Abstract
With the ability to induce rapid and efficient repair of disease-causing mutations, CRISPR/Cas9 technology is ideally suited for gene therapy approaches for recessively and dominantly inherited monogenic disorders. In this study, we have corrected a causal hotspot mutation in exon 6 of the keratin 14 gene (KRT14) that results in generalized severe epidermolysis bullosa simplex (EBS-gen sev), using a double-nicking strategy targeting intron 7, followed by homology-directed repair (HDR). Co-delivery into EBS keratinocytes of a Cas9 D10A nickase (Cas9n), a predicted single guide RNA pair specific for intron 7, and a minicircle donor vector harboring the homology donor template resulted in a recombination efficiency of >30% and correction of the mutant KRT14 allele. Phenotypic correction of EBS-gen sev keratinocytes was demonstrated by immunofluorescence analysis, revealing the absence of disease-associated K14 aggregates within the cytoplasm. We achieved a promising safety profile for the CRISPR/Cas9 double-nicking approach, with no detectable off-target activity for a set of predicted off-target genes as confirmed by next generation sequencing. In conclusion, we demonstrate a highly efficient and specific gene-editing approach for KRT14, offering a causal treatment option for EBS.
Keywords: epidermolysis bullosa simplex, keratin 14, CRISPR/Cas9, homology-directed repair double nicking
Kocher et al. established a CRISPR/Cas9-based gene-editing strategy with the aim to correct a recurrent dominantly inherited KRT14 hotspot mutation causal for the phenotypic manifestation in epidermolysis bullosa simplex (EBS). The homology-directed repair (HDR) was induced via Cas9 nickases in a double-nicking configuration showing an improved safety profile compared with the commonly used wild-type Cas9 nuclease. Treatment of patient keratinocytes and correction of the mutation at genomic level resulted in a full reversion of the disease phenotype.
Introduction
The CRISPR/Cas9 genome-editing technology is capable of modifying genes in a precise and efficient manner, and has become a widely used tool in medical research for the correction of disease-associated mutations. The rescue of a disease phenotype by specific correction of a mutation is executed via a cut-and-paste strategy. Modified from a naturally occurring bacterial defense mechanism, the Cas9 nuclease induces DNA double-strand breaks (DSBs), which activate the cell’s endogenous repair machinery, thus offering the possibility for genetic modifications. The specificity of the CRISPR/Cas9 technology is determined by a single guide RNA (sgRNA), which is designed to be complementary to the targeted genomic region and linked to the Cas9 nuclease, ensuring specifically targeted DSB induction (Figure 1A). Subsequently, cells repair such DSBs either by non-homologous end joining (NHEJ), wherein the DNA ends are randomly ligated, thereby introducing small insertions or deletions, or by homology-directed repair (HDR), where endogenously or externally supplied homologous donor templates are incorporated into the DNA at the site of the DSB.1 Exploiting this technology, genomic sequences can be extensively modified, allowing the introduction or knockout of genes, the creation of distinct deletions or insertions, or the specific correction of any disease-associated mutation.2, 3, 4
Figure 1.
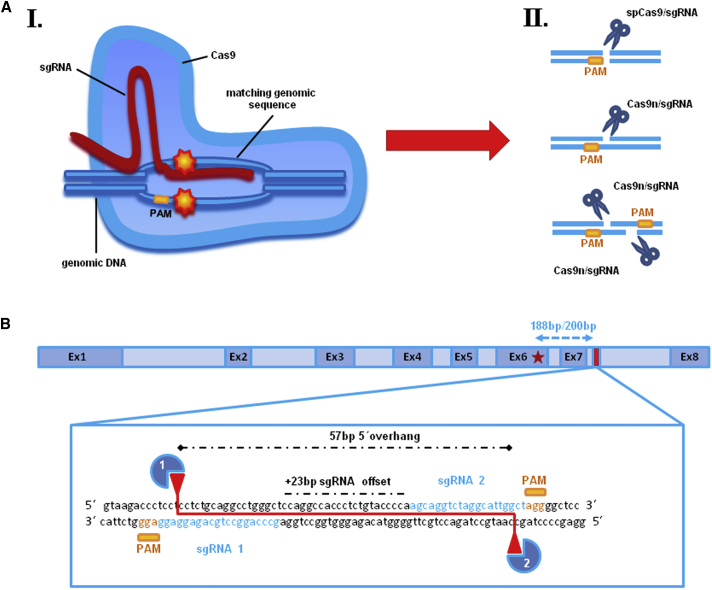
CRISPR/Cas9 Technology: Design of sgRNA Pair for Double-Nicking within KRT14 Intron 7
(A) Schematic depiction of the CRISPR/Cas9 mechanism. (AI) The specificity of the CRISPR/Cas9 technology is determined by a single guide RNA (sgRNA), which is designed to be complementary to the targeted genomic region and linked to the Cas9 nuclease, ensuring specific and directed genomic DSB induction. Such DSBs are repaired by cellular mechanisms such as non-homologous end joining (NHEJ) or homology-directed repair (HDR), where endogenously or externally supplied homologous donor sequences are specifically incorporated into the DNA at the site of the DSB. (II) Cas9 nuclease induces DNA double-strand breaks (DSBs), which activate the cell’s endogenous repair machinery. A modified Cas9 nuclease (Cas9n), in which the nuclease domain RuvC is inactivated,5 causes a single-strand nick. The combination of two single guide RNAs with the Cas9n causes one nick on each strand of the target sequence, thus inducing a DSB at the desired gene locus. (B) A pair of computationally predicted sgRNAs, targeting opposite strands of KRT14 intron 7, 188 and 200 nt downstream of the missense mutation (c.1231G>A) within exon 6 (red star), with an sgRNA offset of +23 bp, mediates DSBs with 57-bp-long 5′ overhangs.
Nevertheless, the likelihood of off-target effects, as seen when using the wild-type CRISPR/Cas9 system derived from streptococcus pyogenes (spCas9), remains an important issue to be addressed. One way to improve the targeting specificity, and to further shift the preferred repair mechanism from the unspecific NHEJ to the more specific HDR, is to utilize two distinct sgRNAs in combination with a modified Cas9 nuclease (Cas9 D10A nickase [Cas9n]), which, by virtue of its mutated RuvC nuclease domain, instead causes paired nicks, one on each strand of the target sequence, which is tantamount to inducing a DSB at the desired gene locus (Figure 1A). Recently, Ran et al.5 successfully performed double-nicking-mediated gene targeting with an efficient offset distance of up to 100 bp between the protospacer adjacent motif (PAM)-distal (5′) ends of both sgRNA sequences, which revealed a significant decrease of off-target activity compared with the spCas9, because of the fact that potential off-target single nicks are more likely to be repaired without causing deletions or insertions.5, 6, 7 The chance of unspecific binding of both sgRNAs in close proximity to each other at any location other than the desired locus, and thereby inducing a DSB, is unlikely, which can be confirmed via next generation sequencing (NGS) of potential off-target regions. It is an essential pre-clinical requirement for any specific CRISPR/Cas9 molecule to ensure high on-target activity in combination with negligible off-target activity.
The CRISPR/Cas9 technology has enormous potential in the development of gene therapies for monogenic disorders. Epidermolysis bullosa simplex (EBS) is a rare genetic condition that is caused primarily by mutations in either the keratin 14 (KRT14) or keratin 5 (KRT5) gene. The intermediate filament proteins encoded by these genes are expressed in keratinocytes within the basal epidermal layer, and deleterious mutations in either impair cellular stability, resulting in cytolysis even upon minor external friction, thereby leading to blister formation. EBS is the most frequently occurring subtype of EB, with an incidence of approximately 1 case per 25,000 live births.8, 9, 10 In the generalized-severe subtype of EBS (EBS-gen sev), the vast majority of mutations are either missense or small in-frame insertions or deletions (indels),11 acting in an autosomal dominant manner. These mutations mainly affect the helix initiation motif or the helix termination motif of the keratin rod domain, which play crucial roles in the formation of the heterodimers resulting in intermediate filament instability and the characteristic presence of cytoplasmic aggregates containing mutant keratin.10 Considering the dominant negative effect of mutant keratins on the intermediate filament cytoskeleton, therapies for the dominantly inherited EBS-gen sev subtype are limited and cannot be addressed by simple gene-replacement strategies. Rather, causal therapies must not only supply a wild-type gene, but also downregulate the disease-causing allele.
