Abstract
5α-Cyprinol 27-sulfate is the major biliary bile salt present in cypriniform fish including the zebrafish (Danio rerio). The current study was designed to identify the zebrafish cytosolic sulfotransferase (Sult) enzyme(s) capable of sulfating 5α-cyprinol and to characterize the zebrafish 5α-cyprinol-sulfating Sults in comparison with human SULT2A1. Enzymatic assays using zebrafish homogenates showed 5α-cyprinol-sulfating activity. A systematic analysis, using a panel of recombinant zebrafish Sults, revealed two Sult2 subfamily members, Sult2st2 and Sult2st3, as major 5α-cyprinol-sulfating Sults. Both enzymes showed higher activities using 5α-cyprinol as the substrate, compared to their activity with DHEA, a representative substrate for mammalian SULT2 family members, particularly SULT2A1. pH-Dependence and kinetics experiments indicated that the catalytic properties of zebrafish Sult2 family members in mediating the sulfation of 5α-cyprinol were different from those of either zebrafish Sult3st4 or human SULT2A1. Collectively, these results imply that both Sult2st2 and Sult2st3 have evolved to sulfate specifically C27-bile alcohol, 5α-cyprinol, in Cypriniform fish, whereas the enzymatic characteristics of zebrafish Sult3 members, particularly Sult3st4, correlated with those of human SULT2A1.
Keywords: Cytosolic sulfotransferase, SULT, sulfation, zebrafish, 5α-cyprinol
1. Introduction
Sulfation is a key conjugation reaction of bile salts in mammals and the reaction is mainly mediated by a hydroxysteroid sulfotransferase, SULT2A1, using 3′-phosphoadenosine 5′-phosphosulfate (PAPS) [1,2] as a sulfate donor. SULT2A1 is a member of the SULT2 family and is known to catalyze the sulfation of a variety of steroid hormones and bile salts [3,4]. Sulfation of bile acids plays an important role in the elimination and detoxification of bile acids, especially the cytotoxic bile acid lithocholic acid (LCA; 3α-hydroxy-5β-cholan-24-oic acid) [2,5,6]. While bile acids are the major end-metabolites in the biosynthesis of cholanoids (bile acids/alcohols) from cholesterol in mammals, birds, reptiles and some teleost fish, bile alcohols have been also known to be major end-metabolites in jawless, cartilaginous, and cypriniform fish, as well as in amphibians [7,8]. A detailed survey of cholanoids in over 1,000 different vertebrate species has revealed that the family contains a great diversity of chemical structures, ranging from C27-bile alcohols in basal vertebrates to C24-bile acids in higher vertebrates [9–11]. The synthesis of cholanoids requires a long series of enzymes, and is a pathway that has become progressively longer in parallel with the evolution of vertebrates. In most teleost fish, especially ray-finned fish, 5β-C24-bile acids, such as cholic acid and chenodeoxycholic acid, are the main cholanoids, as they are in human bile [10]. However, cypriniform fish including the zebrafish commonly used in scientific research exclusively utilize a 5α-C27-bile alcohol, 5α-cyprinol (3α,7α,12α,26,27-pentahydroxy-5α-cholestane) [10, 12]. Most bile salts are found with the A/B ring junction in the 5β-position. The unusual A/B ring junction in cypriniform fish appears to be the result of a special case where there is a double mutation in the bile acid synthesis pathway. The first mutation is a partial loss of function in the mitochondrial sterol 27-hydroxylase (CYP27A1), an enzyme that normally performs three sequential reactions. It first hydroxylates the sterol side-chain at carbon 27, and then progressively oxidizes this carbon, first to an aldehyde and then to the stereospecific 27R carboxylic acid isomer. Only the first step (hydroxylation at C27) has been observed in Cypriniform fish. The second mutation affects the enzyme Δ4-3-oxosteroid 5β-reductase (AKR1D1). This enzyme removes the double bond found in the 3-oxo-Δ4-structure of ring A in the steroid nucleus, and transforms the flat A/B ring junction into the bent 5β-ring configuration [10]. A previous study has shown that 5α-cyprinol (when sulfated at position C-27) retains physiological properties similar to the common taurine conjugated 5β-bile acids cholic acid and chenodeoxycholic acid [13].
In zebrafish, eighteen distinct Sults, including nine Sult1sts, three Sult2sts, five Sult3sts, and one Sult6, have been identified and characterized to date [14,15]. Previous work has shown that zebrafish Sult2 and Sult3 members are capable of catalyzing the sulfation of both 5β-bile acids and a bile alcohol, 5α-petromyzonol. In general, the catalytic activities of these zebrafish Sults are lower than that of human SUT2A1 [16]. Based on this study, we anticipated that members of the zebrafish Sult2 and Sult3 families would also be active for 5α-cyprinol. The zebrafish Sult2 family has been identified as a counterpart of the human SULT2 family (which includes SULT2A1 and SULT2B1) based on their similarity in amino acid sequence [17,18]. The zebrafish Sult3 family has been identified as an ortholog of mammalian arylamine SULT, a SULT3 family member [19–21]. For the 5β-bile acid previously studied, zebrafish Sult3 family members showed higher activities toward steroids and bile acids than did those of the Sult2 family.
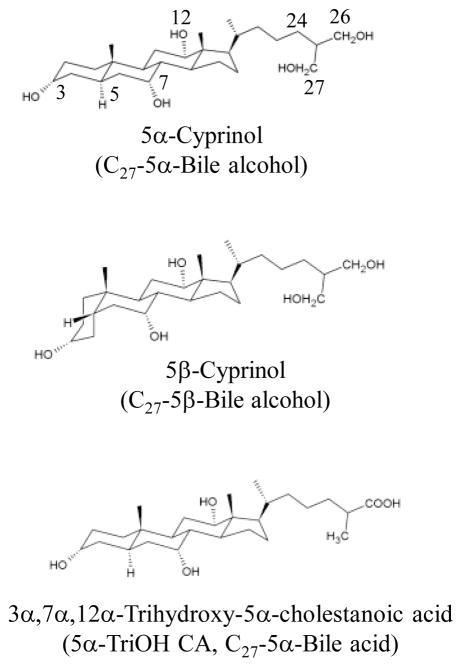
The zebrafish is an excellent model for investigations into the physiological function of sulfation and the SULT enzymes [22,23]. In this paper, we investigated the identities and properties of the zebrafish SULT enzymes involved in the 27-sulfation of the major bile alcohol, 5α-cyprinol, and compare the activities of these enzymes with those of human SULT2A1, using structural isomers of 5α-cyprinol, including 5β-cyprinol and 3α,7α,12α-trihydroxy-5α-cholestanoic acid (5α-triOH CA) (Figure 1). The pH-dependence and kinetics of the sulfation of 5α and 5β-cyprinol by the relevante zebrafish and human SULTs were determined to clarify the effect of C5-configuration of cyprinol on their catalytic activity.
