Abstract
Purpose
We evaluated the association of prostate specific antigen (PSA) and androgen receptor splice variant-7 (AR-V7) transcript levels in patients’ blood with time to treatment failure (TTF) and overall survival (OS) with abiraterone acetate (AA) and/or enzalutamide treatment in castration resistant prostate cancer (CRPC) patients.
Experimental Design
RNA levels of AR-V7 and PSA in peripheral blood collected before treatment were quantified using Droplet Digital-PCR in retrospective cohorts treated with AA (N=81) or enzalutamide (N=51) for CRPC. Multivariable Cox regression adjusted for known prognostic factors was used for analyses.
Results
PSA-transcripts were detected in 57% of AA-treated patients and in 63% of enzalutamide-treated patients. PSA-positive patients had a shorter TTF than PSA-negative patients (adjusted-HR=2.27 (95%CI: 1.26, 4.10) and 2.60 (95%CI: 1.19, 5.69); p-value=0.006 and 0.017 in AA and enzalutamide cohorts, respectively). Patients with a higher-AR-V7 transcript level had a shorter TTF with AA and enzalutamide in univariate analysis (median 8.0 versus 15.6 months, p=0.046 in AA-cohort and 3.6 versus 5.6 months, p=0.050 in enzalutamide-cohort). In multivariable models, the association with TTF remained significant in the enzalutamide-cohort (adjusted-HR=2.02; 95%CI:1.01, 4.05; p=0.048), but statistically insignificant in the AA-cohort. In both cohorts, we observed potential prognostic value of both PSA and AR-V7 RNA expression on OS; patients with detectable PSA transcripts and high AR-V7 predicted the poorest OS.
Conclusions
PSA and AR-V7 transcripts in blood potentially serve as biomarkers predicting TTF and OS with AA or enzalutamide treatment. If validated prospectively, their detection could be facilitated without isolation of circulating tumor cells.
Keywords: AR-V7, prostate cancer, abiraterone acetate, enzalutamide
Introduction
Androgen deprivation therapy (ADT) is the most effective-widely used treatment for advanced hormone sensitive prostate cancer (PCa) (HSPC) patients (1, 2). Most HSPCs initially respond to ADT but eventually progress to castration resistant PCa (CRPC), usually leading to death (3-5). The androgen receptor (AR) remains a principal target in CRPC, which can be sustained by intratumoral androgens from circulating adrenal androgens or from de novo synthesis (6-10). New therapies targeting the AR signaling axis have emerged for CRPC (11, 12). The FDA recently approved several drugs for treatment of metastatic CRPC and two of these drugs target the AR or the androgen synthesis pathway. Abiraterone acetate (AA) is an inhibitor of CYP17A1 that blocks androgen production (13, 14). AA can also be converted to a more active Δ4-abiraterone in vivo, which blocks multiple steroidogenic enzymes and antagonizes AR (15). Enzalutamide is an AR antagonist that binds to the ligand-binding domain of the AR, competing with testosterone and dihydrotestosterone and thereby blocking the translocation of AR to nucleus, inhibiting AR function (16, 17). Even though studies have shown that these drugs prolong overall survival (OS), a significant proportion of patients does not respond to these drugs or develops resistance shortly after the treatment (18-23).
The AR splice variant 7 (AR-V7), which lacks the ligand-binding domain but retains functional transcriptional element binding domains, mediates intracellular AR signaling in a ligand independent manner (24-26). Accumulating evidence has suggested the association of the presence of AR-V7 with CRPC development and resistance to treatment with AA and enzalutamide (27-35). In a small study, Antonarakis et al. (36) reported that detection of AR-V7 mRNA in circulating tumor cells (CTC) from CRPC patients was associated with resistance to AA and enzalutamide. Later, Antonarakis et al. and other groups showed that detection of AR-V7 in CTCs from men with metastatic CRPC did not appear to be associated with primary resistance to taxanes (37, 38). However, a more recent study by Scher et al. demonstrated that the CTC nuclear expression of AR-V7 protein in men with mCRPC is associated with superior survival on taxane therapy over AR-directed therapy in a clinical practice setting (39). Nevertheless, these results suggested that AR-V7 might serve as a biomarker for treatments targeting the AR axis in CRPC.
We previously demonstrated the prognostic value of detection of prostate specific antigen (PSA) transcripts in the blood of men with CRPC (40-42). This study was intended to evaluate the potential application of whole blood AR-V7 and PSA transcript levels as prognostic markers for mCRPC patients treated with AA and enzalutamide. We performed quantitative analysis of AR-V7 and PSA transcripts in blood to determine their association with time to treatment failure (TTF) and OS. Since expression of AR-V7 and PSA are thought to be prostate tumor cell specific, signals detected in the peripheral blood mononuclear cell fraction would presumably represent those from CTCs. Thus we evaluated the expression levels of AR-V7 and PSA in the peripheral blood mononuclear cell fraction using droplet digital PCR (ddPCR) (43, 44), from samples derived pre-abiraterone and/or pre-enzalutamide treatment, in retrospective cohorts of men.
Materials and methods
Patients
Patients were retrospectively identified from Dana-Farber Cancer Institute Prostate Clinical Research Information System (CRIS) database (45). Eligible patients were those who had CRPC treated with AA and/or enzalutamide between April 2010 and May 2015 and had the peripheral blood mononuclear cell fraction-derived RNA available within one year prior to treatment initiation. All patients were consented to an IRB-approved protocol that permits collection of clinical and specimen data. Baseline patient characteristics and treatment history were extracted from the database and medical record review.
