Abstract
As the therapeutic landscape for chronic lymphocytic leukemia (CLL) continues to expand, biological predictors of response to therapy are becoming increasingly important. One such predictive biomarker is the mutational status of the variable region of the immunoglobulin heavy chain (IGHV) gene, which is a powerful predictor of duration of response and overall survival with chemoimmunotherapy (CIT). As this test may influence choice of therapy between CIT and novel agents, it is critical that providers understand how mutational status is determined and the limitations of testing. Here, we describe the details of IGHV mutational status testing, highlighting the appropriate way to interpret this information and best apply it to the care of patients with CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common hematologic malignancy in the Western world, with approximately 19,000 new cases each year in the United States.1 The disease is remarkably heterogeneous, with some patients never requiring treatment and others having rapidly progressive disease despite maximal therapy. The two best established CLL prognostic markers are fluorescence in situ hybridization (FISH) and the mutational status of the variable region of the immunoglobulin heavy chain (IGHV) gene.2,3 Here, we focus on IGHV mutational status as a key prognostic factor that can help to delineate differences in disease outcomes, and increasingly can inform treatment choices.
IGHV Testing in the Clinic
In 1999, two landmark papers were published which identified the importance of somatic mutations in IGHV genes as a prognostic marker in CLL.4,5 In both studies, mutated IGHV correlated with marked improvement in both progression free survival (PFS) and overall survival (OS). Since then, several other studies have validated these results. A recent meta-analysis confirmed an improved PFS for mutated IGHV patients, with a range 9.2 to 18.9 years, compared to unmutated patients, with a range of 1 to 5 years.6 Similarly, OS was found to range from 17.9 to 25.8 years in mutated patients, compared to 3.2 to 10 years in unmutated patients.
In the majority of cases, favorable cytogenetics track with mutated IGHV mutational status and unfavorable cytogenetics track with unmutated IGHV mutational status. Interestingly, in the event of discordant prognostic features, there is evidence that mutational status may have a superior ability to predict overall survival.7,8 Recently, an international prognostic index score that includes IGHV mutation status, along with age, clinical stage, mutations of the tumor suppressor gene TP53, and serum beta-2 microglobulin level, was also found to predict time to first treatment and OS in CLL.9–11 In this weighted model, the relative risk of IGHV mutational status is twice as much as age and clinical stage, and is second only to TP53 mutational status in importance. More simplified prognostic scores, consisting of only IGHV and TP53 mutational status, have similarly been shown to predict survival in patients with CLL.12
In recent years, the value of IGHV mutational status has become most clear in its ability to predict the durability of response to chemoimmunotherapy (CIT). FCR300, the original phase II study of fludarabine, cyclophosphamide and rituximab (FCR) for initial therapy of patients with CLL, demonstrated a PFS of 53.9% for patients with IGHV mutated disease compared to 8.7% in unmutated patients after 12.8 years of follow up.13 The PFS curves for mutated IGHV patients also plateaued, suggesting sustained, long-term remissions, and even cure in a subset of these patients. In the subsequent phase III study, CLL8, the German CLL Study Group showed that IGHV unmutated status, along with TP53 mutations and the presence of deletion 17p, had the strongest negative prognostic impact on PFS and OS.14,15 Additionally, after nearly six years of follow up, more than 83% of IGHV mutated patients were still alive, and median OS was not reached, suggesting that as in FCR300, patients with IGHV mutated CLL benefit significantly from CIT with long-term disease control.
While patients with IGHV unmutated CLL have inferior duration of response to CIT, there does not appear to be a difference in response duration for these patients when treated with recently approved novel agents that target the B cell receptor (BCR) pathway kinases, albeit with shorter follow-up. In fact, responses may actually be more rapid in patients with unmutated IGHV, perhaps due to the greater reliance of such cells on tonic BCR signaling.16 For example, a study of the Bruton’s Tyrosine Kinase (BTK) inhibitor ibrutinib for frontline treatment of CLL reported a 40% rate of complete remission (CR) in patients with unmutated disease, compared to only 6% of patients with mutated disease.17 This discrepancy does not mean that the drug was more effective in unmutated patients; rather, CR in CLL requires complete clearance of lymphocytosis, and patients with unmutated CLL tend to have faster resolution of the lymphocyte redistribution that is characteristic of this class of drugs compared to mutated patients where prolonged lymphocytosis often delays achievement of CR, even though the patients are responding well clinically to treatment.16 Importantly, recently reported data on the 5-year follow-up of patients treated with ibrutinib showed equivalent PFS for mutated and unmutated patients, a remarkable finding that has not been observed in trials of CIT in CLL.18 Similarly, other targeted therapies, including the PI3K-delta inhibitor idelalisib and the BCL-2 antagonist venetoclax achieve equivalently high response rates and PFS for both IGHV mutated and unmutated CLL.19,20
Despite the clear clinical observations regarding the prognostic importance of IGHV mutational status, the mechanism underlying this observation remains incompletely understood. One hypothesis is that IGHV unmutated cells are more prone to undergo apoptosis, and that faster kinetics of CLL cell growth rather than greater resistance to cell death may be responsible for the inferior response durability with traditional CIT.21 Additionally, whole exome sequencing efforts of CLL samples have identified an increased frequency of driver mutations in unmutated as compared to mutated IGHV CLL.22
Given the prognostic implications of IGHV mutational status and the potential of this test to influence treatment, it is important to understand how mutational status is determined and limitations of this testing.
