Abstract
Hepatocytes represent an important target for gene therapy and editing of single-gene disorders. In α-1 antitrypsin (AAT) deficiency, one missense mutation results in impaired secretion of AAT. In most patients, lung damage occurs due to a lack of AAT-mediated protection of lung elastin from neutrophil elastase. In some patients, accumulation of misfolded PiZ mutant AAT protein triggers hepatocyte injury, leading to inflammation and cirrhosis. We hypothesized that correcting the Z mutant defect in hepatocytes would confer a selective advantage for repopulation of hepatocytes within an intact liver. A human PiZ allele was crossed onto an immune-deficient (NSG) strain to create a recipient strain (NSG-PiZ) for human hepatocyte xenotransplantation. Results indicate that NSG-PiZ recipients support heightened engraftment of normal human primary hepatocytes as compared with NSG recipients. This model can therefore be used to test hepatocyte cell therapies for AATD, but more broadly it serves as a simple, highly reproducible liver xenograft model. Finally, a promoterless adeno-associated virus (AAV) vector, expressing a wild-type AAT and a synthetic miRNA to silence the endogenous allele, was integrated into the albumin locus. This gene-editing approach leads to a selective advantage of edited hepatocytes, by silencing the mutant protein and augmenting normal AAT production, and improvement of the liver pathology.
Keywords: humanized liver mouse model, liver xenograft, liver regeneration, α-1 antitrypsin deficiency, nuclease-free genome editing, gene editing, AAV, miRNA, shRNA, RNAi, AAT, AATD, A1AT
Borel et al. describe two studies based on misfolded human α-1 antitrypsin (A1AT). First, when A1AT is expressed in livers of NSG, mice it allows for reproducible engraftment of human hepatocytes. Second, gene editing of hepatocytes to decrease misfolded protein results in expansion of corrected cells and amelioration of liver disease.
Introduction
α-1 antitrypsin deficiency (AATD) is a common genetic disorder that can lead to both liver and lung disease and currently affects an estimated 3.4 million patients worldwide.1 AAT is encoded by SERPINA1 and is primarily secreted by hepatocytes, making it the most abundant serum antiprotease. One of the most common disease variants in AATD is a mutation resulting in a glutamate to lysine (Glu342Lys) substitution known as the PiZ allele or Z-AAT.2 In contrast to the normal PiM allele (M-AAT) the Z-AAT protein is prone to polymerization and consequently is either directed for proteolysis or aggregates in the endoplasmic reticulum of hepatocytes.3 With up to 85% of the AAT protein being retained as polymers or degraded in the liver, it sets the stage for both the loss-of-function (lung) and gain-of-function (liver) diseases observed in AATD patients.
Normally, AAT diffuses into all organs, but its main site of action is the lower respiratory tract, where it protects the alveoli and the surrounding connective tissue matrix from destruction by neutrophil elastase. In this context, AATD patients develop panacinar emphysema owing to years of a protease/antiprotease imbalance leading to alveolar wall degradation and decreases in airway tethering as a consequence of the loss of interstitial elastin.4 The Z-AAT aggregation and polymerization causes liver disease by a toxic gain-of-function mechanism due to accumulation of misfolded protein in the hepatocytes whereby 10%–20% of PiZ homozygote patients suffer from clinical liver disease ranging from fulminant liver failure and cirrhosis to hepatocellular carcinoma.5, 6, 7 Development of these disorders is thought to be a consequence of the intracellular accumulation of Z-AAT polymers in hepatocytes leading to hepatocellular death by apoptosis or other death mechanisms.
Insight into the pathobiology of the liver disease has been derived from the PiZ mouse, which is a transgenic C57BL/6 mouse expressing the human Z-AAT gene at high levels. In this model, polymer accumulation is heterogeneous throughout the liver, and cells with lower Z-AAT protein burdens proliferate to maintain liver mass, leading to cycles of cell death and regeneration that end with the activation of hepatic stellate cells and eventually hepatic fibrosis.8 Consequently, cells with higher Z-AAT protein burden have decreased mitotic index.9
Currently, the treatment of AATD liver disease can only be addressed by liver transplantation, and due to the associated morbidity, it is an option for patients with significant cirrhosis, hepatocellular carcinoma, and liver failure. In contrast, studies have shown a slower progression of the lung disease with weekly intravenous augmentation therapy of purified pooled human plasma AAT.10, 11 However, this long-life therapy engenders considerable costs and lifestyle adjustments; thus, AATD has been a target for gene-augmentation therapy. Strategies for recombinant adeno-associated virus (AAV)-mediated gene augmentation for the lung, and simultaneous gene augmentation with mutant gene reduction for both lung and liver disease have been described.12, 13, 14, 15, 16, 17 However, to our knowledge there are currently no pre-clinical studies taking advantage of the competitive disadvantage of Z-AAT-burdened hepatocytes compared to wild-type murine hepatocytes that were previously documented in the PiZ mouse.9
We therefore hypothesized that the cell death and regeneration cycle in Z-AAT-burdened livers will allow normal donor human hepatocytes or genetically corrected murine hepatocytes to progressively outcompete host hepatocytes expressing the mutant form of AAT. This concept has important clinical implications for cell transplant therapy, but it will also be highly relevant from the perspective of genome editing. Specifically, determining if the genetic correction of hepatocytes in AATD patients can lead to cells with increased engraftment is crucial for this therapeutic approach. Here, we report that the generation of the NOD-scid-gamma-PiZ transgenic mice expressing mutant Z-AAT on the severe immune deficient NSG background allows for the repopulation of the liver by normal donor human hepatocytes as well as by murine hepatocytes corrected by genome editing. The data supports the use of hepatocyte-transplant and genome-editing approaches to treat emphysema and liver disease in patients with AATD. Furthermore, this is a novel, potent, and technically simple approach to generate xeno-repopulation of the liver with normal donor human hepatocytes or with induced pluripotent stem cell-derived hepatic cells, and it may prove a useful tool for the liver regeneration field.