In the present study, we describe a CRISPR/Cas9n-mediated double-nicking strategy for the specific repair of a dominant mutation within exon 6 of the KRT14 gene in a patient keratinocyte line. Double-nicking-mediated DNA cleavage was recently described to be a versatile gene-editing strategy with an improved safety profile in comparison with the commonly used introduction of DSBs by wild-type spCas9 nuclease associated with considerable off-target activity.5, 12, 13 While the usage of the nickase Cas9n is known to facilitate HDR at the target region,14 the combined application of two nickases in a double-nicking configuration further improves its efficiency, which is, in single-nicking approaches, substantially reduced in comparison with the wild-type spCas9.5 We targeted KRT14 intron 7 by simultaneous single-nicking on both DNA strands (double-nicking) and offering a homologous wild-type donor template at the same time. Correction of the mutation via HDR was confirmed, resulting in a full rescue of the EBS phenotype in vitro, as shown by a normalization of the overall KRT14 expression and the absence of K14 aggregates upon heat shock.15
Results
Selection of a Specific Guide RNA Pair for DSB Induction
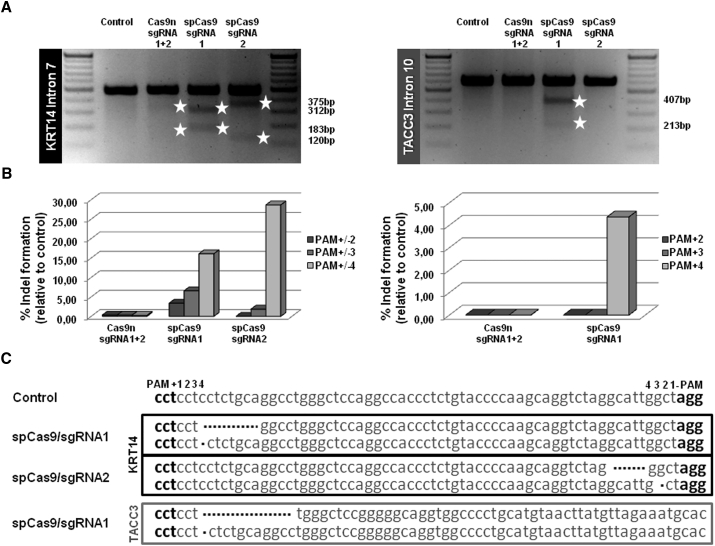
To establish a strategy for correction of a dominantly inherited hotspot missense mutation (c.1231G>A) within exon 6 of the KRT14 gene16, 17 using CRISPR/Cas9 and HDR, we initially evaluated both the wild-type Cas9 nuclease from streptococcus pyogenes (spCas9) and its modified variant D10A (Cas9n) for DNA cleavage efficiency at the selected KRT14 target site. Ran et al.5 successfully performed double-nicking-mediated gene targeting with an efficient offset distance between the PAM-distal (5′) ends of both sgRNA sequences of up to 100 bp. To reduce the risk of nuclease-mediated mutagenesis of coding regions, we decided to screen for sgRNAs targeting an intronic region next to the mutation. Therefore, we screened intron 6 for suitable sgRNAs, which could further be used for our double-nicking approach. Unfortunately, we could not identify a suitable sgRNA pair for intron 6. However, we were able to predict a pair of suitable sgRNAs that bind with an offset of 23 bp to a region within KRT14 intron 7 located 188–200 nt downstream of the causal mutation.18 sgRNA1 and sgRNA2 was the closest sgRNA pair downstream of the mutation that fulfilled the current criteria for efficient double-nicking.5 Co-delivery of sgRNA1 and sgRNA2 in combination with Cas9n into the target cells is predicted to induce DSBs with 57-bp-long 5′ overhangs (Figure 1B). We analyzed the functionality of our selected sgRNAs by delivering the respective spCas9/sgRNA expression plasmids into HEK293 cells and performing a T7 endonuclease I (T7EI) assay on the PCR-amplified KRT14 target region.19, 20 T7EI recognizes and cleaves DNA at sites of base pair mismatches, such as those formed upon NHEJ repair of DSBs. As such, the T7EI assay serves as an indirect test for DSB and NHEJ induction. Depending on the nuclease cutting site within KRT14 intron 7, the PCR-amplified target region was fragmented into two pieces of 312 and 183 bp (spCas9/sgRNA1) and 375 and 120 bp (spCas9/sgRNA2), respectively, confirming the ability of both sgRNAs to direct wild-type spCas9 to the target site for induction of DSBs. In contrast, when co-delivering sgRNA1 and sgRNA2 in combination with the modified Cas9n into HEK293 cells (double-nicking approach), we observed minimal levels of T7EI-mediated cleavage (Figure 2A, left panel), suggesting a general low efficiency of DSB induction via this approach. We confirmed these results by NGS of the PCR-amplified KRT14 region, which revealed a general NHEJ induction efficiency of up to 16% for the spCas9/sgRNA1 and 30% for the spCas9/sgRNA2 at locations immediately adjacent to the PAM sequence (+2 to +4 nt) (Figure 2B, left panel), which was reduced to undetectable levels in the double-nicking approach.
Figure 2.
Specificity Test of Selected sgRNAs Using T7EI Assay and NGS Analysis
(A) Upon delivery of the respective spCas9/sgRNA (sgRNA1 and sgRNA2) expression plasmids into HEK293 cells, the target region was PCR-amplified using a primer pair binding specifically to KRT14 exon 6 and intron 7. The resulting PCR products were cleaved by T7EI treatment into two fragments, 312 and 183 bp for sgRNA1 and 375 and 120 bp for sgRNA2, thus confirming the on-target functionality of both sgRNAs. In the double-nicking approach, both sgRNAs were transfected in combination with Cas9n, resulting in the desired T7EI digestion pattern detectable via agarose gel analysis. The resulting digest pattern is a mixed pattern derived from both sgRNAs (fragments between 375 and 312 bp, and fragments between 183 and 120 bp). The predicted off-target region for sgRNA1 was PCR-amplified using a primer pair binding specifically within TACC3 intron 10. The resulting PCR products were cleaved by T7EI treatment into two fragments of 407 and 213 bp confirming the predicted off-target activity of sgRNA1 when combined with the wild-type spCas9. (B) Relative cutting efficiencies of different Cas9 enzymes were determined by next generation sequencing. Using a custom-designed panel for on/off-target analysis of the KRT14 target region and predicted off-target regions of the selected sgRNAs, the relative amount of indel formation at intron 7 of KRT14 (left panel) and intron 10 of TACC3 (right panel) was analyzed. The graphs show the relative amount of indels compared with untreated control. (C) Sequences of NGS analysis showing mainly small deletions of 1–20 bp for each spCas9/sgRNA at the KRT14 and TACC3 locus.
Specificity Test Revealed No Off-Target Effects for the Double-Nicking Approach
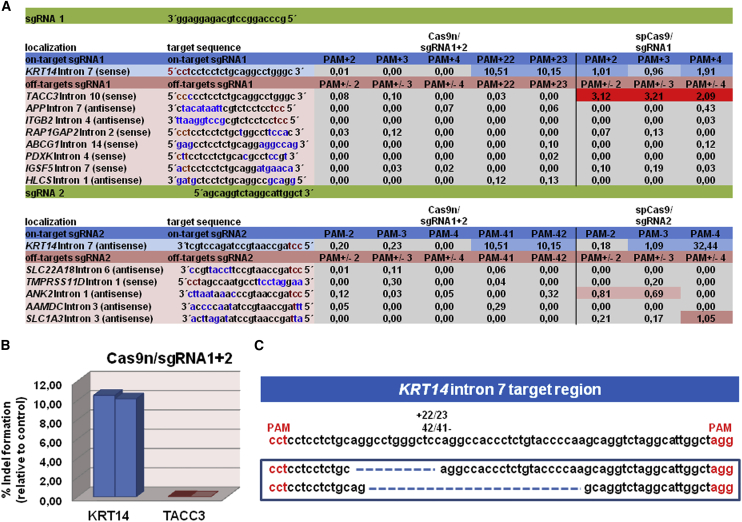
The double-nicking strategy utilizing two separate sgRNAs in combination with Cas9n has recently been described to result in reduced off-target activity in comparison with sgRNA/wild-type spCas9 approaches.5 To confirm this, we analyzed the DNA cleavage activity of these respective nucleases at several potential off-target regions, identified by BLAST analysis, for sgRNA1 and sgRNA2 (Table S1; Figure S1). Of note, sgRNA1 is highly homologous to a region within intron 10 of TACC3, differing by only 1 nt within the predicted PAM sequence, making this locus a potential target for this sgRNA. Indeed, DNA DSB induction by spCas9/sgRNA1 at this site was confirmed by T7EI assay showing digestion of the amplified TACC3 region into two fragments of 407 and 213 bp upon T7EI treatment (Figure 2A, right panel). sgRNA2, which shares no homology to TACC3, was unable to direct spCas9 to induce DNA DSBs within this locus as demonstrated by the T7EI assay, and cells treated with the double-nicking constructs were also negative in this assay (Figure 2A, right panel). NGS analysis further verified the generation of deletions within intron 7 of KRT14 and intron 10 of TACC3 caused by spCas9/sgRNA1, which differed neither in position nor in deletion size (Figure 2C). However, indels were undetectable for Cas9n/sgRNA1 and sgRNA2 at the TACC3 locus, underscoring the specificity and safety advantage of the double-nicking approach, which requires both sgRNAs to bind in close proximity to each other to generate a DSB. For all other predicted off-target genomic loci, we observed no or, if any, <3% (as for RAP1GAP2 intron 2) indel formation by either spCas9/sgRNA (Table S1). All values (% indels of total reads) were at or near the baseline calculated from NGS data obtained from untreated HEK293 cells.
