Prahl et al., 1997; 1998), providing further evidence that conditions should allow microbial Mn reduction.
Fig. 1.
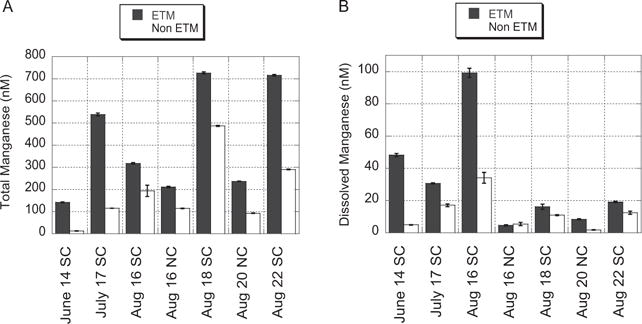
Measured values for (A) total manganese and (B) dissolved manganese during and c. 2 h before or after an ETM event in the South Channel (SC) or North Channel (NC) of the Columbia River estuary in 2007. Data represent an average of triplicate samples and bars represent standard error.
Measurements of ascorbate-reducible Mn were made to determine if Mn oxides were present during ETM events. All samples taken in June, July and August 2007 in the North and South channel showed a significant increase in total manganese concentrations (dissolved + ascorbate-released Mn) during an ETM event, relative to samples taken approximately 2 h either before or after the corresponding ETM event (Fig. 1A). These results indicate that Mn oxides are present and concentrations are enhanced during an ETM event relative to before or after an event. In fact, total Mn (including dissolved and ascorbate-released) had a more significant relationship with turbidity compared with dissolved Mn alone (Fig. S3B). Dissolved Mn showed very little relationship with turbidity when graphed across all ETM events sampled, as demonstrated by a relatively small slope (0.28) and low R value. This is, at least in part, due to the wide variation in NTU values measured during an ETM event (from 4 to 80). Such variation in NTU values could be due to seasonal differences, or possibly differences in sampling depth or location relative to the centre of the ETM (e.g. sampling on the fringes of the ETM where Mn may be enhanced more than particulate matter). Both dissolved Mn and ascorbate-released Mn (particulate Mn oxides) showed a strong correlation with turbidity for a single ETM event and associated non-ETM samplings. Few studies of Mn oxides in the Columbia River have been conducted previously. However, at least one study found that particulate matter in the Columbia River was enriched in manganese oxides, and that particulate Mn is slow settling and may be suspended higher in the water column and for longer than fast-settling particles rich in detrital material (Covert, 2001). This may explain why high values for total and ascorbate-released Mn were obtained despite low NTU values during the August 2007 samplings (Fig. S2). We can infer from these biogeochemical analyses that microbially mediated Mn oxidation is occurring. These results support other studies that have implicated microbial processes in producing high rates of Mn(II) oxidation in other rivers and estuaries, including the lower Mississippi River (Shiller and Stephens, 2005), the Chesapeake Bay (Moffett, 1994) and the Newport River estuary (Sunda and Huntsman, 1987).
Culture-dependent studies
Culturing of Mn oxidizers from serial dilutions of river water in MnA1 media resulted in high numbers of Rhodobacter spp. isolates from multiple dilutions (10−2 to 10−5). Seventy-six of the 77 isolates, whose SSU rRNA gene fragments were successfully sequenced, belonged to a clade of Rhodobacter somewhat related to Pseudorhodobacter ferrugineus (95% identity across ~1400 bp; isolates CR07-8, -44, -62 and -97 in Fig. 2), and the other isolate (CR07-5 in Fig. 2) was related (96% identity) to Rhodobacter vinaykumaraii. We noted that a large percentage (up to c. 30%) of colonies on low-nutrient agar tested positive for Mn oxides using the leucoberbelin blue (LBB) colorimetric test (Tebo et al., 2006); however, none of the colonies tested on rich media, such as K medium, tested LBB positive (data not shown). Similarly, most probable number (MPN) results across a variety of different carbon sources in low-nutrient agar gave similar numbers for all culturable heterotrophic bacteria (2.4 × 103 to 1.1 × 104) and culturable heterotrophic Mn(II) oxidizers (3.6 × 102 to 3.8 × 104, Fig. 3). MPN counts of Mn oxidizers in Mn(III)-pyrophosphate glycerol media were significantly lower (21 cells ml−1, Fig. 3), suggesting that some, but not all culturable bacteria can produce oxides from the Mn(III) intermediate. At least 15% of the Rhodobacter isolates were capable of producing Mn oxides on Mn(III)-pyrophosphate glycerol plates, although not all isolates were tested (data not shown). MPN counts of Mn oxidizers were dramatically reduced (down to 13 cells ml−1) in 50% natural seawater K medium, suggesting that the culturable Mn-oxidizing community in the Columbia River either was outcompeted on rich media, or showed a preference for or adaptation to more oligotrophic conditions (Fig. 3). It is less likely that cells were unable to oxidize Mn in K medium, since all of the Mn-oxidizing isolates tested were capable of oxidation in this medium (data not shown). Total numbers of heterotrophic bacteria did not change significantly on either type of medium (Fig. 3).
Fig. 2.
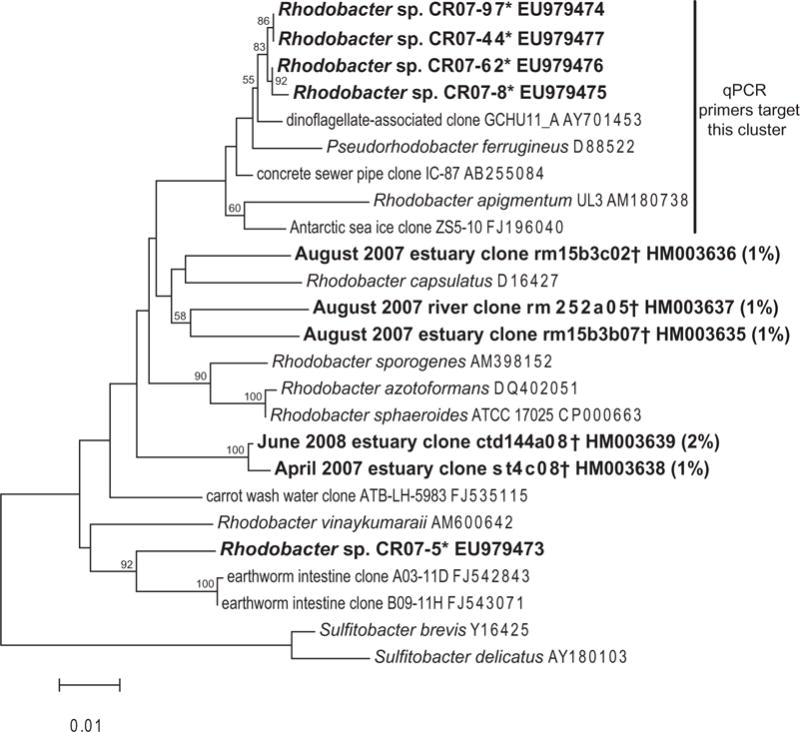
Neighbour-joining tree inferring the phylogenetic relationship between cultured strains* (asterisks) and those found in clone libraries† (daggers) in the Columbia River in this study. Alignments were created using the on-line SILVA aligner and then exported from ARB. Dendrogram was created using PHYLIP. The percentage of the sequences from each library that represented a particular OTU is given in parentheses: actual sequence abundance ranged from 1/150 to 2/80. Bootstrapping values are shown for nodes that were supported >50% of the time. Nodes with bootstrapping values >60 were also supported with maximum-likelihood analysis (data not shown). Sulfitobacter species were used as the outgroup. The cluster of Rhodobacter spp. targeted by the qPCR primers is marked by a bar.
Fig. 3.