Results
Wild-Type Mouse Hepatocytes Have an Increased Engraftment as Compared to PiZ Mouse Hepatocytes in the NSG-PiZ Mouse
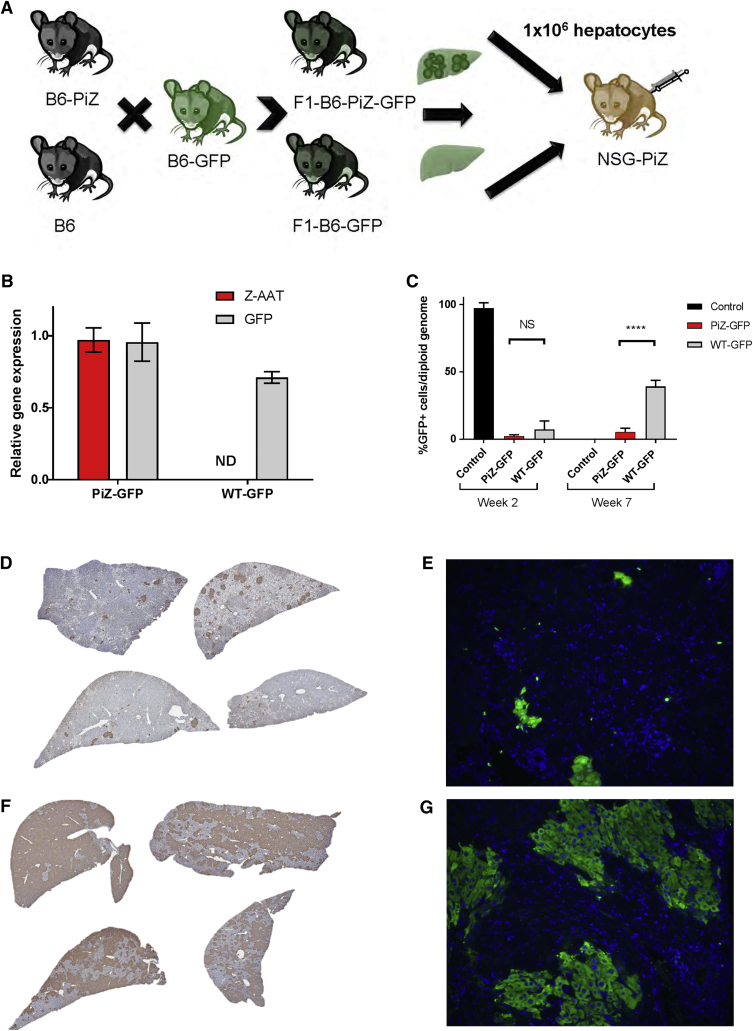
It has been reported that Z-AAT globule-containing murine hepatocytes have an increased incidence of apoptosis and that this leads to a proliferative disadvantage of these cells when compared to murine hepatocytes with lower or no Z-AAT globule loads.8, 9 In order to confirm this hypothesis and determine whether normal donor human hepatocytes could regenerate a PiZ liver, we used the PiZ mouse that expresses the human Z-AAT gene to create two new strains. First, the PiZ mouse was backcrossed onto the NOD-scid IL2rgnull (NSG) background.18 The severely immunocompromised NSG-PiZ strain allows these mice to host human as well as allogeneic mouse hepatocytes without rejection. Second, we generated the hepatocyte donor mice by crossing the transgenic C57BL/6-Tg(CAG-EGFP)1Osb mouse strain with either a wild-type C57BL/6 or a C57BL/6-PiZ transgenic mouse. These crosses generated C57BL/6-Tg(CAG-EGFP)1Osb-PiZ F1 litters hemizygous for GFP with (PiZ-GFP) or without (wild-type, WT-GFP) the PiZ mutant human α-1 antitrypsin (AAT) gene (Figure 1A). As shown in Figure 1B, hepatocytes isolated from the PiZ-GFP mice expressed both the human Z-AAT and GFP genes whereas the WT-GFP donors only expressed GFP. Hepatocytes from these mice were then used to engraft male NSG-PiZ mice by intrasplenic injection of 1 × 106 PiZ-GFP or WT-GFP donor hepatocytes. Eight weeks post-injection, livers from recipient NSG-PiZ mice were examined for donor repopulation by digital droplet PCR (ddPCR) as well as by immunohistology. The quantitative ddPCR data confirms that that the level of donor hepatocyte repopulation in the NSG-PiZ mouse is dependent on whether the donor hepatocytes express the Z-AAT or not. The results in Figure 1C show that on average 40% of the hepatocytes of the NSG-PiZ livers were GFP positive when mice were engrafted with wild-type GFP-expressing normal hepatocytes as compared to only 5% when the donor hepatocytes were expressing GFP and Z-AAT (PiZ-GFP). These results were confirmed by immunohistochemical (IHC) and immunofluorescent (Figures 1D–1G) staining of host livers where WT-GFP hepatocytes engrafted at a significantly higher proportion as compared to the PiZ-GFP hepatocytes. GFP IHC staining and positive pixel count threshold analysis of the images revealed repopulation varied by lobes between 32% and 48% (Table S1), but overall the positive pixel count analysis for histology sections supports the ddPCR results. This data confirms the hypothesis that wild-type murine hepatocytes have an increased regenerative capacity as compared to Z-AAT containing hepatocytes and moreover that the Z-AAT-burdened host liver is undergoing sufficient turnover to provide wild-type donor hepatocytes with the ability to repopulate it at significant levels.
Figure 1.
Wild-Type Mouse Hepatocytes Have an Increased Engraftment as Compared to PiZ Mouse Hepatocytes in the NSG-PiZ Mouse
(A) Experimental outline, GFP-expressing hepatocytes from PiZ-GFP or wild-type (WT)-GFP donor mice were isolated and engrafted into male NSG-PiZ mice by intrasplenic injection of 1 × 106 cells per mouse. The recipient mice were euthanized 8 weeks post-engraftment, and liver tissue was used for analysis. (B) Human Z-AAT and GFP gene expression were determined by qRT-PCR in donor mice (mean ± SEM, n = 3). Statistical significance of the difference between donor cell types was determined by a two-tailed, unpaired t test. ****Statistically significant difference; NS, not significant. (C) The percentage of GFP+ cells per diploid genome was determined by ddPCR performed on genomic DNA from NSG-PiZ mouse liver engrafted with PiZ-GFP or WT-GFP hepatocytes (mean ± SEM; controls, n = 2; PiZ-GFP, n = 3; WT-GFP, n = 4). Genomic DNA from WT-GFP donor liver was used as a positive control. Presence of GFP+ cells is shown by (D and F) immunohistochemistry for all animals and (E and G) immunofluorescence for one representative animal in NSG-PiZ mice engrafted with (D and E) PiZ-GFP hepatocytes and (F and G) WT-GFP hepatocytes.
Normal Human Hepatocytes Readily Repopulate NSG-PiZ Mouse Livers
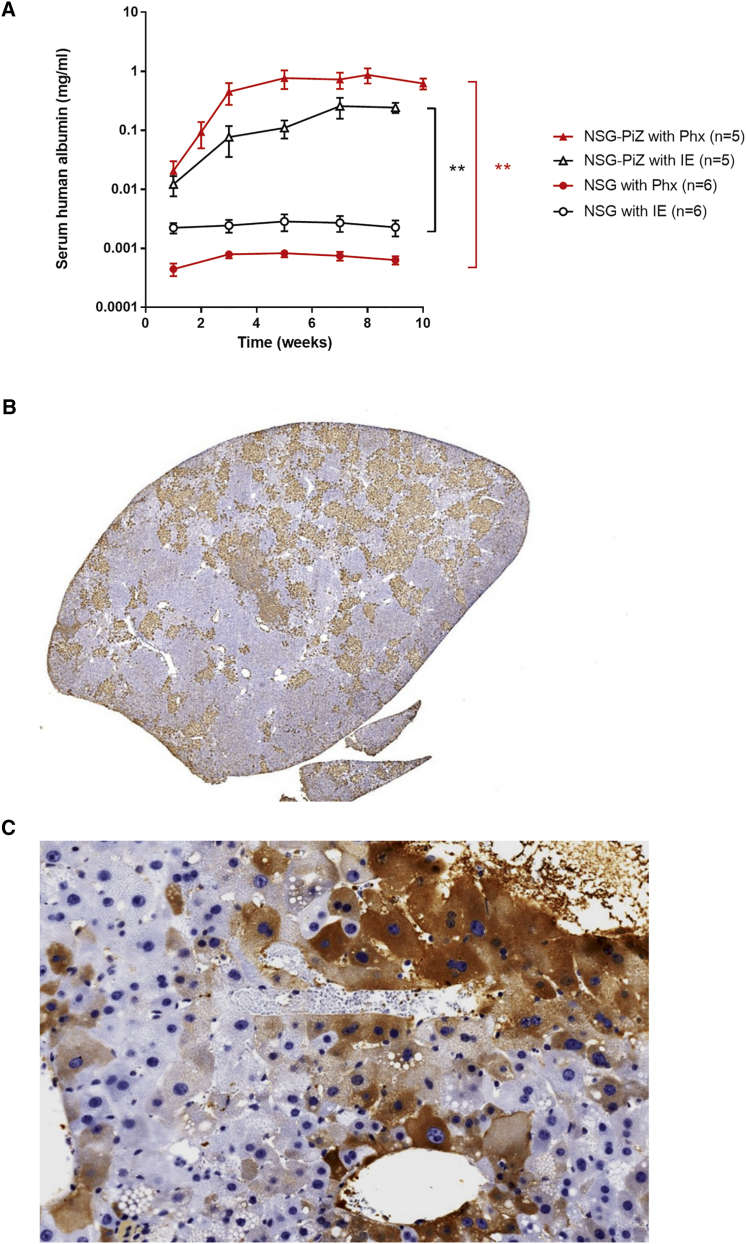
Given the increased fitness of normal mouse hepatocytes in NSG-PiZ mice, we next sought to determine if this mouse strain could sustain a human hepatocyte xenograft. To test this, NSG-PiZ and NSG control mice were injected intrasplenically with 1 × 106 mature human hepatocytes. Human albumin levels were monitored prospectively in mouse sera samples as a surrogate for human liver repopulation. Serum human albumin levels in Figure 2A confirm that liver chimerism is possible in these mice and that the level of human hepatocyte repopulation achieved in NSG-PiZ mouse livers expressing the Z-AAT is dramatically increased over that achieved in NSG mouse livers. This again confirms the role for the Z-AAT burden in creating a niche whereby wild-type hepatocytes are able to expand preferentially. Given that the human hepatocytes also benefited from an increased engraftment in the NSG-PiZ mice, we hypothesized that increasing hepatocyte turnover may lead to increased repopulation. To test this, we repeated the same experiments with an added partial hepatectomy at the time of the intrasplenic injections. Given that in mice the original liver mass is regenerated in about 7 days after a 70% partial hepatectomy,19 we hypothesized that during this proliferative phase we may get increased repopulation. As shown in Figure 2B, partial hepatectomy leads to a 5- to 10-fold increase in circulating serum human albumin levels. Interestingly, in the NSG mice, hepatectomy caused a decrease in repopulation. This is likely due to the advantage of wild-type-proliferating mouse hepatocytes over human hepatocytes in the context of a mouse liver. IHC staining for human albumin in the NSG-PiZ mouse livers (Figure 2C) confirmed that human hepatocytes are also able to repopulate a significant portion of the NSG-PiZ livers.