Biology of the Immunoglobulin Heavy Chain Genes
The adaptive immune system is designed to provide dynamic protection against a wide array of potential pathogens. During normal B-cell maturation, chromosomal recombination of the V (variable), D (diversity) and J (junctional) segments form the V region of the heavy and light immunoglobulin chains. These recombination events account for the tremendous immunologic diversity that is fundamental to the humoral immune response. The addition or removal of nucleotides at the junctions of these segments creates further sequence complexity. While the entire variable region influences immunoglobulin function, three complementarity-determining regions (CDRs) contribute most to antibody specificity. CDR1 and CDR2 are located within the V segment, and CDR3, the most variable region, includes some of the V, all of the D and part of the J region (Figure 1).23,24
Figure 1.
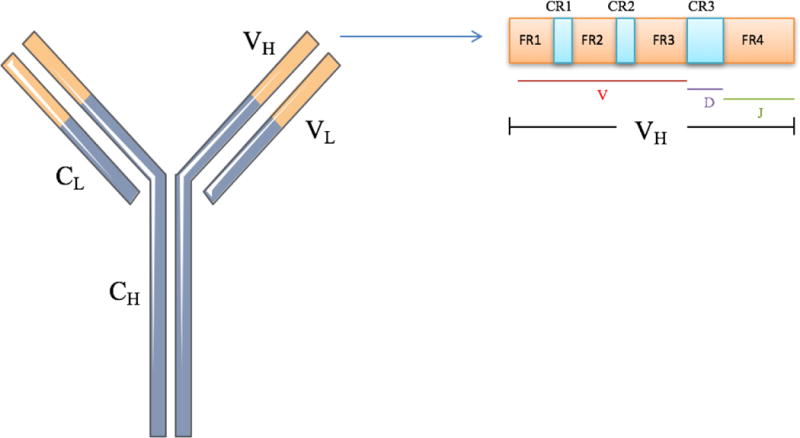
Schematic of an immunoglobulin molecule. The variable regions for the heavy (VH) and light (VL) are depicted in orange. The constant regions of the heavy (CH) and light chain (CL) are depicted in purple. Within the VH region, consisting of V, D, and J segments, are the framework regions (FR1-4) and the complementarity-determining regions (CDR1-3).
When a mature B-cell comes into contact with an antigen, further diversification takes place through a process of somatic hypermutation, typically within the germinal center. Somatic hypermutation introduces random nucleotide changes into the V genes, selecting for B-cells that produce immunoglobulins with the highest degree of selectivity, a process known as affinity maturation. Cells that fail to go through the affinity maturation process subsequently undergo apoptosis. As germline genes used to encode the V region have been mapped, it is possible to determine whether the malignant B-cell clone has undergone somatic hypermutation.
What specimens to Test
As lymphocytes are present in both peripheral blood and bone marrow, either source can be used for testing. Samples should be collected in tubes containing either ethylenediaminetetraacetic acid (EDTA) or trisodium citrate/pyridoxal 5'-phosphate/Tris (CPT). Heparin should be avoided, as it has the potential to interfere with DNA polymerase and reverse transcriptase.25 A minimum of 3 mL of peripheral blood or 1 mL of bone marrow is typically required.