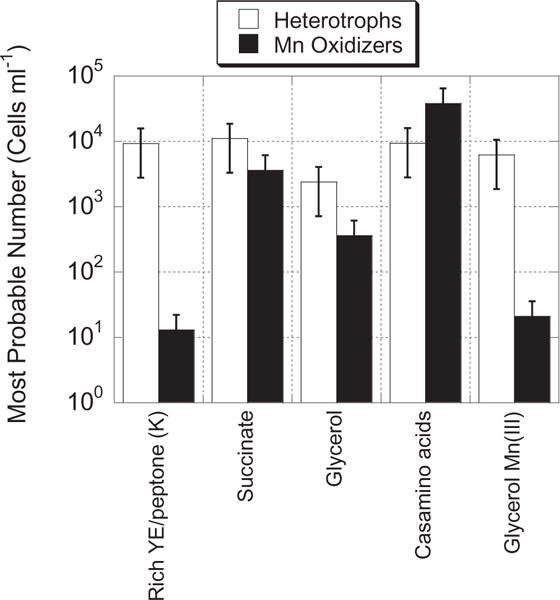
Most probable number (MPN) counts for detectable heterotrophic bacteria versus heterotrophic Mn-oxidizing bacteria on three different carbon substrates, on rich K medium, or with Mn(III) provided as a source of electrons. MPN dilutions were made from ETM water collected in July 2007. Data represent an average of quadruplicate samples and bars represent the upper and lower 95% confidence intervals.
Quantification of Mn reducers (Fig. 4B) was estimated at 2.7 × 104 cells ml−1 in the June ETM and 2.8 × 104 cells ml−1 in the July ETM using MPN cell counts. Further, multiple isolates of a Shewanella sp. (asterisks, Figs 5 and S4) were cultured from the highest dilution of 10−5 and likely represent an important group in the Columbia River. Corroborating these data are molecular phylogenetic analyses of the Columbia River water samples, where sequences clustering with the isolated Shewanella strains were detected in 1/100 and 1/190 clones in libraries from samples collected in April 2007 and August 2007 respectively (daggers, Figs 5 and S4). Both culturing (Nealson et al., 1991; DeLong et al., 1997; Ziemke et al., 1997; Brettar et al., 2001; Ivanova et al., 2003) and molecular techniques (DiChristina and DeLong, 1993; Brown and Bowman, 2001; Bowman and McCuaig, 2003) have demonstrated presence and/or abundance of Shewanella species in various environments. However, there is often a discrepancy between strains that are retrieved by culturing and those that are retrieved by molecular techniques (Suzuki et al., 1997), sometimes termed the great plate count anomaly (Amann et al., 1995). Here we have managed to culture strains with high identity (98% by SSU rRNA sequence) to sequences retrieved from clone libraries. The sequences from the cultured strains (A1 and Gly) also cluster most closely with uncultured clones from the Columbia River, than with other top blast hits, either cultured or uncultured (Figs 5 and S4). These results indicate that at least one of the more abundant Shewanella phylotypes in this environment is readily culturable. The use of a lower-nutrient medium and cultivation from high dilutions may have also contributed to our success in culturing an environmentally relevant strain.
Fig. 4.
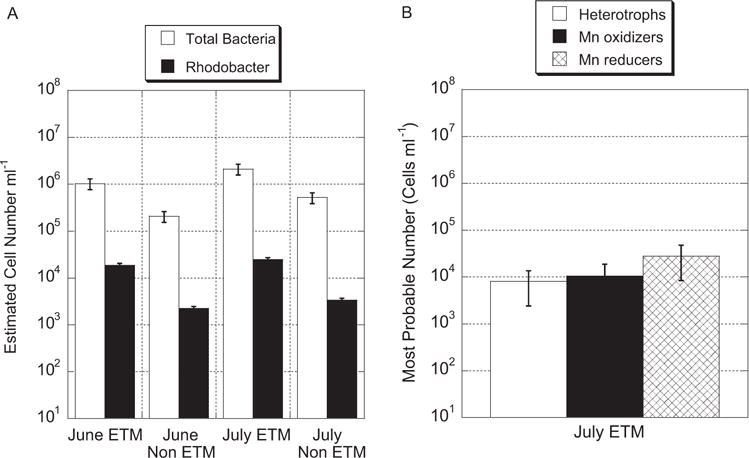
A. Real-time qPCR quantification of 16S rRNA gene copy number of total Bacteria and Rhodobacter spp. in Columbia River estuary samples during both ETM and non-ETM events. The y-axis represents the 16S rRNA gene copy number normalized by three rRNA operon copies as an estimate of cell number (see Experimental procedures and Fogel et al., 1999). Data represent an average of triplicate samples and bars represent standard error.
B. Most probable number (MPN) counts for heterotrophic bacteria, Mn-oxidizing bacteria (including Rhodobacter sp.) averaged across all carbon substrates, and for Mn-reducing bacteria (including Shewanella sp.) for samples taken in July 2007. Data represent an average of quadruplicate samples and bars represent the upper and lower 95% confidence intervals.
Fig. 5.
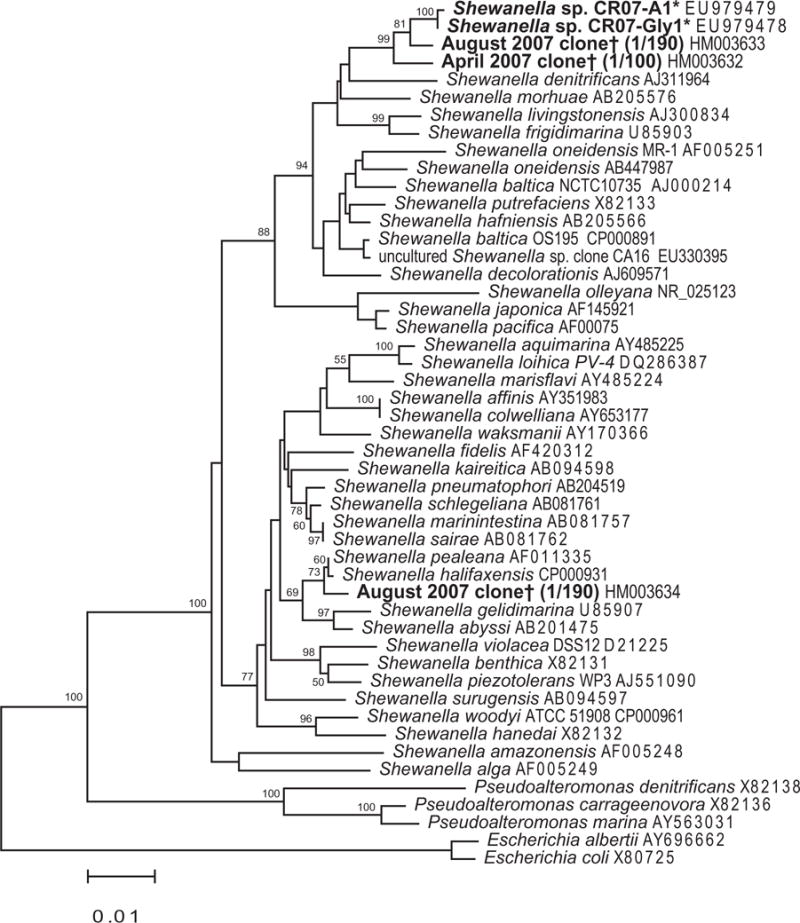
Neighbour-joining tree inferring the phylogenetic relationship between Mn(IV)-reducing and Mn(II)-oxidizing Shewanella sp. cultured strains* (asterisks) and those found in clone libraries† (daggers) in the Columbia River in this study. Top blast hits to the isolates were included, as well as a broad range of described species. Alignments were created using the on-line SILVA aligner and then exported from ARB. Dendrogram was created using PHYLIP. Bootstrapping values are shown for nodes that were supported at least 50% of the time. The dendrogram was only weakly supported with maximum-likelihood analysis, and many branches had low bootstrapping values in both analyses (Fig. S4). Escherichia species were used as the outgroup.
While these Shewanella sp. were isolated for their capability to reduce Mn(IV) oxides, they are also capable of oxidizing Mn(II) slowly on MnA1 medium supplemented with casamino acids, or more rapidly when succinate is used (empirical observation, rates not measured). Shewanella species have been isolated from estuarine waters previously (Das and Caccavo, 2000; Skerratt et al., 2002; Kan et al., 2006). The more recent discoveries of Mn(II) oxidation by Shewanella strains VS-7 and VS-58 58 isolated from Vailulu’u Seamount (Staudigel et al., 2006) and by other environmental Shewanella strains (C.R. Anderson, R.E. Davis, N.S. Bandolin, A.M. Baptista and B.M. Tebo, in preparation) could have important implications for electron shuttling and carbon cycling in diverse environments such as Vailulu’u Seamount and the Colum-bia River. Taken together, the culture-dependent MPN data presented here suggest that a large percentage of the culturable population is capable of Mn cycling, either oxidation or reduction, and that Rhodobacter spp. and Shewanella sp. are among the most abundant.