Thus, our results demonstrate that the double-nicking Cas9n approach is less efficient in inducing DSBs than the wild-type spCas9/sgRNA approaches. However, because of the reduced off-target activity of the double-nicking approach, which is an important prerequisite for any future clinical application, we decided to proceed with the Cas9n/sgRNA1 and sgRNA2 configuration for subsequent gene correction studies in EBS keratinocytes.
HDR-Mediated Gene Repair Using an MC System
To achieve HDR of the causal heterozygous mutation (c.1231G>A) within exon 6 of KRT14 in a patient-derived EBS human keratinocyte line (EBS hKc), we first generated a minicircle (MC) vector that carries the homologous donor template extending from intron 3 to the 3′ UTR of KRT14. The MC vector lacks any bacterial backbone, resulting in an overall vector size reduction from ∼8,000 (parental vector) to ∼3,500 bp (MC) (Figure S2A). This results in increased transfection efficiency into cells when compared with standard plasmids (Figure S2B), which is expected to be associated with an enhanced HDR efficiency because of an increased availability of HDR donor templates. The MC included engineered restriction sites for EcoRI and NheI within intron 7, which were included to facilitate the quantification of recombination events. Additionally, a GFP-IRES-blasticidin selection cassette under the control of the EF1 promoter was incorporated downstream of the KRT14 3′ UTR for initial selection of transfected cells (Figure 3). The MC was co-transfected into EBS hKc cells together with spCas9/sgRNA1, spCas9/sgRNA2, or Cas9n/sgRNA1 and sgRNA2. To more accurately determine HDR efficiency achieved with our double-nicking approach compared with the spCas9 approaches and to exclude differences due to different transfection efficiencies, we pre-selected our treated cell population with blasticidin, to maintain a homogeneous cell population carrying the transfected vectors. We achieved a transfection efficiency of ∼30% into EBS hKc cells as estimated by the analysis of GFP expression via flow cytometry. Following blasticidin selection, ∼90% of EBS hKc were GFP-positive (Figure S2C). For analysis of HDR-mediated integration of donor template sequences, PCR amplification of genomic DNA from treated patient cells was performed using a forward primer that specifically binds the integrated restriction sites EcoRI and NheI and a reverse primer binding downstream of the KRT14 3′ UTR, in a region not present in the MC vector. Thus, amplification of the MC donor template is excluded by this strategy. We observed a PCR product of the expected size of 893 bp under all conditions tested, revealing the accurate, HDR-mediated integration of these restriction sites within intron 7 of KRT14 (Figure 4A). In order to more accurately assess the HDR efficiency achieved under the different gene-editing conditions tested, we performed a separate PCR using primers specifically amplifying the KRT14 region spanning exon 6 to 60 nt downstream of the 3′ UTR, which again excludes amplification of the MC vector and results in a product of 1,189 bp. Digestion of this PCR product (1,189 bp) with EcoRI generated the expected cleavage products of 874 and 315 bp that were readily detectable only in the double-nicking approach, indicating a higher HDR efficiency compared with the spCas9/sgRNA approaches (Figure 4B).
Figure 3.
HDR of KRT14 Using an MC-Based System
Co-transfection into EBS hKc of the respective Cas9/sgRNA combination (spCas9-sgRNA1, spCas9-sgRNA2, or both Cas9n-sgRNA1 and Cas9n-sgRNA2) and the MC donor plasmid, carrying the homology donor template spanning from intron 3 to the 3′ UTR of KRT14 and a GFP-IRES-blasticidin under the control of an EF1 promoter, led to DSB induction within intron 7 of KRT14 and subsequently to the HDR-mediated introduction of the silent mutations (gray stars) and two restriction sites provided by the MC donor vector, and the repair of the dominant disease-causing mutation (black star).
Figure 4.
Integration and HDR Analysis of CRISPR/Cas9-Edited EBS hKc
After co-transfection of the respective Cas9/sgRNA combinations (spCas9-sgRNA1, spCas9-sgRNA2, or both Cas9n-sgRNA1 and Cas9n-sgRNA2) and the MC donor plasmid into EBS hKc, we performed two rounds of blasticidin selection. The resulting blasticidin-resistant cells were then analyzed for site-specific integration of the donor template at the genomic level. (A) Integration-specific PCR using a forward primer binding the restriction sites EcoRI and NheI (white box) provided by the MC and a reverse primer binding downstream to the 3′ UTR of the KRT14 gene resulted in a PCR product of 893 bp. (B) EcoRI digestion to estimate the relative recombination efficiency in the treated cell pool. We performed a PCR using a forward primer binding to exon 6 and a reverse primer binding a sequence downstream of KRT14 3′ UTR. The PCR resulted in a specific product of 1,189 bp. EcoRI digestion generated the expected fragments of 874 and 315 bp, only visible in the Cas9n 1- and 2-treated EBS hKc. (C) Schematic depiction of the primer binding sites used for amplification of the integration-specific PCR fragments. Gray arrows indicate a forward primer binding to the restriction sites EcoRI and NheI integrated via HDR. Black arrows indicate forward and reverse primers binding to endogenous KRT14. Black star: missense mutation (c.1231G>A) in exon 6; gray stars: silent mutations.
HDR Efficiency upon Double-Nicking-Induced DNA Cleavage
To evaluate the relative HDR efficiency in blasticidin-selected EBS hKc treated with the double-nicking constructs, the PCR-amplified KRT14 target region was subcloned into an appropriate subcloning DNA vector for transformation into competent bacterial cells and analysis of resulting colonies. To increase the subcloning efficiency, we first performed a nested PCR on the initial integration-specific product (1,189 bp) spanning exon 6 to 60 nt downstream of the 3′ UTR, which excludes amplification of the MC vector, reducing the size of PCR product to be subcloned to 507 bp (Figure 5A). Colony PCR of single colonies and EcoRI restriction digestion of the amplified DNA resulted in the correct digestion pattern in 11 of 32 colonies, demonstrating an HDR efficiency of approximately 34% (Figure 5A, right panel).
Figure 5.
Analysis of HDR Efficiency
(A) PCR analysis of genomic DNA from CRISPR/Cas9-treated EBS hKc using a forward primer binding to exon 6 and a reverse primer binding a sequence downstream of KRT14. To minimize the size of the PCR product and to facilitate subcloning, we performed a nested PCR using a forward primer binding to exon 6 and a reverse primer binding to intron 7, resulting in a specific product of 507 bp. An EcoRI restriction digest confirmed the presence of recombined KRT14 alleles in treated EBS cells. Upon bacterial transformation, single colonies were picked for colony PCR and amplified product digested with EcoRI to identify recombined alleles. (B) Sequence analysis of one representative single clone, containing a modified wild-type allele (green arrow: wild-type sequence at mutation site [position 178] within exon 6), is shown. The allele additionally contains the introduced restriction sites NheI/EcoRI and the silent mutations (blue stars). Sequence alignment was performed with the software “Multiple sequence alignment with hierarchical clustering.”21
For a more detailed analysis of the editing processes induced by the double-nicking approach, we picked a total of 96 bacterial colonies and performed sequence analysis on the resulting colony PCR products. This revealed an HDR efficiency of up to 32% (Figure 6B), in keeping with results from the restriction digestion assay above. Silent mutations introduced into both homologous KRT14 arms allowed us to assess sites preferentially used for recombination, which seem to be located within the 5′ region of the KRT14 donor template (Figure 6C). Sequencing analysis further revealed NHEJ indel formation of 47%, which can be subdivided into 38% deletions and 9% insertions. Finally, we observed a ratio of wild-type /mutation allele of 63/33. Assuming a wild-type/mutation allele ratio of 50/50 in untreated patient keratinocytes, we calculated a mutation correction efficiency of approximately 16% with the double-nicking approach (Figure 6A).