Figure 1.
Stereochemical structures of 5α- and 5β-cyprinol and 3α,7α,12α-Trihydroxy-5α-cholestanoic acid (5α-TriOH CA). 5α-cyprinol and 5α-TriOH CA possess a planar structure due to the 5α-configuration at A/B ring junction, in contrast to 5β-cyprinol which has a bent orientation at the C5 position.
2. Materials and Methods
2.1. Materials
5α-Cyprinol and 5β-cyprinol were generated by deconjugating their sulfate salts purified from carp gallbladder as previously described [24]. 3α,7α,12α-trihydroxy-5α-cholestanoic acid (5α-triOH CA) were prepared as previously described [25]. Dehydroepiandrosterone (DHEA), adenosine 5′-triphosphate (ATP), sodium dodecyl sulfate (SDS), dithiothreitol (DTT), dimethyl sulfoxide (DMSO), sodium acetate, 2-morpholinoethanesulfonic acid (Mes), 3-(N-morpholino)propanesulfonic acid (Mops), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), 3-([1,1-dimethyl-2-hydroxyethyl]amino)-2-hydroxypropanesulfonic acid (Ampso), 3-(cyclohexylamino)-2-hydroxy-1-propanesulfonic acid (Capso), 3-(cyclohexylamino)-1-propanesulfonic acid (Caps) were from Sigma Chemical Company. Carrier-free sodium [35S]sulfate was from American Radiolabeled Chemicals. Silica gel thin-layer chromatography (TLC) plates were from Merck Millipore. All other chemicals were of the highest grade commercially available.
2.2. Preparation of zebrafish homogenate
Adult zebrafish (strain AB) were obtained from the Zebrafish International Resource Center at the University of Oregon (Eugene, OR, USA; P40 RR012546 from NIH-NCRR). The fish were kept in tanks containing buffered water (pH 7.2) at 28 °C, and were fed daily live brine shrimp naupli and Tetramin dried flake food (Tetra, Blacksburg, VA, USA). The day-night cycle was maintained at 14:10 h, and spawning and fertilization were stimulated by the onset of first light. Marbles were used to cover the bottom of the spawning tank to protect newly laid eggs and facilitate their retrieval for study. Fertilized zebrafish embryos were collected from the bottom of the tank by siphoning with a disposable pipette. The eggs were placed in Petri dishes and washed thoroughly with E3 water (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4). 50 zebrafish larvae at 7 dpf were chilled in ice-cold water for 20 min and then homogenized by glass tissue grinder in 50 μl homogenate buffer (20 mM Hepes-NaOH, pH7.5, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 1 mM PMSF, protease inhibitor cocktail). The homogenate was subjected to a centrifuge at 10,000 × g for 20 min and collected supernatant was stored at −80°C.
2.3. Enzymatic assay with zebrafish homogenate
The sulfating activity of zebrafish homogenate was assayed using [35S]PAPS as the sulfonate group donor. [35S]PAPS was synthesized from ATP and carrier-free [35S]sulfate using the recombinant human bifunctional PAPS synthase as previously described [26]. The standard assay mixture, in a final volume of 20 μl, contained 50 mM Mops buffer at pH 7.0, 14 μM [35S]PAPS (15 Ci/mmol), 1 mM DTT, 50 mM NaF, and 10 μM substrate, 5α-cyprinol. The substrates tested were prepared in DMSO. The reaction was started by the addition of 2.0 μg homogenate, allowed to proceed for 15 min at 28°C and terminated by heating at 100°C for 3 min. The precipitates formed were cleared by centrifugation at 16,000 × g for 5 min, and the supernatant was subjected to the analysis of [35S]sulfated product by TLC with ethyl acetate/acetonitrile/n-butanol/isopropanol/formic acid (2:2:2:4:1; by volume) as the solvent system. Afterwards, the TLC plate was dried and the radioactive spots located by autoradiography, excised from the plate, and eluted in 0.5 ml H2O in a glass vial. 4.5 ml of Ecolume scintillation liquid was then added to each vial and counted for the radioactivity of sulfur-35 using a liquid scintillation counter. The cpm count obtained was calculated into pmol of sulfated product formed based on the specific activity of the [35S]PAPS prepared for the corresponding reactions and expressed in pmol of sulfated product/min/mg protein. Kinetic assay was performed using varying concentrations of 5α-cyprinol under the above-mentioned condition and the parameters were calculated from substrate inhibition kinetics using GraphPad Prism5 software and non-linear regression.
2.4. Enzymatic assay with purified recombinant SULTs
The sulfating activity of purified recombinant SULT enzymes was assayed using [35S]PAPS as sulfonate group donor. Human SULT2A1 and eighteen zebrafish Sult enzymes were expressed using pGEX-2TK or pMALc5x prokaryotic expression and purified as previously described [17–21, 27]. The enzymatic assay was performed under the following conditions. The standard assay mixture, in a final volume of 20 μl, contained 50 mM Mops buffer at pH 7.0, 14 μM [35S]PAPS (15 Ci/mmol), 1 mM DTT and 50 μM substrate. This substrate concentration (50 μM) was selected for screening the cyprinol-sulfating activity of the zebrafish Sults since it has been successfully used for the identification of the bile acid-sulfating zebrafish Sults in our previous study [16]. The reaction was started by the addition of 1.0 μg enzyme, allowed to proceed for 10 min at 37°C for human SULT and at 28°C for zebrafish Sults and terminated by heating at 100°C for 3 min. TLC analysis of sulfated product was carried out under the above mentioned procedure. The pH-dependent assays were performed with different pH buffers (50 mM sodium acetate at 4.5 or 5.5; Mes at 5.5, 6.0, or 6.5; Mops at 6.5 or 7.0; Hepes at 7.0, 7.5, or 8.0; Ampso at 8.0, 8.5 or 9.0; Capso 9.0, 9.5, or 10.0; and Caps at 10.0, 10.5, 11.0). For the kinetic studies on the sulfation of 5α-cyprinol and 5β-cyprinol by the relevant Sults, enzyme assays were performed with varying concentrations of the substrates and 50 mM Mops at pH 7.0. Kinetic parameters were calculated from the Michaelis-Menten and substrate inhibition equation using GraphPad Prism5 software and non-linear regression.