Culture-independent studies
Quantitative real-time PCR (qPCR) of total bacterial SSU rRNA genes in June and July, respectively, were estimated to represent 3.09 × 106 and 6.36 × 106 cells ml−1 during an ETM and 6.22 × 105 and 1.56 × 106 cells ml−1 during a non-ETM event (Fig. 4A). Quantification of SSU rRNA genes of a select clade of Rhodobacter spp., including the most abundant isolates, yielded in June and July, respectively, 5.63 × 104 and 7.43 × 104 during an ETM and 6.77 × 103 and 1.01 × 104 during a non-ETM event (Fig. 4A). Numbers of either Bacteria or Rhodobacter spp. were four- to eightfold greater during an ETM event supporting studies that found higher numbers of particle-attached bacteria and increased microbial activity during ETM events (Crump and Baross, 1996; 2000; Crump et al., 1998; 1999). The data presented here also demonstrated that Rhodobacter spp. relative to total Bacteria represented approximately 1% of the population for samples taken either during or before or after an ETM event. Interestingly, these data corroborate data from clone libraries which found that approximately 1–2% of the clones were Rhodobacter spp. related to Rhodobacter capsulatus and R. sphaeroides (Fig. 2, daggers). It was also interesting that numbers of Rhodobacter spp. (including culturable species) as determined by qPCR very closely approximated those found for Mn oxidizers in MPN analyses (Fig. 4). Additionally, culturable bacteria as determined by MPNs (3.8 × 103 to 3.8 × 104) represented roughly 1% or less of the total bacterial population as determined by qPCR (roughly 106). This falls well within the range of culturable bacteria given in the literature from 0.01% to 0.1% (Kogure et al., 1979), up to at least 60% (Button et al., 1993). Numbers were comparable to that in a similar study (C.R. Anderson, R.E. Davis, N.S. Bandolin, A.M. Baptista and B.M. Tebo, in preparation) of the Columbia River plume off the coast of Oregon that found 103–105 bacteria per ml of plume water. That same study also detected Rhodobacter sp. in T-RFLP electropherograms, but not in MPNs using nutrient-rich K medium, or on agar plates made with nutrient-rich K, M or Lept medium (C.R. Anderson, R.E. Davis, N.S. Bandolin, A.M. Baptista and B.M. Tebo, in preparation), again suggesting that these strains prefer oligotrophic medium. Overall, this study demonstrates high concentrations of Mn oxides and Mn-oxidizing bacteria. We suggest that at least some of the Mn oxides present are a result of rapid Mn cycling in the water column. Future studies should measure rates of Mn oxidation inside and outside of the ETM.
Several studies have found Rhodobacter spp. in marine/estuarine environments including the Columbia River (Crump et al., 1999). Studies by Zhao and colleagues (2009) and Kan and colleagues (2007) found that both Roseobacter and Rhodobacter (P. ferrugineus) populations are important in the Chesapeake bay with both groups accounting for 18% of the population in a March 2003 clone library. Waidner and Kirchman examined the presence (Waidner and Kirchman, 2005) and abundance (Waidner and Kirchman, 2008) of photosynthesis genes and found that pufM genes most closely related to the Rhodobacter genus were abundant and ubiquitous in the Delaware estuary, a system somewhat analogous to that of the Columbia River estuary. Stevens and colleagues (2005) evaluated 265 clones from 12 different studies of microbial diversity in the Wadden Sea, Germany and found at least two clones in the Rhodobacter clade of the Rhodobacter group (AY145556 and AY332128). A large percentage (c. 20%) of the clones they evaluated fell in the Roseobacter clade. Not all of the aforementioned studies were conducted in the estuarine tidal flats, some were conducted in more saline/marine environments of the Wadden Sea. Overall, our study supports a mounting body of literature suggesting that members of the Rhodobacter clade are abundant and widespread in estuarine environments (Waidner and Kirchman, 2008; Zhao et al., 2009), whereas members of the Roseobacter clade are usually the more dominant group in marine environments (Buchan et al., 2005). Members of the Roseobacter clade have been known to oxidize Mn(II) and have been found in various environments including the deep-sea (Templeton et al., 2005) and coastal estuaries (Hansel and Francis, 2006). To our knowledge, this is the first time that members of the Rhodobacter clade (relating to the genus Rhodobacter) have been implicated in manganese(II) oxidation. We can infer from in vitro analyses that Rhodobacter spp. and/or Shewanella spp. may be involved in Mn cycling in situ, and molecular data support this hypothesis by demonstrating that both sequence types are abundant. Sequences related to cultured Shewanella spp. were abundant in clone libraries, and qPCR results indicate an abundance of sequences related to cultured Rhodobacter spp. Unfortunately there is no molecular marker available to determine the diversity and/or quantity of manganese oxidizers in the Columbia River. The gene for MofA has been used (Siering and Ghiorse, 1997), and perhaps moxA may be used as a proxy for Mn oxidizer diversity in future experiments.
Interestingly, all of the Rhodobacter sp. isolates that were tested from the largest cluster contained the gene for MoxA (Fig. 6), whereas Rhodobacter sp. CR07-5 did not. MoxA is a multicopper oxidase that is thought to be involved in Mn oxidation, either directly or indirectly. In fact Rhodobacter sp. CR07-5 is a poor oxidizer, and oxidizes Mn(II) more sporadically and more weakly than the other strains. While MoxA may not be solely responsible for Mn oxidation (i.e. it may not be the Mn oxidase), it may be involved in mediating Mn oxidation, or it may occur as part of an Mn oxidase complex. Thus, it is interesting that the strains with the strongest Mn oxidation appear to have the gene. It is important to note, however, that some strains containing moxA homologues, such as Sulfitobacter sp. EE-36, have tested negative for manganese oxidation (Dick, 2006). More work would be needed to determine the role that moxA plays in Mn oxidation, and to determine the diversity of moxA genes present in the Columbia River. Anderson and colleagues (C.R. Anderson, R.E. Davis, N.S. Bandolin, A.M. Baptista and B.M. Tebo, in preparation) have detected manganese oxidase enzymes in the Columbia River, but have not examined the diversity of those proteins.
Fig. 6.
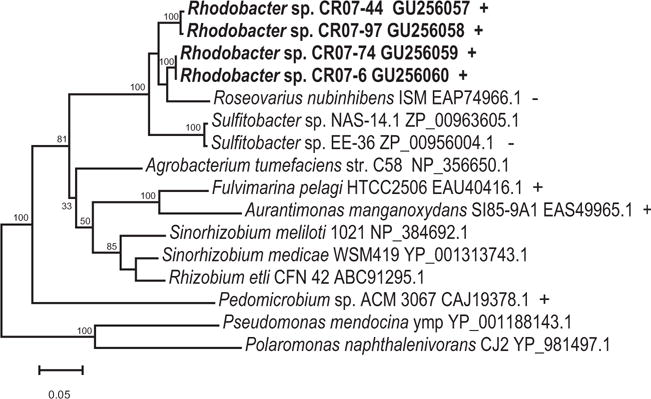
Neighbour-joining tree demonstrating the phylogenetic relationship between putative multicopper oxidase genes that are widely dispersed among Alphaproteobacteria. Alignments were created using CLUSTALX. Dendrogram was created using PHYLIP. Bootstrapping values are shown for nodes that were supported >70% of the time, or for nodes supported by both neighbour-joining and maximum-likelihood analyses. All nodes with bootstrapping values >70% were supported in both analyses (data not shown). Strains capable of Mn(II) oxidation are denoted with a plus (+), and those known to be incapable are denoted with a minus (−). The remaining strains have not been tested, or the result is unknown. Pseudomonas mendocina and Polaromonas naphthalenivorans were used as the outgroups.
Overall, our molecular, microbiological and chemical-analytical results build on previous studies and suggest that microbially produced particulate Mn oxides, and Mn cycling bacteria are abundant in ETM waters and that Mn cycling is intensified during ETM events. Further, we speculate that Mn oxide particles and associated oxidizers may create anaerobic microenvironments in the ETM that allow for Mn reducers and contribute to the degradation of particulate organic matter. This and other studies (Gebhardt et al., 2005; Shiller and Stephens, 2005) indicate that the rapid cycling of trace metals such as Fe and Mn in the ETM plays an important role in biogeochemical and organic matter cycling and in supporting the dynamic estuarine food web.
