Abstract
Human cytosolic sulfotransferase 1A1 (SULT1A1) is considered to be one of the most important SULT isoforms for metabolism, detoxification, and carcinogenesis. This theory is driven by observations that SULT1A1 is widely expressed in multiple tissues and acts on a wide range of phenolic substrates. SULT1A1 is subject to functional common copy number variation (CNV) including deletions or duplications. However, it is less clear how SULT1A1 CNV impacts health and disease. To better understand the biological role of SULT1A1 in human health, we genotyped CNV in 14,275 Marshfield Clinic patients linked to an extensive electronic health record. Since SULT1A1 is linked to steroid metabolism, select serum steroid hormones were measured in 100 individuals with a wide spectrum of SULT1A1 CNV genotypes. Furthermore, comprehensive phenome-wide association studies (PheWAS) were conducted using diagnostic codes and clinical text data. For the first time, individuals homozygous null for SULT1A1 were identified in a human population. Thirty-six percent of the population carried >2 copies of SULT1A1 whereas 4% had ≤ 1 copy. Results indicate SULT1A1 CNV was negatively correlated with estrone-sulfate to estrone ratio predominantly in males (E1S/E1; p=0.03, r=−0.21) and may be associated with increased risk for common allergies. The effect of SULT1A1 CNV on circulating estrogen metabolites was opposite to the predicted CNV-metabolite trend based on enzymatic function. This finding, and the potential association with common allergies reported herein, warrants future studies.
Keywords: PheWAS, TextWAS, allergy, estrogen, sulfation, metabolism
1. Introduction
Human cytosolic sulfotransferase 1A1 (SULT1A1) is one of four genes in the SULT1A subfamily (SULT1A1, 1A2, 1A3, and 1A4) that is mapped to the short arm of chromosome 16 [1–4]. This chromosomal region is rich in repetitive sequences and segmental duplications [3, 5], as demonstrated by a large ancestral duplication containing SULT1A3/SULT1A4 [3] and a highly polymorphic copy number variant (CNV) resulting in the complete deletion and duplication of SULT1A1 [6]. SULT1A1 is hypothesized to be one of the most important SULT isoforms. This assumption is driven by observations that SULT1A1 is widely expressed in multiple human tissues, and biochemical experiments demonstrate that SULT1A1 activity plays an important role in the metabolism, bioactivation, and detoxification of procarcinogens, medications, and steroid hormones, including estrogens [7–9].
In addition to the biochemical properties of SULT1A1, early functional genetic studies have identified coding and regulatory single-nucleotide polymorphisms (SNPs) that are associated with SULT1A1 transcription, translation, and enzyme activity [10–14]. Many of these SNPs have been associated with a wide spectrum of clinical phenotypes [15]. For example, variant SULT1A1*2 has been reported to be associated with cancer risk in various organs and tissues in different ethnic groups [16–21], but other research groups have found conflicting evidence [22]. Since these studies often fail to consider the functional CNV [6, 23, 24], interpreting the relevance of these associations is challenging. Furthermore, SULT1A1 CNV may have an impact on SNP genotype quality when there can be between 0–6 copies of SULT1A1 with varying dosages of SNP alleles. A few studies have begun to assess the relevance of SULT1A1 CNV with clinical phenotypes [5, 10, 14, 25–27] and metabolism. These studies are often limited in scope, sample size, and do not capture extreme CNV genotypes [23, 24].
Given the observed importance of CNV in SULT1A1 activity, biochemical relevance of SULT1A1 with respect to metabolism, and potential role in human health, we conducted a large-scale screen of SULT1A1 CNV in 14,725 patients of European descent linked to an extensive electronic health record (EHR). To better understand the potential metabolic impact of SULT1A1, we selected 100 individuals with a wide range of SULT1A1 genotypes (0–6 copies) and correlated those genotypes with serum estrogen levels. To further assess the potential clinical relevance of CNV, we associated SULT1A1 genotype with over 30,000 phenotypes by a phenome-wide association study (PheWAS) and a text-wide association study (TextWAS). This research builds on the relevance of SULT1A1 CNV in estrogen metabolism, identified individuals who are homozygous deleted for this important enzyme, and describes how SULT1A1 CNV may be associated with phenotypes linked to common allergies.
2. Materials and methods
2.1 Population
All subjects studied came from Marshfield Clinic’s Personalized Medicine Research Project (PMRP) and have been described previously [28–31]. PMRP is a homogenous cohort of adult patients who are predominantly Caucasian, 77% claiming German ancestry, and have on average over 30 years of EHR data. In total, all individuals over the current age of 40 (14,275 participants) were genotyped for SULT1A1 copy number. The electronic medical records of individuals with rare variants of SULT1A1 and 100 randomly selected individuals taking Nasacort for nasal allergies were reviewed manually by a trained study coordinator. This study was approved by the Marshfield Clinic Institutional Review Board. Written and informed consent was acquired for all participants prior to study enrollment.