Technical Aspects and Limitations of IGHV Testing
Given the importance of IGHV mutation status on prognostication, investigators of the European Research Initiative on CLL (ERIC) developed a consensus recommendation on the minimum requirements for reliable and reproducible IGHV mutational testing.25 Currently, IGHV mutational status is determined by amplifying the IGHV transcript by polymerase chain reaction (PCR), sequencing the gene through Sanger sequencing, and comparing the transcript to known germline genes available in immunoglobulin databases.25,26
According to ERIC recommendations, both complementary DNA (cDNA) and genomic DNA (gDNA) are suitable for analysis. cDNA has the advantage of preferentially identifying functional rearrangements. However, the use of gDNA avoids the need for reverse transcription and can be more easily performed on stored samples. Either leader primers, which allow for sequencing of the entire IGHV region, or primers specific to framework-region 1 (FR1), can be used. While FR1 primers require approximation of the percentage of identity to the closest germline gene, they are more efficient than leader primers. If a borderline result is obtained, the lab can utilize both primers for a more comprehensive evaluation.
The percentage of identity is calculated using the ratio of the number of mutations within the IGHV region and the length in nucleotides of the most homologous germline IGHV gene found in immunoglobulin databases, including international immunogenetics information (IMGT), V-Base, and GenBank. Insertions and deletions are counted as one mutation, irrespective of the number of extra or missing nucleotides. Also, if an FR1 primer is used, the 5’ nucleotides equal to the primer length are excluded.
Given the different techniques available for IGHV testing, there is a potential for discrepant results across institutions. There is also a risk that a transcript other than the primary clonal transcript will be amplified and that certain primers miss subclones. Framework-region primers also do not yield a full-length transcript, and can potentially lead to an inaccurate calculation of percent similarity to germline. In addition, there are differences in the immunoglobulin databases, consisting of germline immunoglobulin genes and polymorphisms, as well as variations in the software programs used to calculate the percentage of nucleotide mutations.
More recently, targeted exome or genome next generation sequencing (NGS) has emerged as an alternative method to reliably sequence the entire V-D-J sequence, including the highly variable CDR3.27 Comparisons of NGS-based testing to traditional methods suggests that NGS may provide more accurate results. In addition, NGS allows for identification of multiple productive clonally unrelated IGHV rearrangements, which were observed in 24% of patients with CLL in one study.28
Application of Testing
Mutation rates of ≥2% difference from germline are considered mutated, while unmutated disease has a <2% mutation rate. The 98% homology cutoff was chosen to exclude potential polymorphic variant sequences.4,5 It is important that providers be mindful that this is a mathematical cutoff, especially when interpreting borderline mutational values. Identification of more than one clone, where one is mutated while the other is unmutated, also appears to have an intermediate prognosis as compared to mutated alone or unmutated alone.28,29 In the event of discordant clones, additional prognostic markers can also help guide interpretation and prognostication. For example, concurrent ZAP70 overexpression or 11q deletion by FISH are more likely to be associated with unmutated disease.30,31
It should be noted that unlike FISH testing, which can change over time through a process of clonal evolution, IGHV mutational status is generally considered to be a stable prognostic marker, and as such serial testing is generally not required. However, changes in mutational status have been reported, possibly due to the emergence of subclones.32 Since IGHV mutational status provides useful prognostic information that can influence counseling or patient follow up, we generally send it soon after a diagnosis of CLL is made. However, IGHV status is not typically a major factor in the decision of when to initiate therapy. Rather, initiation of therapy continues to be guided by the 2008 International Workshop on CLL (IWCLL) criteria, in which only patients with advanced stage or symptomatic disease warrant therapy.33 IGHV mutational status can influence choice of therapy, with mutated IGHV patients being better candidates for chemoimmunotherapy, while unmutated IGHV patients are more likely to benefit from a novel agent based approach. Ongoing prospective studies will provide further evidence for the utility of this approach.
Summary Table.
What is the test?
IGHV mutational testing assesses the percentage of sequence variability between the V region of the immunoglobulin heavy chain gene in a clonal CLL population compared to the homologous germline V region sequence.
How is it measured?
IGHV mutational status is determined by amplifying the expressed, clonal IGHV transcript by PCR, sequencing the gene through Sanger sequencing and comparing the transcript to known germline sequence.
What are the normal range values?
Mutated IGVH is classified as ≥2% mutated when compared to germline. Unmutated disease is <2% mutated compared to germline.
What conditions or types of conditions is it used for?
IGHV testing is a useful prognostic tool for patients with CLL. IGHV status may also have prognostic implications in other lymphomas, such as follicular and mantle cell lymphoma.34,35
What tests are helpful to do with it for a more complete picture?
In addition to IGHV testing, other prognostic testing such as FISH cytogenetics should be performed for patients with CLL. FISH can identify chromosomal abnormalities which may have prognostic and treatment implications. For example, deletions of chromosome 17p or 11q are associated with an adverse clinical outcome, while deletions of 13q, suggest a more favorable clinical course.2
How does it impact treatment?