Figure 2.
Normal Human Hepatocytes Readily Repopulate NSG-PiZ Mouse Livers
Male NSG-PiZ and NSG mice were engrafted with 1 × 106 mature human hepatocytes, and serum was collected biweekly post-engraftment for 10 weeks for (A) quantification of human albumin levels by ELISA after either intrasplenic engraftment alone (IE) or intrasplenic engraftment with partial hepatectomy (PHx); data are represented as mean ± SEM. **Statistically significant differences between groups (two-way ANOVA). Liver tissue was harvested at 10 weeks post-PHx and stained for human albumin (B and C); one representative animal is shown.
Z-AAT Burden in NSG-PIZ Mice Decreases with Age, and Hepatocyte Repopulation Correlates with Z-AAT Levels
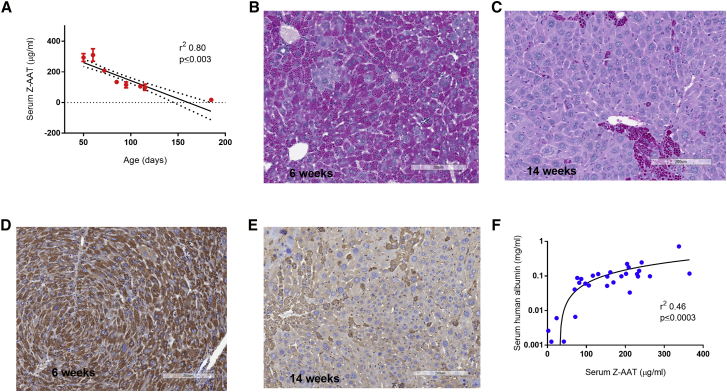
It has been established that in PiZ transgenic mice Z-AAT globule-containing hepatocytes have a higher incidence of apoptosis and that these cells tend to get outcompeted by hepatocytes with lower Z-AAT globule loads.8 This phenomenon is likely the underlying basis for the competitively advantaged repopulation of wild-type murine or human hepatocytes in NSG-PiZ mice. To more clearly define the consequences of this normal hepatocyte turnover in the absence of donor hepatocytes, we investigated whether this natural turnover in NSG-PiZ mice would also lead to a decrease in the Z-AAT globule load over time. A prospective analysis of serum Z-AAT levels in these mice shows a clear and strong inverse correlation (r2 = 0.80, p ≤ 0.003, two-tailed) between the age of the mice and the Z-AAT serum levels (Figure 3A). As NSG-PiZ mice age, the serum Z-AAT levels decrease, which is also reflected histologically by the decrease in the globule-containing status of the hepatocytes between 6 and 14 weeks of age. This is clearly visualized by both a diastase-resistant periodic acid-Schiff (PASD; Figures 3B and 3C) and human AAT staining of livers at different ages (Figures 3D and 3E), consistently with previous reports.20, 21 This data suggests that in the absence of engrafted wild-type hepatocytes, the liver in NSG-PiZ mice still exerts enough selective pressure to favor survival and expansion of hepatocytes that produce and accumulate less Z-AAT. Interestingly, as the mice age there are some hepatocytes that continue to accumulate Z-AAT globules, and it appears that even though they are scarcer, their globules tend to be larger (Figure 3C). As the Z-AAT burden decreases with the age of the mice, we would expect that the proliferative advantage of the normal human engrafted hepatocytes would also be diminished. To confirm if this was the case, we performed a retrospective analysis comparing the average human albumin level of a given mouse to the Z-AAT serum level at the time of engraftment. Our analysis revealed a moderate yet highly significant positive correlation (r2 = 0.46, p ≤ 0.0003) between Z-AAT serum levels and the levels of human liver chimerism as determined by average human albumin serum levels (Figure 3F).
Figure 3.
Z-AAT Burden in NSG-PiZ Mice Decreases with Age and Hepatocyte Repopulation Correlates with Z-AAT Levels
(A) Correlation between the age of NSG-PiZ mice and their serum Z-AAT levels. Serum was collected and Z-AAT levels were quantified by ELISA. Data are plotted as mean ± SEM; n per group is as follows: 50 days, n = 10; 60 days, n = 9; 72 days, n = 27; 85 days, n = 12; 95 days, n = 17; 110 days, n = 7; 115 days, n = 3; 185 days, n = 5. (B–E) Liver tissue from NSG-PiZ mice was stained for PASD (B and C) or human AAT (D and E) at 6 weeks of age (B and D) or 14 weeks of age (C and E); results are presented for one representative animal. (F) Correlation between serum Z-AAT levels of the NSG-PiZ hosts at the time of engraftment and subsequent level of repopulation indicated by serum human albumin 7 weeks post-engraftment. Serum was collected at engraftment and 7 weeks post-engraftment, and Z-AAT and human albumin levels (respectively) were quantified by ELISA for 30 animals.
Optimization of the Human Liver Xenograft Model
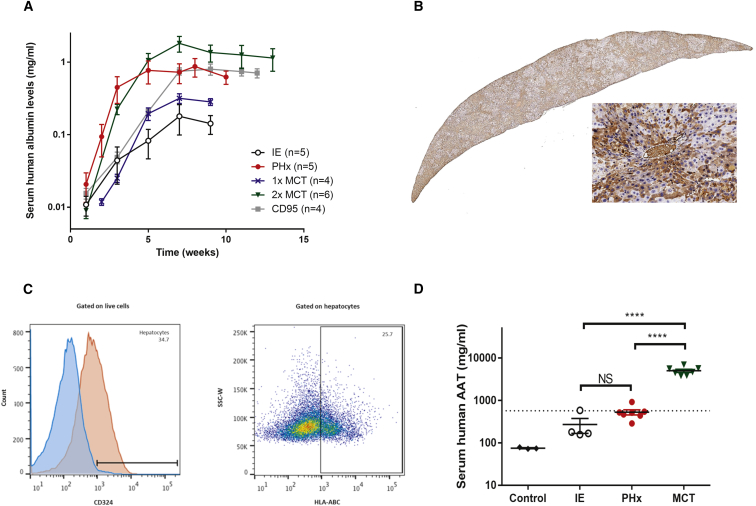
To investigate alternatives to the partial hepatectomy for increased human hepatocyte repopulation, we tested two approaches that have been previously used in the liver xenograft field, namely monocrotaline (MCT) and a mouse-specific anti-Fas antigen (CD95) antibody. MCT is a pyrrolizidine plant alkaloid with known endothelial toxicity in the lung, liver, and kidney and has been shown to increase cell repopulation in the liver.22 The anti-CD95 antibody recognizes mouse Fas and leads to cytolytic activity of cells expressing mouse Fas by inducing apoptosis of mouse hepatocytes and other cells.23 We explored various protocols employing these two liver injury models to create a “two hit” protocol that can be easily and reproducibly performed without the need for the partial hepatectomy. Repopulation was significantly increased with both of these interventions and achieved levels comparable or higher than with the partial hepatectomy (Figure 4A). We administered either a single or two doses MCT (50 mg/kg) prior to engraftment, with the highest levels achieved when mice were given two MCT doses (Figure 4A). The kinetics of repopulation following a single intraperitoneal injection of the anti-CD95 antibody at the time of hepatocyte delivery was just as efficient as the partial hepatectomy as determined by the serum human albumin levels (Figure 4A). Staining of the MCT- and CD95-treated mouse livers for human albumin confirmed the repopulation of the human hepatocytes (Figure 4B). Furthermore, a quantitative flow-cytometry analysis using an anti-HLA antibody confirms that at least 25% of all hepatocytes in the MCT-treated mice are of human origin (Figure 4C). Human specificity of the anti-HLA antibody was demonstrated by absence of staining of strictly murine hepatocytes (Figure S1). The FDA-accepted therapeutic serum threshold for protein augmentation of human AAT is currently set at 11 μM, equivalent to 572 μg/mL. The results from the xenograft experiments indicate for the first time that this threshold can be achieved with a simple hepatocyte cell therapy approach. Quantification of total human AAT levels after human hepatocyte transplantation in the NSG-PiZ mice shows that it is possible to achieve this therapeutic threshold and that it may be of clinically relevant interest when combined with a partial hepatectomy (Figure 4D).