Experimental procedures
Sample collection and storage
Water samples from the Columbia River estuary (Fig. S1) were collected in duplicate or triplicate in sterile 50 ml Falcon tubes onboard either the R/V Forerunner (in June and July 2007), the R/V Barnes (in August 2007) or the R/V Wecoma (in August 2007 and all other samplings). For cruises on the R/Vs Barnes and Forerunner, samples were retrieved using an air pump, and were coordinated with a conductivity, temperature and depth recorder (CTD). Samples for clone libraries were collected in April 2007, August 2007 and June 2008 on the R/V Wecoma. For these cruises, samples were retrieved using an SBE Carousel sampler with twelve 10 l Niskin sampling bottles. Cast data were collected using a SeaBird brand CTD equipped with a transmissometer, fluorometer, thermometer, photosynthetically available radiation (PAR) light sensor, O2 probe and altimeter. Microbial cells for molecular assays were concentrated onto triplicate or quadruplicate Sterivex filters (Millipore, Billerica, MA) onboard the ship. The volume filtered was recorded, and was approximately 600–900 ml per filter. Filters were stored shipboard in a freezer containing dry ice and were transferred to a −80°C freezer in the laboratory until further processing. Water for culturing and MPN assays was stored at 4°C in sterile Falcon tubes until the samples were transported to the laboratory (usually within 24–48 h). Sub-samples of water for ICP analysis were either filtered or unfiltered and acidified with 3% nitric acid.
Culturing and MPNs
A new medium, called MnA1 medium, was designed and used for isolation of Mn(II) oxidizers from dilutions of Columbia River water. Constituents were the same as J medium (Tebo et al., 2006), but with 40% natural seawater, and the addition of 10 mg l−1 yeast extract, a mix of MnA1 trace metals, modified from Holden and colleagues (2001) to include the following final concentrations (in μM): 8.5 disodium nitrilotriacetic acid, 1.5 KCl, 1.0 AlK(SO4)2 × 12H2O, 0.4 CoCl2 × 6H2O, 0.4 CuSO4 × 5H2O, 16.0 H3BO3, 0.2 H2WO4, 0.4 KBr, 0.6 KI, 0.25 LiCl, 6.0 MnSO4, 0.8 Na2MoO4 × 2H2O, 0.6 Na2SeO3 × 2H2O, 0.4 NiCl2 × 6H2O, 0.3 VOSO4, 7.5 ZnCl2 and 10.0 FeCl3 × 6H2O, as well as one of each of the following: 10 mM glycerol, 10 mM succinate, 10 mM glycolate or 0.5 g l−1 casamino acids. To test for oxidation of Mn(III), Mn(III)-pyrophosphate was used instead of 100 μM MnCl2. Mn(III)-pyrophosphate was modified from Klewicki and Morgan (1998) by adding each of the following in the order listed and mixing thoroughly by inversion between ingredients: 2.0 ml of 100 mM Na pyrophosphate-HCl (pH 7.5), 4.0 ml of 1 M Hepes pH 7.5, 320 μl of 0.1 M MnCl2 and 400 μl of 20 mM Mn(VII) as potassium permanganate. After mixing, the solution was incubated on a rotary shaker overnight and filter-sterilized prior to use. For MPN analyses, 225 ml of MnA1 medium or K medium made with 40% seawater (Tebo et al., 2006) were added to each well in a 96-well culturing plate. Twenty-five microlitres of inoculum was added along a dilution of 10−1 to 10−8, so that the top half of the plate represented quadruplicates for analysis of metabolic activity, and the bottom half of the plate represented quadruplicate dilutions for detection of Mn oxides. Positive controls were inoculated in duplicate in column 12 with Aurantimonas manganoxydans strain SI85-9A1. The plates were incubated for 4 weeks in the dark at room temperature, at which time 50 μl of 0.04% leucoberbelin blue (LBB) (Tebo et al., 2006) was added to the wells for detection of Mn oxides in the top four rows, and 50 μl of 0.3% Iodonitotetrazolium salt (INT), a redox indicator, was added to the bottom four rows as a proxy for cell growth/activity. The solutions were allowed to react overnight and were scored as positive (strongly coloured red for INT or blue for LBB) or negative (weak or colourless) by visual observation. Controls were conducted at the beginning of the experiment to test for background reaction with LBB. All dilutions were negative for LBB, indicating that there were not sufficient quantities of Mn oxide to react with LBB prior to incubation. MPN estimates were made using a downloadable MPN calculator (http://www.i2workout.com/mcuriale/mpn/index.html). For Mn reducers, serial dilutions were conducted in triplicate in shake tubes, or media with low-melt agarose (Adrian et al., 2000). Shake tubes contained the following: 50% natural seawater and 20 mg ml−1 low-melt agarose that was heated in a microwave, dispensed in the glove box, autoclaved, flushed with ultra high purity N2, and supplemented with the following (final concentrations): 10 mM Hepes pH 7.75, 5 mM KHCO3, 0.025 g l−1 yeast extract, J medium vitamin solution (Tebo et al., 2006), 1 mM synthetic Mn oxides (Tebo et al., 2006), Balch trace metals (Balch et al., 1979) and 10 mM acetate. After inoculation, tubes were inverted, solidified briefly on ice and incubated at room temperature. The Mn oxides were suspended evenly and Mn reduction resulted in clearings in the agarose. Material from the highest dilutions was further diluted into shake agar tubes and/or spread onto solid MnA1 agar plates for isolation and phylogenetic identification. Amplicons from colony PCR reactions were screened prior to cloning and full-length sequencing.
Biogeochemical analyses of Mn
Unfiltered samples were reacted with ascorbic acid (10%) overnight as a proxy for total Mn (dissolved plus ascorbate-reducible); available Mn oxide concentration was calculated by subtracting the dissolved concentrations (as defined by passing through a 0.2 μm filter) from total Mn. All samples for analysis (dissolved and total) were centrifuged, transferred to new sterile 15 ml Falcon tubes, and Mn concentrations were determined by inductively coupled plasma-optical emission spectroscopy (ICPOES) on a Perkin–Elmer Optima 2000 instrument and were adjusted for changes in dilution due to acid addition.
DNA extraction, cloning and sequencing
Filters containing microbial cells for DNA extraction were cracked open using pliers to allow the inner cylinder containing the filter to be removed. The filter was cut loose from the cylinder using a sterile scalpel and was gently folded and cut into tiny pieces using sterile scissors. The filter pieces were allowed to fall directly into bead beating tubes from the FastDNA Spin Kit for Soil (MP Biomedical Sciences, Solon, OH), and extraction was carried out according to the manufacturer’s instructions. DNA concentration was determined using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Pure cultures of Shewanella sp. and Rhodobacter sp. were grown in liquid K medium (Tebo et al., 2006), pelleted, and genomic DNA was extracted using the UltraClean® Microbial DNA Isolation Kit (MoBioLaboratories, Carlsbad, CA). The SSU rRNA gene was then amplified (from either metagenomic DNA or DNA from isolates) using the 27F and 1492R primers (Lane, 1990). PCR products from three to six PCR reactions were pooled for metagenomic DNA, and amplicons were cloned into TOPO TA pCR®2.1 vectors (Invitrogen, Carlsbad, CA). Plasmid DNA from transformants was extracted using the QIAprep Spin Miniprep kit (Qiagen, Valencia, CA), and screened using the M13 forward (−20) primer. Ninety-six-well plates of glycerol stocks were sent to Washington University (Saint Louis, MO) for sequencing of clone libraries. Sequences of clones clustering with Rhodobacter or Shewanella were deposited into GenBank with Accession No. HM003632–HM003639. For the isolates, one clone was selected from each isolate and was fully sequenced with primers M13 forward (−20), M13 reverse (−27), 357f (5′-CCTACGGGAGGCAGCAG), 515r (5′-TTACCGCGGC KGCTGRCAC), 926f (5′-AAACTYAAAKGAATWGRCGG) and 1098r (5′-GGGTYKCGCTCGTTGC). Sequences were deposited in GenBank with Accession No. EU979473–EU979479. Alignments were created using the on-line SILVA aligner and then exported from release 94 (Pruesse et al., 2007) of the SILVA database for ARB (Ludwig et al., 2004) along with other sequences of interest. Phylogenetic trees were constructed using neighbour-joining and maximum-likelihood methods in the PHYLIP software package (Felsenstein, 2004). The percentage of sequences from each library that represented a particular OTU is given in parentheses: actual sequence abundance ranged from 1/150 to 2/80.