Although IGHV mutation status is a powerful prognostic tool, the decision to initiate therapy continues to be based on the 2008 IWCLL guidelines, in which only patients with advanced or symptomatic disease should be treated. However, IGHV status can facilitate counseling and appropriate follow-up intervals. IGHV status may also influence what type of therapy is started in the frontline setting, as CLL patients with IGHV mutated disease can have highly durable responses to CIT. In contrast, patients with unmutated IGHV should be considered for novel targeted therapies, which appear to be equally efficacious irrespective of IGHV status.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 3.Davids MS, Vartanov A, Werner L, Neuberg D, Dal Cin P, Brown JR. Controversial fluorescence in situ hybridization cytogenetic abnormalities in chronic lymphocytic leukaemia: new insights from a large cohort. Br J Haematol. 2015;170(5):694–703. doi: 10.1111/bjh.13498. [DOI] [PubMed] [Google Scholar]
- 4.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–1847. [PubMed] [Google Scholar]
- 5.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–1854. [PubMed] [Google Scholar]
- 6.Parikh SA, Strati P, Tsang M, West CP, Shanafelt TD. Should IGHV status and FISH testing be performed in all CLL patients at diagnosis? A systematic review and meta-analysis. Blood. 2016;127(14):1752–1760. doi: 10.1182/blood-2015-10-620864. [DOI] [PubMed] [Google Scholar]
- 7.Gladstone DE, Swinnen L, Kasamon Y, et al. Importance of immunoglobulin heavy chain variable region mutational status in del(13q) chronic lymphocytic leukemia. Leuk Lymphoma. 2011;52(10):1873–1881. doi: 10.3109/10428194.2011.585529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gladstone DE, Blackford A, Cho E, et al. The importance of IGHV mutational status in del(11q) and del(17p) chronic lymphocytic leukemia. Clin Lymphoma Myeloma Leuk. 2012;12(2):132–137. doi: 10.1016/j.clml.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International CLLIPIwg. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. Lancet Oncol. 2016;17(6):779–790. doi: 10.1016/S1470-2045(16)30029-8. [DOI] [PubMed] [Google Scholar]
- 10.Molica S, Shanafelt TD, Giannarelli D, et al. The chronic lymphocytic leukemia international prognostic index predicts time to first treatment in early CLL: Independent validation in a prospective cohort of early stage patients. Am J Hematol. 2016;91(11):1090–1095. doi: 10.1002/ajh.24493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentile M, Shanafelt TD, Rossi D, et al. Validation of the CLL-IPI and comparison with the MDACC prognostic index: analysis of 1364 newly diagnosed patients. Blood. 2016 doi: 10.1182/blood-2016-07-728261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delgado J, Doubek M, Baumann T, et al. Chronic lymphocytic leukemia: A prognostic model comprising only two biomarkers (IGHV mutational status and FISH cytogenetics) separates patients with different outcome and simplifies the CLL-IPI. Am J Hematol. 2017;92(4):375–380. doi: 10.1002/ajh.24660. [DOI] [PubMed] [Google Scholar]
- 13.Thompson PA, Tam CS, O'Brien SM, et al. Fludarabine, cyclophosphamide, and rituximab treatment achieves long-term disease-free survival in IGHV-mutated chronic lymphocytic leukemia. Blood. 2016;127(3):303–309. doi: 10.1182/blood-2015-09-667675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer K, Bahlo J, Fink AM, et al. Long-term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: updated results of the CLL8 trial. Blood. 2016;127(2):208–215. doi: 10.1182/blood-2015-06-651125. [DOI] [PubMed] [Google Scholar]
- 15.Brown JR, Hallek MJ, Pagel JM. Chemoimmunotherapy Versus Targeted Treatment in Chronic Lymphocytic Leukemia: When, How Long, How Much, and in Which Combination? American Society of Clinical Oncology educational book American Society of Clinical Oncology Meeting. 2016;35:e387–398. doi: 10.1200/EDBK_159018. [DOI] [PubMed] [Google Scholar]
- 16.Guo A, Lu P, Galanina N, et al. Heightened BTK-dependent cell proliferation in unmutated chronic lymphocytic leukemia confers increased sensitivity to ibrutinib. Oncotarget. 2016;7(4):4598–4610. doi: 10.18632/oncotarget.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byrd JC, Furman RR, Coutre SE, et al. Three-year follow-up of treatment-naive and previously treated patients with CLL and SLL receiving single-agent ibrutinib. Blood. 2015;125(16):2497–2506. doi: 10.1182/blood-2014-10-606038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Brien SFR, Coutre S. Five-Year Experience with Single-Agent Ibrutinib in Patients with Previously Untreated and Relapsed/Refractory Chronic Lymphocytic Leukemia/Small Lymphocytic Leukemia. Blood. 2016;123 (abstract 233) [Google Scholar]