2.2 Copy number variation genotyping
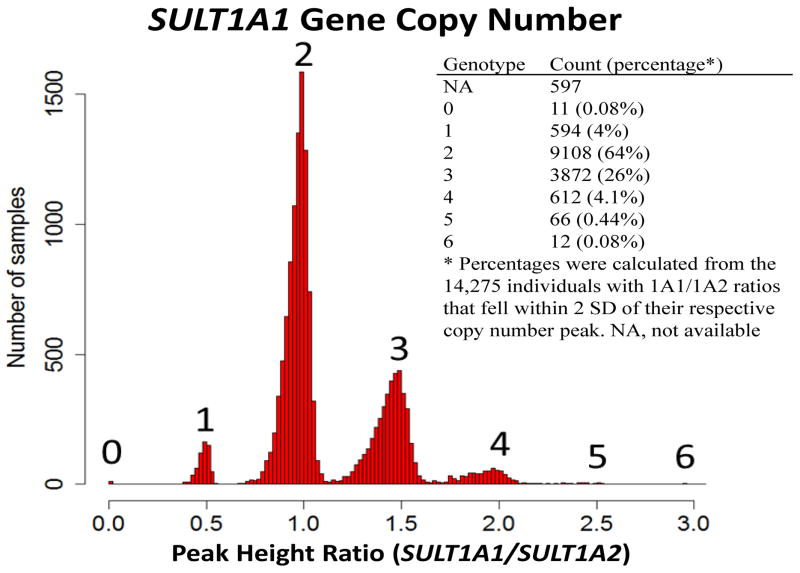
Fluorescent-based semi-quantitative PCR was used to genotype SULT1A1 CNV [6]. A set of PCR primers were designed to co-amplify a 212 bp fragment within exons 2 and 3 of SULT1A1 (NM_177534) and a 208 bp fragment within exons 3 and 4 of SULT1A2 (NM_177528). SULT1A2 was used as an internal two copy control. PCR products were analyzed on an ABI3130 DNA analyzer (Foster City, CA). Copy number was estimated by calculating the height ratio of the 212 bp amplicon of SULT1A1 to the reference 208 bp amplicon of SULT1A2. Samples with a ratio of 0, ~0.5, ~1.0, ~1.5, ~2.0, ~2.5, and ~3.0 were defined as having a SULT1A1 copy number of 0, 1, 2, 3, 4, 5, and 6, respectively (Figure 1). Assuming there are four possible alleles for SULT1A1 CNV (0–3 copies), and seven observed CNV genotypes (0–6 copies), an expectation-maximization algorithm was utilized to estimate allele frequencies. The initial allele frequency was set at 0.25 for all alleles and the model converged after 13 iterations with a sum of all errors across the alleles of 5.6E-7.
Figure 1. Histogram of SULT1A1/SULT1A2 ratios for all individuals genotyped.
Each peak with 1A1/1A2 ratios of 0, ~0.5, ~1.0, ~1.5, ~ 2.0, ~2.5, and ~3.0 define those individuals with 0, 1, 2, 3, 4, 5, and 6 copies of SULT1A1, respectively.
2.3 Steroid measurements
2.3.1 Sample collection
To understand the potential biological involvement of SULT1A1 activity on estrogen metabolism, we quantified estrogen metabolites in serum from 100 PMRP patients. This included 50 males between the ages of 40 to 50 and 50 premenopausal females between the ages of 30 to 40; age is defined as age at blood draw. All individuals with rare CNV genotypes in their respective age group (0, 5 and 6 copies) were selected whereas 10 randomly selected individuals from each of the remaining common copy number categories (1–4 copies) were selected for analysis. No individual was pregnant within a year of blood draw, on hormone replacement therapy, or had a history of breast or prostate cancer.
2.3.2 Sample processing and steroid measurements
All sample processing and liquid chromatography–mass spectrometry (LC-MS) analysis was performed by researchers blinded to sample identity and CNV genotype. Measurement of the products of SULT1A1 estrogen metabolites, specifically free estrone (E1) and sulfated estrone (E1S) were conducted. As a physiological negative control, free dehydroepiandrosterone (DHEA) and sulfated dehydroepiandrosterone (DHEA-S) were analyzed, as these compounds are generated primarily by adrenal synthesis and undergo sulfation by SULT2A1 [32].
Steroid levels were measured on an Ultimate 3000 quaternary UHPLC coupled to a Q Exactive Plus mass spectrometer operating in the negative ion mode (conjugates) and positive mode (unconjugated steroids). LC-MS Optima grade solvents (water, methanol, acetonitrile, and acetic acid) were purchased from Fisher Scientific (Pittsburg, PA). DHEA-S, E1S, E1, DHEA as well as stable isotope-labeled standards [2H5]-DHEA-S and [2H4]-E1S were purchased from Sigma (St. Louis, MO) and were tested before analysis for cross-contamination for hormones measured here. Girard P reagent was from Tokyo Chemical Industry Company, LTD (Tokyo, Japan). Stable isotope-labeled [13C3]-E1 98% purity and [2H5]-DHEA 97% purity were from Cambridge Isotope Labs (Andover, MA). Double charcoal stripped human serum from Golden West Biologicals, Inc. (Temecula, CA, USA) was used as a surrogate matrix for calibrators and quality control and contained no detectable levels of any analytes.
Levels of unconjugated steroids were measured using a validated and previously published protocol of Girard P derivatization for keto-steroids [33, 34]. For conjugated steroid analysis, internal standard solutions containing 10 pg/μL [2H5]-DHEA-S and 10 pg/μL [2H4]-E1S in methanol were added (20 μL) to each sample of serum (100 μL). These samples were diluted with 320 μL of methanol, vortexed, and centrifuged at 15,000 × g for 10 minutes to precipitate insoluble components. Supernatants were transferred to new microcentrifuge tubes and evaporated to dryness under nitrogen then re-suspended in 100 μL solution of 95:5 water:methanol. Following resuspension, samples were vortexed, centrifuged at 15,000 × g for 5 minutes, and transferred to injection vials (90 μL total sample). A 10 μL aliquot of sample with 20 pg of each internal standard was injected on the column and analyzed via LC-MS/HRMS. LC separation of derivatized sample components was performed using a Waters XBridge C18 column (3.5 μm particle size, 2.1 × 150 mm) stored at 40ºC in a column heater with a two solvent gradient where solvent A was water with 0.2 mM ammonium fluoride and solvent B was methanol. The LC gradient was set at 0.2 ml/min flow 5% B for 1 min increasing in solvent concentration to 50% B at 5 min, with an increase in flow rate and solvent concentration to 0.225 ml/min and 90% B at 20 min and holding these parameters to 25 min, followed by re-equilibration at starting conditions from 26 to 30 min. MS analysis was performed using alternating full scan with data independent analysis at a 1 m/z isolation widow looped three times based on appearance of [M-H]− of each analyte and internal standard for a total of 18 scans. Peak integration was performed from the full scan at a 5 ppm window with a confirming ion from the MS/HRMS scan (10 ppm window) using the matched stable isotope labeled internal standard. Standard curves were linear within the range of the samples and quality control samples falling within 20% coefficient of variation across the analytical runs. The limits of quantitation (LOQ) were conservatively set at 10 times the lowest non-zero standard curve point since the signal intensity was zero in multiple analyte channels for the matrix blanks. LOQs for selected hormones were as follows: E1: 0.75 pg/mL, DHEA: 3.75 pg/mL, E1S: 0.24 ng/mL, and DHEA-S: 15.6 ng/mL. Selected samples were re-injected over the storage time during analysis to confirm stability within 5% of the original values. One sample was lost during processing due to defective glassware.