Sample collection and RNA extraction
The peripheral blood mononuclear cell fraction were isolated by density separation over Ficoll-Hypaque (GE Healthcare) from 8 mL of whole blood collected in Vacutainer cell preparation tubes (BD, Franklin Lakes, NJ) with EDTA within 2 hours after blood drawn (46). Samples generated from patients were then blind-coded. Total RNA from the peripheral blood mononuclear cell fraction was extracted with Trizol reagent (Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol. The resulting RNA was precipitated in 70% ethanol and 0.3 M sodium acetate, and stored at −80°C until use. RNA was further purified by phenol/chloroform extraction and re-suspended in nuclease-free water prior to experiments. Quality of purified RNA samples was verified on 2 % agarose E-gels (Life Technologies, Carlsbad, CA) and no visible signs of degradation were shown; RNA concentrations were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
cDNA synthesis
Reverse transcription (RT) was conducted using iScript advanced cDNA synthesis Kit (Bio-Rad, Hercules, CA) with random hexamer primers as per manufacturer's instruction. Briefly, ~2.5 μg of total RNA sample was subjected to RT-PCR in the 15 μL reaction system; and incubated at 42°C for 30 min and 85°C for 5 min. cDNA was stored at −20°C until use for ddPCR.
Droplet digital polymerase chain reaction (ddPCR)
DdPCR (QX200, Bio-Rad, Hercules, CA, USA) was used in this study. Droplet generation, PCR reactions, and detection were carried out according to the manufacturer's instruction (43, 47). Briefly, the reactions were performed in 20-μL reaction volume that consisted of 10 μL of 2× ddPCR Supermix for probes (No dUTP) (Bio-Rad), 1 μL of gene-specific primers (900 nM) and probes (250 nM) and 2 μL of the cDNA sample. Each reaction mix was converted to droplets with the QX200 droplet generator (Bio-Rad). Droplet-partitioned samples were then transferred to a 96-well plate, sealed and cycled in a C1000 Touch thermal cycler (Bio-Rad) under the following cycling protocol: 95 °C for 10 min, followed by 40 cycles of 94 °C for 30 s, 60°C for 60 s, and a 10-min incubation at 98 °C. The cycled plates were then read on a Bio-Rad QX200 droplet reader. At least two negative control wells with no cDNA template were included in every run. The data analysis was performed with QuantaSoft droplet reader software v1.7.4 (Bio-Rad). The target mRNA concentrations were calculated using Poisson statistics (44) and the background was corrected based on the data of the no template control. Absolute transcript levels were initially presented as copies per μL and converted to copies per μg RNA based on the input amount of RNA.
Primers and probes for the full length AR (AR-FL), AR-V7 and PSA are provided in supplemental data.
Statistical analysis
The cutoff for high AR-V7 was set at the upper tertile of the expression value normalized to the amount of input-RNA since similar results (TTF or OS) were observed between the lowest and middle tertile so they were combined. Samples with detectable PSA transcripts were considered PSA positive. Patient disease characteristics and prior treatments were evaluated by descriptive statistics; Fisher’s exact and Wilcoxon rank sum tests were used to compare the expression status of AR-V7 (high versus low) or PSA (positive or negative) for categorical and continuous characteristics.
TTF was defined as time from treatment initiation until the date of drug discontinuation for any reason, censored at the date of last follow-up for patients who were still on therapy. OS was defined as time from treatment initiation to death from any cause, censored at the date of last follow-up for patients who were still alive. The distributions of TTF and OS were estimated using the Kaplan-Meier method, with 95% confidence intervals (CIs); their associations with AR-V7 and PSA expression status were evaluated using the log-rank test or the Wald chi-square test in multivariable Cox regression model adjusted for known prognostic factors. The multivariable model was constructed by including variables with p<0.10 in univariate analysis no formal model selection was used. For patients treated with AA, we included baseline PSA (dichotomized at the median), albumin< lower limit of normal (yes versus no), ECOG performance status (0 versus >0), prior use of docetaxel (yes versus no) as covariates. Similar covariates were used for those treated with enzalutamide except “prior use of AA (yes versus no)” was used as a covariate instead of albumin. Statistical analyses were performed with SAS version 9.4 (SAS Institute, Cary, NC) and all statistical tests were two-sided.
Results
Detection of AR-V7 and PSA in the peripheral blood mononuclear cell fraction by ddPCR
We first conducted a series of experiments intended to optimize conditions for the detection of AR-V7 and PSA by ddPCR. The absolute quantification and linearity detection of the transcripts were validated by using a series of diluted plasmid DNAs; cross-reaction between AR-V7 and full-length androgen receptor (AR-FL) were not detected (Fig. S1). To document its sensitivity, AR-V7 transcripts were determined in DU145 (an AR negative and PSA negative prostate cancer cell line) cells spiked with different numbers of 22RV1 cells, a prostate-cancer cell line known to express both AR-FL and AR-V7. As shown in Fig. S2, AR-V7 transcripts were detectable in samples with one 22RV1 cell to 104 DU145 cells. We measured the level of AR-FL in the peripheral blood mononuclear cell fraction and found expectedly that high levels of expression of AR-FL, potentially masking the detection of tumor derived AR-FL. After the validation of linearity of detection, we applied the assay to patient samples. As examples, droplet plots and histogram of 6 representative samples and the negative control are shown in Fig. S3.