Figure 4.
Optimization of the Human Liver Xenograft Model
(A) Repopulation rate was assessed after various challenges. The challenges compared were as follows: intrasplenic engraftment alone (IE, white circles), intrasplenic injection with partial hepatectomy (PHx, red circles), a single intraperitoneal dose of 50 mg/kg monocrotaline (MCT) at 7 days pre-engraftment (1× MCT, blue crosses), a double intraperitoneal dose of 50 mg/kg MCT at 14 and 7 days pre-engraftment (2× MCT, green triangles), and a dose of 1 μg CD95 intravenously (i.v.) at the time of engraftment (CD95, gray squares). Serum was collected biweekly post-engraftment, and human albumin levels were quantified as a measure of repopulation. Data are plotted as mean ± SEM. **Statistically significant differences between groups. (B) Liver tissue was harvested at 10 weeks post-engraftment from MCT-treated mice and stained for human albumin; one representative animal is shown. (C) Hepatocytes of human origin were quantified by flow cytometry. Hepatocytes were gated as CD324+ cells, and human hepatocytes are defined as cells expressing the human HLA-ABC marker within the CD324+ subset. One representative animal is shown. Percentages are indicated in the histogram and dot plot. (D) Serum human AAT levels were quantified by ELISA in various treatment groups at 10 weeks post-engraftment. Data are plotted as mean ± SEM. The dotted line denotes the therapeutic threshold at 572 μg/mL. ****Statistically significant difference between groups (two-way unpaired t test).
Selective Expansion of Genome-Edited Hepatocytes
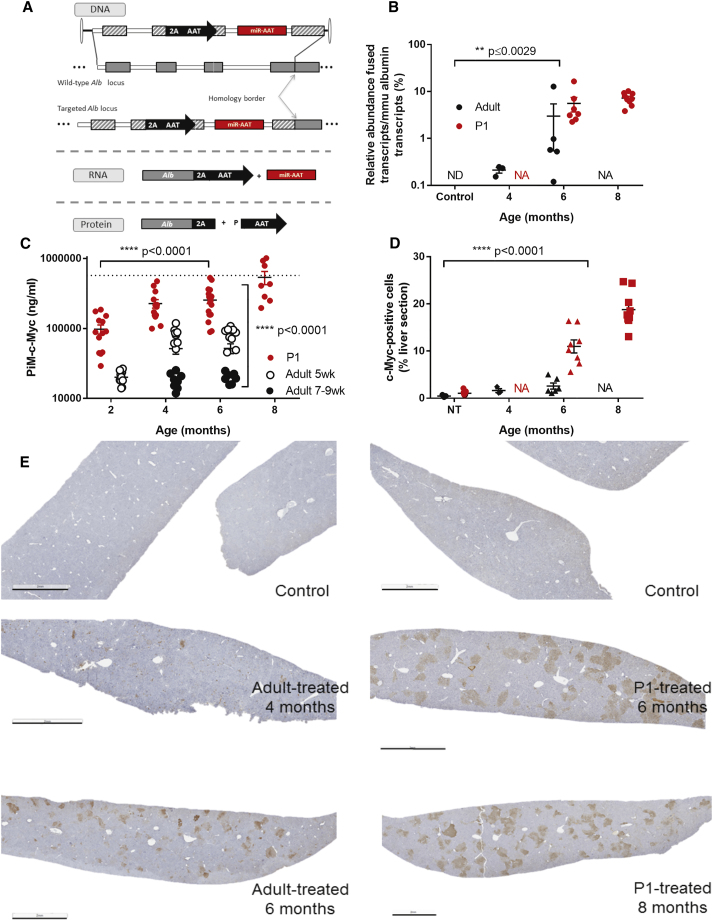
Adults and P1 B6-PiZ mice were injected systemically with either saline or AAV-GeneRide-DualFunctionAAT. GeneRide-DualFunctionAAT is a promoterless cassette containing a 2A-peptide sequence followed by a de-targeted, c-Myc-tagged human AAT sequence, and an artificial miRNA targeting Z-AAT.14 The cassette is flanked by two homology arms (1.1–1.3 kb) that are complementary to the C57BL/6 Alb locus and allow homologous recombination (HR). Following HR, Alb and AAT will be co-transcribed as a single Alb-AAT mRNA—which will be referred to here as “fused transcript”—and will lead to the production of two proteins through ribosomal skipping (Figure 5A). To measure the expression imparted by the genome-edited alleles, we quantified by ddPCR the relative abundance of the fused mRNA transcript as compared to the normal albumin transcript in a subset of mice at 4, 6, and 8 months post-injection. The fused transcript is only detected in the treated animals (Figure 5B). In adult-treated mice, at 4 months the ratio was 0.2%, while at 6 months it was increased to 3%; in P1-treated mice, at 6 months the ratio of 5.6%, and at 8 months it was increased to 7.2% (Figure 5B; one-way ANOVA, p value ≤ 0.0029), supporting expansion of the genome-edited hepatocytes as function of time. These results were further confirmed by measuring the c-Myc-tagged AAT in serum (M-AAT-c-Myc). Mice were bled prior to injection and biweekly thereafter, which allowed for monitoring of M-AAT-c-Myc serum levels by ELISA. The levels of M-AAT-c-Myc increase on average 10-fold over time when between 2 and 6 months in adult-treated mice, and such increase over time is also observed in P1-treated mice (Figure 5C). This difference in expression over time is statistically significant (two-way ANOVA, p < 0.0001), further supporting expansion of the genome-edited hepatocytes as a function of time. P1-treated animals present at all time points higher M-AAT-c-Myc levels than age-matched adult-treated animals (Figure 5C). This difference between adult-treated and P1-treated is statistically significant (two-way ANOVA, p value < 0.0001). This was further confirmed in a subset of livers stained for c-Myc at 4, 6, and 8 months post-injection where the number of positive cells was quantified (Figure 5D). The staining shows clusters of hepatocytes throughout the entire liver (Figure 5E), and the size of the clusters increases between early and late time points for both the adult-treated and the P1-treated animals. This qualitative observation is supported by a quantitative threshold analysis (Figure 5D), which estimates the percentage of c-Myc positive pixel counts at 1.6% at 4 months and 2.6% at 6 months for the adult-treated mice and 11% at 6 months and 18.8% at 8 months for the P1-treated animals. At 6 months of age, the P1-treated animals present levels approximately 4-fold higher than adult-treated animals. The differences observed were statistically significant both between treatment groups (two-way ANOVA, p value < 0.0001) and over time (two-way ANOVA, p value < 0.0001).
Figure 5.