2.3.3 Correlation analysis of SULT1A1 CNV and estrogen metabolism
Since all metabolite levels were highly skewed, (|skew| > 0.6 checked by manual observation of histograms of hormone levels, with the exception of DHEA-S/DHEA in men only), Spearman rank correlations were used to examine the relationships among free compounds, conjugated analytes, paired free/conjugate ratios, and copy number stratified by sex. To determine if there was a significant difference between compound levels by copy number, the data was analyzed using non-parametric Kruskal-Wallis tests stratified by sex. Less than 2% of free steroids (E1, DHEA) had values that fell below the limit of detection. All statistical analyses were performed using SAS version 9.3.
2.4 Association methods
Because SULT1A1 genotype may be involved in estrogen metabolism and human health, a PheWAS was conducted. The phenome was defined by ICD9 coding extracted from patient EHR data using standard methods as described previously [29–31]. Individuals whose medical records contained ICD9 codes inclusive of three levels of resolution defined by ICD9 code suffix (for example, ICD9 720, 720.8, 720.89) were designated as a case for a particular condition, whereas individuals with no record of the broadest code (e.g., 720) were classified as controls. Due to privacy concerns, only those phenotypes that were observed >9 times within the cohort were assessed. Utilizing this approach, there were 6,910 phenotypes extracted from the EHR. Special attention was given toward two codes defining breast cancer (ICD9 174, malignant neoplasm of breast; and ICD9 233.0, carcinoma in situ of breast) in females only and 115 codes that define adverse drug events (E930–E949, E850–E858, except E850.1, E854.1) [35]. As an alternative to PheWAS, a TextWAS was conducted with methodologies reported previously [29]. Briefly, all clinical notes were broken down into four possible combinations of word strings which included unigrams (one word), bigrams (two adjacent words), trigrams (three adjacent words), and quad-grams (four adjacent words). All word strings were then cross referenced with the National Library of Medicine’s unified Medical Language System medical dictionary. In total, the text-based phenome consisted of 23,382 clinically relevant terms (word strings). Individuals with a given word string were considered cases for that word string while all others were considered controls. Logistic regression using CNV as a continuous variable was conducted in both PheWAS and TextWAS. Sex and EHR length were included in the analysis as covariates. The p-value of CNV was generated using Wald statistics. All associations were conducted by Plink v1.9 (http://pngu.mgh.harvard.edu/purcell/plink/) [36] and R i386 3.1.0 (http://www.R-project.org/) [37].
3. Results
3.1 Correlation between SULT1A1 CNV and estrogen sulfation
SULT1A1 CNV ranged from zero to six copies in the study population. Among the 14,872 individuals genotyped, approximately 4% could not be definitively defined due to 1A1/1A2 ratios falling between copy number bins. Of the remaining 14,275 subjects, 605 individuals (4%) carried less than two copies, 9,108 individuals (64%) had two copies, and 4,562 individuals (32%) carried three or more copies of SULT1A1. The frequency for the common CNV is similar to previous reports [6]. For the first time, 11 individuals (0.08%) were identified as homozygous deleted and 12 individuals (0.08%) had six copies of SULT1A1 (Figure 1). Given there were seven observed genotypes (0–6 copies), it was assumed that there were predominantly 4 possible alleles of SULT1A1 on any given chromosome (0–3 copies). Under this assumption, the frequency of the null allele was estimated at 2.6% while the frequencies for two and three copies of SULT1A1 alleles were estimated at 17% and 1.2%, respectively.
To assess the physiological significance of SULT1A1 CNV with respect to estrogen metabolism, E1 and E1S serum levels were measured from 50 males and 50 females, including those with extreme CNV genotypes. Estrogen levels from one female were identified as an extreme outlier and were excluded from further analysis. Spearman correlation analysis of SULT1A1 CNV against measurements of E1 and E1S in all individuals and after adjusting for sex indicated that SULT1A1 CNV was not individually associated with E1 or E1S levels; however, SULT1A1 CNV was significantly correlated with E1S/E1 levels (p= 0.035, r=−0.21). Further analyses were performed with data from females and males separately. In males, SULT1A1 CNV was significantly correlated with E1S/E1 levels (p=0.025, r=−0.32). This association was not as pronounced in females but had a similar direction of effect (p=0.15, r=−0.21) (Table 1). E1S levels in males were significantly different among different copy number groups (p= 0.012). As expected, no significant associations were detected between SULT1A1 CNV and levels of DHEA and DHEA-S.
Table 1.