2.5. Mass spectrometry (MS) and nuclear magnetic resonance (NMR) analyses of sulfated 5α-cyprinol
Enzymatic sulfation of 5α-cyprinol was performed with zebrafish Sult2st3 and nonradioactive PAPS as the sulfate donor based on the above-mentioned procedure. The final reaction mixture was applied to a Waters Sep-Pak C18 cartridge. The bound 5α-cyprinol and its sulfated metabolite were eluted using 80% methanol, and the eluate was dried using a SpeedVac concentrator. The eluate reconstituted into 50% methanol/1% formic acid was analyzed by direct-infusion electrospray ion-trap mass spectrometry in the negative ion mode using a Thermo-Finnigan LCQ DECA XP Plus mass spectrometer. Ionization conditions employed were: spray voltage, 5 kV; sheath gas flow rate, 40; capillary temperature, 315 °C, and the infusion flow rate, 5 μL/min. For NMR analysis, the eluate from Waters Sep-Pak C18 cartridge was further loaded into Oasis WAX column. The bound 5α-cyprino sulfate was eluted into 80% methanol/1% NH4OH and dried using a SpeedVac concentrator. The isolated sample was dissolved in CD3OD, respectively, (Aldrich) and analyzed using a Bruker Advance 600 instrument (600 MHz) (Bruker) with 1H-NMR, 1H-1H COSY, and HSQC modes. The data obtained are summarized in Table S1.
3. Results
3.1. Sulfation of 5α-cyprinol by zebrafish homogenate
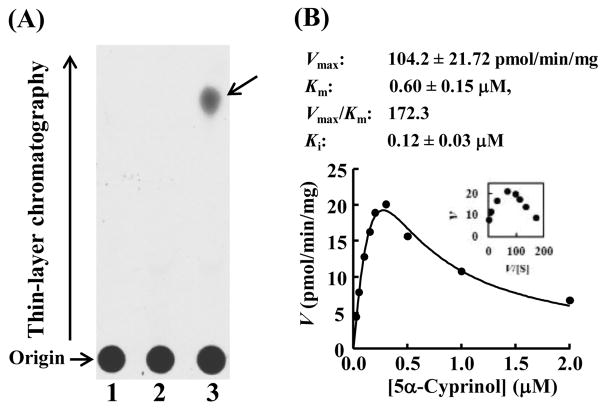
Zebrafish homogenate at 7 dpf was used to detect the in vivo 5α-cyprinol-sulfating activity, performed with 35S-labeled PAPS. As shown in Figure 2, zebrafish homogenate showed the sulfating activity toward 5α-cyprinol. The saturation curve analysis with 5α-cyprinol as a substrate exhibited the substrate inhibition kinetics for 5α-cyprinol sulfation by zebrafish homogenate (Figure 2 (B)), with Km and Vmax/Km values of 0.60 μM and 172.3 pmol/min/mg/μM, respectively. These results indicated clearly the capacity of zebrafish homogenate in sulfating 5α-cyprinol.
Figure 2.
Zebrafish homogenate exhibits a significant 5α-cyprinol-sulfating activity. (A) Enzymatic sulfation of cyprinol was performed with 7-dpf zebrafish homogenate under assay condition as described in Materials and Methods. The figure shows the autoradiograph of the TLC analysis for the reaction product without substrate (lane 1), without homogenate (lane 2), or with substrate and homogenate (lane 3). (B) Kinetic analysis for the sulfation of 5α-Cyprinol by zebrafish homogenate. The fitted curve was generated using substrate inhibition kinetics. An Eadie-Hofstee plot is inserted in the fitting curve. Kinetic parameters compiled in this figure were calculated based on the substrate inhibition kinetics. The data are calculated mean ± SD derived from three experiments.
3.2. Sulfation of 5α-cyprinol and its structural analogs by the zebrafish Sults and human SULT2A1
Systematic enzyme assays were carried out in order to identify the zebrafish Sult enzyme(s) responsible for the sulfation of 5α-cyprinol and investigate the structure-activity relationship of the sulfation reaction using 5β-cyprinol, 5α-triOH CA, and DHEA. Human SULT2A1 was also used in the enzymatic assay with those four substrates for comparison. All members of zebrafish Sult2 and Sult3 families showed activity toward 5α-cyprinol (Table 1). Among them, Sult2st2 and Sult2st3 were the most active, compared to the other zebrafish Sults. Although both Sult2st2 and Sult2st3 effectively sulfated both 5α-cyprinol and 5β-cyprinol (two isomers with opposite ring A/B junctions), neither were active in sulfating 5α-triOH CA. The activity of both enzymes using DHEA as substrate was much less than that shown for 5α- or 5β-cyprinol. In contrast to Sult2st2 and Sult2st3, zebrafish Sult3 enzymes showed only weak activities toward the 5α-cyprinol, 5β-cyprinol, and 5α-triOH CA. In contrast to the Sult2 enzymes, zebrafish Sult3 enzymes were quite active with DHEA as a substrate. Human SULT2A1, on the other hand, exhibited moderate activity with 5α-cyprinol and 5α-triOH as substrates, and a much higher activity with DHEA as substrate. Human SULT2A1 was more active with 5α-cyprinol than with 5β-cyprinol, indicating that SULT2A1 is readily capable of catalyzing the sulfation of 5α-configured bile salts, with structures not normally encountered in human bile.
Table 1.