In total, we assayed 171 pre-treatment samples: 102 were AA treated and 69 were enzalutamide treated patients. In both cohorts, samples were removed if a) they were from the same patient (N=11 and 12), b) they were ineligible (i.e. sample was collected post therapy initiation; AA or enzalutamide was used in the adjuvant setting; patients had been treated with prior AA before sample collection in the AA cohort; or outcome data was not assessable) after we systemically reviewed clinical data (N=7 and 5), or c) the housekeeping TBP gene expression was extremely low (N=3 and 1) in the AA and enzalutamide cohort, respectively. The final number was 81 and 51 in the AA and enzalutamide cohort respectively. The number of transcripts was normalized to the amount of input RNA prior to the analysis.
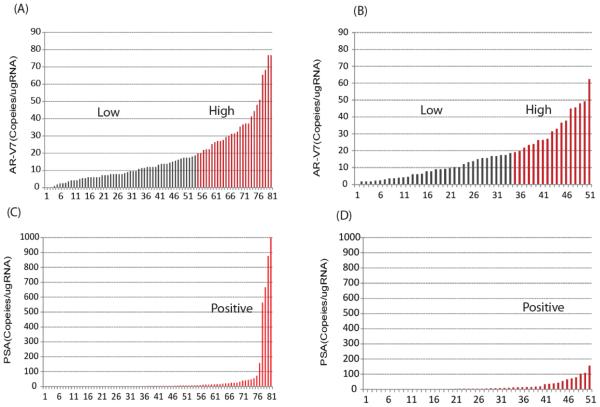
The overall distributions of AR-V7 and PSA transcripts detected in the AA and the enzalutamide cohorts are shown in Fig. 1. PSA transcripts were detected in 46 (57%) of AA-treated patients and in 32 (63%) of enzalutamide treated patients. AR-V7 transcripts were detected in greater than 95% of patients in both cohorts. The distribution of AR-V7 expression level was similar in the AA- and enzalutamide-treated patients (median and interquartile range: 13.2(7.2 26.4) and 13.8(6.0, 24.0) copies/ugRNA, respectively). In the AA cohort, among 27 patients with high AR-V7 expression (defined as the top tertile, i.e. >19 copies/ugRNA), 21 (78%) patients demonstrated PSA transcripts and 6 (22%) patients did not. Similarly, in the enzalutamide cohort, the majority of patients with high AR-V7 patients were also positive for PSA transcripts (77%).
Figure 1. The distribution of AR-V7 and PSA transcript levels in the peripheral blood mononuclear cell fraction of samples.
AR-V7 transcripts in (A) AA and (B) enzalutamide cohort; PSA transcripts in (C) AA and (D) enzalutamide cohort. X axis represents the number of samples; Y axis represents the normalized transcription numbers.
Patient characteristics and outcomes
Patient and disease characteristics were presented in Table 1. Eighty one were AA-treated patients and 51 were enzalutamide-treated patients. 25 (30.9%) of patients in the AA cohort received prior docetaxel. In the enzalutamide cohort, 23 (45.1%) patients received prior docetaxel and 35 (68.8%) also received prior AA for CRPC or HSPC. Median follow-up was 29.7 (range: 3.6+, 47.5) and 23.9 (range: 0.9+, 48.3) months in the AA and enzalutamide cohorts, respectively. In the AA cohort, median TTF and OS were 10.3 (95% CI: 8.0, 19.9) and 34.4 (95% CI: 25.5, 38.7) months respectively. In the enzalutamide cohort, median TTF and OS were only 3.7 (95% CI: 2.8,6.0) months and 21.4 (15.1,35.3) months, respectively. These differences in outcomes were likely due to baseline characteristics including high prior treatment with AA in the enzalutamide treated cohort.
Table 1.
Patient and disease characteristics
| AA cohort (N=81) |
Enzalutamide cohort (N=51) |
|||
|---|---|---|---|---|
| N | % | N | % | |
| At diagnosis of cancer | ||||
| Gleason | ||||
| 6 or less | 7 | 8.6 | 8 | 15.7 |
| 7 | 27 | 33.3 | 16 | 31.4 |
| 8-10 | 40 | 49.4 | 21 | 41.2 |
| Unknown | 7 | 8.6 | 6 | 11.8 |
| At treatment initiation | ||||
| ECOG performance status | ||||
| 0 | 62 | 76.5 | 28 | 54.9 |
| 1 | 9 | 11.1 | 16 | 31.4 |
| ≥2 | 5 | 6.2 | 5 | 9.8 |
| Unknown | 5 | 6.2 | 2 | 3.9 |
| Presence of metastasis | 79 | 97.5 | 50 | 98.0 |
| Low albumin(<LLN)* | 5 | 6.8 | 7 | 14.3 |
| Prior use of Docetaxel** | 25 | 30.9 | 23 | 45.1 |
| Prior use of AA** | - | - | 35 | 68.6 |
| Prior use of Enzalutamide** | 4 | 4.9 | - | - |
| Age, years (median) | 68.3 | IQR:62-74 Range:46-89 |
69.0 | IQR:63-74 Range:50-88 |
| PSA,ng/mL (median) | 16.4 | IQR:4.8-52.1 Range: 0.1-972.1 |
45.5 | IQR:7.4-126.6 Range: 0.3-1148.4 |
|
Months from sample collection
to treatment start |
||||
| ≤3 | 33 | 40.7 | 19 | 37.3 |
| 3~6 | 20 | 24.7 | 13 | 25.5 |
| >6 | 28 | 34.6 | 19 | 37.3 |
evaluable N=74 and 49 for AA and enzalutamide cohort, respectively
treatment for metastasis or as a secondary hormone for castration-resistant prostate
Association of PSA and AR-V7 expression with patient characteristics
Median serum PSA value was higher in patients with detectable PSA transcripts (62.7 versus 8.5 ng/mL, p=0.051) in the enzalutamide cohort; but this trend was not statistically significant in the AA cohort (median 18.2 versus 16.4, p=0.357). Serum PSA value was not associated with patient AR-V7 expression status in both cohorts (p-value=0.502 and 0.811, respectively). In addition, PSA and AR-V7 transcript status in the peripheral blood mononuclear cell fraction was not associated with patients’ other baseline characteristics, such as ECOG performance status and albumin (p values > 0.30, data not shown). However, we observed that patients who had started chemotherapy at time of sample collection more likely had high AR-V7 or positive PSA transcripts than those who had not received chemotherapy in the AA cohort. Similar trend was also observed in the enzalutamide cohort (Table S1).