Figure 6.
HDR Efficiency of the MC Double-Nicking Approach
96 subcloned nested PCR fragments were sequenced and analyzed. (A) Ratio between wild-type and mutated alleles. (B) Ratio between NHEJ (insertions and deletions) and HDR. n = absolute numbers of analyzed sequences. (C) HDR analysis. The position of sequence modifications (P) was defined according to the KRT14 target sequence exon 6-intron 6-exon 7-intron 7 (exon 6: P1-P221; intron 6: P222-P315; exon 7: P316-P362; intron 7: P363-P923). wRS, with restriction sites (exon 6: P1-P221; intron 6: P222-P315; exon 7: P316-P362; intron 7: P363-P935).
Off-Target Site Analysis in EBS Keratinocytes
To reconfirm the specificity of our double-nicking approach, we performed NGS on genomic DNA isolated from treated and blasticidin-selected EBS keratinocyte bulk populations. NGS of the PCR-amplified on-target KRT14 region revealed a general NHEJ induction efficiency of up to 10% for our double-nicking approach, 2% for the spCas9/sgRNA1 and 32% for spCas9/sgRNA2, respectively (Figure 7A). The reduced number of NHEJ events detectable in the respective Cas9/gRNA-treated cell populations is due to the presence of the MC vector at the time point of sequence analysis, influencing the evaluation of the results (Figure 7). The NHEJ rates are therefore underestimated compared with NHEJ rates we obtained by the subcloning strategy (Figure 6) wherein amplification of the MC was excluded by the PCR strategy. To confirm the reduced off-target activity of the Cas9n in comparison with the wild-type spCas9 nuclease, we analyzed the DNA cleavage activity of the respective nucleases at the potential off-target regions for sgRNA1 and sgRNA2 (Figure 7A) as already described above. NGS analysis revealed deletions within intron 7 of KRT14 and intron 10 of TACC3 in cells treated with spCas9/sgRNA1, as expected. Additionally, NGS analysis of two predicted off-targets for sgRNA2 indicated possible DNA cleavage activity. However, in cells treated with the double-nicking constructs, only indels within intron 7 of KRT14, but not within any predicted off-target region, including intron 10 of TACC3, were detectable, again emphasizing the improved safety profile of our double-nicking approach (Figure 7B). Treatment of cells with the double-nicking constructs resulted in deletions located between the binding sites of both sgRNAs, but further away from the PAM sequences, and they were larger in size compared with deletions resulting from spCas9/sgRNAs (Figure 7C).
Figure 7.
Specificity Test of Selected sgRNAs in an MC-Treated EBS hKc Cell Line Using NGS
(A) DNA cleavage activities of sgRNAs at on-target and off-target regions are represented as % indels relative to untreated control. The cutoff for cleavage activity was set to 0.5%. Colored boxes indicate cleavage activity of the respective Cas9/sgRNAs at on-target and off-target regions. On-target and off-target region sequences are listed, with red letters indicating the PAM position, blue letters indicating mismatches between the sgRNA on-target binding sites and the potential off-target regions, and black letters indicating matches between on-target and off-target regions. (B) Graphs show the relative amount of indels (PAM +22/23) within KRT14 intron 7 and TACC3 intron 10, caused by double-nicking compared with untreated control. (C) Sequences of NGS analysis showing frequent deletions for the Cas9n/sgRNA1 and Cas9n/sgRNA2 configuration.
Corrected EBS Single-Cell Clones Exhibit Reversion of Disease Phenotype
Our results thus far demonstrate an HDR efficiency of up to 32% and a mutation correction efficiency of approximately 16% in pre-selected EBS keratinocytes by our double-nicking approach. In order to demonstrate that correction of the causal mutation results in the reversion of the EBS disease phenotype, we first isolated single cells from the blasticidin-selected EBS bulk population. From 384 single cells initially isolated, we were able to maintain and expand a total of 17 clonal populations. 12 clones were screened via western blot analysis for keratin 14 (K14) expression, resulting in the identification of 5 clones showing restoration of normal K14 protein levels, 7 of which exhibited increased K14 levels characteristic of the parental EBS hKc cell line (data not shown). We chose two separate clones, clones 16 and 17, with opposite K14 profiles, for further analysis. PCR amplification of the KRT14 target region from isolated genomic DNA and synthesized cDNA of the selected cell clones revealed correction of the mutation at genomic level and RNA level for single-cell clone 17, whereas there was no correction for single-cell clone 16 (Figures S3A and S3B). Both cell clones showed signs of CRISPR/Cas9 HDR-mediated gene editing. This was evident in the introduction of silent mutations engineered into the MC donor template. We also amplified the off-target region within intron 10 of TACC3 and could again demonstrate the absence of indel formation within this predicted off-target site (Figure S3C).
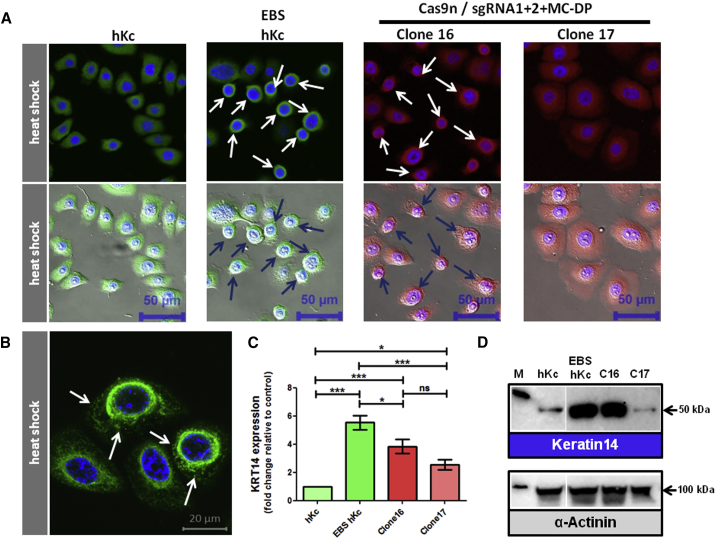
EBS-gen sev keratinocytes accumulate K14 because of a positive autocrine feedback loop involving interleukin-1β (IL-1β) and the stress-related JNK pathway.15, 22 The incorporation of mutant K14 proteins into the intermediate filament network renders the network unstable and sensitive to minor stress, such as heat shock, resulting in its collapse into aggregates, which accumulate in the cytoplasm. Thus, correction of the causal mutation should restore intermediate filament stability and normalize K14 expression within the cell. In line with this, heat shock assays demonstrate the collapse of the intermediate filament network into aggregates in parental EBS hKc cells and in clone 16, which is completely absent in clone 17 (Figures 8A and 8B). Furthermore, whereas parental EBS hKc cells and the uncorrected clone 16 showed increased KRT14 expression compared with wild-type hKcs, the corrected clone 17 showed a normalized KRT14 expression pattern (Figures 8C and 8D). Thus, correction of the disease-causing mutation, as achieved here by HDR and CRISPR/Cas9 technology, results in the functional reversion of the EBS-gen sev-associated phenotype.
Figure 8.
CRISPR/Cas9-Mediated Reversion of Disease Phenotype in EBS Single-Cell Clones
(A) Immunofluorescence staining using antibodies directed against K14 upon 30 min of heat stress (43°C) followed by 15 min recovery time at 37°C in CRISPR/Cas9-treated EBS keratinocytes. Human keratinocytes and EBS keratinocytes served as controls. K14 staining showed a full reversion of the disease-associated K14 aggregation in Cas9n-sgRNA1 and Cas9n-sgRNA2/MC-DP-treated single-cell clone 17. EBS hKc and the uncorrected Cas9n-sgRNA1 and Cas9n-sgRNA2/MC-DP-treated single-cell clone 16 showed the characteristic K14 aggregates around the nucleus and the periphery of the cytoplasm (arrows), which are not visible in human wild-type keratinocytes (hKc) and Cas9n/sgRNA1 and 2/MC-DP repaired EBS patient keratinocytes (scale bar, 50 μm). (B) Typical cytokeratin 14 aggregate formation (white arrows) in EBS cells upon heat stress (scale bar, 20 μm). (C) mRNA expression levels of KRT14 in wild-type hKc, CRISPR/Cas9-treated, and nontreated EBS patient keratinocytes via semi-quantitative real-time PCR. Expression levels were normalized to GAPDH. Relative expression levels were shown as fold change. The gene expression levels in human wild-type keratinocytes (hKc) were set to 1. Nine different runs with duplicates were analyzed using three different RNA samples of each cell line. Statistics were performed using the one-way ANOVA-Tukey’s multiple comparison test. (D) Western blot analysis on whole cell extracts revealed a normalization of K14 for clone 17 at protein level compared with untreated EBS hKc and clone 16, which showed no correction of the causal mutation. α-Actinin was included as loading control.