Specific activities of the zebrafish Sults and human SULTs with 5α-cyprinol and its structural analogs as substratesa
| Enzymes | Specific activity (nmol/min/mg) | |||
|---|---|---|---|---|
|
| ||||
| 5α-cyprinol | 5β-cyprinol | 5α-triOH CA | DHEA | |
| zfSult2st1 | 1.60 ± 0.07 | 1.52 ± 0.10 | N.D.b | 1.45 ± 0.04 |
| zfSult2st2 | 10.14 ± 0.68 | 10.08 ± 0.06 | N.D. | 0.90 ± 0.01 |
| zfSult2st3 | 27.86 ± 0.20 | 26.42 ± 0.76 | N.D. | 0.11 ± 0.01 |
| zfSult3st1 | 0.64 ± 0.03 | 0.04 ± 0.01 | N.D. | 0.34 ± 0.01 |
| zfSult3st2 | 0.77 ± 0.06 | 0.35 ± 0.03 | 0.14 ± 0.01 | 16.72 ± 0.12 |
| zfSult3st3 | 0.44 ± 0.03 | 0.22 ± 0.01 | 0.07 ± 0.01 | 12.53 ± 0.05 |
| zfSult3st4 | 1.43 ± 0.03 | 1.15 ± 0.04 | 0.26 ± 0.01 | 26.88 ± 0.21 |
| zfSult3st5 | 0.68 ± 0.07 | 0.60 ± 0.01 | 0.09 ± 0.01 | 5.35 ± 0.01 |
| hSULT2A1 | 4.71 ± 0.20 | 0.34 ± 0.01 | 1.94 ± 0.04 | 13.94 ± 0.47 |
Specific activity corresponds to nmol substrate sulfated/min/mg purified enzyme. Results shown represent mean ± standard deviation derived from three separate assays.
ND refers to activity not detected. The detection limit of this assay was estimated to be ~0.01 nmol/min/mg protein.
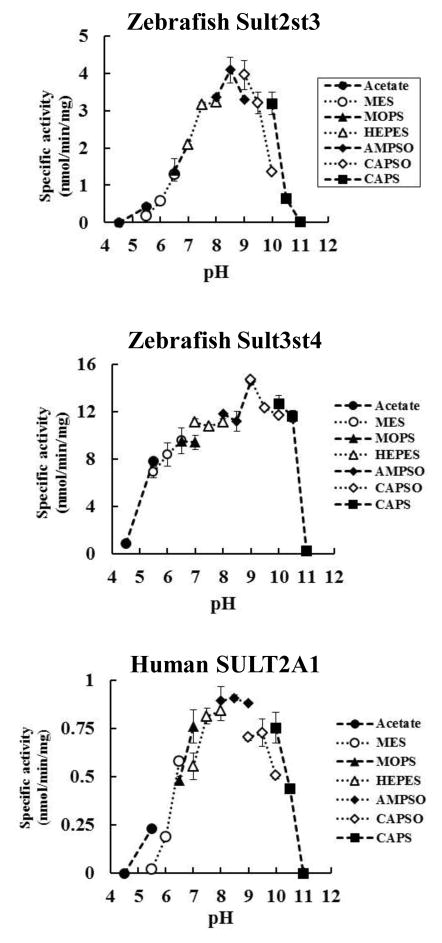
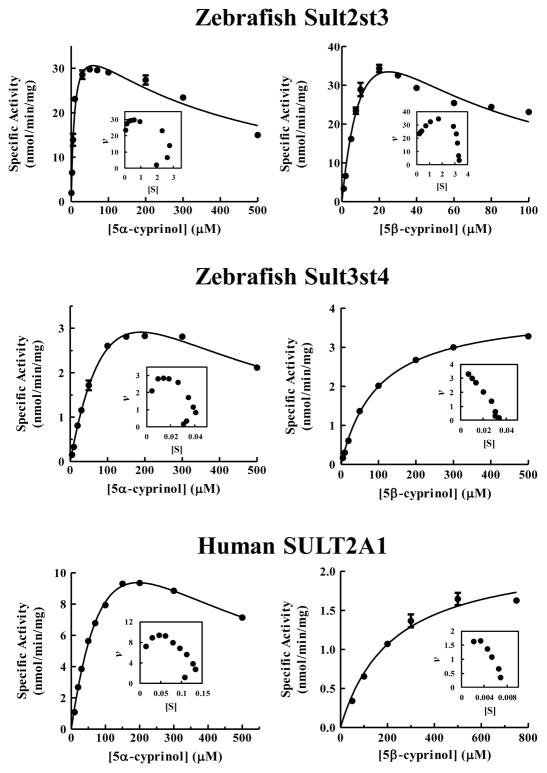
3.3 pH-Dependence and kinetic properties of the sulfation of 5α-cyprinol by zebrafish Sults and human SULT2A1
To further characterize the sulfation of 5α-cyprinol by the relevant zebrafish Sults and human SULT2A1, pH-dependence and kinetic properties were analyzed. Zebrafish Sult2st3 and Sult3st4 and human SULT2A1 were tested in the pH-dependent assay. As shown in Figure 3, zebrafish Sult2st3 was active over a pH range from 6.0 to 10.5, with a distinct optimal activity detected at pH 8.5. In contrast, zebrafish Sult3st4 exhibited a broad pH optimum spanning 6.0–10.5 with an optimal activity at pH 9.5. Similar to zebrafish Sult3st4, human SULT2A1 exhibited a broad pH optimum spanning 7.5–9.5. Zebrafish Sult2st1, Sult2st2, Sult2st3, and Sult3st4 and human SULT2A1 were analyzed in the kinetics study. To investigate the effect of cyprinol’s A/B ring junction on the kinetic properties, 5β-cyprinol was also used as a substrate. Substrate saturation curves of zebrafish Sult2st3 and Sult3st4 and human SULT2A1 are shown in Figure 4. The saturation curve analysis demonstrated that the sulfation of both of 5α-cyprinol and 5β-cyprinol by zebrafish Sult2st2 and Sult2st3 could be fitted to substrate inhibition kinetics, which were further confirmed by a linear Eadie-Hofstee plot (Figure 4). In contrast to these two zebrafish Sults, zebrafish Sult2st1 showed Michaelis–Menten kinetics for the sulfation of both 5α-cyprinol and 5β-cyprinol (data not shown). Zebrafish Sult3st4 and human SULT2A1 showed substrate inhibition kinetics for 5α-cyprinol and Michaelis–Menten kinetics for 5β-cyprinol (Figure 4). Table 2 shows the kinetic constants determined for the sulfation of 5α-cyprinol and 5β-cyprinol by the five enzymes tested. The Km values of zebrafish Sult2st1, Sult2st2, and Sult2st3 were much lower than those of human SULT2A1 and zebrafish Sult3st4. Furthermore, the catalytic efficiencies of all members of zebrafish Sult2 subfamily with 5α-cyprinol were much higher than those of human SULT2A1 and zebrafish Sult3st4 based on the calculated Vmax/Km values. The kinetic parameters of zebrafish Sult2 members with 5α-cyprinol as a substrate appeared to be comparable to those with 5β-cyprinol, except for the Ki values. These results suggest that zebrafish Sult2 members are the primary enzymes responsible for the sulfation of 5α-cyprinol and the human orthologous isoform, SULT2A1, is far less efficient in sulfating 5α-cyprinol.