Association of PSA and AR-V7 transcript level in the peripheral blood mononuclear cell fraction with TTF
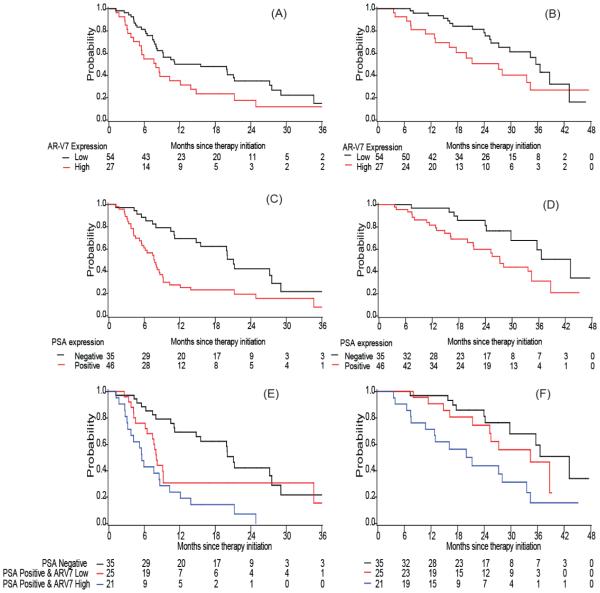
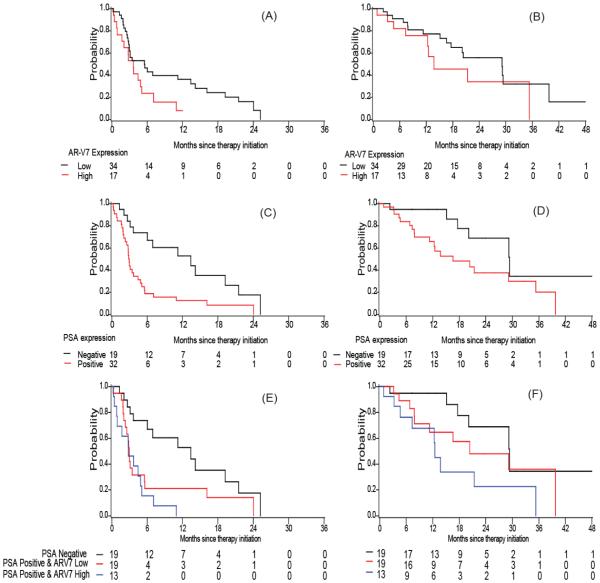
PSA transcript positive patients had a shorter TTF than PSA-negative patients; the adjusted hazard ratio was 2.27 (95% CI: 1.26, 4.10) in the AA cohort and 2.60 (95% CI: 1.19, 5.69) in the enzalutamide cohort (adjusted p=0.006 and p=0.017 respectively). Patients with high AR-V7 expression tended to have a shorter TTF: median 8.0 versus 15.6 months in the AA cohort (log-rank p=0.046) and median 3.6 versus 5.6 months in the enzalutamide cohort (log-rank p=0.050). In multivariable analysis when adjusted for the above mentioned covariates, AR-V7 remained significant in the enzalutamide cohort (adjusted HR=2.02 (95% CI: 1.01, 4.05), p=0.048), but not in the AA cohort (adjusted HR=1.31 (95% CI: 0.74, 2.32), p=0.353), as shown in Table 2, Table 3, Fig. 2 A, C. E and Fig. 3 A, C, E). For both cohorts, the effect of PSA and AR-V7 transcripts on TTF appeared additive. Those with both PSA positive and high AR-V7 expression had the shortest TTF compared to those with no PSA transcript detected or those who were PSA transcript positive but low AR-V7 expression (median 21.1, 8.1 and 5.6 months, adjusted HR=1.00, 2.04 (95% CI: 1.03,4.02) and 2.58 (95% CI: 1.29, 5.15), respectively in the AA cohort; median 13.4, 3.0 and 2.8 months, adjusted HR=1.00, 2.02 (95% CI: 0.85,4.82) and 3.70 (95% CI:1.52,8.96) in the enzalutamide cohort, Table 2 and 3).
Table 2.