Real-time quantitative PCR
Primers specific for bacteria were examined for target diversity using the Probe Match tool on the RDP webpage (Cole et al., 2009), and for other traits using Primer Express® software (Applied Biosystems™, Foster City, CA). The forward primer 338f 5′-TCCTACGGGAGGCAGCAGT (Nadkarni et al., 2002) and the reverse primer 518r 5′-ATTACCGC GGCTGCTGG (Einen et al., 2008) were chosen as they gave broad coverage and a good amplicon size. Primers specific for Rhodobacter spp. were designed using ARB probe design (Ludwig et al., 2004), the Silva release 94 database (Pruesse et al., 2007), Primer Express® software (Applied Biosystems™, Foster City, CA), the Probe Match tool on the RDP webpage (Cole et al., 2009), and by taking into consideration optimal primer characteristics (Nadkarni et al., 2002). Both the forward primer 391f (5′TAGCCATGCCGCGTGATC) and the reverse primer 536r (5′AACGCTAGCCCCCTCCG) matched the largest clade of cultured Rhodobacter spp. and had only three common non-target sequences in the database. The primer set was challenged against non-target sequences (with 2 basepair mismatches or more) from clone libraries. At 62°C non-target clones were not amplified, whereas Rhodobacter sp. clones were amplified. Amplifications were performed on triplicate samples at two dilutions (0.1 ng μl−1 and 0.2 ng μl−1) in Maxima™ SYBR Green qPCR Master Mix (Fermentas, Glen Burnie, MD) containing 0.2 μM of each primer and were carried out using an Applied Biosystems™ 7300 Real-Time PCR System (Foster City, CA) with the following amplification protocol: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C with 1 min at either 60°C for bacteria or 62°C for Rhodobacter spp. After amplification, a dissociation protocol was added to verify dissociation profiles. Plasmid DNA containing the SSU rRNA gene from Rhodobacter sp. CR07-74 was used in the range of 109–102 target copies per microlitre to generate calibration curves for quantification as described previously (Fey et al., 2004). Copy number results were expressed on a basis of amount of DNA extracted per ml of water filtered. Because many bacteria contain more than one SSU rRNA gene copy in the genome, it is common to divide the number of copies in the sample, as determined by qPCR, by the average number of copies per genome to obtain a more accurate estimate of genome copy number, as a proxy for cell number (Fogel et al., 1999). The Ribosomal RNA Operon Copy Number Database (Kaplenbach et al., 2001) was used to estimate the number of SSU rRNA genes copies for all bacteria (4.06), and for Rhodobacterales (2.69), according to the database on 2 August 2010. Since dividing by these numbers would yield a higher number of Rhodobacterales relative to total Bacteria, copy numbers of both groups were normalized by an average of three copies per genome as a more conservative estimate of Rhodobacter species.
moxA-type genes
Primers for moxA-type multicopper oxidase genes were designed using a CLUSTALX multiple sequence alignment of a range of multicopper oxidase-containing Alphaproteobacteria, and using the SciTools available at Integrated DNA technologies (http://www.idtdna.com). Forward primer MoxAr 5′-AAC ATG CCG CCC ATY TCG A and reverse primer MoxAf 5′-AGA TGG CSA TGG GSA TGA TG were chosen and both of these matched eight of the mox sequences in the alignment. Reaction mixtures were carried out in 50 μl of reactions containing AmpliTaq Gold® (Applied Biosystems™, Foster City, CA) reaction buffer (1×) and DNA Polymerase (1.5 U), 0.4 μM of each primer, 800 μM dNTPs, 0.2 mg ml−1 Bovine Serum Albumin (BSA, New England Biolabs, Ipswich, MA) and 20 ng of DNA. The programme consisted of 3 min at 95°C, 30 cycles of 95°C for 1 min, 52°C for 1.5 min and 72°C for 3 min, followed by a final elongation step at 72°C for 10 min. The PCR products were screened on a 1% agarose gel in TBE buffer. Genomic DNA from A. manganoxydans strain SI85-9A1 was used as a control and all strains gave a band around 650 bp except Rhodobacter sp. CR07-5. All strains, including the control, had some smearing and gave multiple bands even after optimization of MgCl and temperature. Therefore, bands were cut out and purified using a QIAquick Gel Extraction Kit (Qiagen, Valencia, CA), prior to sequencing. moxA sequences were generated using the MoxAf and MoxAr primer set and were deposited in GenBank (Accession No. GU256057–GU256060). The sequences for Rhodobacter sp. CR07-44 and Rhodobacter sp. CR07-62 were identical across 423 bp. However, the sequencing reads for Rhodobacter sp. CR07-62 were incomplete, and were thus omitted from the tree. Multicopper oxidase predicted amino acid sequences were aligned using CLUSTALX (Thompson et al., 1997) and phylogenetic trees were constructed using neighbour-joining and maximum-likelihood methods in the PHYLIP software package (Felsenstein, 2004).
Supplementary Material
Fig. S1. Hindcast models of bottom salinity in the Columbia River estuary during sampling. Models were downloaded and modified from the Model Browser created by the Center for Coastal Margin and Prediction Program (http://www.stccmop.org/), a multi-institutional National Science Foundation Science and Technology Center.
A and B. June samples taken (A) during an ETM with turbidity at 50 ntu and salinity at 19 psu, and (B) after an ETM with turbidity at 8 ntu and salinity at 25 psu.
C and D. July samples taken (C) during an ETM with turbidity at 70 ntu and salinity at 5 psu, and (D) after an ETM with turbidity at 6 ntu and salinity at 9 psu. For reference, Astoria (represented by a polygon) and Youngs Bay are labelled in the first panel.
Fig. S2. Changes in total, dissolved and ascorbate-released manganese with depth (A) during and (B) before an ETM event in the South Channel of the Columbia River in July 2007.
Fig. S3. Relationship between turbidity and manganese (A) during and after an ETM event in the South Channel in July 2007, and (B) regression analysis of total manganese versus turbidity and dissolved manganese(II) versus turbidity for all deep water samples (c. 1 m from the bottom, or 10–18 m depth) collected in the South Channel on June, July or August 2007 cruises on the Columbia River.
Fig. S4. Maximum-likelihood tree inferring the phylogenetic relationship between Mn(IV)-reducing and Mn(II)-oxidizing Shewanella sp. cultured strains* (asterisks) and those found in clone libraries† (daggers) in the Columbia River in this study. Top blast hits to the isolates were included, as well as a broad range of described species. Alignments were created using the on-line SILVA aligner and then exported from ARB. Dendrogram was created using PHYLIP. Bootstrapping values are shown for nodes that were supported at least 50% of the time. The number of the sequences from each library that represented a particular sequence/OTU is given in parentheses. The dendrogram was only weakly supported with neighbour-joining analysis, and many branches had low bootstrapping values in both analyses (Fig. 5). Escherichia species were used as the outgroup.
Acknowledgments
The authors would like to thank the captains and crews of the R/Vs Barnes, Forerunner and Wecoma for assistance with sampling. We thank the chief scientists, especially Lydie Herfort and Byron Crump, as well as computer analysts that gave us insight into predicting and capturing samples during ETM events. We thank Craig Anderson for helpful discussion and input in the writing of this manuscript, and Rick Davis for helpful advice, and assistance with phylogenetic analyses and linux. We are appreciative of assistance from Ninian Blackburn, Brenda Broers, Andrew Bauman and Gnana Sutha Siluvai in conducting ICP-OES analyses. Partial support was provided through the following National Science Foundation, Division of Ocean Sciences grants: 0424602 awarded to Antonio Baptista and 0935270 awarded to S. Bräuer. Support was also provided by an Oregon Opportunity grant awarded to B. Tebo and Grant No. P42ES010337 from the National Institute of Environmental Health Sciences (NIEHS).