Selective Expansion of Genome-Edited Hepatocytes
Adults or P1 PiZ mice were injected systemically with (A) recombinant AAV8 vector packaged with a promoterless, dual-function cassette containing an artificial miRNA that targets human AAT and a miRNA-detargeted human AAT sequence with a c-Myc tag. The cassette is flanked by 1.1–1.3 kb homology arms to the albumin locus. Transgene expression was evaluated by several methods. (B) Albumin-α-1 antitrypsin-fused transcript/murine albumin transcript ratio by ddPCR. **Statistically significant differences over time (two-way ANOVA). (C) M-AAT-c-Myc serum levels by ELISA, normalized to the average of the age-matched control group. ****Statistically significant differences both over time and between groups (two-way ANOVA). (D) c-Myc immunohistochemistry, quantified by threshold analysis with (E) representative animals shown for each group (scale bar, 2 mm). (D) ****Statistically significant differences over time (two-way ANOVA). For all graphs, data are represented as mean ± SEM.
Genome Editing Improves Liver Phenotype
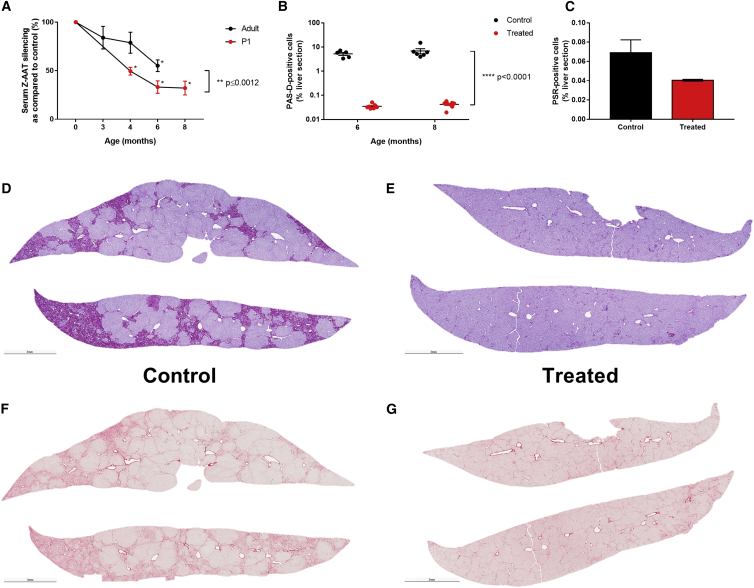
The Z-AAT liver pathology is caused by polymerization of the Z-AAT protein and accumulation of those polymers in globules. Clinically the patients present with liver fibrosis, which may lead to cirrhosis and hepatocellular carcinoma. Levels of serum Z-AAT were evaluated by ELISA, and the percentage of Z-AAT silencing increases over time (Figure 6A) and is statistically significant starting at 6 months in adult-treated mice (two-tailed unpaired t test, p ≤ 0.0002) and starting at 4 months in P1-treated mice (two-tailed unpaired t test, p ≤ 0.0001). Consistent with the M-AAT-c-Myc data, silencing is higher in the P1-treated animals than in the adult-treated animals (two-way ANOVA, p ≤ 0.0012). Interestingly, the magnitude of Z-AAT silencing was unexpected given that it far exceeds the amount of genome-edited hepatocytes. This finding has important implications and suggests that not all hepatocytes produce equal amounts of Z-AAT, and perhaps, as suggested by our data, the highest degree of proliferative advantage for the GeneRide (and thus Z-AAT silencing) may be in those hepatocytes that continue to produce and retain high amounts of Z-AAT. Alternatively, this could point to the inverted terminal repeat (ITR)-driven expression of the anti-AAT miRNA in unedited hepatocytes. Finally, the liver phenotype was further characterized in P1-treated mice, at 6 months and 8 months post-treatment. A periodic acid-Schiff-diastase (PAS-D) staining was performed on liver sections from those mice, in order to assess the amount of Z-AAT globules that appear as magenta spheres. As quantified in Figure 6B, the percentage of PAS-D+ cells is significantly reduced by more than 100-fold at both 6 and 8 months post-treatment (two-way ANOVA, p ≤ 0.0001). A Picro Sirius Red (PSR) staining was performed on liver sections from the male animals, in order to assess the amount of collagen fibers, which will appear in red, as a measure of liver fibrosis. Quantification of PSR+ cells showed a reduction of over 40% at 8 months post-treatment (Figure 6C, two-tailed unpaired t test, non-significant). Representative images are shown in Figures 6D and 6E for the PAS-D staining and in Figures 6G and 6H for the PSR staining.
Figure 6.
Genome Editing Improves Liver Phenotype
Z-AAT serum levels are decreased, as shown in (A) by ELISA, normalized to the average of the age-matched control group and presented as percentage of silencing. *Statistically significant differences between treated and control groups at a given time point (unpaired t test); **statistically significant differences between treatment groups (two-way ANOVA). Liver phenotype was further analyzed at 6 and 8 months post-treatment in the P1-injected group, by (B) PASD immunohistochemistry at 6 and 8 months, quantified by threshold analysis. ****Statistically significant differences between groups (two-way ANOVA). (C) Picro Sirius immunohistochemistry at 8 months, quantified by threshold analysis; (D and E) representative animals for the PASD staining (D, control; E, treated); (F and G) representative animals for the PSR staining (F, control; G, treated). For all graphs, data are represented as mean ± SEM.
Taken all together, the data supports the hypothesis that HR occurred in cells throughout the liver, that the genome-edited hepatocytes then selectively expanded, and that this expansion results in an increase of serum M-AAT with a concomitant decrease of Z-AAT over time. This results in a marked improvement of the liver pathology.
Discussion
The current report demonstrates an in vivo survival advantage of hepatocytes in which WT-AAT is expressed, as compared with those expressing the common human missense mutant AAT, Z-AAT, in line with a previous study in PiZ mice demonstrating that cell therapy could partially improve the liver phenotype.24 Similarly, here we show for the first time that gene-edited PIZ hepatocytes can also be corrected to lower the Z-AAT burden and provide competitive survival advantage to those hepatocytes, thereby normalizing serum AAT levels and improving the toxicity in the liver imparted by the misfolded Z-AAT. Disorders in which corrected cells possess a competitive survival advantage have been viewed as optimal candidates for gene and cell therapy. In cases such as severe combined immune deficiency (SCID) due to adenosine deaminase (ADA) deficiency, the survival advantage of corrected cells appears to contribute to the robust clinical efficacy observed despite modest initial transduction efficiency in early trials.25 A survival advantage in genetically corrected hepatocytes has been demonstrated in the case of hereditary tyrosinemia type 1 due to fumarylacetoacetate hydrolase (FAH)-deficiency.26 The latter could present an opportunity for cell or gene therapy or genome editing of this disease,27 if safety and efficacy prove to provide an advantage over small-molecule therapy with 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC). In the case of AAT deficiency, no therapy currently exists to correct or prevent liver disease, and attempts to use intramuscular gene augmentation to secrete WT-AAT and treat lung disease have been hampered by suboptimal serum levels at the doses of vector used to date.15 Additionally, transplantation of WT-AAT hepatocytes in an AAT-deficient patient with end-stage liver disease previously resulted in an increase in AAT serum levels from 12–19 mg/dL to 290 mg/dL.28
The competitive advantage for repopulation of non-PiZ-expressing hepatocytes in the NSG-PiZ recipient also presents numerous opportunities to use this system as an experimental tool in both xenograft and isograft settings. The Fah-deficient mouse strain has frequently been used for a similar purpose. The NSG-PiZ mouse may represent an alternative human hepatocyte repopulation model in which the uncorrected animals breed and survive easily without maintenance drug therapy. The human xenograft version of this system may be used as an in vivo model to identify optimal vectors for transduction of human hepatocytes, which often differ from mouse hepatocytes in their tropism for specific AAV capsid variants.29 Similarly, human hepatocyte xenografts are a useful tool for in vivo directed evolution studies.30
This study validates the NSG-PiZ mouse model as a platform to test human hepatocyte cell-based therapy for liver disease and genome editing for AATD. Besides disease-specific applications, the NSG-PiZ mouse model offers an in vivo tool to evaluate the liver repopulation ability of any cells with hepatocytic potential, including human induced pluripotent stem cell-derived hepatic cells. Overall, this study provides an important additional liver disease mouse model to the liver regeneration field, which is needed to investigate ways to treat human liver disease.