Time to treatment failure (TTF) and overall survival (OS) from treatment initiation (AA cohort)
| Time to treatment failure | Overall survival | |||||
|---|---|---|---|---|---|---|
| N/N events |
Median. month |
Adjusted Hazard Ratio* |
N/N events |
Median, month |
Adjusted Hazard Ratio* |
|
| All patients (N=81) | 81/58 | 10.3 (8.0,19.9) |
- | 81/35 | 34.4 (25.5,38.7) |
- |
| By AR-V7 expression | ||||||
| Low | 54/36 | 15.6 (8.1,21.3) |
1.00 (reference) |
54/19 | 35.6 (27.3,43.2) |
1.00 (reference) |
| High (top 33th percentile) | 27/22 | 8.0 (4.2,12.1) |
1.31
(0.74,2.32) |
27/16 | 27.2 (13.0,34.4) |
1.73
(0.83,3.60) |
| P-value | 0.046 | 0.353 | 0.060 | 0.145 | ||
| By PSA expression | ||||||
| Negative | 35/20 | 21.1 (14.8,27.6) |
1.00 (reference) |
35/10 | 43.2 (29.8,NR) |
1.00 (reference) |
| Positive | 46/38 | 7.7 (5.5,9.1) |
2.27
(1.26,4.10) |
46/25 | 27.3 (20.0,34.4) |
1.84
(0.81,4.19) |
| P-value | 0.002 | 0.006 | 0.019 | 0.146 | ||
| By PSA and ARV7 status | ||||||
| PSA negative | 35*/20 | 21.1 (14.8,27.6) |
1.00 (reference) |
35/10 | 43.2 (29.8,NR) |
1.00 (reference) |
| PSA positive and low ARV7 | 25/18 | 8.1 (6.5,9.2) |
2.04
(1.03,4.02) |
25/10 | 34.4 (21.4,NR) |
1.30
(0.50,3.37) |
| PSA positive and high ARV7 | 21/20 | 5.6 (3.2,8.6) |
2.58
(1.29,5.15) |
21/15 | 21.3 (10.8,33.6) |
2.82
(1.11,7.18) |
| P-value | 0.0001 | 0.018 | 0.004 | 0.071 | ||
adjusted for variables with p<0.10 in univariate analysis, including baseline PSA (dichotomized at the median), Low albumin (yes versus no), ECOG PS (0 versus >0), prior use of docetaxel (yes versus no).
Table 3.
Time to treatment failure (TTF) and overall survival (OS) from treatment initiation (Enzalutamide cohort)
| Time to treatment failure | Overall survival | |||||
|---|---|---|---|---|---|---|
| N/N event s |
Median. month |
Adjusted Hazard Ratio* |
N/N event s |
Median, month |
Adjusted Hazard Ratio* |
|
| All patients (N=51) | 51/43 | 3.7 (2.8,6.0) |
51/25 | 21.4 (15.1,35.3) |
||
| By AR-V7 expression | ||||||
| Low | 34/28 | 5.6 (2.9,13.4) |
1.00 (reference) |
34/16 | 29.1 (16.6,39.8) |
1.00 (reference) |
| High | 17/15 | 3.6 (0.9,5.1) |
2.02
(1.01,4.05) |
17/9 | 13.8 (7.4,35.3) |
2.08
(0.83,5.24) |
| P-value | 0.050 | 0.048 | 0.242 | 0.119 | ||
| By PSA expression | ||||||
| Negative | 19/13 | 13.4 (3.6,21.5) |
1.00 (reference) |
19/6 | 29.4 (17.6,NR) |
1.00 (reference) |
| Positive | 32/30 | 2.9 (2.0,4.5) |
2.60
(1.19,5.69) |
32/19 | 16.6 (7.9,35.3) |
1.61
(0.63,4.17) |
| P-value | 0.003 | 0.017 | 0.053 | 0.322 | ||
| By PSA and ARV7 status | ||||||
| PSA negative | 19/13 | 13.4 (3.6,21.5) |
1.00 (reference) |
19/6 | 29.4 (17.6,NR) |
1.00 (reference) |
| PSA positive and low ARV7 | 19/17 | 3.0 (2.0,5.6) |
2.02
(0.85,4.82) |
19/10 | 20.4 (7.8,39.8) |
1.08
(0.37, 3.14) |
| PSA positive and high ARV7 | 13/13 | 2.8 (0.9,4.9) |
3.70
(1.52,8.96) |
13/9 | 12.5 (4.6,35.3) |
3.08
(1.02,9.30) |
| P-value | 0.003 | 0.015 | 0.045 | 0.074 | ||
adjusted for variables with p<0.10 in univariate analysis, including baseline PSA (dichotomized at the median), ECOG PS (0 versus >0), prior use of docetaxel (yes versus no), prior use of AA (yes versus no)
Figure 2. Kaplan Meier plots in AA cohort.
Plots of TTF according to (A) AR-V7 expression (C) PSA expression (E) combination of PSA and AR-V7 expression; Plots of OS according to (B) AR-V7 expression (D) PSA expression (F) combination of PSA and AR-V7 expression in AA cohort.
Figure 3. Kaplan Meier plots in Enzalutamide cohort.
Plots of TTF according to (A) AR-V7 expression (C) PSA expression (E) combination of PSA and AR-V7 expression; Plots of OS according to (B) AR-V7 expression (D) PSA expression (F) combination of PSA and AR-V7 expression in Enzalutamide cohort.
Association of PSA and AR-V7 expression with OS
In both cohorts, we observed that patients with high AR-V7 expression had a shorter overall survival (OS) (median OS: 35.6 versus 27.2 month in the AA cohort and 29.1 versus 13.8 months in the enzalutamide cohort). Similarly, median OS was shorter among men with detectable PSA transcripts than among those with negative PSA transcripts (Table 2 and 3; Fig. 2 and 3, panels B, C, F). However, these observed associations became insignificant in the multivariable models adjusted for other baseline factors, most likely due to the limited number of deaths (i.e. 35 deaths in the AA cohort and 25 deaths in the enzalutamide cohort) in these analyses (HR ranging 1.6~2.1, p>0.05). In addition, we observed an additive effect of PSA transcript detection and AR-V7 expression in the prediction for OS (Fig. 2F and 3F). When compared to those with negative PSA or those with PSA positive but low AR-V7 expression, those with both positive PSA transcripts and high AR-V7 transcripts had the worst OS (median 43.2, 34.4 and 21.3 months, adjusted HR=1.00, 1.30 (95% CI: 0.50,3.37) and 2.82 (95% CI: 1.11, 7.18), respectively in the AA cohort; median 29.4, 20.4 and 12.5 months, adjusted HR=1.00, 1.08 (95% CI: 0.37,3.14) and 3.08 (95% CI:1.02,9.30) in the enzalutamide cohort, as shown in Table 2 and 3).