Discussion
The generalized severe simplex subtype of EBS has an inflammatory phenotype caused by a single heterozygous mutation within either of the basal keratins that results in functionally impaired protein that interferes with the correct assembly of the intermediate filament network in basal keratinocytes.15, 22 As highlighted by the few clinical trials and case reports that have been published so far,23, 24, 25, 26, 27, 28 there are no established targeted/molecular treatment options for EBS, and current standard care for EBS-gen sev patients still focuses on wound care and symptomatic treatments. In general, therapeutic strategies for genetic diseases target either the genetic cause underlying a specific disorder or disease-specific pathophysiological pathways. For EBS, both causal and symptomatic treatment strategies have been investigated during the last decade. In order to identify potential targets for novel treatments, Wally et al.15, 29 investigated the EBS-gen sev pathomechanism, identifying the pro-inflammatory interleukin-1β (IL-1β)-mediated stress pathway to be constitutively activated. Disrupting IL-1β signaling with antibodies or the small molecule diacerein resulted in a reversion of the IL-1β-related gene expression signature toward the wild-type signature. Translation of this approach into a clinical setting resulted in a significant amelioration of the disease phenotype, i.e., a reduction of blister numbers, in EBS-gen sev patients.15, 29 While diacerein represents a specifically targeted, symptomatic, and potentially short-term accessible treatment option that has already taken steps forward into clinics, causal treatment approaches, such as spliceosome-mediated pre-mRNA trans-splicing (SMaRT), are still under development.15 Causal treatment of monogenic diseases requires sustained replacement or correction of the affected gene or mRNA. In addition, considering the dominant negative effect of mutant keratins on the intermediate filament network, therapies for EBS-gen sev are limited, and conventional gene replacement strategies are not applicable. Even though progress has been made in the field of ex vivo gene therapy for other subtypes of EB (junctional and recessive dystrophic EB), for the dominantly inherited EBS-gen sev, this therapeutic approach is currently not available30, 31, 32 because dominant negative interference cannot be overcome simply by providing a wild-type cDNA. Rather a significant reduction of mutated K14 determines the success of the treatment as it is the incorporation of this faulty molecule into the intermediate filament network that, upon stress, subsequently triggers filament aggregation.22, 33, 34, 35 Accordingly, success has also been achieved using short inhibitory RNAs (siRNAs) and therapeutic trans-splicing, both of which result in reduction in levels of the faulty transcript.22, 35 Permanent deletion or repair of the causal mutation at genomic level, using current state-of-the-art gene-editing tools such as TALEN or CRISPR/Cas9, would circumvent the need for repeated administration of transiently acting drugs. In this respect, Petek et al.36 have already demonstrated that targeted alteration of a disease-causing KRT14 allele using a gene-targeting vector with promoter trap design results in restoration of the intermediate filament cytoskeleton of EBS keratinocytes in a xenograft mouse model.
The potential to repair any mutation that causes a severe disease phenotype renders the CRISPR/Cas9 system an attractive and broadly applicable gene-editing tool. CRISPR/Cas9 constructs can be easily generated and adapted for almost any genomic region of interest. The main difficulty concern regarding a future clinical application is to predict off-target activity of the nuclease. Until now, low HDR efficiencies accompanied by high risks of off-target effects have impeded its way into the clinic as a curative treatment for monogenetic disorders.12 Indeed, Fu et al.13 showed frequent off-target mutagenesis induced by the commonly used spCas9 in human cells, exhibiting the need for both extensive sgRNA pre-selection and very sensitive off-target analysis tools. Therefore, recent studies have focused on the improvement of HDR efficiency and the safety profile of the commonly used spCas9 nucleases. Ran et al.5 described a method to increase HDR rates and to reduce the frequency of off-target events by using the Cas9 mutant D10A (Cas9n), which preferably induces single-strand breaks (nicks) within the DNA, in a double-nicking configuration. Targeting opposite DNA strands separately to induce what is tantamount to DSBs within the desired region reduces the chance of unwanted indel formation elsewhere in the genome, a common side event observed with the wild-type spCas9.5, 12 In this study, we demonstrate the feasibility of this approach in the HDR-mediated correction of a dominant hotspot mutation within the KRT14 gene, which is associated with the EBS-gen sev phenotype. The site chosen for DSB induction was determined by the presence of PAM sequences on both DNA strands spaced 23 bp apart and is located ∼200 bp downstream of the mutation within intron 7 of KRT14. Even though the relatively large distance to the mutation is expected to reduce the general repair efficiency, we chose the double-nicking strategy because of safety concerns. Besides the auspicious safety profile of the Cas9n nickase, targeting an intron reduces the risk for nuclease-mediated mutagenesis of coding sequences.
Using this approach, we achieved HDR efficiencies of up to 32% in pre-selected EBS patient keratinocytes accompanied by an overall mutation correction rate of approximately 16%. Furthermore, corrected single-cell clones showed a restored K14 expression pattern. A heat shock assay confirmed successful correction at the functional level, as demonstrated by the absence of the characteristic EBS-gen sev K14 aggregates, indicating restoration of the stability of the intermediate filament network. The double-nicking approach is not only an example for what can be accomplished with CRISPR/Cas9, but also shows proof-of-principle for the repair of dominant mutations.
A major aspect of our study is the development of a safe repair strategy. Additional to commonly used T7EI assays and surveyor assays, we performed an extended off-target analysis via NGS. The off-target analysis via T7EI assay and NGS indicates adverse effects caused by the wild-type spCas9/sgRNA1 and wild-type spCas9/sgRNA2, respectively, at predicted target regions highly homologous to the respective sgRNA binding sequence. However, this was not detectable in the double-nicking approach, rendering this the safest and most feasible of all strategies tested here for the specific gene editing of KRT14. Although the frequency of DSB induction was significantly lower than in the single cutting approaches using the spCas9 nuclease, HDR efficiency was increased, making this strategy auspicious for future investigations in primary patient keratinocytes into which plasmid delivery is a key issue. In this respect, further improvements of the double-nicking strategy described here can include a direct Cas9 RNA or protein delivery into the cells, reducing the time of nickase activity, and thereby further reducing the frequency of off-target events. However, prior to translating our approach into the clinics, the risk of adverse off-target activity has to be extensively addressed by NGS. Specifically, whole-genome sequencing can further increase the sensitivity of the off-target analysis as it has already been shown for CRISPR/Cas9- and TALEN-edited human induced pluripotent stem cells (IPSCs) by Smith et al.37
The possibility to apply this gene-editing tool as an ex vivo gene therapy for patients suffering from blistering skin diseases further improves its safety profile. The ex vivo gene therapy will rely on the correction of autologous stem cells isolated from patients, which are further expanded to epidermal sheets and transplanted back. Modified stem cells can be clonally selected upon treatment, thereby increasing the ability to assess the safety profile of the approach significantly.38, 39 Epidermal sheets can be expanded from gene-corrected cells and subsequently transplanted back onto the patient, successfully demonstrating the high therapeutic potential of ex vivo gene therapy in the skin.30, 31, 32 Considering that the mutational hotspots for EBS-gen sev are located in exons 1 and 6 of the KRT14 gene, it is possible to treat the cells of multiple individuals from different families with a small set of designed sgRNA pairs, simplifying the therapeutic approach in this patient group. A CRISPR/Cas9n ex vivo gene therapy treatment would offer a safe, effective, and, compared with other treatment options, permanent repair of the EBS skin. In many cases, EBS-gen sev is severe in childhood, but improves with age. However, specific sites of the body, such as the soles of the feet, are particularly prone to blistering, causing substantial pain throughout life. Ex vivo gene therapy could provide an additional and permanent treatment option for this patient population.
In summary, we here present an efficient and specific gene-editing approach for KRT14 repair, which can be further translated into a clinical setting offering a causal treatment for patients suffering from severe skin blistering diseases.
Materials and Methods
Selection and Cloning of sgRNA Specific for Intron 7 of KRT14
Putative sgRNA target sites within intron 7 of the KRT14 gene were predicted using the online prediction platform CHOPCHOP.18 The sgRNAs were designed (sgRNA1: 5′- ATCCGCCCAGGCCTGCAGAGGAGG-3′/5′-AAACCCTCCTCTGCAGGCCTGGGC-3′ and sgRNA2: 5′-ATCCAGCAGGTCTAGGCATTGGCT-3′/5′-AAACAGCCAATGCCTAGACCTGCT-3′) and cloned as double-stranded oligonucleotides into either the CMV-T7-hspCas9-T2A-GFP-H1-gRNA CAS740G-1 linearized SmartNuclease vector or the CMV-T7-hspCas9-nickase-T2A-GFP-H1-gRNA CAS790G-1 linearized SmartNickase vector (System Bioscience, Palo Alto, CA, USA) according to the manufacturer’s protocol.