Future prospects for liver-directed cell and gene therapy of AAT deficiency include innovations for in vivo or ex vivo genome editing with nucleotide-directed endonucleases, such as CRISPR/Cas9 and Cpf1. Many of the questions to be addressed regarding both safety and efficacy depend on targeting human, as opposed to murine, hepatocytes. For instance, studies of off-target effects of genome editing are only valid in the context of the human genome. Likewise, the potential for vector carcinogenesis due to recombinant AAV (rAAV) integration cannot be readily extrapolated between species. This is particularly true for liver-directed rAAV therapy, in which vector-related hepatocellular carcinoma has been tracked to the Rian locus in murine cells. Because of the dissimilarity in sequence between the murine and human loci, the model presented here could potentially be of value in addressing these and other related questions in future studies.
Materials and Methods
Animals
NOD.Cg-PrkdcscidIl2rgtm1wjl/SzJ (NSG) and NSG-Tg(SERPINA1*E342K)#Slcw/Sz (NSG-PiZ) mice were obtained from the research colonies of Leonard D. Shultz at The Jackson Laboratory. FVB.Cg-Tg(SERPINA1E342K*)#Slcw mice carrying the PiZ allele of the serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (Homo sapiens) transgene were kindly provided by David Perlmutter (University of Pittsburgh School of Medicine). This transgenic model possesses the Z variant (PiZ) allele of the human AAT (SERPINA1*) transgene, allowing for human Z-AAT expression. The PiZ allele31 was backcrossed five generations by a marker-assisted speed congenic approach to the NSG strain background. NSG-Tg(SERPINA1*E342K)#Slcw/Sz was abbreviated as NSG-PiZ mice. The PiZ transgenic hemizygous mice were intercrossed to fix the PiZ allele to homozygosity. NSG-PiZ homozygotes were raised in a specific pathogen-free (SPF) barrier mouse room at The Jackson Laboratory and maintained by intercrossing homozygous breeders. NSG-PiZ homozygotes were shipped to UMMS for hepatocyte engraftment. The mice used for engraftment experiments were maintained on sulfamethoxazole/trimethoprim in drinking water on alternate weeks as previously described.32 Mice were housed with 12 hr light/dark cycles on Bed-o’cob 1/4” bedding (Andersons), and fed a standard laboratory diet. Mice were not fasted before any of the procedures. All procedures were carried out during the light cycle.
The B6-PiZ mice used for the genome-editing experiments have a high transgene copy number (>10).
Animal Procedures
All animal procedures were performed according to the guidelines indicated for rodent surgery on the Institutional Animal Care and Use Committee webpage and by the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School. Aseptic techniques and sterile instruments were used during all procedures. All recipient animals were males, age 6–10 weeks, and they were anesthetized by isoflurane inhalation in a closed ventilation chamber.
Intrasplenic Injection
An incision was made on mouse left paralumbar fossa in order to expose the spleen. One million primary human hepatocytes (Bioreclamation IVT) suspended in 50 μL of Hanks Balanced Saline solution were injected into the inferior pole of the spleen, using a 25G needle connected to a 1/3 mL syringe. The exposed spleen was returned to the abdominal cavity, and the incision was closed with an absorbable suture (4-0 Vicryl). Vet Bond was applied topically to seal the skin incision.
Partial Hepatectomy
A median-line incision was made to expose the liver. The median lobe of the liver and the left lateral lobe were securely tied off with 4-0 silk Blake braided suture and excised. Two-thirds of the total liver was removed by partial hepatectomy. The remaining liver was gently returned to the abdominal cavity. Finally, the opposing sections of the abdominal wall were aligned at the ventral midline, and an absorbable suture (4-0 Vicryl) was applied to close the incision. The skin incision was sealed with the topical application of Vet Bond.
Vector Injection for Genome Editing
For the treatment of adult mice, males between 5 and 9 weeks of age received an intravenous injection of 1E12 gc of vector in 200 μL total volume. For the treatment of P1 mice, both males and females were injected intraperitoneally with 2E11 gc of vector. Animals were bled biweekly via sub-mandibular puncture as per our protocol, and serum was stored at −80°C until time of analysis. At endpoint, animals were euthanized by CO2 asphyxiation followed by cervical dislocation as per our protocol. Liver tissue was harvested, consistent sections were fixed for histology, and the remaining tissue was snap-frozen for downstream molecular analysis.
Liver Perfusion/Primary Hepatocyte Isolation
Mice were euthanized and an abdominal incision was made to expose the abdominal cavity, and the viscera were arranged in order to expose the inferior vena cava. A catheter needle was used to puncture the inferior vena cava and inserted ∼1 cm into the inferior vena cava. Suture silks were applied to secure the catheter. Pump tubing was attached to the end of the catheter. The portal vein was sliced to allow outflow. The diaphragm was opened, and the inferior vena cava was clamped off above the liver. To perfuse the liver, liver perfusion medium (Life Technologies) was first pumped through the circulatory system for 50 mL, followed by liver digest medium (Life Technologies) for 50 mL. The liver was removed and immersed in ice-cold hepatocyte wash medium (HWM, Life Technologies) allowing for the hepatocytes to diffuse into solution. The cell suspension was drawn up and pipetted through 70 μm mesh and was centrifuged at 50 g for 3min at 4°C. After spinning down the cells, 10 mL of HWM and 10 mL 90% Percoll (GE Healthcare) in phosphate-buffered saline (PBS; Ca2+, Mg2+ free) were added to the solution to remove non-viable cells. The hepatocyte solution was washed three times with 30 mL of HWM.
ELISA
The mouse sera were collected biweekly pre- and post-engraftment and tested for total AAT, Z-AAT, c-Myc, and human albumin by ELISA. For all assays, high binding 96-well plates (Immulon4, Dynatech Laboratories) were coated with the relevant capture antibody in 100 μL Voller’s buffer overnight at 4°C. Unless otherwise indicated, all subsequent incubations were done at room temperature for 1 hr, and plates were washed in between steps with PBS 0.05% Tween 20.
Human AAT ELISA
Blocking was done with 1% milk. Capture antibody is human-specific goat anti-AAT (1:500, Cat#55111, MP Biomedicals); detection antibody is goat anti-hAAT (horseradish peroxidase [HRP]; 1:5,000, Cat#ab191350, Abcam). Standard is AAT, (Cat# 16-16-011609, Athens Research and Technology).
Z-AAT ELISA
Blocking was done with 1% milk. Capture antibody is a custom antibody (available through http://www.umassmed.edu/muellerlab/contact/reagent-request); detection antibody is described above.
c-Myc ELISA
All incubations were done at 37°C. Blocking was done with 5% BSA. Capture antibody is (1:1,000, Cat# ab19234, Abcam).
Human Albumin ELISA
Human albumin levels were evaluated post-engraftment by using the human Albumin ELISA quantitation set (Bethyl Labs) as described by the manufacturer. For all assays, after incubation with the HRP-conjugated antibody, 3,3',5,5'-tetramethylbenzidine (TMB) peroxidase substrate (KPL) was applied, and the reactions were stopped by adding 2 N H2SO4 (Fisher Scientific) after 15–30 min. Plates were read at 450 nm on a VersaMax microplate reader (Molecular Devices).
Immunohistochemistry
Mouse liver was serially sectioned 4 μm.
Human Albumin Staining
Sections were de-paraffinized and treated by Citra (Biogenex) at 98°C for 30 min. Background Sniper (Biocare Medical) was applied to reduce non-specific background staining. Sections were incubated with rabbit anti-human albumin (1:4,000, Cat#ab2604, Abcam) for 1 hr. Stain was visualized using Mach2 Goat × Rabbit HRP polymer (Biocare Medical), the 3,3'-diaminobenzidine (DAB) chromagen (Biocare Medical) and Churukian-Allison-Tacha (CAT) hematoxylin counterstain (Biocare Medical).