Correlation between SULT1A1 CNV and Estrogen sulfation
| Gender (sample size) |
Metabolite | Categories of CNV (Mean±SD) | r (p-value) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CP0 (n=3F, 2M) |
CP1 (n=10F, 10M) |
CP2 (n=10F, 10M) |
CP3 (n=10F, 10M) |
CP4 (n=10F, 10M) |
CP5 (n=6F, 6M) |
CP6 (n=0F, 2M) |
|||
| Male (n=50) | E1 (pg/mL serum) | 25.18±15.34 | 25.34±18.57 | 48.02±45.49 | 52.03±46.30 | 48.71±28.53 | 41.68±21.88 | 27.55±6.07 | 0.23 (0.11) |
| E1S (ng/mL serum) | 2.92±0.36 | 2.82±0.77 | 2.72±1.44 | 4.03±1.38 | 2.21±0.77 | 2.51±0.77 | 1.17±0.021 | −0.22 (0.11) | |
| E1S/E1 | 147.81 ± 104.27 | 182.61±137.54 | 80.63±45.73 | 142.38±114.90 | 83.12±80.11 | 75.84±43.85 | 43.57±8.84 | −0.32 (0.025) | |
| DHEA (pg/mL serum) | 24614.97±2953.30 | 21104.87±9556.66 | 37211.83±26096.38 | 30202.69±22522.43 | 31417.76±24561.34 | 32065.90±8646.91 | 40043.00±1870.94 | 0.26 (0.06) | |
| DHEAS (ng/mL serum) | 2491.77±511.21 | 3740.32±5343.70 | 13158.48±24639.23 | 8316.33±14730.70 | 2826.07±3308.61 | 3952.67±3909.26 | 2804.18±448.25 | −0.005 (0.97) | |
| DHEAS/DHEA | 100.71± 8.69 | 284.73± 613.71 | 345.25± 434.25 | 256.08± 322.08 | 119.67± 147.13 | 118.24± 104.96 | 70.37± 14.48 | −0.20 (0.16) | |
| Female (n=49) | E1 | 68.61±39.60 | 57.86±52.49 | 74.08±46.69 | 77.69±45.60 | 175.07±186.32 | 84.89±86.97 | NA | 0.19 (0.20) |
| E1S | 4.38±3.06 | 2.72±1.53 | 2.88±0.48 | 3.05±0.93 | 3.56±2.29 | 2.78±0.92 | NA | 0.03 (0.85) | |
| E1S/E1 | 70.74 ±34.23 | 300.85 ±481.30 | 55.63±39.31 | 68.99±66.23 | 40.17±45.87 | 92.57±84.91 | NA | −0.21 (0.15) | |
| DHEA | 19644.86±9322.50 | 47853.93±38728.78 | 32841.50±18937.03 | 33987.81±23845.05 | 46801.92±54419.97 | 16745.35±6075.99 | NA | −0.17(0.25) | |
| DHEAS | 927.29±243.11 | 3353.70±3109.54 | 1751.68±1036.42 | 1114.11±506.36 | 1458.83±779.25 | 1167.37±381.91 | NA | −0.17 (0.26) | |
| DHEAS/DHEA | 54.33± 30.07 | 76.54± 61.26 | 67.60± 44.77 | 39.52± 17.04 | 44.52± 25.88 | 79.81± 40.99 | NA | −0.04 (0.77) | |
| Female + Male (n=99) | E1 | 51.23±37.54 | 40.64±40.72 | 61.05±46.82 | 64.86±46.62 | 115.21±148.05 | 61.32±61.43 | 27.55±6.07 | 0.14 (0.17) |
| E1S | 3.79±2.32 | 2.77±1.18 | 2.80±1.05 | 3.54±1.25 | 2.88±1.80 | 2.65±0.82 | 1.17±0.021 | −0.10 (0.31) | |
| E1S/E1 | 101.57 ± 71.32 | 238.25±338.39 | 68.13±43.44 | 105.69±98.73 | 60.51±66.26 | 84.20±62.62 | 43.57±8.84 | −0.21 (0.035) | |
| DHEA | 21632.90±7283.24 | 34479.40±30692.70 | 35026.66±22304.34 | 32095.25±22657.93 | 39109.84±41843.38 | 24405.63±10713.59 | 40043.00±1870.94 | 0.04 (0.71) | |
| DHEAS | 1553.08±910.58 | 3547.01±4259.76 | 7154.90±17452.69 | 4715.22±10796.22 | 2142.45±2442.32 | 2560.02±3021.36 | 2804.18±448.25 | −0.08 (0.43) | |
| DHEAS/DHEA | 72.88± 33.41 | 180.64± 437.72 | 206.43± 332.50 | 142.10± 242.06 | 82.09± 109.81 | 99.03± 78.58 | 70.37± 14.48 | −0.08 (0.41) | |
Abbreviations: CP0, CP1, CP2, CP3, CP4, CP5, and CP6 stands for the number of copy number variants 1–6 for SULT1A1; r (p-value), Spearman’s rank correlation coefficient and p-value.
3.2 Association analysis
SNPs in SULT1A1 have been associated with numerous phenotypes with varying and often conflicting results [16–22, 38]. Few studies have assessed the functionally relevant CNV. To understand the potential clinical impact of SULT1A1 CNV, we leveraged extensive EHR data linked to all 14,275 individuals. Initial focus was directed toward the 11 individuals who were homozygous null for SULT1A1. After manual chart review, none of these individuals had any common or unusual phenotypic patterns. Therefore, it is unlikely the homozygous null genotype results in an unusual congenital abnormality. Because SULT1A1 CNV was associated with baseline E1S/E1 ratios and SNPs in SULT1A1 have previously been associated with breast cancer [20, 21], we further evaluated the relevance of SULT1A1 CNV to breast cancer risk by analyzing the medical records of females diagnosed with ICD9 174, malignant neoplasm of breast (640 cases) and ICD9 233.0, carcinoma in situ of breast (210 cases). In this population, neither ICD9 code was associated with SULT1A1 CNV (p=0.24 and 0.69, respectively).