Figure 3.
Zebrafish Sult2st3, zebrafish Sult3st4, and human SULT2A1 exhibit differential pH profiles in the sulfation of 5α-cyprinol. The enzymatic assays were performed using different pH-buffer systems as indicated under standard assay conditions as described in Materials and Methods. The data are calculated mean ± SD derived from three experiments.
Figure 4.
Zebrafish Sult2st3, but not zebrafish Sult3st4 and human SULT2A1, displays similar kinetics in the sulfation of 5α- and 5β-cyprinol. Figures in the left panels show the kinetic assay with 5α-cyprinol and in the right panels the kinetics with 5β-cyprinol. The fitting curves were generated using Michaelis-Menten or substrate inhibition kinetics. Eadie-Hofstee plots are inserted under each fitting curve. The data are calculated mean ± SD derived from three experiments.
Table 2.
Kinetic constant of the sulfation of cyprinol by and the relevant zebrafish Sults and human SULT 2A1a
| Vmax (nmol/min/mg) | Km (μM) | Vmax/Km | Ki (μM) | |
|---|---|---|---|---|
| zfSult2st1 | ||||
| 5α | 1.57 ± 0.05 | 1.56 ± 0.24 | 1.00 | - |
| 5β | 1.78 ± 0.05 | 1.38 ± 0.19 | 1.29 | - |
| zfSult2st2 | ||||
| 5α | 24.73 ± 5.40 | 7.73 ± 2.67 | 3.20 | 27.43 ± 11.16 |
| 5β | 22.84 ± 1.10 | 7.15 ± 0.67 | 3.19 | 58.15 ± 5.74 |
| zfSult2st3 | ||||
| 5α | 40.34 ± 1.31 | 9.47 ± 0.86 | 4.26 | 373.9 ± 38.0 |
| 5β | 75.57 ± 7.71 | 15.77 ± 2.48 | 4.79 | 39.78 ± 6.70 |
| zfSult3st4 | ||||
| 5α | 9.59 ± 0.89 | 217.0 ± 26.0 | 0.04 | 164.9 ± 22.2 |
| 5β | 3.97 ± 0.05 | 98.9 ± 2.89 | 0.04 | - |
| hSULT2A1 | ||||
| 5α | 26.73 ± 1.61 | 181.0 ± 14.6 | 0.15 | 210.8 ± 19.9 |
| 5β | 2.24 ± 0.09 | 221.3 ± 23.1 | 0.01 | - |
Results represent means ± SD derived from three determinations.
3.4. Structure of 5α-cyprinol sulfated product
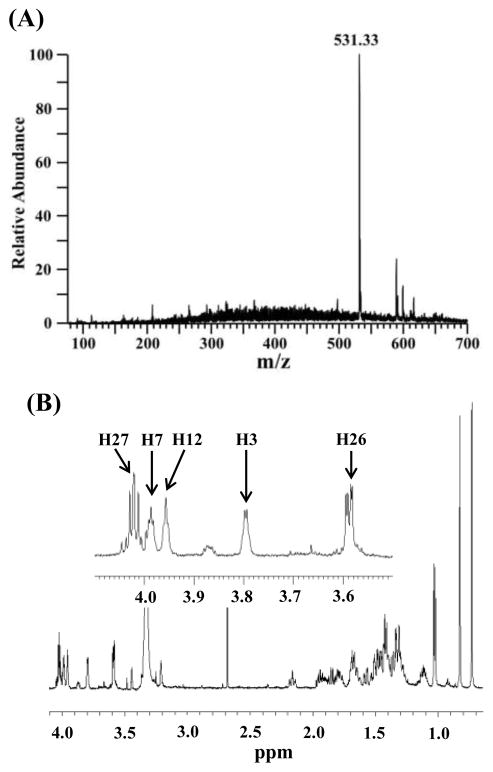
Following the sulfation of 5α-cyprinol with the zebrafish enzymes, the structure of the sulfated product was examined using mass spectrometry and NMR. Mass spectrometry analysis of the reaction product with Sult2st3 showed a prominent ion at m/z 531.33, equivalent to a deprotonated mono-sulfated pentahydroxy C27 bile alcohol ion (Figure 5A). Further MS/MS analysis was performed but showed no fragmentation (data not shown). The 1H-NMR spectrum (600 MHz, CD3OD) for the sulfated 5α-cyprinol clearly showed the five types of protons, at 3α-OH-bearing carbon (3.79 ppm), 7α-OH-bearing carbon (3.98 ppm), 12α-OH-bearing carbon (3.93 ppm), 26-OH-bearing carbon (3.59 ppm), and 27-O-sulfate-bearing carbon (4.02 ppm) (Figure 5B). All assigned protons were supported by H1-H1 COSY and H1-C13 HSQC analyses (Figure S1 and Table S1). The spectrum obtained in this study was similar to that of the NMR data previously reported for 5α–cyprinol 27-O-sulfate [28].
Figure 5.