Subgroup and sensitivity analysis
All samples for this study were collected within one year prior to treatment initiation. We suspected that the time of the blood draw may potentially impact the level of AR-V7 and PSA expression and confound the observed association. Thus we conducted a subgroup analysis by restricting the samples to those with blood draw within 6 months prior to treatment initiation. The results suggested that there was no significant difference in the groups (HRs ranging 1.3~2.5) but there was limited statistical power for the subgroup analysis (Table S2).
In addition, we noticed that a few patients (6 in the AA cohort and 4 in the enzalutamide cohort) who had no PSA transcripts detected had high AR-V7 transcripts, though the majority (77%) of patients with high AR-V7 was also positive for PSA transcripts. It is possible that for these cases, AR-V7 might be mainly derived from non-prostate cells. If we consider that these patients may have been misclassified, by placing them in the low AR-V7 group, we found that the association of AR-V7 with TTF and OS became stronger (for TTF: adjusted HR =1.93 (95% CI: 1.04,3.55) and 2.52 (95% CI: 1.2, 5.15); for OS: adjusted HR=2.47 (95% CI: 1.12,5.41) and 2.95 (95% CI:1.17,7.5), in the AA and enzalutamide cohort respectively).
Discussion
We had previously shown that PSA detected by RT-PCR was predictive of OS (40-42). Under the assumption that tumor specific transcripts in blood are most likely derived from CTCs, we directly analyzed the tumor specific RNAs, AR-V7 and PSA, in the peripheral blood mononuclear cell fraction, thus by-passing the isolation of CTCs. Our data showed that the detection of PSA transcripts in the peripheral blood mononuclear cell fraction predicted TTF in CRPC patients who were treated with AA or enzalutamide, with adjustment of serum PSA, albumin, ECOG performance status, prior use of docetaxel, and/or prior use of AA. The association of AR-V7 expression with the TTF was statistically significant in univariable analyses but insignificant in multivariable analysis although trends remained. Both the transcript level of PSA and AR-V7 in the peripheral blood mononuclear cell fraction were associated with shorter OS (HR ranging 1.6~2.1) and the prognostic effect was additive. These results demonstrate the potential application of PSA and AR-V7 transcripts in the peripheral blood mononuclear cell fraction as biomarkers for predicting OS and potentially TTF on AA or enzalutamide treatment.
Among 132 samples analyzed, we found that almost all of the samples had detectable AR-V7, while only ~60% of samples had detectable PSA. There was some degree of correlation as most patients with high AR-V7 also had positive PSA expression. We noticed that a few patients had no detectable PSA transcripts but high AR-V7 transcripts. It is possible that some CTCs express AR-V7 but not PSA. Another possibility is that in some cases, the AR-V7 may originate from non-prostate cells as noted in the report by Takeuchi et al. (48). Our re-grouping study supported the possibility that in some cases, AR-V7 in the peripheral blood mononuclear cell fraction may not be derived from CTCs and this event may potentially alter the assessment of AR-V7 alone when using this methodology.
For this study, we defined high AR-V7 expression as the upper tertile of expression level (i.e. ≥19 copies/ugRNA). The analysis was initially conducted with three tertile groups, and subsequently condensed into two groups as we observed similar results (TTF and OS) among those with low and intermediate AR-V7 expression (data not shown). In addition, classifying 33% of patients to the high expression group was consistent with the reported detection rate (ranging 20~40%) in circulating tumor cells from patients treated with AA or enzalutamide (37, 38).
The limitation of this study includes its retrospective nature and a wide range of time window for the sample collection. However, we observed a consistent trend when the analysis was restricted to samples collected within 6 months prior to therapy initiation. In addition, due to limited sample size and absent data issue, we could not include some important covariates, such as LDH, hemoglobin, AP, which have been shown to be of prognostic value for patients treated on AA. Lastly, we could not analyze treatment response based on PSA and/or radiographic criteria due to incomplete data issue. Also, as no data was available from untreated patients as the control group, it was not possible to investigate the interaction between treatment and these biomarkers to assess its potential predictive value, in addition to its prognostic role.
It is interesting to note that Reig et al. recently has reported that the detection of TMPRSS2-ERG fusion transcripts in the peripheral blood mononuclear cell fraction predicted a lower PSA progression free survival to docetaxel or cabazitaxel treatments (49). Since the TMPRSS2-ERG fusion is driven by AR and unique to PCa cells, we would consider the detection of TMPRSS2-ERG fusion transcript in our future validation studies. This additional control may help eliminate issues relating to signals derived from non-tumor cells. Currently, we are performing validation analyses in cohorts obtained from other institutions.
In conclusion, this study suggests that PSA and AR-V7 transcripts detected in the peripheral blood mononuclear cell fraction may potentially serve as biomarkers predicting TTF and OS with AA or enzalutamide treatment. The combination of PSA and AR-V7 signals may provide a more accurate assessment. If validated prospectively, the direct detection of AR-V7 and PSA transcripts in the peripheral blood mononuclear cell fraction could be a simple and powerful prognostic tool without the need for CTC isolation.