Generation of the MC Donor Plasmid
The KRT14 homology donor template was PCR-amplified in two fragments for ease of cloning into the MN511A1 vector (System Bioscience, Palo Alto, CA, USA). The first fragment was amplified using an intron-3-specific forward primer (5′-GATCTCTAGAGTGAGAACTATATGGAAAAGTCAGCTTAAAAGAAATGC-3′) and an intron-7-specific reverse primer (5′- GATCGCTAGCAGGGTCTTACCATCTCTTGAGGATTGCGATCCAGAGG-3′) and cloned into the MN511A1 vector via the XbaI and NheI sites incorporated within the primers. The resulting 1,173 bp fragment incorporates silent mutations within exon 7. The second fragment (805 bp) was amplified using an intron-7-specific forward primer, which incorporates three silent mutations (5′- GATCGAATTCCATCATCTGCAGGCCTGGGCTCCAGGCCACCCTCTGTACCCCAAGCAGGTCTAGGCATTGGCTACGGGCTCCGTG-3′) and a KRT14 3′ UTR-specific reverse primer (5′-GATCGGATCCTTATGCAACTCAG ATAATGAAGCTGTATTGATTG-3′), and subsequently cloned into EcoRI and BamHI restriction sites. Correct cloning was confirmed by sequence analysis. The resulting homology donor template encompasses the entire KRT14 region from intron 3 to the 3′ UTR and incorporates NheI and EcoRI sites within intron 7, which facilitates detection of the recombined allele. The IRES-blasticidin cassette was cloned downstream of the KRT14 3′ UTR according to Peking et al.40 For production of MC DNA, the MC-Easy-Minicircle DNA production kit (System Bioscience, Palo Alto, CA, USA) was used according to the manufacturer’s protocol.
Transient Transfection of Donor Plasmids
The embryonic kidney cell line HEK293 (Stratagene, La Jolla, CA, USA) was cultivated in DMEM supplemented with 10% fetal bovine serum (FBS) and 100 U/mL penicillin/100 μg/mL streptomycin (Biochrom, Berlin, Germany). Human EBS keratinocytes (EBDM-2) carrying a heterozygous mutation (c.1231G>A) in exon 6 of KRT14 and normal hKcs were cultivated in keratinocyte serum-free medium (SFM) GIBCO (Invitrogen, Paisley, UK) supplemented with bovine pituitary extract (BPE), epidermal growth factor (EGF) according to manufacturer’s protocol, and 100 U/mL penicillin/100 μg/mL streptomycin. All cell lines were maintained at 37°C and 5% CO2 in a humidified incubator.
Transient transfections of HEK293AD cells were performed using jetPEI reagent (Polyplus-transfection SA, Strasbourg, France), whereas transient transfections of EBS patient keratinocytes were performed using Xfect transfection reagent (Takara Bio Europe, Saint-Germain-en-Laye, France) according to the manufacturer’s protocol. HEK293AD cells were cultivated in 60 mm plates and transfected with the respective Cas9/sgRNA-expressing plasmids (5 μg). Human EBS keratinocytes were transfected with the respective Cas9/sgRNA-expressing plasmids and the MC donor template in a ratio of 2:1 (4 μg DP and 2 μg sgRNA) or in the case of double-nicking in a ratio of 2:1:1 (4 μg DP and 2 μg of each sgRNA). Antibiotic selection of transfected EBS keratinocytes was performed when cells reached 60% confluency, by supplementing the medium with 10 μg/mL blasticidin (Invivogen, San Diego, CA, USA). Untransfected EBS keratinocytes cells were used as a negative control. For transfection efficiency tests of our different donor plasmids into EBS keratinocytes, cells were cultured in six-well plates and transiently transfected with 4.5 μg of plasmid using Xfect transfection reagent.
Flow Cytometric Analysis and Fluorescence-Activated Cell Sorting
Transfection efficiency was evaluated 48 hr post-transfection via flow cytometric analysis using a Beckman Coulter FC500 FACS (fluorescence-activated cell sorting) analyzer (Beckman Coulter, Brea, CA, USA). GFP (expressed from Cas9/sgRNA vector)-expressing cells were analyzed using the Kaluza Flow Cytometry Analysis Software (Beckman Coulter).
Isolation of Single-Cell Clones
To obtain a homogeneous cell population upon genome editing, we isolated single-cell clones using single-cell dilution. Cells were counted using a TC20 cell counter (Bio-Rad, Hercules, CA, USA) and manually seeded to obtain 0.5 cell per well of a 96-well plate in co-culture with 3T3-J2 mouse fibroblasts feeder cells (5 × 102 cells/cm2) growth arrested with 4 μg/mL mitomycin C (Roche, Basel, Switzerland). We initially isolated 384 single-cell clones from the blasticidin-selected bulk keratinocyte population that had been treated with Cas9n/MC-DP (MC donor plasmid) and sgRNA1 and sgRNA2. Of these, only 17 clonal lines could be expanded up to T75 flask format for protein expression studies.
T7EI Assay
Mismatches upon Cas9/sgRNA-mediated DSB induction and subsequent NHEJ at the desired genomic locus in HEK293AD cells were evaluated via T7EI assays (New England Biolabs, Frankfurt, Germany) on genomic DNA using a KRT14 exon-6-specific forward primer (5′-CAGGAGATGATTGGCAGCGTGG-3′) and a KRT14 intron-7-specific reverse primer (5′-AAGCAAGAGGTGGGGGCTGCC-3′) for the PCR. TACC3 off-target region for sgRNA1 was amplified using a TACC3 intron-10-specific forward primer (5′-GTGCAGACAGGGTGGTTCCG-3′) and a TACC3 intron-10-specific reverse primer (5′-TCCCGGTCACCTGCTCTGAC-3′). The digest was performed according to the manufacturer’s protocol.
Integration and HDR Efficiency Analysis
For analysis of site-specific integration and recombination, genomic DNA of pre-selected Cas9n/MC-DP and sgRNA1- and sgRNA2-treated keratinocytes was used. The target region for site-specific integration was PCR-amplified using a forward primer (5′-GATGGTAA GACCCTGCTAGCGAATTCCCT-3′) binding to the multiple cloning site of the MC-DP and a reverse primer (5′-GATGCTTCTCCCACTTTCTCCCC-3′) binding to the endogenous 3′ downstream sequence of the KRT14 gene. For estimation of the HDR efficiency, we PCR-amplified the KRT14 target region using a forward primer binding to KRT14 exon 6 (5′-CAGGAGATGATTGGCAGCGTGG-3′) and again a reverse primer binding to the endogenous 3′ downstream sequence (5′-GATGCTTCTCCCACTTTCTCCCC-3′) of the KRT14 gene, resulting in a PCR product of 1,189 bp, which was subsequently digested with EcoRI restriction enzyme (Thermo Fisher). To minimize the size of the PCR product and facilitate subcloning, we performed a nested PCR using a forward primer binding to exon 6 (5′-CAGGAGATGATTGGCAGCGTGG-3′) and a reverse primer binding to intron 7 (5′-AAGCAAGAGGTGGGGGCTGCC-3′). For subcloning of the PCR product, the StrataClone PCR Cloning Kit (Aligent Technologies, Santa Clara, CA, USA) was used according to the manufacturer’s protocol. Single clones were analyzed via EcoRI digestion and Sanger sequencing.
Semiquantitative Real-Time PCR
Total RNA was isolated from cultured keratinocytes using an innuPREP RNA Mini Kit (Analytik Jena, Jena, Germany). cDNA was synthesized from 2 μg of total RNA, using iScript cDNA Synthesis Kit (Bio-Rad). 100 ng of cDNA and 15 pmol of each primer were added to each reaction. Reactions were prepared as master mixes, and two reactions were analyzed per sample. Semi-quantitative real-time PCR was performed to analyze mRNA expression levels of KRT14. For amplification of the transcripts, the following primers were used: keratin-14: KRT14_fw: 5′-GAGGATATGGTGGTGGCCTTGGTGCTGG-3′ and KRT14_rv: 5′- GATCGGATCCTTGGTGCGGAAGTCATCCG-3′. PCR conditions were 3 min at 95°C followed by 50 cycles of 30 s at 95°C, 30 s at 64°C, and 30 s at 72°C. Semi-quantitative real-time PCR was performed using the Bio-Rad iCycler System and Bio-Rad iQ SYBR Green Supermix Kit (Bio-Rad). At least 9 different runs with duplicates were analyzed using 3 different RNA samples of each cell line. Statistics were performed using the one-way ANOVA-Tukey’s multiple comparison test. Transcript levels were calculated after normalization to glyceraldehyde 3-phosphate dehydrogenase (GAPDH; GAPDH_fw: 5′- GCCAACGTGTCAGTGGTGGA-3′; GAPDH_rv: 5′- CACCACCCTGTTGCTGTAGCC-3′).