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Acinas SG, Anton J, Rodriguez-Valera F. Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol. 1999;65:514–522. doi: 10.1128/aem.65.2.514-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian L, Szewzyk U, Wecke J, Gorisch H. Bacterial dehalorespiration with chlorinated benzenes. Nature. 2000;408:580–583. doi: 10.1038/35046063. [DOI] [PubMed] [Google Scholar]
- Aguilar-Islas AM, Bruland KW. Dissolved manganese and silicic acid in the Columbia River plume: a major source to the California current and coastal waters off Washington and Oregon. Mar Chem. 2006;101:233–247. [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CR, Dick GJ, Chu ML, Cho JC, Davis RE, Bräuer SL, Tebo BM. Aurantimonas manganoxydans, sp. nov. and Aurantimonas litoralis, sp. nov.: Mn(II)-oxidizing representatives of a globally distributed clade of Alphaproteobacteria from the order Rhizobiales. Geomicrobiol J. 2009a;26:189–198. doi: 10.1080/01490450902724840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CR, Johnson HA, Caputo N, Davis RE, Torpey JW, Tebo BM. Mn(II) oxidation is catalyzed by heme peroxidases in ‘Aurantimonas manganoxydans’ strain SI85-9A1 and Erythrobacter sp. strain SD-21. Appl Environ Microbiol. 2009b;75:4130–4138. doi: 10.1128/AEM.02890-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista AM, Zhang YL, Chawla A, Zulauf M, Seaton C, Myers EP, et al. A cross-scale model for 3D baroclinic circulation in estuary-plume-shelf systems: II. Application to the Columbia River. Cont Shelf Res. 2005;25:935–972. [Google Scholar]
- Baptista A, Howe B, Freire J, Maier D, Silva CT. Scientific exploration in the era of ocean observatories. Comput Sci Eng. 2008;10:53–58. [Google Scholar]
- Barnes C, Duxbury A, Morse B. Circulation and selected properties of the Columbia River effluent at sea. In: Pruter A, Alverson D, editors. The Columbia River Estuary and Adjacent Ocean Waters. Seattle, WA, USA: University of Washington Press; 1972. pp. 41–80. [Google Scholar]
- Bencharit S, Ward MJ. Chemotactic responses to metals and anaerobic electron acceptors in Shewanella oneidensis MR-1. J Bacteriol. 2005;187:5049–5053. doi: 10.1128/JB.187.14.5049-5053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JP, McCuaig RD. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl Environ Microbiol. 2003;69:2463–2483. doi: 10.1128/AEM.69.5.2463-2483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettar I, Moore ERB, Hofle MG. Phylogeny and abundance of novel denitrifying bacteria isolated from the water column of the central Baltic Sea. Microb Ecol. 2001;42:295–305. doi: 10.1007/s00248-001-0011-2. [DOI] [PubMed] [Google Scholar]
- Brouwers GJ, de Vrind JPM, Corstjens PLAM, Cornelis P, Baysse C, de Vrind-de Jong EW. CumA, a gene encoding a multicopper oxidase, is involved in Mn2+ oxidation in Pseudomonas putida GB-1. Appl Environ Microbiol. 1999;65:1762–1768. doi: 10.1128/aem.65.4.1762-1768.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwers GJ, Corstjens PLAM, de Vrind JPM, Verkamman A, de Kuyper M, de Vrind-de Jong EW. Stimulation of Mn2+ oxidation in Leptothrix discophora SS-1 by Cu2+ and sequence analysis of the region flanking the gene encoding putative multicopper oxidase MofA. Geomicrobiol J. 2000a;17:25–33. [Google Scholar]
- Brouwers GJ, Vijgenboom E, Corstjens PLAM, de Vrind JPM, de Vrind-de Jong EW. Bacterial Mn2+-oxidizing systems and multicopper oxidases: an overview of mechanisms and functions. Geomicrobiol J. 2000b;17:1–24. [Google Scholar]
- Brown MV, Bowman JP. A molecular phylogenetic survey of sea-ice microbial communities (SIMCO) FEMS Microbiol Ecol. 2001;35:267–275. doi: 10.1111/j.1574-6941.2001.tb00812.x. [DOI] [PubMed] [Google Scholar]
- Bruland KW, Lohan MC, Aguilar-Islas AM, Smith GJ, Sohst B, Baptista A. Factors influencing the chemistry of the near-field Columbia River plume: nitrate, silicic acid, dissolved Fe, and dissolved Mn. J Geophys Res. 2008;113:C00B02. [Google Scholar]
- Buchan A, Gonzalez JM, Moran MA. Overview of the marine Roseobacter lineage. Appl Environ Microbiol. 2005;71:5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burla M. PhD Thesis. Beaverton, OR: Division of Environmental and Biomolecular Systems, Oregon Health & Science University; 2009. The Columbia River estuary and plume: natural variability, anthropogenic change and physical habitat for salmon. [Google Scholar]
- Button DK, Schut F, Quang P, Martin R, Robertson BR. Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corstjens PLAM, de Vrind JPM, Goosen T, de Vrind-de Jong EW. Identification and molecular analysis of the Leptothrix discophora SS-1 mofA gene, a gene putatively encoding a manganese-oxidizing protein with copper domains. Geomicrobiol J. 1997;14:91–108. [Google Scholar]
- Cottrell MT, Kirchman DL. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl Environ Microbiol. 2000;66:5116–5122. doi: 10.1128/aem.66.12.5116-5122.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covert PA. An examination of the form and variability of manganese oxide in Columbia River suspended material. Chemical Oceanography, College of Oceanic and Atmospheric Sciences; Corvallis, OR: Oregon State University; Corvallis, OR: 2001. p. 81. [Google Scholar]
- Crump BC, Baross JA. Particle-attached bacteria and heterotrophic plankton associated with the Columbia River estuarine turbidity maxima. Mar Ecol Prog Ser. 1996;138:265–273. [Google Scholar]
- Crump BC, Baross JA. Characterization of the bacterially-active particle fraction in the Columbia River estuary. Mar Ecol Prog Ser. 2000;206:13–22. [Google Scholar]
- Crump BC, Baross JA, Simenstad CA. Dominance of particle-attached bacteria in the Columbia River estuary, USA. Aquat Microb Ecol. 1998;14:7–18. [Google Scholar]
- Crump BC, Armbrust EV, Baross JA. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol. 1999;65:3192–3204. doi: 10.1128/aem.65.7.3192-3204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang HY, Lovell CR. Numerical dominance and phylotype diversity of marine Rhodobacter species during early colonization of submerged surfaces in coastal marine waters as determined by 16S ribosomal DNA sequence analysis and fluorescence in situ hybridization. Appl Environ Microbiol. 2002;68:496–504. doi: 10.1128/AEM.68.2.496-504.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Caccavo F. Dissimilatory Fe(III) oxide reduction by Shewanella alga BrY requires adhesion. Curr Microbiol. 2000;40:344–347. doi: 10.1007/s002849910068. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Franks DG, Yayanos AA. Evolutionary relationships of cultivated psychrophilic and barophilic deep-sea bacteria. Appl Environ Microbiol. 1997;63:2105–2108. doi: 10.1128/aem.63.5.2105-2108.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiChristina TJ, DeLong EF. Design and application of rRNA-targeted oligonucleotide probes for the dissimilatory iron- and manganese-reducing bacterium Shewanella putrefaciens. Appl Environ Microbiol. 1993;59:4152–4160. doi: 10.1128/aem.59.12.4152-4160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick G. PhD Thesis. San Diego, CA, USA: Marine Biology Research Division, Scripps Institute of Oceanography, University of California; 2006. Microbial manganese(II) oxidation: biogeochemistry of a deep-sea hydrothermal plume, enzymatic mechanism and genomic perspectives; p. 190. [Google Scholar]
- Dick GJ, Torpey JW, Beveridge TJ, Tebo BA. Direct identification of a bacterial Manganese(II) oxidase, the multicopper oxidase MnxG, from spores of several different marine Bacillus species. Appl Environ Microbiol. 2008a;74:1527–1534. doi: 10.1128/AEM.01240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick GJ, Podell S, Johnson HA, Rivera-Espinoza Y, Bernier-Latmani R, McCarthy JK, et al. Genomic insights into Mn(II) oxidation by the marine alphaproteobacterium Aurantimonas sp. strain SI85-9A1. Appl Environ Microbiol. 2008b;74:2646–2658. doi: 10.1128/AEM.01656-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einen J, Thorseth IH, Ovreas L. Enumeration of archaea and bacteria in seafloor basalt using real-time quantitative PCR and fluorescence microscopy. FEMS Microbiol Lett. 2008;282:182–187. doi: 10.1111/j.1574-6968.2008.01119.x. [DOI] [PubMed] [Google Scholar]