Supplementary Material
Statement of translational relevance:
Our study indicated that the quantity of prostate specific antigen (PSA) and AR-V7 transcripts detected in the blood is inversely associated with time to treatment failure (TTF) and overall survival (OS) in castration resistant prostate cancer patients who were treated with abiraterone acetate and/or enzalutamide treatment. This result suggests that PSA and AR-V7 transcripts in patients’ blood may potentially serve as biomarkers predicting TTF and OS for treatments targeting at androgen receptor axis in castration resistant prostate cancer patients. The combination of PSA and AR-V7 signals likely provides a more accurate assessment. If validated prospectively, the direct detection of AR-V7 and PSA transcripts in the blood could be a simple and powerful prognostic tool without the need for the isolation of circulating tumor cells.
Acknowledgements
The authors thank all the participants for their assistance with the study. We would like to thank Dr. Tara Ellison, Eric Miller and other experts from Bio‐Rad Laboratories for their great technical assistant on ddPCR platform. The study was supported by the Dana-Farber Prostate Cancer SPORE P50CA090381.
Footnotes
Conflicts of Interest: The authors disclose no potential conflicts of interest.
References
- 1.Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin. 1972;22:232–40. doi: 10.3322/canjclin.22.4.232. [DOI] [PubMed] [Google Scholar]
- 2.Messing EM, Manola J, Sarosdy M, Wilding G, Crawford ED, Trump D. Immediate hormonal therapy compared with observation after radical prostatectomy and pelvic lymphadenectomy in men with node-positive prostate cancer. N Engl J Med. 1999;341:1781–8. doi: 10.1056/NEJM199912093412401. [DOI] [PubMed] [Google Scholar]
- 3.Nelson WG, De Marzo AM, Isaacs WB. Prostate cancer. N Engl J Med. 2003;349:366–81. doi: 10.1056/NEJMra021562. [DOI] [PubMed] [Google Scholar]
- 4.Ross RW, Xie W, Regan MM, Pomerantz M, Nakabayashi M, Daskivich TJ, et al. Efficacy of androgen deprivation therapy (ADT) in patients with advanced prostate cancer: association between Gleason score, prostate-specific antigen level, and prior ADT exposure with duration of ADT effect. Cancer. 2008;112:1247–53. doi: 10.1002/cncr.23304. [DOI] [PubMed] [Google Scholar]
- 5.Klotz L, Toren P. Androgen deprivation therapy in advanced prostate cancer: is intermittent therapy the new standard of care? Curr Oncol. 2012;19:S13–21. doi: 10.3747/co.19.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:8253–61. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 7.Mostaghel EA, Nelson PS. Intracrine androgen metabolism in prostate cancer progression: mechanisms of castration resistance and therapeutic implications. Best Pract Res Clin Endocrinol Metab. 2008;22:243–58. doi: 10.1016/j.beem.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bluemn EG, Nelson PS. The androgen/androgen receptor axis in prostate cancer. Curr Opin Oncol. 2012;24:251–7. doi: 10.1097/CCO.0b013e32835105b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan X, Cai C, Chen S, Chen S, Yu Z, Balk SP. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene. 2014;33:2815–25. doi: 10.1038/onc.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan CJ, Tindall DJ. Androgen receptor rediscovered: the new biology and targeting the androgen receptor therapeutically. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:3651–8. doi: 10.1200/JCO.2011.35.2005. [DOI] [PubMed] [Google Scholar]
- 11.Longo DL. New therapies for castration-resistant prostate cancer. N Engl J Med. 2010;363:479–81. doi: 10.1056/NEJMe1006300. [DOI] [PubMed] [Google Scholar]
- 12.Attar RM, Takimoto CH, Gottardis MM. Castration-resistant prostate cancer: locking up the molecular escape routes. Clin Cancer Res. 2009;15:3251–5. doi: 10.1158/1078-0432.CCR-08-1171. [DOI] [PubMed] [Google Scholar]
- 13.Jarman M, Barrie SE, Llera JM. The 16,17-double bond is needed for irreversible inhibition of human cytochrome p45017alpha by abiraterone (17-(3-pyridyl)androsta-5, 16-dien-3beta-ol) and related steroidal inhibitors. J Med Chem. 1998;41:5375–81. doi: 10.1021/jm981017j. [DOI] [PubMed] [Google Scholar]
- 14.O'Donnell A, Judson I, Dowsett M, Raynaud F, Dearnaley D, Mason M, et al. Hormonal impact of the 17alpha-hydroxylase/C(17,20)-lyase inhibitor abiraterone acetate (CB7630) in patients with prostate cancer. Br J Cancer. 2004;90:2317–25. doi: 10.1038/sj.bjc.6601879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Bishop AC, Alyamani M, Garcia JA, Dreicer R, Bunch D, et al. Conversion of abiraterone to D4A drives anti-tumour activity in prostate cancer. Nature. 2015;523:347–51. doi: 10.1038/nature14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–90. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wadosky KM, Koochekpour S. Therapeutic Rationales, Progresses, Failures, and Future Directions for Advanced Prostate Cancer. International journal of biological sciences. 2016;12:409–26. doi: 10.7150/ijbs.14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Attard G, Reid AH, Yap TA, Raynaud F, Dowsett M, Settatree S, et al. Phase I clinical trial of a selective inhibitor of CYP17, abiraterone acetate, confirms that castration-resistant prostate cancer commonly remains hormone driven. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:4563–71. doi: 10.1200/JCO.2007.15.9749. [DOI] [PubMed] [Google Scholar]