Human AAT Staining
Slides were de-paraffinized with xylene and re-hydrated through decreasing concentrations of ethanol to water, including an intermediary step to quench endogenous peroxidase activity (3% hydrogen peroxide in methanol). Slides were transferred to 1× Tris-buffered saline (TBS). For enzyme-induced antigen retrieval, sections were heated for 5 min at 37°C, while submerged in Digest-all Trypsin (Invitrogen). Slides were subsequently rinsed in 1× TBS and incubated with a universal protein blocker Sniper (Biocare Medical), for 15 min at room temperature (RT). Slides were rinsed in 1× TBS and incubated for 60 min with a primary rabbit antibody that recognizes human (and not mouse) AAT (1:600, Cat#KDI-A1ATRYPabr, Fitzgerald). Slides were rinsed in 1× TBS, followed by application of conjugated secondary antibody (Mach 2 goat anti-rabbit horse radish peroxidase-conjugated, Biocare Medical) for 30 min at RT. Detection of AAT was achieved by incubating slides in 3′3′ diaminobenzidine (Biocare Medical) for 1.5 min at RT. Slides were counterstained with hematoxylin for 30 s and mounted with Cytoseal XYL (Richard-Allen Scientific).
PAS-D Staining
Schiff’s reagent was pre-warmed at RT for at least 30 min. Sections were de-paraffinized and rehydrated. Amylase was applied on the slides for 20 min at RT and rinsed with water. Sections were incubated in 0.5% Periodic acid for 10 min and rinsed. Schiff’s reagent was applied to the slides for 15 min. Then sections were washed in lukewarm tap water for 5 min. Hematoxylin counterstain was applied for 15 s and blue nuclei for 1 min. One slide without amylase treatment acted as the control.
PSR Staining
PSR staining was performed using the Picrosirius Red Stain Kit (Cat#24901-250, Polysciences Inc.).
Threshold Analyses
Analyses were carried out blindly by an experienced technician at the University of Florida Pathology Core.
Model Optimization
Animals were pre-treated with 50 mg/kg MCT (Oakwood Chemical) intraperitoneally 7 days or 14 and 7 days prior to engraftment or treated at the time of engraftment with 1 μg purified NA/LE hamster anti-mouse CD95 antibody (first apoptosis signal [FAS], BD PharMingen) intravenously.
Flow Cytometry Assay
After dissociation, cells were stained on ice for 30 min with anti-mouse/human CD324 (Biolegend) and anti-HLA-ABC (BD Biosciences) antibodies to determine human hepatocyte percentage. Cells were then washed twice with 1× PBS and resuspended in 1× PBS supplemented with 0.5% FBS and 2 mM EDTA. Cells were acquired using FACS-LSRII flow cytometer (BD Biosciences) and analyzed with FlowJo software (version 10.1; Tree Star Inc.).
ddPCR Assay
Genomic DNA was isolated (Gentra Puregene kit, QIAGEN) from flash-frozen liver and ddPCR (Bio-Rad) was performed according to the manufacturer’s recommendations using 50 ng of DNA as input and TaqMan assays detecting GFP and RPP30.
Statistics
Regression analysis was followed by a two-tailed Pearson R test (Figure 4). Effects of time and treatment were assessed through a repeated-measure two-way ANOVA (Figure 6C). Statistical significance of the difference between age-matched control and treated groups was determined with a paired t test (Figure 6F). Statistical significance was defined as p < 0.05.
Author Contributions
F.B., L.D.S., D.L.G., M.A.B., T.R.F., and C.M. designed experiments; F.B., Q.T., G.G., C.G., and Z.W. performed experiments; A.B., M.A.K., and C.M. generated the GeneRide construct; L.D.S., D.L.G., and M.A.B. generated the NSG-PiZ strain; F.B., Q.T., T.R.F., and C.M. wrote the manuscript.
Conflicts of Interest
A.B. is co-founder and chief science officer of LogicBio Therapeutics and is listed as an inventor on relevant patent applications. M.A.K. is a founder and advisor to LogicBio Therapeutics. LogicBio Therapeutics has licensed GeneRide technology from Stanford University. None of this work was funded by LogicBio Therapeutics. D.L.G. and M.A.B. are consultants and receive research support from The Jackson Laboratory. T.R.F. is a paid consultant and scientific advisor to Dimension Therapeutics (in which he holds equity) and Editas Medicine and a founder of AGTC but has donated his equity to the Alpha-1 Foundation. C.M. and T.R.F. are co-founders of Apic Bio, which has licensed the dual-function vector for AAT from UMMS. None of this work was funded by Apic Bio. C.M. and T.R.F. are inventors on patents for technologies described within the manuscript and may be entitled to royalty payments in the future. The other authors (F.B., Q.T., G.G., C.G., Z.W., and L.D.S.) declare no competing financial interests.
Acknowledgments
We thank Ann Fu at the University of Florida Molecular Pathology Core for her assistance with histology and the UMMS Flow Cytometry Core Facility, David Perlmutter, and Ira Fox for providing L.D.S. with the STOCK-PiZ mice. This research was supported by the Office of the Director of the NIH (1R24OD018259-01 to D.L.G., C.M., M.A.B., and L.D.S.), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the NIH (1R01DK098252-01 to C.M. and T.R.F.), the National Heart, Lung, and Blood Institute (NHLBI) of the NIH (1P01HL131471-01 and 1P01HL131471-01 to C.M. and R01 HL064274 to M.K.), and an Alpha-1 Foundation postdoctoral fellowship (to F.B.) and research grant (to C.M.). L.D.S. was supported by a Cancer Core grant (CA034196) to The Jackson Laboratory. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. None of this work was funded by LogicBio Therapeutics.
Footnotes
Supplemental Information includes one figure and one table and can be found with this article online at https://doi.org/10.1016/j.ymthe.2017.09.020.
Contributor Information
Michael A. Brehm, Email: michael.brehm@umassmed.edu.
Christian Mueller, Email: chris.mueller@umassmed.edu.