- Emerson D, Ghiorse WC. Isolation, cultural maintenance, and taxonomy of a sheath-forming strain of Leptothrix discophora and characterization of manganese-oxidizing activity associated with the sheath. Appl Environ Microbiol. 1992;58:4001–4010. doi: 10.1128/aem.58.12.4001-4010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. phylip (Phylogeny Inference Package) Version 3.68 Distributed by the author. Department of Genome Sciences, University of Washington; Seattle, WA, USA: 2004. [Google Scholar]
- Fey A, Eichler S, Flavier S, Christen R, Hofle MG, Guzman CA. Establishment of a real-time PCR-based approach for accurate quantification of bacterial RNA targets in water, using Salmonella as a model organism. Appl Environ Microbiol. 2004;70:3618–3623. doi: 10.1128/AEM.70.6.3618-3623.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel GB, Collins CR, Li J, Brunk CF. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb Ecol. 1999;38:93–113. doi: 10.1007/s002489900162. [DOI] [PubMed] [Google Scholar]
- Frolov S, Baptista AM, Zhang YL, Seaton C. Estimation of ecologically significant circulation features of the Columbia River estuary and plume using a reduced-dimension Kalman filter. Cont Shelf Res. 2009;29:456–466. [Google Scholar]
- Gebhardt AC, Schoster F, Gaye-Haake B, Beeskow B, Rachold V, Unger D, Ittekkot V. The turbidity maximum zone of the Yenisei River (Siberia) and its impact on organic and inorganic proxies. Estuar Coast Shelf Sci. 2005;65:61–73. [Google Scholar]
- Gelfenbaum G. Suspended sediment response to semidiurnal and fortnightly tidal variations in a mesotidal estuary – Columbia River, USA. Mar Geol. 1983;52:39–57. [Google Scholar]
- Geszvain K, Tebo BM. Identification of a two-component regulatory pathway essential for Mn(II) oxidation in Pseudomonas putida GB-1. Appl Environ Microbiol. 2010;76:1224–1231. doi: 10.1128/AEM.02473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Rappé MS. Evolution, diversity, and molecular ecology of marine prokaryotes. In: Kirchman DL, editor. Microbial Ecology of the Oceans. New York, USA: Wiley; 2000. pp. 47–84. [Google Scholar]
- Gonzalez JM, Simo R, Massana R, Covert JS, Casamayor EO, Pedros-Alio C, Moran MA. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl Environ Microbiol. 2000;66:4237–4246. doi: 10.1128/aem.66.10.4237-4246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabemann I, Uncles RJ, Krause G, Stephens JA. Behaviour of turbidity maxima in the Tamar (UK) and Weser (FRG) estuaries. Estuar Coast Shelf Sci. 1997;45:235–246. [Google Scholar]
- Gralnick JA, Vali H, Lies DP, Newman DK. Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc Natl Acad Sci USA. 2006;103:4669–4674. doi: 10.1073/pnas.0505959103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel CM, Francis CA. Coupled photochemical and enzymatic Mn(II) oxidation pathways of a planktonic Roseobacter-like bacterium. Appl Environ Microbiol. 2006;72:3543–3549. doi: 10.1128/AEM.72.5.3543-3549.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hau HH, Gralnick JA. Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol. 2007;61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- Holden JF, Takai K, Summit M, Bolton S, Zyskowski J, Baross JA. Diversity among three novel groups of hyperthermophilic deep-sea Thermococcus species from three sites in the northeastern Pacific Ocean. FEMS Microbiol Ecol. 2001;36:51–60. doi: 10.1111/j.1574-6941.2001.tb00825.x. [DOI] [PubMed] [Google Scholar]
- Ivanova EP, Sawabe T, Zhukova NV, Gorshkova NM, Nedashkovskaya OI, Hayashi K, et al. Occurrence and diversity of mesophilic Shewanella strains isolated from the north-west Pacific Ocean. Syst Appl Microbiol. 2003;26:293–301. doi: 10.1078/072320203322346155. [DOI] [PubMed] [Google Scholar]
- Jay DA, Musiak JD. Particle trapping in estuarine tidal flows. J Geophys Res. 1994;99:20445–20461. [Google Scholar]
- Jay DA, Smith JD. Circulation, density distribution and neap-spring transitions in the Columbia River estuary. Prog Oceanogr. 1990;25:81–112. [Google Scholar]
- Kan JJ, Wang K, Chen F. Temporal variation and detection limit of an estuarine bacterioplankton community analyzed by denaturing gradient gel electrophoresis (DGGE) Aquat Microb Ecol. 2006;42:7–18. [Google Scholar]
- Kan J, Suzuki MT, Wang K, Evans SE, Chen F. High temporal but low spatial heterogeneity of bacterioplankton in the Chesapeake Bay. Appl Environ Microbiol. 2007;73:6776–6789. doi: 10.1128/AEM.00541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplenbach J, Saxman P, Cole J, Schmidt T. RRNDB: the ribosomal RNA operon copy number database. Nucleic Acids Res. 2001;29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepkay P, Nealson KH. Growth of a manganese-oxidizing Pseudomonas sp. in continuous culture. Arch Microbiol. 1987;148:63–67. [Google Scholar]
- Klewicki JK, Morgan JJ. Kinetic behavior of Mn(III) complexes of pyrophosphate, EDTA, and citrate. Environ Sci Technol. 1998;32:2916–2922. [Google Scholar]
- Klinkhammer GP, McManus J. Dissolved manganese in the Columbia River estuary: production in the water column. Geochim Cosmochim Acta. 2001;65:2835–2841. [Google Scholar]
- Klinkhammer GP, Chin CS, Wilson C, Rudnicki MD, German CR. Distributions of dissolved manganese and fluorescent dissolved organic matter in the Columbia River estuary and plume as determined by in situ measurement. Mar Chem. 1997;56:1–14. [Google Scholar]
- Kogure K, Simidu U, Taga N. A tentative direct microscopic method for counting living marine bacteria. Can J Microbiol. 1979;25:415–420. doi: 10.1139/m79-063. [DOI] [PubMed] [Google Scholar]
- Lane DS. 16S and 23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. New York, USA: John Wiley; 1990. pp. 115–148. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett JW. A radiotracer study of cerium and manganese uptake onto suspended particles in Chesapeake Bay. Geochim Cosmochim Acta. 1994;58:695–703. [Google Scholar]
- Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiol-SGM. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- Nealson KH, Myers CR. Microbial reduction of manganese and iron: new approaches to carbon cycling. Appl Environ Microbiol. 1992;58:439–443. doi: 10.1128/aem.58.2.439-443.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson KH, Saffarini D. Iron and manganese in anerobic respiration: environmental significance, physiology, and regulation. Annu Rev Microbiol. 1994;48:311–343. doi: 10.1146/annurev.mi.48.100194.001523. [DOI] [PubMed] [Google Scholar]
- Nealson K, Scott J. Ecophysiology of the genus Shewanella. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes: Proteobacteria: Gamma Subclass. New York, USA: Springer-Verlag; 2006. pp. 1133–1151. [Google Scholar]
- Nealson KH, Myers CR, Wimpee BB. Isolation and identification of manganese-reducing bacteria and estimates of microbial Mn(IV)-reducing potential in the Black Sea. Deep Sea Res. 1991;38:S907–S920. [Google Scholar]
- Poulain AJ, Newman DK. Rhodobacter capsulatus catalyzes light-dependent Fe(II) oxidation under anaerobic conditions as a potential detoxification mechanism. Appl Environ Microbiol. 2009;75:6639–6646. doi: 10.1128/AEM.00054-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahl FG, Small LF, Eversmeyer B. Biogeochemical characterization of suspended particulate matter in the Columbia River estuary. Mar Ecol Prog Ser. 1997;160:173–184. [Google Scholar]