- 19.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368:138–48. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taplin ME, Montgomery B, Logothetis CJ, Bubley GJ, Richie JP, Dalkin BL, et al. Intense androgen-deprivation therapy with abiraterone acetate plus leuprolide acetate in patients with localized high-risk prostate cancer: results of a randomized phase II neoadjuvant study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:3705–15. doi: 10.1200/JCO.2013.53.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravi P, Mateo J, Lorente D, Zafeiriou Z, Altavilla A, Ferraldeschi R, et al. External validation of a prognostic model predicting overall survival in metastatic castrate-resistant prostate cancer patients treated with abiraterone. European urology. 2014;66:8–11. doi: 10.1016/j.eururo.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 23.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–97. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 24.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–13. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–77. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J, Van der Steen T, Tindall DJ. Are androgen receptor variants a substitute for the full-length receptor? Nat Rev Urol. 2015;12:137–44. doi: 10.1038/nrurol.2015.13. [DOI] [PubMed] [Google Scholar]
- 27.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–62. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73:483–9. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. 2010;107:16759–65. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hornberg E, Ylitalo EB, Crnalic S, Antti H, Stattin P, Widmark A, et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One. 2011;6:e19059. doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fenner A. Prostate cancer: unravelling AR splice variant signalling in CPRC. Nat Rev Urol. 2012;9:410. doi: 10.1038/nrurol.2012.139. [DOI] [PubMed] [Google Scholar]
- 33.Ciccarese C, Santoni M, Brunelli M, Buti S, Modena A, Nabissi M, et al. AR-V7 and prostate cancer: The watershed for treatment selection? Cancer Treat Rev. 2016;43:27–35. doi: 10.1016/j.ctrv.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–30. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Z, Chen S, Sowalsky AG, Voznesensky OS, Mostaghel EA, Nelson PS, et al. Rapid induction of androgen receptor splice variants by androgen deprivation in prostate cancer. Clin Cancer Res. 2014;20:1590–600. doi: 10.1158/1078-0432.CCR-13-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–38. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Nakazawa M, et al. Androgen Receptor Splice Variant 7 and Efficacy of Taxane Chemotherapy in Patients With Metastatic Castration-Resistant Prostate Cancer. JAMA oncology. 2015;1:582–91. doi: 10.1001/jamaoncol.2015.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onstenk W, Sieuwerts AM, Kraan J, Van M, Nieuweboer AJ, Mathijssen RH, et al. Efficacy of Cabazitaxel in Castration-resistant Prostate Cancer Is Independent of the Presence of AR-V7 in Circulating Tumor Cells. European urology. 2015;68:939–45. doi: 10.1016/j.eururo.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Scher HI, Lu D, Schreiber NA, Louw J, Graf RP, Vargas HA, et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA oncology. 2016 Jun 4; doi: 10.1001/jamaoncol.2016.1828. dio: 10.1001/jamaoncol.2016.1828. [Epub ahead ofprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kantoff PW, Halabi S, Farmer DA, Hayes DF, Vogelzang NA, Small EJ. Prognostic significance of reverse transcriptase polymerase chain reaction for prostate-specific antigen in men with hormone-refractory prostate cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2001;19:3025–8. doi: 10.1200/JCO.2001.19.12.3025. [DOI] [PubMed] [Google Scholar]
- 41.Halabi S, Small EJ, Hayes DF, Vogelzang NJ, Kantoff PW. Prognostic significance of reverse transcriptase polymerase chain reaction for prostate-specific antigen in metastatic prostate cancer: a nested study within CALGB 9583. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2003;21:490–5. doi: 10.1200/JCO.2003.04.104. [DOI] [PubMed] [Google Scholar]
- 42.Ross RW, Manola J, Hennessy K, Galsky M, Scher H, Small E, et al. Prognostic significance of baseline reverse transcriptase-PCR for prostate-specific antigen in men with hormone-refractory prostate cancer treated with chemotherapy. Clin Cancer Res. 2005;11:5195–8. doi: 10.1158/1078-0432.CCR-05-0431. [DOI] [PubMed] [Google Scholar]
- 43.McDermott GP, Do D, Litterst CM, Maar D, Hindson CM, Steenblock ER, et al. Multiplexed target detection using DNA-binding dye chemistry in droplet digital PCR. Anal Chem. 2013;85:11619–27. doi: 10.1021/ac403061n. [DOI] [PubMed] [Google Scholar]
- 44.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–5. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oh WK, Hayes J, Evan C, Manola J, George DJ, Waldron H, et al. Development of an integrated prostate cancer research information system. Clin Genitourin Cancer. 2006;5:61–6. doi: 10.3816/CGC.2006.n.019. [DOI] [PubMed] [Google Scholar]
- 46.Nguyen HC, Xie W, Yang M, Hsieh CL, Drouin S, Lee GS, et al. Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. The Prostate. 2013;73:346–54. doi: 10.1002/pros.22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dodd DW, Gagnon KT, Corey DR. Digital quantitation of potential therapeutic target RNAs. Nucleic acid therapeutics. 2013;23:188–94. doi: 10.1089/nat.2013.0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeuchi T, Okuno Y, Hattori-Kato M, Zaitsu M, Mikami K. Detection of AR-V7 mRNA in whole blood may not predict the effectiveness of novel endocrine drugs for castration-resistant prostate cancer. Res Rep Urol. 2016;8:21–5. doi: 10.2147/RRU.S98877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reig O, Marin-Aguilera M, Carrera G, Jimenez N, Pare L, Garcia-Recio S, et al. TMPRSS2-ERG in Blood and Docetaxel Resistance in Metastatic Castration-resistant Prostate Cancer. European urology. 2016 Feb 29; doi: 10.1016/j.eururo.2016.02.034. pii: S0302-2838(16)00212-8.dio: 10.1016/j.eururo.2016.02.034.[Epub ahead of print] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.