Supplemental Information
References
- 1.de Serres F.J. Worldwide racial and ethnic distribution of alpha1-antitrypsin deficiency: summary of an analysis of published genetic epidemiologic surveys. Chest. 2002;122:1818–1829. doi: 10.1378/chest.122.5.1818. [DOI] [PubMed] [Google Scholar]
- 2.Brantly M., Nukiwa T., Crystal R.G. Molecular basis of alpha-1-antitrypsin deficiency. Am. J. Med. 1988;84(Suppl 6A):13–31. doi: 10.1016/0002-9343(88)90154-4. [DOI] [PubMed] [Google Scholar]
- 3.Lomas D.A., Evans D.L., Finch J.T., Carrell R.W. The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- 4.Larsson C. Natural history and life expectancy in severe alpha1-antitrypsin deficiency, Pi Z. Acta Med. Scand. 1978;204:345–351. doi: 10.1111/j.0954-6820.1978.tb08452.x. [DOI] [PubMed] [Google Scholar]
- 5.Sharp H.L. The current status of alpha-1-antityrpsin, a protease inhibitor, in gastrointestinal disease. Gastroenterology. 1976;70:611–621. [PubMed] [Google Scholar]
- 6.Nemeth A., Strandvik B. Liver disease in children with alpha 1-antitrypsin deficiency without neonatal cholestasis. Acta Paediatr. Scand. 1982;71:1001–1005. doi: 10.1111/j.1651-2227.1982.tb09563.x. [DOI] [PubMed] [Google Scholar]
- 7.Dawwas M.F., Davies S.E., Griffiths W.J., Lomas D.A., Alexander G.J. Prevalence and risk factors for liver involvement in individuals with PiZZ-related lung disease. Am. J. Respir. Crit. Care Med. 2013;187:502–508. doi: 10.1164/rccm.201204-0739OC. [DOI] [PubMed] [Google Scholar]
- 8.Lindblad D., Blomenkamp K., Teckman J. Alpha-1-antitrypsin mutant Z protein content in individual hepatocytes correlates with cell death in a mouse model. Hepatology. 2007;46:1228–1235. doi: 10.1002/hep.21822. [DOI] [PubMed] [Google Scholar]
- 9.Ding J., Yannam G.R., Roy-Chowdhury N., Hidvegi T., Basma H., Rennard S.I., Wong R.J., Avsar Y., Guha C., Perlmutter D.H. Spontaneous hepatic repopulation in transgenic mice expressing mutant human α1-antitrypsin by wild-type donor hepatocytes. J. Clin. Invest. 2011;121:1930–1934. doi: 10.1172/JCI45260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman K.R., Burdon J.G., Piitulainen E., Sandhaus R.A., Seersholm N., Stocks J.M., Stoel B.C., Huang L., Yao Z., Edelman J.M., McElvaney N.G., RAPID Trial Study Group Intravenous augmentation treatment and lung density in severe α1 antitrypsin deficiency (RAPID): a randomised, double-blind, placebo-controlled trial. Lancet. 2015;386:360–368. doi: 10.1016/S0140-6736(15)60860-1. [DOI] [PubMed] [Google Scholar]
- 11.McElvaney N.G., Burdon J., Holmes M., Glanville A., Wark P.A., Thompson P.J., Hernandez P., Chlumsky J., Teschler H., Ficker J.H., RAPID Extension Trial Group Long-term efficacy and safety of α1 proteinase inhibitor treatment for emphysema caused by severe α1 antitrypsin deficiency: an open-label extension trial (RAPID-OLE) Lancet Respir. Med. 2017;5:51–60. doi: 10.1016/S2213-2600(16)30430-1. [DOI] [PubMed] [Google Scholar]
- 12.Mueller C., Chulay J.D., Trapnell B.C., Humphries M., Carey B., Sandhaus R.A., McElvaney N.G., Messina L., Tang Q., Rouhani F.N. Human Treg responses allow sustained recombinant adeno-associated virus-mediated transgene expression. J. Clin. Invest. 2013;123:5310–5318. doi: 10.1172/JCI70314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller C., Flotte T.R. Gene-based therapy for alpha-1 antitrypsin deficiency. COPD. 2013;10(Suppl 1):44–49. doi: 10.3109/15412555.2013.764978. [DOI] [PubMed] [Google Scholar]
- 14.Mueller C., Tang Q., Gruntman A., Blomenkamp K., Teckman J., Song L., Zamore P.D., Flotte T.R. Sustained miRNA-mediated knockdown of mutant AAT with simultaneous augmentation of wild-type AAT has minimal effect on global liver miRNA profiles. Mol. Ther. 2012;20:590–600. doi: 10.1038/mt.2011.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flotte T.R., Trapnell B.C., Humphries M., Carey B., Calcedo R., Rouhani F., Campbell-Thompson M., Yachnis A.T., Sandhaus R.A., McElvaney N.G. Phase 2 clinical trial of a recombinant adeno-associated viral vector expressing α1-antitrypsin: interim results. Hum. Gene Ther. 2011;22:1239–1247. doi: 10.1089/hum.2011.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiuchiolo M.J., Kaminsky S.M., Sondhi D., Mancenido D., Hollmann C., Crystal R.G. Phase I/II study of intrapleural administration of a serotype rh.10 replication-deficient adeno-associated virus gene transfer vector expressing the human α1-antitrypsin cDNA to individuals with α1-antitrypsin deficiency. Hum. Gene Ther. Clin. Dev. 2014;25:112–133. doi: 10.1089/humc.2014.2513. [DOI] [PubMed] [Google Scholar]
- 17.Chiuchiolo M.J., Kaminsky S.M., Sondhi D., Hackett N.R., Rosenberg J.B., Frenk E.Z., Hwang Y., Van de Graaf B.G., Hutt J.A., Wang G. Intrapleural administration of an AAVrh.10 vector coding for human α1-antitrypsin for the treatment of α1-antitrypsin deficiency. Hum. Gene Ther. Clin. Dev. 2013;24:161–173. doi: 10.1089/humc.2013.168. [DOI] [PubMed] [Google Scholar]
- 18.Li S., Ling C., Zhong L., Li M., Su Q., He R., Tang Q., Greiner D.L., Shultz L.D., Brehm M.A. Efficient and targeted transduction of nonhuman primate liver with systemically delivered optimized AAV3B vectors. Mol. Ther. 2015;23:1867–1876. doi: 10.1038/mt.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michalopoulos G.K. Liver regeneration. J. Cell. Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piccolo P., Annunziata P., Soria L.R., Attanasio S., Barbato A., Castello R., Carissimo A., Quagliata L., Terracciano L.M., Brunetti-Pierri N. Down-regulation of hepatocyte nuclear factor-4α and defective zonation in livers expressing mutant Z α1-antitrypsin. Hepatology. 2017;66:124–135. doi: 10.1002/hep.29160. [DOI] [PubMed] [Google Scholar]
- 21.Teckman J.H., An J.K., Loethen S., Perlmutter D.H. Fasting in alpha1-antitrypsin deficient liver: constitutive [correction of consultative] activation of autophagy. Am. J. Physiol. Gastrointest. Liver Physiol. 2002;283:G1156–G1165. doi: 10.1152/ajpgi.00041.2002. [DOI] [PubMed] [Google Scholar]
- 22.Shah M., Patel K., Sehgal P.B. Monocrotaline pyrrole-induced endothelial cell megalocytosis involves a Golgi blockade mechanism. Am. J. Physiol. Cell Physiol. 2005;288:C850–C862. doi: 10.1152/ajpcell.00327.2004. [DOI] [PubMed] [Google Scholar]
- 23.Ogasawara J., Watanabe-Fukunaga R., Adachi M., Matsuzawa A., Kasugai T., Kitamura Y., Itoh N., Suda T., Nagata S. Lethal effect of the anti-Fas antibody in mice. Nature. 1993;364:806–809. doi: 10.1038/364806a0. [DOI] [PubMed] [Google Scholar]
- 24.Baligar P., Kochat V., Arindkar S.K., Equbal Z., Mukherjee S., Patel S., Nagarajan P., Mohanty S., Teckman J.H., Mukhopadhyay A. Bone marrow stem cell therapy partially ameliorates pathological consequences in livers of mice expressing mutant human α1-antitrypsin. Hepatology. 2017;65:1319–1335. doi: 10.1002/hep.29027. [DOI] [PubMed] [Google Scholar]
- 25.Biasco L., Scala S., Basso Ricci L., Dionisio F., Baricordi C., Calabria A., Giannelli S., Cieri N., Barzaghi F., Pajno R. In vivo tracking of T cells in humans unveils decade-long survival and activity of genetically modified T memory stem cells. Sci. Transl. Med. 2015;7:273ra13. doi: 10.1126/scitranslmed.3010314. [DOI] [PubMed] [Google Scholar]
- 26.Paulk N.K., Wursthorn K., Wang Z., Finegold M.J., Kay M.A., Grompe M. Adeno-associated virus gene repair corrects a mouse model of hereditary tyrosinemia in vivo. Hepatology. 2010;51:1200–1208. doi: 10.1002/hep.23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strom S.C., Fisher R.A., Thompson M.T., Sanyal A.J., Cole P.E., Ham J.M., Posner M.P. Hepatocyte transplantation as a bridge to orthotopic liver transplantation in terminal liver failure. Transplantation. 1997;63:559–569. doi: 10.1097/00007890-199702270-00014. [DOI] [PubMed] [Google Scholar]
- 29.Ling C., Lu Y., Kalsi J.K., Jayandharan G.R., Li B., Ma W., Cheng B., Gee S.W., McGoogan K.E., Govindasamy L. Human hepatocyte growth factor receptor is a cellular coreceptor for adeno-associated virus serotype 3. Hum. Gene Ther. 2010;21:1741–1747. doi: 10.1089/hum.2010.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azuma H., Paulk N., Ranade A., Dorrell C., Al-Dhalimy M., Ellis E., Strom S., Kay M.A., Finegold M., Grompe M. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat. Biotechnol. 2007;25:903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlson J.A., Rogers B.B., Sifers R.N., Finegold M.J., Clift S.M., DeMayo F.J., Bullock D.W., Woo S.L. Accumulation of PiZ alpha 1-antitrypsin causes liver damage in transgenic mice. J. Clin. Invest. 1989;83:1183–1190. doi: 10.1172/JCI113999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shultz L.D., Lyons B.L., Burzenski L.M., Gott B., Chen X., Chaleff S., Kotb M., Gillies S.D., King M., Mangada J. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.