Sulfation of 5α-cyprinol by zebrafish Sult2st3 occurs specifically at the 27-hydroxyl group. (A) Negative mode ESI-Mass spectrometry profile of sulfated 5α-cyprinol. (B) 1H-NMR spectrum of the sulfated 5α-cyprinol. Assigned protons, H3, H7, H12, H26, H27 are referred to the 5α-cyprinol in Figure 1.
4. Discussion
Bile salts, including bile acids and bile alcohols, is known to vary in the structure among different vertebrates. While many species of teleost fish utilize taurine conjugated C24-5β-bile acids, zebrafish uniquely synthesize a C27-5α-bile alcohol, 5α-cyprinol, present in bile as the sulfate, 5α-cyprinol 27-O-sulfate [10,12]. The primitive nature of this bile alcohol is an unusual finding, given even the earliest teleost fish contain a bile acid synthetic pathway capable of converting cholesterol into an advanced C24 bile acid like cholic acid [29,30]. Previously, it was shown that some human SULTs, particularly SULT2A1, are capable of sulfating C24-5β-bile acids, while the relevant zebrafish Sults are less efficient at sulfating C24-5β-bile acids [16]. In this study, zebrafish larva homogenate actively sulfated 5α-cyprinol with a low Km value (Figure 2). This suggests that zebrafish Sults are evolutionally better suited for sulfating bile salt structures with the A/B ring junctions in the flat trans position.
The current study also identified the zebrafish Sult enzyme(s) responsible for the sulfation of 5α-cyprinol and compared the catalytic properties of these enzymes with SULT2A1, the enzyme responsible for the sulfation of bile acids in humans. Of the eight 5α-cyprinol-sulfating zebrafish identified, Sult2st2 (Km 7.73 μM) and Sult2st3 (Km 9.47 μM) had the highest activity with 5α-cyprinol as substrate (Figure 4 and Table 2). The Km value (0.60 μM) determined for the homogenate, however, was much lower than either Sult2st2 or Sult2st3 and closer in value to Sult2st1 (1.56 μM). Previous ontogenic expression analysis has shown that both Sult2st2 and Sult2st3 were fully expressed at 1-week post fertilization and that their expression continued into the adult stage [18]. These observations, therefore, suggested that either or both Sult2st2 and Sult2st3 (but not Sult2st1) are the major sulfating enzymes for 5α-cyprinol.
Interestingly, both Sult2st2 and Sult2st3 apparently did not distinguish between 5α-cyprinol and 5β-cyprinol, two bile salts with opposite A/B ring junctions (Table 1 and 2). Zebrafish Sult2 family members are expected to be involved in the sulfation of hydroxysteroids like DHEA and corticosterone [18]. However, the specific activities of zebrafish Sult2 members toward hydroxysteroids are much lower (and the Km values much larger) than those with cyprinol. Significantly, none of the zebrafish Sult2 members examined were capable of sulfating C24-5β-bile acids, including chenodeoxycholic acid and lithocholic acid [16], two bile salts not encountered in zebrafish bile. The enzymatic properties of Sult2st2 and Sult2st3 match the profile of bile salts in the zebrafish, and suggest that their primary function in the fish is to sulfate 5α-cyprinol. Our observations of the effective DHEA-sulfating activities of zebrafish Sult3 members are consistent with previous studies showing high catalytic activity of zebrafish Sult3 members for 3β-hydroxysteroids but not for C24-5β-bile acids [16, 19–21]. Zebrafish Sult3 members are relatively comparable in their substrate preferences to human SULT2A1, being capable of sulfating C24-5β-bile acids [16]. However, it should be pointed out that a previous study showed that Sult3st1 exhibited specific sulfating activity toward ursodeoxycholic acid [16], while Sult3st1 displayed low sulfating activities toward other C24-5β-bile acids, cyprinol, and DHEA. Amino acid sequence analysis indicated that zebrafish Sult3st1 shares relatively low amino acid sequence identities (50–55%) with other zebrafish Sult3 family members and can in fact be classified into a separate subfamily [19–21], which may underscore the unique enzymatic property of Sult3st1. Zebrafish Sult3 members are also expected to be similar to mammalian SULT3 family members (based on amino acid sequence homologies), which are capable of catalyzing arylamine compounds [19–21]. All mammalian SULT enzymes have in common three flexible loops which form the substrate-binding pocket and two of the loops are highly variable in amino acid sequence, implying the role of these loops in substrate selectivity and specificity [31]. Crystal structure analysis of human SULT2A1 have shown that these three flexible loops (short loop-1 in SULT2A1) and a loop between 2nd-β-sheet and 1st-α-helix (Pro14-Ser20) are involved in the recognition of a representative substrate, DHEA [32,33]. The three dimensional structures of zebrafish Sults are not yet available, but amino acid sequence alignments show significant differences within the four loops, for zebrafish Sult2 members, human SULT2A1, zebrafish Sult3 members, and mouse SULT3A1 (Figure 6).
Figure 6.
Substrate recognition sites of zebrafish Sult2 and Sult3 family members differ significantly from those of human SULT2A1 and mouse Sult3A1. Alignment of amino acid sequences of members of SULT2 and SULT3 family were performed with the Clustal Omega tool. Identical and similar residues conserved among the enzymes analyzed are drawn in black and gray. Four loop structures involved in the substrate recognition are shown in the figure.
A previous study has reported that Sult2st3 showed the strongest activity toward corticosterone among hydroxysteroids tested [18]. It appears that the catalytic pocket of Sult2st3 is wide enough to accept structurally dissimilar sterol side-chains, for example, C21 (in corticosterone) and C27 (as in α- and β-cyprinol), a flexibility that may have led to the subsequent evolutionary selection of this enzyme for the sulfation of 5α-cyprinol.
In our assay, a small amount of 5α-cyprinol was sulfated using human SULT2A1. This enzyme is known to be capable of sulfating both the 3α- and 3β-hydroxyl groups of C24 5β-bile acids [2,16], although the catalytic activity of human SULT2A1 toward 5β-bile acids, such as lithocholic acid, is much higher than what we observed with 5α-cyprinol. In addition, the enzyme has been implicated in the sulfation of 24-hydroxycholesterol at C24 [34], and the steroid drug tibolone (7α-methylnoretynodrel) at C21 [35]. We did not examine the position of the sulfate in α-cyprinol after the reaction with human SULT2A1, but the mixed ability of SULT2A1 to sulfate numerous positions on the steroid skeleton leaves enough uncertainty to believe that the sulfate could be found on either C3 or C27.