Given SULT1A1’s reported involvement in the metabolism of numerous drugs [9], we further evaluated 115 ICD9 codes that define adverse drug events [35]. The strongest association was between SULT1A1 CNV and “Anticoagulants causing adverse effects in therapeutic use” (ICD9 E934.2, p=0.0024, OR=0.64 [0.48–0.85] (Supplementary Table 1). Given the number of tests, this association was not statistically significant (p<4.3E-4 assuming α<0.05, 115 tests/phenotypes).
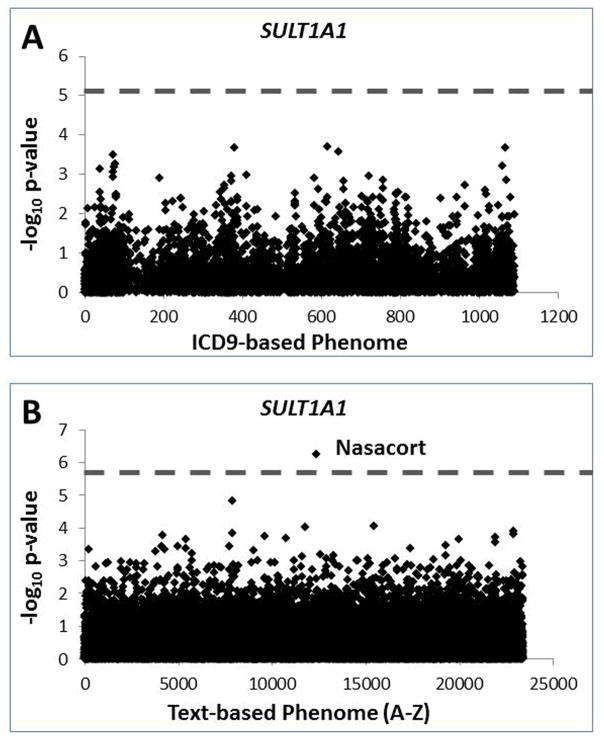
To further assess the impact of SULT1A1 CNV on thousands of phenotypes, a PheWAS was conducted. Based on ICD9 codes to define cases and controls [29–31], no phenotype passed a conservative Bonferroni threshold (p<7.2E-6, assuming α < 0.05 and 6910 tests/phenotypes). The top associations included ICD9 616.3 defining abscess of Bartholin’s gland (p= 0.00020, OR=2.0[1.4–2.9]) followed by ICD9 379.92 defining swelling or mass of eye (p= 0.00021, OR=1.7[1.3–2.3]) (Figure 2, Supplementary Table 2); the relevance of these association is uncertain. Because it has been demonstrated that clinical text data can provide complementary data to ICD9 coding and provide additional phenotypic specificity [29], we conducted a TextWAS that associated 23,382 medical terms with SULT1A1 CNV genotype. In this analysis, the top association was for the term “Nasacort” (1,455 cases; p=5.8E-7, OR 0.8[0.73–0.87]) (Figure 2, Supplementary Table 2). This association passed a conservative experiment-wise Bonferroni threshold (p<2.2E-6 assuming α<0.05, 23,382 tests/phenotypes), and further passed a study-wise Bonferroni threshold when considering the total number of tests from both the TextWAS and PheWAS (p<1.7E-6 assuming α<0.05, 30,292 tests/phenotypes) (Figure 2). Nasacort is an over-the-counter nasal spray containing triamcinolone, an adrenocortical steroid commonly used to treat nasal allergy symptoms such as sinusitis and rhinitis. An additional top TextWAS result included “Claritin,” another over-the-counter drug frequently taken to treat common allergy symptoms (p=3.7E-4 OR 0.9[0.85–0.95]; Supplemental Table 2). Manual chart review of 100 randomly selected individuals with “Nasacort” indicated that none of the patients had any adverse drug reactions though approximately 15% and 42% with Nasacort documented were in reference to the treatment of sinusitis and rhinitis, respectively. The term “allergic rhinitis” extracted from TextWAS data was moderately associated with SULT1A1 genotype (p=0.028) whereas the ICD9 code defining rhinitis (ICD9 477) from PheWAS data had a suggestive association (p=0.057).
Figure 2. Manhattan plot for (A) PheWAS and (B) TextWAS of SULT1A1 CNV.
Dotted lines represent thresholds for statistical significance for the respective experiment.
4. Discussion
SULT1A1 is proposed to be involved in a variety of pathophysiologic processes such as drug metabolism, cancer, hormone regulation, and neurotransmitter biology [5]. Multiple coding and promoter variants have been associated with human disease with varying results. Many of these studies were conducted without considering the common CNV, even when functional studies suggest SULT1A1 CNV may have a significant influence on enzyme activity [6, 10]. Only recently have other research groups begun to assess the impact of SULT1A1 CNV on disease risk and estrogen metabolism.