To analyze correction of the mutation and accurate integration of the MC sequence at RNA level, we used a forward primer binding to the exon 5-exon 6 junction (5′-AGCTCAGCATGAAAGCATCCCT-3′) and a reverse primer binding to exon 8 (5′-CATCGTGCACATCCATGACCTTGGTG-3′) of KRT14. The resulting PCR fragment was confirmed by sequence analysis.
Protein Isolation and Western Blot Analysis
2.5 × 105 EBS keratinocytes were cultivated in keratinocyte SFM GIBCO (Invitrogen, Paisley, UK) and grown to 70%–80% confluence. Whole cell lysates were generated by lysing the cell pellet in radioimmunoprecipitation assay (RIPA) buffer (Santa Cruz Biotechnology, Heidelberg, Germany). Whole cell lysates were centrifuged at full speed at 4°C for 20 min, and supernatant was frozen at −20°C until usage.
Protein samples were denatured for 5 min at 95°C in 4× loading buffer (0.25 M Tris-HCl; 8% SDS; 30% glycerol; 0.02% bromphenol blue; 0.3 M β-mercaptoethanol [pH 6.8]). The procedure of western blotting was performed as previously described.40, 41 Ponceau red (Sigma-Aldrich, St. Louis, MO, USA) staining of the nitrocellulose membrane was performed after electro-blotting. Nitrocellulose membrane was blocked with blocking reagent (Roche Diagnostics, Mannheim, Germany) diluted 1:10 in Tris-buffered saline with 0.2% Tween (TBS-T) for 1 hr at room temperature (RT). For detection of K14, a mouse anti-cytokeratin 14 antibody (LL001:sc53253) (Santa Cruz Biotechnology, Heidelberg, Germany) diluted 1:400 in TBS-T and blocking reagent was used and incubated at 4°C overnight. A polyclonal α-actinin antibody (H-300:sc15335) served as loading control, diluted 1:1,000 in TBS-T and blocking reagent. As secondary antibodies, either the horseradish peroxidase (HRP) Envision+-labeled anti-mouse or anti-rabbit antibody (Dako, Santa Clara, CA, USA) diluted 1:200 in TBS-T was used. Nitrocellulose membrane was incubated for 1 hr at RT. Protein bands were visualized using the Immobilon Western Chemiluminescent HRP Substrate (Merck, Darmstadt, Germany) and the ChemiDoc XRS Imager (Bio-Rad, Hercules, CA, USA).
Immunofluorescence Staining of Cytokeratin 14 after Heat Shock
2.5 × 105 keratinocytes were seeded into 35 mm glass-bottom dishes (MatTek In Vitro Life Science Laboratories, Bratislava, Slovak Republic) and grown to 50%–70% confluency. Heat shock was performed in a water bath at 43°C for 30 min, followed by a 15 min recovery time at 37°C and 5% CO2 in a humidified incubator. Cells were fixed with 4% formaldehyde solution (Sigma-Aldrich) overnight at RT. The next day cells were treated with an antigen retrieval buffer (1 mM EDTA and 0.05% Tween in PBS [pH 8]) at 95°C for 40 min in a water bath. Permeabilization and staining of the cells were performed in a single step. The mouse anti-cytokeratin 14 antibody (LL001:sc53253) was used in a 1:100 dilution, diluted in 0.3% Triton X, blocking reagent (1:10), and TBS-T. As secondary antibody, either Alexa Fluor 488 goat anti-mouse IgG (Invitrogen, Paisley, UK) (1:400 in TBS-T) or Alexa Fluor 594 goat anti-mouse IgG (Invitrogen, Paisley, UK) (1:400 in TBS-T) was used for 1 hr at RT. DAPI was diluted 1:4,000 in TBS-T and added to the cells for 10 min. Cells were analyzed using an inverted microscope system, which includes the laser scanning confocal microscope Zeiss LSM 700 and the Axio Observer Z1 (Carl Zeiss, Oberkochen, Germany).
Off-Target Prediction
Off-target prediction was performed using the BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Both sgRNAs were compared with the genomic and transcript databases to identify potential off-target loci.
NGS
We designed a comprehensive AmpliSeq panel to perform NGS analyses on the Ion Torrent Personal Genome Machine (PGM) platform. A setup of a 400 bp customized panel was chosen to cover the cutting sites of the single gRNA constructs within the on-target region of the KRT14 gene, as well as cutting sites within predicted off-target regions; primers are listed in Table S2. A mean vertical coverage of at least 5,000 reads was planned. The library preparation, template preparation, and the sequencing run were performed according to manufacturer’s protocols (Thermo Fisher Scientific/Life Technologies, Carlsbad, CA, USA). Data analysis was implemented on the Integrative Genome Viewer (IGV). We attained a mean coverage of about 4,072 reads for the EBS cell line and about 1,602 reads for HEK293 cells with our customized panel.
Author Contributions
Conceptualization, T.K. and U.K.; Methodology, T.K., U.K., J.P.H., and E.M.M.; Investigation, T.K., S.H., P.P., T.L., and M.A.; Validation, A.K.; Writing – Original Draft, T.K., P.P., and U.K.; Writing – Review & Editing, J.R., J.W.B., and J.P.H.; Resources, V.W., A.K., and M.A.; Supervision, U.K., E.M.M., and J.R.
Acknowledgments
The authors thank Franz Zimmermann for his suggestions and critical scientific input. Special thanks to Univ.-Prof. Dr. Eva Rohde and Karin Roider, MSc, of the Spinal Cord Injury and Tissue Regeneration Center Salzburg (SCI-TReCS) for providing their Core Facility for Microscopy. This work was supported by DEBRA Austria.
Footnotes
Supplemental Information includes three figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.ymthe.2017.08.015.