5. Conclusion
The present study has revealed Sult2st2 and Sult2st3 as the major 5α-cyprinol-sulfating Sults in zebrafish that sulfate 5α-cyprinol at C27 position. Whereas zebrafish Sult2 family members are active in the sulfation of bile alcohols, Sult3 family members sulfate 3β-hydroxysteroids such as pregnenolone and DHEA. Although the catalytic properties of zebrafish Sult3 family members are not the same as those of mammalian SULT3 enzymes, they are relatively comparable to that of human SULT2A1.
Supplementary Material
Highlights.
Zebrafish homogenates are capable of sulfating 5α-cyprinol with a high affinity.
Zebrafish Sult2 and Sult3 members exhibit strong 5α-cyprinol-sulfating activities.
Kinetic studies indicate that zebrafish Sult2 members are more catalytically efficient toward cyprinol than zebrafish Sult3 members.
The catalytic property of human SULT2A1 is comparable to that of zebrafish Sult3 members, but not zebrafish Sult2 members.
Acknowledgments
Funding: This work was supported in part by a National Institutes of Health grant R03HD071146. M.D.K. received support from K08-GM074238 from the National Institutes of Health.
Abbreviations
- SULT
cytosolic sulfotransferase
- PAPS
3′-phosphoadenosine-5′-phosphosulfate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alnouti Y. Bile Acid sulfation: a pathway of bile acid elimination and detoxification. Toxicol Sci. 2009;108:225–246. doi: 10.1093/toxsci/kfn268. [DOI] [PubMed] [Google Scholar]
- 2.Hofmann AF. Detoxification of lithocholic acid, a toxic bile acid: relevance to drug hepatotoxicity. Drug Metab Rev. 2004;36:703–722. doi: 10.1081/dmr-200033475. [DOI] [PubMed] [Google Scholar]
- 3.Radominska A, Comer KA, Zimniak P, Falany J, Iscan M, Falany CN. Human liver steroid sulphotransferase sulphates bile acids. Biochem J. 1990;272:597–604. doi: 10.1042/bj2720597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otterness DM, Wieben ED, Wood TC, Watson RWG, Madden BJ, Mccormick DJ, Weinshilboum RM. Human liver dehydroepiandrosterone sulfotransferase: molecular cloning and expression of cDNA. Mol Pharmacol. 1992;41:865–872. [PubMed] [Google Scholar]
- 5.Kitada H, Miyata M, Nakamura T, Tozawa A, Honma W, Shimada M, Nagata K, Sinal CJ, Guo GL, Gonzalez FJ, Yamazoe Y. Protective role of hydroxysteroid sulfotransferase in lithocholic acid-induced liver toxicity. J Biol Chem. 2003;278:17838–17844. doi: 10.1074/jbc.M210634200. [DOI] [PubMed] [Google Scholar]
- 6.Saini SP, Sonoda J, Xu L, Toma D, Uppal H, Mu Y, Ren S, Moore DD, Evans RM, Xie W. A novel constitutive androstane receptor-mediated and CYP3A-independent pathway of bile acid detoxification. Mol Pharmacol. 2004;65:292–300. doi: 10.1124/mol.65.2.292. [DOI] [PubMed] [Google Scholar]
- 7.Haslewood GA. Bile salt evolution. J Lipid Res. 1967;8:535–550. [PubMed] [Google Scholar]
- 8.Hofmann AF, Hagey LR, Krasowski MD. Bile salts of vertebrates: structural variation and possible evolutionary significance. J Lipid Res. 2010;51:226–246. doi: 10.1194/jlr.R000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagey LR, Vidal N, Hofmann AF, Krasowski MD. Evolutionary diversity of bile salts in reptiles and mammals, including analysis of ancient human and extinct giant ground sloth coprolites. BMC Evol Biol. 2010;10:133. doi: 10.1186/1471-2148-10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagey LR, Møller PR, Hofmann AF, Krasowski MD. Diversity of bile salts in fish and amphibians: evolution of a complex biochemical pathway. Physiol Biochem Zool. 2010;83:308–321. doi: 10.1086/649966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagey LR, Vidal N, Hofmann AF, Krasowski MD. Complex evolution of bile salts in birds. Auk. 2010;127:820–831. doi: 10.1525/auk.2010.09155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reschly EJ, Ai N, Ekins S, Welsh WJ, Hagey LR, Hofmann AF, Krasowski MD. Evolution of the bile salt nuclear receptor FXR in vertebrates. J Lipid Res. 2008;49:1577–1587. doi: 10.1194/jlr.M800138-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goto T, Holzinger F, Hagey LR, Cerrè C, Ton-Nu HT, Schteingart CD, Steinbach JH, Shneider BL, Hofmann AF. Physicochemical and physiological properties of 5alpha-cyprinol sulfate, the toxic bile salt of cyprinid fish. J Lipid Res. 2003;44:1643–1651. doi: 10.1194/jlr.M300155-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Liu TA, Bhuiyan S, Liu MY, Sugahara T, Sakakibara Y, Suiko M, Yasuda S, Kakuta Y, Kimura M, Williams FE, Liu MC. Zebrafish as a model for the study of the phase II cytosolic sulfotransferases. Curr Drug Metab. 2010;11:538–546. doi: 10.2174/138920010791636158. [DOI] [PubMed] [Google Scholar]
- 15.Kurogi K, Liu TA, Sakakibara Y, Suiko M, Liu MC. The use of zebrafish as a model system for investigating the role of the SULTs in the metabolism of endogenous compounds and xenobiotics. Drug Metab Rev. 2013;45:431–440. doi: 10.3109/03602532.2013.835629. [DOI] [PubMed] [Google Scholar]