Our results suggest that increasing copies of SULT1A1 are inversely associated with E1S/E1 levels in males and, to a lesser degree, in females. Interestingly, SULT1A1 copy number has been previously associated with male breast cancer risk [26]. Our associations may agree with Moyer, et al. who reported that increased copies of SNP alleles driven by the SULT1A1 CNV are associated with lower E2S/E2 levels. These associations were only observed in women treated with oral conjugated equine estrogen [24]. In combination with Moyer et al., our findings seem counterintuitive since increased copies of SULT1A1 would result in increased SULT1A1 enzyme activity and should result in increased levels of E1S and E2S levels. It may be hypothesized that intracellular estrogen levels have an inverse relationship to circulating estrogens and that SULT1A1 CNV may influence this dichotomy. It should be noted that SULT1E1 has been reported to be the primary conjugating enzyme for estrogen sulfation at physiological concentrations with two reported genetic variants that influence enzyme activity, but SULT1E1 genetics may not have large influences on estrogen metabolism variability given reported functional variants are predominantly rare (minor allele frequency </= 1%) [39]. Although our estrogen metabolite experiments were the first to evaluate extreme CNV genotypes, larger population studies with sufficient power to evaluate both the CNV and other potential functional variants in SULT1A1 [16, 17, 22] in different ethnicities are required. This may be particularly relevant when evaluating extreme duplication events, which are more common in populations with African compared to European ancestries [6, 10]. Such studies should consider standardized sample collection to account for circadian rhythms and can stratify by therapeutic drugs such as aromatase inhibitors or tamoxifen, in combination with further functional studies, to better understand the relationship between SULT1A1 genotype and estrogen metabolism. Likewise, further studies on other substrates of SULT1A1 would be highly informative to understanding the paradoxical relationship between CNV and metabotype.
To identify individuals with extreme CNV genotypes for the estrogen experiments required widespread screening. For the first time, individuals who were homozygous null for SULT1A1 were observed but these individuals presented with no overt phenotype. This finding corresponds to phenotypic analysis of SULT1A1 mouse knockouts that are viable and have no outwardly apparent phenotype. However, the absence of functional SULT1A1 enzyme in mouse knockouts may have an influence on the reduction of DNA adducts [40, 41], warranting future investigation into the effects of extreme SULT1A1 CNV.
Because all individuals genotyped were linked to extensive phenotypic data, we conducted the first PheWAS/TextWAS for SULT1A1. Based on ICD9 coding, no association passed statistical significance and there was no evidence for association with female breast cancer risk. Although ICD9 coding has repeatedly been shown to be effective in defining case-control groups for thousands of phenotypes [15, 30, 42], ICD9 coding is primarily applied for billing purposes in the United States and can be limited in the phenotypes it captures. To address these limitations, it has been shown that clinical text data can complement ICD9 coding and provide additional phenotypic information [29]. In our TextWAS, we identified a statistically significant association with the term “Nasacort” (p=5.83E-7). As described previously, Nasacort, also known as triamcinolone, is an over-the-counter medication often used to treat allergy symptoms including rhinitis. Although text data can often provide additional phenotypic information not acquired by ICD9 coding, it can be more difficult to interpret text data without context. In relation to Nasacort, this challenge may be further exacerbated given many patients likely self-medicate with this over-the-counter drug. Association data from text and ICD9 coding related to rhinitis may support SULT1A1 CNV involvement in risk for symptoms related to common allergies. Future disease specific studies that consider both CNV and SNP genotyping will be required to better understand this potential relationship.
5. Conclusions
In conclusion, this study is the first large-scale screen of SULT1A1 CNV that identified individuals that were null for SULT1A1. Metabolic analysis of circulating hormones in the context of SULT1A1 CNV identified a significant inverse relationship with E1/E1S concentration primarily in males. Further PheWAS/TextWAS results suggest SULT1A1 CNV may be related to the treatment of common allergies. These results build on the growing importance of SULT1A1 CNV on metabolism and human health.
Supplementary Material
PheWAS results of ICD9 codes for adverse drug events with SULT1A1 CNV.
Top clinical terms and ICD9 codes associated with SULT1A1 CNV (p<0.001) by TextWAS and PheWAS, respectively.
Acknowledgments
We wish to thank Dr. Emily A Andreae for her assistance in editing this manuscript.
Funding
This work was supported by the generous donors of Marshfield Clinic, National Institutes of Health National Center for Advancing Translations Sciences [UL1TR000427], National Human Genome Research Institute [U01HG006389], National Institute of General Medical Science [R01GM114128], National Library of Medicine [K22LM011938], and National Institute of Environmental Health Sciences [K22ES026235].