- Prahl FG, Small LF, Sullivan BA, Cordell J, Simenstad CA, Crump BC, Baross JA. Biogeochemical gradients in the lower Columbia River. Hydrobiologia. 1998;361:37–52. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge JP, Lin M, Larsen EI, Fegan M, McEwan AG, Sly LI. A multicopper oxidase is essential for manganese oxidation and laccase-like activity in Pedomicrobium sp. ACM 3067. Environ Microbiol. 2007;9:944–953. doi: 10.1111/j.1462-2920.2006.01216.x. [DOI] [PubMed] [Google Scholar]
- Shiller AM. Dissolved trace elements in the Mississippi River: seasonal, interannual, and decadal variability. Geochim Cosmochim Acta. 1997;61:4321–4330. [Google Scholar]
- Shiller AM, Stephens TH. Microbial manganese oxidation in the lower Mississippi River: methods and evidence. Geomicrobiol J. 2005;22:117–125. [Google Scholar]
- Siering PL, Ghiorse WC. PCR detection of a putative manganese oxidation gene (mofA) in environmental samples and assessment of mofA homology among diverse manganese-oxidizing bacteria. Geomicrobiol J. 1997;14:109–125. [Google Scholar]
- Skerratt JH, Bowman JP, Nichols PD. Shewanella olleyana sp. nov., a marine species isolated from a temperate estuary which produces high levels of polyunsaturated fatty acids. Int J Syst Evol Microbiol. 2002;52:2101–2106. doi: 10.1099/00207713-52-6-2101. [DOI] [PubMed] [Google Scholar]
- Small LF, Prahl FG. A particle conveyor belt process in the Columbia River estuary: evidence from chlorophyll a and particulate organic carbon. Estuaries. 2004;27:999–1013. [Google Scholar]
- Staudigel H, Hart SR, Pile A, Bailey BE, Baker ET, Brooke S, et al. Vailulu’u seamount, Samoa: life and death on an active submarine volcano. Proc Natl Acad Sci USA. 2006;103:6448–6453. doi: 10.1073/pnas.0600830103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens H, Stubner M, Simon M, Brinkhoff T. Phylogeny of Proteobacteria and Bacteroidetes from oxic habitats of a tidal flat ecosystem. FEMS Microbiol Ecol. 2005;54:351–365. doi: 10.1016/j.femsec.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Sunda WG, Huntsman SA. Microbial oxidation of manganese in a North Carolina estuary. Limnol Oceanogr. 1987;32:552–564. [Google Scholar]
- Sunda WG, Kieber DJ. Oxidation of humic substances by manganese oxides yields low-molecular-weight organic substrates. Nature. 1994;367:62–64. [Google Scholar]
- Suzuki MT, Rappe MS, Haimberger ZW, Winfield H, Adair N, Strobel J, Giovannoni SJ. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MT, Preston CM, Beja O, Torre JR, Steward GF, DeLong EF. Phylogenetic screening of ribosomal RNA gene-containing clones in bacterial artificial chromosome (BAC) libraries from different depths in Monterey Bay. Microb Ecol. 2004;48:473–488. doi: 10.1007/s00248-004-0213-5. [DOI] [PubMed] [Google Scholar]
- Tebo BM. The Ecology and Ultrastructure of Marine Manganese Oxidizing Bacteria. San Diego, CA, USA: University of California; 1983. [Google Scholar]
- Tebo BM, Bargar JR, Clement B, Dick G, Murray KJ, Parker D, et al. Biogenic manganese oxides: properties and mechanisms of formation. Annu Rev Earth Planet Sci. 2004;32:287–328. [Google Scholar]
- Tebo BM, Johnson HA, McCarthy JK, Templeton AS. Geomicrobiology of manganese(II) oxidation. Trends Microbiol. 2005;13:421–428. doi: 10.1016/j.tim.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Tebo BM, Clement B, Dick GJ. Biotransformations of manganese. In: Hurst CJ, Crawford RL, Garland JL, Lipson DA, Mills AL, Stetzenbach LD, editors. Manual of Environmental Microbiology. Washington, DC, USA: ASM Press; 2006. pp. 1223–1238. [Google Scholar]
- Templeton AS, Staudigel H, Tebo BM. Diverse Mn(II)-oxidizing bacteria isolated from submarine basalts at Loihi Seamount. Geomicrobiol J. 2005;22:127–139. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouwborst RE, Clement BG, Tebo BM, Glazer BT, Luther GW. Soluble Mn(III) in suboxic zones. Science. 2006;313:1955–1957. doi: 10.1126/science.1132876. [DOI] [PubMed] [Google Scholar]
- de Vrind JPM, Boogerd FC, de Vrind-de Jong EW. Manganese reduction by a marine Bacillus species. J Bacteriol. 1986;167:30–34. doi: 10.1128/jb.167.1.30-34.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Waasbergen LG, Hildebrand M, Tebo BM. Identification and characterization of a gene cluster involved in manganese oxidation by spores of the marine Bacillus sp. strain SG-1. J Bacteriol. 1996;178:3517–3530. doi: 10.1128/jb.178.12.3517-3530.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Döbler I, Biebl H. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol. 2006;60:255–280. doi: 10.1146/annurev.micro.60.080805.142115. [DOI] [PubMed] [Google Scholar]
- Waidner LA, Kirchman DL. Aerobic anoxygenic photosynthesis genes and operons in uncultured bacteria in the Delaware River. Environ Microbiol. 2005;7:1896–1908. doi: 10.1111/j.1462-2920.2005.00883.x. [DOI] [PubMed] [Google Scholar]
- Waidner LA, Kirchman DL. Diversity and distribution of ecotypes of the aerobic anoxygenic phototrophy gene pufM in the Delaware estuary. Appl Environ Microbiol. 2008;74:4012–4021. doi: 10.1128/AEM.02324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Baptista AM. SELFE: a semi-implicit Eulerian-Lagrangian finite-element model for cross-scale ocean circulation. Ocean Model. 2008;21:71–96. [Google Scholar]
- Zhang YL, Baptista AM, Myers EP. A cross-scale model for 3D baroclinic circulation in estuary-plume-shelf systems: I. Formulation and skill assessment. Cont Shelf Res. 2004;24:2187–2214. [Google Scholar]
- Zhao YL, Wang K, Budinoff C, Buchan A, Lang A, Jiao NZ, Chen F. Gene transfer agent (GTA) genes reveal diverse and dynamic Roseobacter and Rhodobacter populations in the Chesapeake Bay. ISME J. 2009;3:364–373. doi: 10.1038/ismej.2008.115. [DOI] [PubMed] [Google Scholar]
- Ziemke F, Brettar I, Hofle MG. Stability and diversity of the genetic structure of a Shewanella putrefaciens population in the water column of the central Baltic. Aquat Microb Ecol. 1997;13:63–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Hindcast models of bottom salinity in the Columbia River estuary during sampling. Models were downloaded and modified from the Model Browser created by the Center for Coastal Margin and Prediction Program (http://www.stccmop.org/), a multi-institutional National Science Foundation Science and Technology Center.
A and B. June samples taken (A) during an ETM with turbidity at 50 ntu and salinity at 19 psu, and (B) after an ETM with turbidity at 8 ntu and salinity at 25 psu.
C and D. July samples taken (C) during an ETM with turbidity at 70 ntu and salinity at 5 psu, and (D) after an ETM with turbidity at 6 ntu and salinity at 9 psu. For reference, Astoria (represented by a polygon) and Youngs Bay are labelled in the first panel.
Fig. S2. Changes in total, dissolved and ascorbate-released manganese with depth (A) during and (B) before an ETM event in the South Channel of the Columbia River in July 2007.
Fig. S3. Relationship between turbidity and manganese (A) during and after an ETM event in the South Channel in July 2007, and (B) regression analysis of total manganese versus turbidity and dissolved manganese(II) versus turbidity for all deep water samples (c. 1 m from the bottom, or 10–18 m depth) collected in the South Channel on June, July or August 2007 cruises on the Columbia River.
Fig. S4. Maximum-likelihood tree inferring the phylogenetic relationship between Mn(IV)-reducing and Mn(II)-oxidizing Shewanella sp. cultured strains* (asterisks) and those found in clone libraries† (daggers) in the Columbia River in this study. Top blast hits to the isolates were included, as well as a broad range of described species. Alignments were created using the on-line SILVA aligner and then exported from ARB. Dendrogram was created using PHYLIP. Bootstrapping values are shown for nodes that were supported at least 50% of the time. The number of the sequences from each library that represented a particular sequence/OTU is given in parentheses. The dendrogram was only weakly supported with neighbour-joining analysis, and many branches had low bootstrapping values in both analyses (Fig. 5). Escherichia species were used as the outgroup.