- 16.Kurogi K, Krasowski MD, Injeti E, Liu MY, Williams FE, Sakakibara Y, Suiko M, Liu MC. A comparative study of the sulfation of bile acids and a bile alcohol by the Zebra danio (Danio rerio) and human cytosolic sulfotransferases (SULTs) J Steroid Biochem Mol Biol. 2011;127:307–314. doi: 10.1016/j.jsbmb.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugahara T, Yang YS, Liu CC, Pai TG, Liu MC. Sulphonation of dehydroepiandrosterone and neurosteroid: molecular cloning, expression, and functional characterization of a novel zebrafish SULT2 cytosolic sulphotransferase. Biochem J. 2003;375:785–791. doi: 10.1042/BJ20031050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasuda S, Liu MY, Yang YS, Snow R, Takahashi S, Liu MC. Identification of novel hydroxysteroid-sulfating cytosolic SULTs, SULT2 ST2 and SULT2 ST3, from zebrafish: cloning, expression, characterization, and developmental expression. Arch Biochem Biophys. 2006;455:1–9. doi: 10.1016/j.abb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Yasuda T, Yasuda S, Williams FE, Liu MY, Sakakibara Y, Bhuiyan S, Snow R, Carter G, Liu MC. Characterization and ontogenic study of novel steroid-sufating SULT3 sulfotransferases from zebrafish. Mol Cell Endocrinol. 2008;294:29–36. doi: 10.1016/j.mce.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Yasuda S, Burgess M, Yasuda T, Liu MY, Bhuiyan S, Williams FE, Kurogi K, Sakakibara Y, Suiko M, Liu MC. A novel hydroxysteroid-sulfating cytosolic sulfotransferase, SULT3 ST3, from zebrafish: identification, characterization, and ontogenic study. Drug Metab Lett. 2009;3:217–227. doi: 10.2174/187231209790218154. [DOI] [PubMed] [Google Scholar]
- 21.Mohammed YI, Kurogi K, Shaban AA, Xu Z, Liu MY, Williams FE, Sakakibara Y, Suiko M, Bhuiyan S, Liu MC. Identification and characterization of zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5. Aquat Toxicol. 2012;112–113:11–18. doi: 10.1016/j.aquatox.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Z, Williams FE, Liu MC. Developmental toxicity of dextromethorphan in zebrafish embryos/larvae. J Appl Toxicol. 2011;31:157–163. doi: 10.1002/jat.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crittenden F, Thomas HR, Parant JM, Falany CN. Activity Suppression Behavior Phenotype in SULT4A1 Frameshift Mutant Zebrafish. Drug Metab Dispos. 2015;43:1037–1044. doi: 10.1124/dmd.115.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goto T, Holzinger F, Hagey LR, Cerrè C, Ton-Nu HT, Schteingart CD, Steinbach JH, Shneider BL, Hofmann AF. Physicochemical and physiological properties of 5alpha-cyprinol sulfate, the toxic bile salt of cyprinid fish. J Lipid Res. 2003;44:1643–1651. doi: 10.1194/jlr.M300155-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Ogawa S, Mitamura K, Ikegawa S, Krasowski MD, Hagey LR, Hofmann AF, Iida T. Chemical synthesis of the (25R)- and (25S)-epimers of 3α,7α,12α-trihydroxy-5α-cholestan-27-oic acid as well as their corresponding glycine and taurine conjugates. Chem Phys Lipids. 2011;164:368–377. doi: 10.1016/j.chemphyslip.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Yanagisawa K, Sakakibara Y, Suiko M, Takami Y, Nakayama T, Nakajima H, Takayanagi K, Natori Y, Liu MC. cDNA cloning, expression, and characterization of the human bifunctional ATP sulfurylase/adenosine 5′-phosphosulfate kinase enzyme. Biosci Biotechnol Biochem. 1998;62:1037–1040. doi: 10.1271/bbb.62.1037. [DOI] [PubMed] [Google Scholar]
- 27.Suiko M, Sakakibara Y, Liu MC. Sulfation of environmental estrogen-like chemicals by human cytosolic sulfotransferases. Biochem Biophys Res Commun. 2000;267:80–84. doi: 10.1006/bbrc.1999.1935. [DOI] [PubMed] [Google Scholar]
- 28.Asakawa M, Noguchi T, Seto H, Furihata K, Fujikura K, Hashimoto K. Structure of the toxin isolated from carp (Cyprinus carpio) bile. Toxicon. 1990;28:1063–1069. doi: 10.1016/0041-0101(90)90144-v. [DOI] [PubMed] [Google Scholar]
- 29.Tokarz J, Möller G, de Angelis MH, Adamski J. Zebrafish and steroids: what do we know and what do we need to know? J Steroid Biochem Mol Biol. 2013;137:165–173. doi: 10.1016/j.jsbmb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Tokarz J, Möller G, Angelis MH, Adamski J. Steroids in teleost fishes: A functional point of view. Steroids. 2015;103:123–144. doi: 10.1016/j.steroids.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 31.Tibbs ZE, Rohn-Glowacki KJ, Crittenden F, Guidry AL, Falany CN. Structural plasticity in the human cytosolic sulfotransferase dimer and its role in substrate selectivity and catalysis. Drug Metab Pharmacokinet. 2015;30:3–20. doi: 10.1016/j.dmpk.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Pedersen LC, Petrotchenko EV, Negishi M. Crystal structure of SULT2A3, human hydroxysteroid sulfotransferase. FEBS letters. 2000;475:61–64. doi: 10.1016/s0014-5793(00)01479-4. [DOI] [PubMed] [Google Scholar]
- 33.Rehse PH, Ming Z, Sheng-Xiang L. Crystal structure of human dehydroepiandrosterone sulphotransferase in complex with substrate. Biochem J. 2002;364:165–171. doi: 10.1042/bj3640165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cook IT, Duniec-Dmuchowski Z, Kocarek TA, Runge-Morris M, Falany CN. 24-hydroxycholesterol sulfation by human cytosolic sulfotransferases: formation of monosulfates and disulfates, molecular modeling, sulfatase sensitivity, and inhibition of liver x receptor activation. Drug Metab Dispos. 2009;37:2069–2078. doi: 10.1124/dmd.108.025759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falany JL, Macrina N, Falany CN. Sulfation of tibolone and tibolone metabolites by expressed human cytosolic sulfotransferases. J Steroid Biochem Mol Biol. 2004;88:383–391. doi: 10.1016/j.jsbmb.2004.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.