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- 1.Her C, Raftogianis R, Weinshilboum RM. Human phenol sulfotransferase STP2 gene: molecular cloning, structural characterization, and chromosomal localization. Genomics. 1996;33:409–420. doi: 10.1006/geno.1996.0216. [DOI] [PubMed] [Google Scholar]
- 2.Her C, Szumlanski C, Aksoy IA, Weinshilboum RM. Human jejunal estrogen sulfotransferase and dehydroepiandrosterone sulfotransferase: immunochemical characterization of individual variation. Drug Metab Dispos. 1996;24:1328–1335. [PubMed] [Google Scholar]
- 3.Hildebrandt MA, Salavaggione OE, Martin YN, Flynn HC, Jalal S, Wieben ED, Weinshilboum RM. Human SULT1A3 pharmacogenetics: gene duplication and functional genomic studies. Biochem Biophys Res Commun. 2004;321:870–878. doi: 10.1016/j.bbrc.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 4.Raftogianis RB, Her C, Weinshilboum RM. Human phenol sulfotransferase pharmacogenetics: STP1 gene cloning and structural characterization. Pharmacogenetics. 1996;6:473–487. doi: 10.1097/00008571-199612000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Bradley ME, Benner SA. Phylogenomic approaches to common problems encountered in the analysis of low copy repeats: the sulfotransferase 1A gene family example. BMC Evol Biol. 2005;5:22. doi: 10.1186/1471-2148-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hebbring SJ, Adjei AA, Baer JL, Jenkins GD, Zhang J, Cunningham JM, Schaid DJ, Weinshilboum RM, Thibodeau SN. Human SULT1A1 gene: copy number differences and functional implications. Hum Mol Genet. 2007;16:463–470. doi: 10.1093/hmg/ddl468. [DOI] [PubMed] [Google Scholar]
- 7.Adjei AA, Weinshilboum RM. Catecholestrogen sulfation: possible role in carcinogenesis. Biochem Biophys Res Commun. 2002;292:402–408. doi: 10.1006/bbrc.2002.6658. [DOI] [PubMed] [Google Scholar]
- 8.Coughtrie MW. Sulfation through the looking glass--recent advances in sulfotransferase research for the curious. Pharmacogenomics J. 2002;2:297–308. doi: 10.1038/sj.tpj.6500117. [DOI] [PubMed] [Google Scholar]
- 9.Glatt H, Meinl W. Pharmacogenetics of soluble sulfotransferases (SULTs) Naunyn Schmiedebergs Arch Pharmacol. 2004;369:55–68. doi: 10.1007/s00210-003-0826-0. [DOI] [PubMed] [Google Scholar]
- 10.Yu X, Dhakal IB, Beggs M, Edavana VK, Williams S, Zhang XX, Mercer K, Ning B, Lang NP, Kadlubar FF, Kadlubar S. Functional genetic variants in the 3′-untranslated region of sulfotransferase isoform 1A1 (SULT1A1) and their effect on enzymatic activity. Toxicol Sci. 2010;118:391–403. doi: 10.1093/toxsci/kfq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ning B, Nowell S, Sweeney C, Ambrosone CB, Williams S, Miao X, Liang G, Lin D, Stone A, Ratnasinghe DL, Manjanatha M, Lang NP, Kadlubar FF. Common genetic polymorphisms in the 5′-flanking region of the SULT1A1 gene: haplotypes and their association with platelet enzymatic activity. Pharmacogenet Genomics. 2005;15:465–473. doi: 10.1097/01.fpc.0000166823.74378.79. [DOI] [PubMed] [Google Scholar]
- 12.Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM. Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem Biophys Res Commun. 1997;239:298–304. doi: 10.1006/bbrc.1997.7466. [DOI] [PubMed] [Google Scholar]
- 13.Raftogianis RB, Wood TC, Weinshilboum RM. Human phenol sulfotransferases SULT1A2 and SULT1A1: genetic polymorphisms, allozyme properties, and human liver genotype-phenotype correlations. Biochem Pharmacol. 1999;58:605–616. doi: 10.1016/s0006-2952(99)00145-8. [DOI] [PubMed] [Google Scholar]
- 14.Yu X, Kubota T, Dhakal I, Hasegawa S, Williams S, Ozawa SS, Kadlubar S. Copy number variation in sulfotransferase isoform 1A1 (SULT1A1) is significantly associated with enzymatic activity in Japanese subjects. Pharmgenomics Pers Med. 2013;6:19–24. doi: 10.2147/PGPM.S36579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hebbring SJ. The challenges, advantages and future of phenome-wide association studies. Immunology. 2014;141:157–165. doi: 10.1111/imm.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang G, Miao X, Zhou Y, Tan W, Lin D. A functional polymorphism in the SULT1A1 gene (G638A) is associated with risk of lung cancer in relation to tobacco smoking. Carcinogenesis. 2004;25:773–778. doi: 10.1093/carcin/bgh053. [DOI] [PubMed] [Google Scholar]
- 17.Bamber DE, Fryer AA, Strange RC, Elder JB, Deakin M, Rajagopal R, Fawole A, Gilissen RA, Campbell FC, Coughtrie MW. Phenol sulphotransferase SULT1A1*1 genotype is associated with reduced risk of colorectal cancer. Pharmacogenetics. 2001;11:679–685. doi: 10.1097/00008571-200111000-00006. [DOI] [PubMed] [Google Scholar]
- 18.Ozawa S, Katoh T, Inatomi H, Imai H, Kuroda Y, Ichiba M, Ohno Y. Association of genotypes of carcinogen-activating enzymes, phenol sulfotransferase SULT1A1 (ST1A3) and arylamine N-acetyltransferase NAT2, with urothelial cancer in a Japanese population. Int J Cancer. 2002;102:418–421. doi: 10.1002/ijc.10728. [DOI] [PubMed] [Google Scholar]
- 19.Nowell S, Ratnasinghe DL, Ambrosone CB, Williams S, Teague-Ross T, Trimble L, Runnels G, Carrol A, Green B, Stone A, Johnson D, Greene G, Kadlubar FF, Lang NP. Association of SULT1A1 phenotype and genotype with prostate cancer risk in African-Americans and Caucasians. Cancer Epidemiol Biomarkers Prev. 2004;13:270–276. doi: 10.1158/1055-9965.epi-03-0047. [DOI] [PubMed] [Google Scholar]
- 20.Tang D, Rundle A, Mooney L, Cho S, Schnabel F, Estabrook A, Kelly A, Levine R, Hibshoosh H, Perera F. Sulfotransferase 1A1 (SULT1A1) polymorphism, PAH-DNA adduct levels in breast tissue and breast cancer risk in a case-control study. Breast Cancer Res Treat. 2003;78:217–222. doi: 10.1023/a:1022968303118. [DOI] [PubMed] [Google Scholar]