- 19.Sharman J, Coutre SE, Furman RR, et al. Second interim analsis of a phase 3 study of idelalisib (ZYDELIG) plus rituximab (R) for relapsed chronic lymphocytic leukemia (CLL): efficacy analysis in patient subpopulations with Del(17p) and other adverse prognostic factors. Blood. 2014;124 (abstr 133) [Google Scholar]
- 20.Roberts AW, Davids MS, Pagel JM, et al. Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. The New England journal of medicine. 2016;374(4):311–322. doi: 10.1056/NEJMoa1513257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davids MS, Deng J, Wiestner A, et al. Decreased mitochondrial apoptotic priming underlies stroma-mediated treatment resistance in chronic lymphocytic leukemia. Blood. 2012;120(17):3501–3509. doi: 10.1182/blood-2012-02-414060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525–530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calis JJ, Rosenberg BR. Characterizing immune repertoires by high throughput sequencing: strategies and applications. Trends in immunology. 2014;35(12):581–590. doi: 10.1016/j.it.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahota SS, Babbage G, Zojer N, Ottensmeier CH, Stevenson FK. Determining Mutational Status of Immunoglobulin V Genes in Chronic Lymphocytic Leukemia. In: Illidge T, Johnson PWM, editors. Lymphoma: Methods and Protocols. Totowa, NJ: Humana Press; 2005. pp. 129–144. [DOI] [PubMed] [Google Scholar]
- 25.Ghia P, Stamatopoulos K, Belessi C, et al. ERIC recommendations on IGHV gene mutational status analysis in chronic lymphocytic leukemia. Leukemia. 2007;21(1):1–3. doi: 10.1038/sj.leu.2404457. [DOI] [PubMed] [Google Scholar]
- 26.Szankasi P, Bahler DW. Clinical laboratory analysis of immunoglobulin heavy chain variable region genes for chronic lymphocytic leukemia prognosis. The Journal of molecular diagnostics : JMD. 2010;12(2):244–249. doi: 10.2353/jmoldx.2010.090091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blachly JS, Ruppert AS, Zhao W, et al. Immunoglobulin transcript sequence and somatic hypermutation computation from unselected RNA-seq reads in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2015;112(14):4322–4327. doi: 10.1073/pnas.1503587112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatopoulos B, Timbs A, Bruce D, et al. Targeted deep sequencing reveals clinically relevant subclonal IgHV rearrangements in chronic lymphocytic leukemia. Leukemia. 2016 doi: 10.1038/leu.2016.307. [DOI] [PubMed] [Google Scholar]
- 29.Heyman B, Volkheimer AD, Weinberg JB. Double IGHV DNA gene rearrangements in CLL: influence of mixed-mutated and -unmutated rearrangements on outcomes in CLL. Blood Cancer J. 2016;6(7):e440. doi: 10.1038/bcj.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiestner A, Rosenwald A, Barry TS, et al. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood. 2003;101(12):4944–4951. doi: 10.1182/blood-2002-10-3306. [DOI] [PubMed] [Google Scholar]
- 31.Trbusek M, Malcikova J, Smardova J, et al. Inactivation of p53 and deletion of ATM in B-CLL patients in relation to IgVH mutation status and previous treatment. Leukemia. 2006;20(6):1159–1161. doi: 10.1038/sj.leu.2404195. [DOI] [PubMed] [Google Scholar]
- 32.Osman A, Gocke CD, Gladstone DE. Change in IgHV Mutational Status of CLL Suggests Origin From Multiple Clones. Clin Lymphoma Myeloma Leuk. 2017;17(2):97–99. doi: 10.1016/j.clml.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 33.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berget E, Molven A, Lokeland T, Helgeland L, Vintermyr OK. IGHV gene usage and mutational status in follicular lymphoma: Correlations with prognosis and patient age. Leuk Res. 2015;39(7):702–708. doi: 10.1016/j.leukres.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Navarro A, Clot G, Royo C, et al. Molecular subsets of mantle cell lymphoma defined by the IGHV mutational status and SOX11 expression have distinct biologic and clinical features. Cancer Res. 2012;72(20):5307–5316. doi: 10.1158/0008-5472.CAN-12-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]