Supplemental Information
References
- 1.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.March O.P., Reichelt J., Koller U. Gene editing for skin diseases: designer nucleases as tools for gene therapy of skin fragility disorders. Exp. Physiol. 2017 doi: 10.1113/EP086044. Published online March 17, 2017. [DOI] [PubMed] [Google Scholar]
- 3.Aushev M., Koller U., Mussolino C., Cathomen T., Reichelt J. Traceless targeting and isolation of gene-edited immortalized keratinocytes from epidermolysis bullosa simplex patients. Mol. Ther. Methods Clin. Dev. 2017;6:112–123. doi: 10.1016/j.omtm.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hainzl S., Peking P., Kocher T., Murauer E.M., Larcher F., Del Rio M., Duarte B., Steiner M., Klausegger A., Bauer J.W. COL7A1 editing via CRISPR/Cas9 in recessive dystrophic epidermolysis bullosa. Mol. Ther. 2017 doi: 10.1016/j.ymthe.2017.07.005. Published online July 13, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dianov G.L., Hübscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013;41:3483–3490. doi: 10.1093/nar/gkt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horn H.M., Tidman M.J. The clinical spectrum of epidermolysis bullosa simplex. Br. J. Dermatol. 2000;142:468–472. doi: 10.1046/j.1365-2133.2000.03358.x. [DOI] [PubMed] [Google Scholar]
- 9.Fine J.D., Bruckner-Tuderman L., Eady R.A., Bauer E.A., Bauer J.W., Has C., Heagerty A., Hintner H., Hovnanian A., Jonkman M.F. Inherited epidermolysis bullosa: updated recommendations on diagnosis and classification. J. Am. Acad. Dermatol. 2014;70:1103–1126. doi: 10.1016/j.jaad.2014.01.903. [DOI] [PubMed] [Google Scholar]
- 10.Coulombe P.A., Kerns M.L., Fuchs E. Epidermolysis bullosa simplex: a paradigm for disorders of tissue fragility. J. Clin. Invest. 2009;119:1784–1793. doi: 10.1172/JCI38177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szeverenyi I., Cassidy A.J., Chung C.W., Lee B.T., Common J.E., Ogg S.C., Chen H., Sim S.Y., Goh W.L., Ng K.W. The Human Intermediate Filament Database: comprehensive information on a gene family involved in many human diseases. Hum. Mutat. 2008;29:351–360. doi: 10.1002/humu.20652. [DOI] [PubMed] [Google Scholar]
- 12.Wu Z., Feng G. Progress of application and off-target effects of CRISPR/Cas9. Yi Chuan. 2015;37:1003–1010. doi: 10.16288/j.yczz.15-070. [DOI] [PubMed] [Google Scholar]
- 13.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wally V., Lettner T., Peking P., Peckl-Schmid D., Murauer E.M., Hainzl S., Hintner H., Bauer J.W. The pathogenetic role of IL-1β in severe epidermolysis bullosa simplex. J. Invest. Dermatol. 2013;133:1901–1903. doi: 10.1038/jid.2013.31. [DOI] [PubMed] [Google Scholar]
- 16.Glász-Bóna A., Medvecz M., Sajó R., Lepesi-Benko R., Tulassay Z., Katona M., Hatvani Z., Blazsek A., Kárpáti S. Easy method for keratin 14 gene amplification to exclude pseudogene sequences: new keratin 5 and 14 mutations in epidermolysis bullosa simplex. J. Invest. Dermatol. 2009;129:229–231. doi: 10.1038/jid.2008.223. [DOI] [PubMed] [Google Scholar]
- 17.Kaneko S., Hamada T., Kawano Y., Hashimoto T., Morita E. Missense mutation at the helix termination region in the 2B domain of keratin 14 in a Japanese family with epidermolysis bullosa simplex, generalized, other. Int. J. Dermatol. 2011;50:436–438. doi: 10.1111/j.1365-4632.2010.04702.x. [DOI] [PubMed] [Google Scholar]
- 18.Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42(Web Server issue):W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bloom K., Ely A., Arbuthnot P. A T7 endonuclease I assay to detect Talen-mediated targeted mutation of HBV cccDNA. Methods Mol. Biol. 2017;1540:85–95. doi: 10.1007/978-1-4939-6700-1_8. [DOI] [PubMed] [Google Scholar]
- 20.Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., Rebar E.J. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 21.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wally V., Brunner M., Lettner T., Wagner M., Koller U., Trost A., Murauer E.M., Hainzl S., Hintner H., Bauer J.W. K14 mRNA reprogramming for dominant epidermolysis bullosa simplex. Hum. Mol. Genet. 2010;19:4715–4725. doi: 10.1093/hmg/ddq405. [DOI] [PubMed] [Google Scholar]
- 23.Abitbol R.J., Zhou L.H. Treatment of epidermolysis bullosa simplex, Weber-Cockayne type, with botulinum toxin type A. Arch. Dermatol. 2009;145:13–15. doi: 10.1001/archdermatol.2008.546. [DOI] [PubMed] [Google Scholar]
- 24.Neufeld-Kaiser W., Sybert V.P. Is cyproheptadine effective in the treatment of subjects with epidermolysis bullosa simplex-Dowling-Meara? Arch. Dermatol. 1997;133:251–252. doi: 10.1001/archderm.133.2.251. [DOI] [PubMed] [Google Scholar]
- 25.Veien N.K., Buus S.K. Treatment of epidermolysis bullosa simplex (EBS) with tetracycline. Arch. Dermatol. 2000;136:424–425. doi: 10.1001/archderm.136.3.424-a. [DOI] [PubMed] [Google Scholar]
- 26.Chiaverini C., Fontas E., Vabres P., Bessis D., Mazereeuw J., Charlesworth A., Meneguzzi G., Lacour J.P. Oral erythromycin therapy in epidermolysis bullosa simplex generalized severe. Br. J. Dermatol. 2015;173:563–564. doi: 10.1111/bjd.13672. [DOI] [PubMed] [Google Scholar]
- 27.Retief C.R., Malkinson F.D., Pearson R.W. Two familial cases of epidermolysis bullosa simplex successfully treated with tetracycline. Arch. Dermatol. 1999;135:997–998. doi: 10.1001/archderm.135.8.997. [DOI] [PubMed] [Google Scholar]
- 28.Kerns M.L., Guss L., Fahey J., Cohen B., Hakim J.M., Sung S., Lu R.G., Coulombe P.A. Randomized, split-body, single-blinded clinical trial of topical broccoli sprout extract: assessing the feasibility of its use in keratin-based disorders. J. Am. Acad. Dermatol. 2017;76:449–453.e1. doi: 10.1016/j.jaad.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wally V., Kitzmueller S., Lagler F., Moder A., Hitzl W., Wolkersdorfer M., Hofbauer P., Felder T.K., Dornauer M., Diem A. Topical diacerein for epidermolysis bullosa: a randomized controlled pilot study. Orphanet J. Rare Dis. 2013;8:69. doi: 10.1186/1750-1172-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauer J.W., Koller J., Murauer E.M., De Rosa L., Enzo E., Carulli S., Bondanza S., Recchia A., Muss W., Diem A. Closure of a large chronic wound through transplantation of gene-corrected epidermal stem cells. J. Invest. Dermatol. 2017;137:778–781. doi: 10.1016/j.jid.2016.10.038. [DOI] [PubMed] [Google Scholar]
- 31.Mavilio F., Pellegrini G., Ferrari S., Di Nunzio F., Di Iorio E., Recchia A., Maruggi G., Ferrari G., Provasi E., Bonini C. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat. Med. 2006;12:1397–1402. doi: 10.1038/nm1504. [DOI] [PubMed] [Google Scholar]
- 32.Siprashvili Z., Nguyen N.T., Gorell E.S., Loutit K., Khuu P., Furukawa L.K., Lorenz H.P., Leung T.H., Keene D.R., Rieger K.E. Safety and wound outcomes following genetically corrected autologous epidermal grafts in patients with recessive dystrophic epidermolysis bullosa. JAMA. 2016;316:1808–1817. doi: 10.1001/jama.2016.15588. [DOI] [PubMed] [Google Scholar]
- 33.Cao T., Longley M.A., Wang X.J., Roop D.R. An inducible mouse model for epidermolysis bullosa simplex: implications for gene therapy. J. Cell Biol. 2001;152:651–656. doi: 10.1083/jcb.152.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chamcheu J.C., Navsaria H., Pihl-Lundin I., Liovic M., Vahlquist A., Törmä H. Chemical chaperones protect epidermolysis bullosa simplex keratinocytes from heat stress-induced keratin aggregation: involvement of heat shock proteins and MAP kinases. J. Invest. Dermatol. 2011;131:1684–1691. doi: 10.1038/jid.2011.93. [DOI] [PubMed] [Google Scholar]
- 35.Werner N.S., Windoffer R., Strnad P., Grund C., Leube R.E., Magin T.M. Epidermolysis bullosa simplex-type mutations alter the dynamics of the keratin cytoskeleton and reveal a contribution of actin to the transport of keratin subunits. Mol. Biol. Cell. 2004;15:990–1002. doi: 10.1091/mbc.E03-09-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petek L.M., Fleckman P., Miller D.G. Efficient KRT14 targeting and functional characterization of transplanted human keratinocytes for the treatment of epidermolysis bullosa simplex. Mol. Ther. 2010;18:1624–1632. doi: 10.1038/mt.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith C., Gore A., Yan W., Abalde-Atristain L., Li Z., He C., Wang Y., Brodsky R.A., Zhang K., Cheng L., Ye Z. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15:12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duarte B., Miselli F., Murillas R., Espinosa-Hevia L., Cigudosa J.C., Recchia A., Del Río M., Larcher F. Long-term skin regeneration from a gene-targeted human epidermal stem cell clone. Mol. Ther. 2014;22:1878–1880. doi: 10.1038/mt.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larcher F., Dellambra E., Rico L., Bondanza S., Murillas R., Cattoglio C., Mavilio F., Jorcano J.L., Zambruno G., Del Rio M. Long-term engraftment of single genetically modified human epidermal holoclones enables safety pre-assessment of cutaneous gene therapy. Mol. Ther. 2007;15:1670–1676. doi: 10.1038/sj.mt.6300238. [DOI] [PubMed] [Google Scholar]
- 40.Peking P., Koller U., Hainzl S., Kitzmueller S., Kocher T., Mayr E., Nyström A., Lener T., Reichelt J., Bauer J.W., Murauer E.M. A gene gun-mediated nonviral RNA trans-splicing strategy for Col7a1 repair. Mol. Ther. Nucleic Acids. 2016;5:e287. doi: 10.1038/mtna.2016.3. [DOI] [PubMed] [Google Scholar]
- 41.Tockner B., Kocher T., Hainzl S., Reichelt J., Bauer J.W., Koller U., Murauer E.M. Construction and validation of an RNA trans-splicing molecule suitable to repair a large number of COL7A1 mutations. Gene Ther. 2016;23:775–784. doi: 10.1038/gt.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.