- 21.Zheng W, Xie D, Cerhan JR, Sellers TA, Wen W, Folsom AR. Sulfotransferase 1A1 polymorphism, endogenous estrogen exposure, well-done meat intake, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10:89–94. [PubMed] [Google Scholar]
- 22.Kotnis A, Kannan S, Sarin R, Mulherkar R. Case-control study and meta-analysis of SULT1A1 Arg213His polymorphism for gene, ethnicity and environment interaction for cancer risk. Br J Cancer. 2008;99:1340–1347. doi: 10.1038/sj.bjc.6604683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charoenchokthavee W, Ayudhya DP, Sriuranpong V, Areepium N. Effects of SULT1A1 Copy Number Variation on Estrogen Concentration and Tamoxifen-Associated Adverse Drug Reactions in Premenopausal Thai Breast Cancer Patients: A Preliminary Study. Asian Pac J Cancer Prev. 2016;17:1851–1855. doi: 10.7314/apjcp.2016.17.4.1851. [DOI] [PubMed] [Google Scholar]
- 24.Moyer AM, de Andrade M, Weinshilboum RM, Miller VM. Influence of SULT1A1 genetic variation on age at menopause, estrogen levels, and response to hormone therapy in recently postmenopausal white women. Menopause. 2016;23:863–869. doi: 10.1097/GME.0000000000000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim IW, Han N, Kim MG, Kim T, Oh JM. Copy number variability analysis of pharmacogenes in patients with lymphoma, leukemia, hepatocellular, and lung carcinoma using The Cancer Genome Atlas data. Pharmacogenet Genomics. 2015;25:1–7. doi: 10.1097/FPC.0000000000000097. [DOI] [PubMed] [Google Scholar]
- 26.Palli D, Rizzolo P, Zanna I, Silvestri V, Saieva C, Falchetti M, Navazio AS, Graziano V, Masala G, Bianchi S, Russo A, Tommasi S, Ottini L. SULT1A1 gene deletion in BRCA2-associated male breast cancer: a link between genes and environmental exposures? J Cell Mol Med. 2013;17:605–607. doi: 10.1111/jcmm.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulze J, Johansson M, Thorngren JO, Garle M, Rane A, Ekstrom L. SULT2A1 Gene Copy Number Variation is Associated with Urinary Excretion Rate of Steroid Sulfates. Front Endocrinol (Lausanne) 2013;4:88. doi: 10.3389/fendo.2013.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, Li R, Masys DR, Ritchie MD, Roden DM, Struewing JP, Wolf WA eMerge Team. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hebbring SJ, Rastegar-Mojarad M, Ye Z, Mayer J, Jacobson C, Lin S. Application of clinical text data for phenome-wide association studies (PheWASs) Bioinformatics. 2015;31:1981–1987. doi: 10.1093/bioinformatics/btv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hebbring SJ, Schrodi SJ, Ye Z, Zhou Z, Page D, Brilliant MH. A PheWAS approach in studying HLA-DRB1*1501. Genes Immun. 2013;14:187–191. doi: 10.1038/gene.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye Z, Mayer J, Ivacic L, Zhou Z, He M, Schrodi SJ, Page D, Brillaint MH, Hebbring SJ. Phenome-wide association studies (PheWASs) for functional variants. Eur J Hum Genet. 2015;23:523–529. doi: 10.1038/ejhg.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowell S, Falany CN. Pharmacogenetics of human cytosolic sulfotransferases. Oncogene. 2006;25:1673–1678. doi: 10.1038/sj.onc.1209376. [DOI] [PubMed] [Google Scholar]
- 33.Rangiah K, Shah SJ, Vachani A, Ciccimaro E, Blair IA. Liquid chromatography/mass spectrometry of pre-ionized Girard P derivatives for quantifying estrone and its metabolites in serum from postmenopausal women. Rapid Commun Mass Spectrom. 2011;25:1297–1307. doi: 10.1002/rcm.4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frey AJ, Wang Q, Busch C, Feldman D, Bottalico L, Mesaros CA, Blaire IA, Vachani A, Snyder NW. Validation of highly sensitive simultaneous targeted and untargeted analysis of keto-steroids by Girard P derivatization and stable isotope dilution-liquid chromatography-high resolution mass spectrometry. Steroids. 2016;116:60–66. doi: 10.1016/j.steroids.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Committee UHD. Adverse Events Related to Medical Care, Utah: 1995–99. Salt Lake City, UT: Utah Department of Health; 2001. [Google Scholar]
- 36.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R.C. Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2015. [Google Scholar]
- 38.Moyer AM, Suman VJ, Weinshilboum RM, Avula R, Black JL, Safgren SL, Kuffel MJ, Ames MM, Ingle JN, Goetz MP. SULT1A1, CYP2C19 and disease-free survival in early breast cancer patients receiving tamoxifen. Pharmacogenomics. 2011;12:1535–1543. doi: 10.2217/pgs.11.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adjei AA, Thomae BA, Prondzinski JL, Eckloff BW, Wieben ED, Weinshilboum RM. Human estrogen sulfotransferase (SULT1E1) pharmacogenomics: gene resequencing and functional genomics. Br J Pharmacol. 2003;139:1373–1382. doi: 10.1038/sj.bjp.0705369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sachse B, Meinl W, Glatt H, Monien BH. The effect of knockout of sulfotransferases 1a1 and 1d1 and of transgenic human sulfotransferases 1A1/1A2 on the formation of DNA adducts from furfuryl alcohol in mouse models. Carcinogenesis. 2014;35:2339–2345. doi: 10.1093/carcin/bgu152. [DOI] [PubMed] [Google Scholar]
- 41.Herrmann K, Engst W, Meinl W, Florian S, Cartus AT, Schrenk D, Appel KE, Nolder T, Himmelbauer H, Glatt H. Formation of hepatic DNA adducts by methyleugenol in mouse models: drastic decrease by Sult1a1 knockout and strong increase by transgenic human SULT1A1/2. Carcinogenesis. 2014;35:935–941. doi: 10.1093/carcin/bgt408. [DOI] [PubMed] [Google Scholar]
- 42.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PheWAS results of ICD9 codes for adverse drug events with SULT1A1 CNV.
Top clinical terms and ICD9 codes associated with SULT1A1 CNV (p<0.001) by TextWAS and PheWAS, respectively.