Abstract
The intragenic tumor-suppressor microRNA miR-486-5p is often down-regulated in non-small cell lung cancer (NSCLC) but the mechanism is unclear. This study investigated epigenetic co-regulation of miR-486-5p and its host gene ANK1. MiR-486-5p expression in lung tumors and cell lines was significantly reduced compared to normal lung (p<0.001) and is strongly correlated with ANK1 expression. In vitro, siRNA-mediated ANK1 knockdown in NSCLC cells also reduced miR-486-5p while the DNA methylation inhibitor 5-aza-2′-deoxycytidine induced expression of both. ANK1 promoter CpG island was unmethylated in normal lung but methylated in 45% (118/262) lung tumors and 55% (17/31) NSCLC cell lines. After adjustment for tumor histology and smoking, methylation was significantly more prevalent in adenocarcinoma (101/200, 51%) compared to squamous cell carcinoma (17/62, 27%), p<0.001; HR=3.513 (CI: 1.818–6.788); and in smokers (73/128, 57%) than never-smokers (28/72, 39%), p=0.014; HR=2.086 (CI: 1.157–3.759). These results were independently validated using quantitative methylation data for 809 NSCLC cases from The Cancer Genome Atlas project. Together, our data indicate that aberrant ANK1 methylation is highly prevalent in lung cancer, discriminate tumors by histology and patients’ smoking history, and contributes to miR-486-5p repression.
Keywords: ANK1, miR-486-5p, intronic microRNA, epigenetics, NSCLC
1. Introduction
MicroRNAs (miRs) are small (~22 nucleotides) non-coding RNAs that regulate many fundamental biological processes primarily through inhibiting the translation of their target genes [1]. Some miRs target key oncogenic or tumor-suppressor genes and thereby play a critical role in inhibiting or promoting carcinogenesis, respectively [2]. MiR-486-5p is a tumor-suppressor miR that is often down-regulated across multiple malignancies including lung [3,4], gastric [5], pancreatic [6], and other [7] cancers. In vitro and laboratory animal based gain- or loss-of-function studies on miR-486-5p have confirmed its role in suppressing the growth of lung cancer [8–10]. Reduced miR-486-5p expression strongly contribute to lung cancer progression while recent studies showed that it could be reliably detected in easily accessible samples such as serum and sputum indicating its potential as a prognostic biomarker. The clinical use this and other prognostic miR signatures as disease fingerprints and potential early detection biomarkers including through complementing lung cancer screening by low-dose computed tomography have been demonstrated [11–15]. However, the mechanism(s) leading to this highly prevalent and clinically important miR-486-5p repression remains unclear.
Aberrant promoter CpG island methylation is one of the most common epigenetic modifications that silence tumor-suppressor genes in cancer. Recent publications from our group [16,17] and others [18,19] have shown that some tumor-suppressor miRs are also epigenetically silenced in cancer through promoter hypermethylation and/or histone modification. For intragenic miRs that are located within coding genes, such epigenetic changes in the promoter region of the host gene could simultaneously alter the expression of the gene and the miR. Over 50% of approximately 1900 miRs in the human genome are intragenic, and about a third including miR-486 are located within intronic regions [20,21]. Since some intronic miRs are co-transcribed and consequently co-regulated with their host genes [20–23], we hypothesized that the frequent repression of miR-486-5p in lung cancer could be mediated through aberrant epigenetic modification of its host gene, ANK1.
ANK1 is a large (~230kb) gene within chromosome 8p11.21 that encodes the adapter protein ankyrin-1. Three distinct promoter regions drive tissue-specific expression of ANK1B (brain and muscle), ANK1E (erythroid cells), or ANK1A (ubiquitous) transcript variants [24–26]. MiR-486 is located within the last intron of ANK1 that is common to all transcripts and could be co-transcribed with any or all variants. Indirect evidence in various cell types indicates potential co-transcription and co-regulation of ANK1 and miR-486. GATA1 activates the expression of both ANK1 and miR-486-5p in myeloid leukemia [25,27]. Similarly, MYB (c-myb) binds to the ANK1 promoter and activates coordinated expression of ANK1 and miR-486-3p to promote erythropoiesis [28]. In skeletal muscle cells, MLK (MRTF-A) activates expression of ANK1 and miR-486 [29], while MSTN (myostatin) represses expression of both [30]. These findings support our hypothesis that miR-486-5p down-regulation in lung cancer could be mediated through epigenetic repression of the host gene.
Since the tumor-suppressor function of miR-486-5p and its down-regulation in lung cancer have been extensively studied, the focus of this study was to define the mechanisms leading to this repression. The presence of epigenetic abnormalities across the promoter regions of ANK1 and/or the intronic region surrounding the miR and their role in suppressing ANK1, miR-486-5p, or both in lung cancer were evaluated. First, miR-486-5p expression in lung tumor-normal pairs and cell lines was determined using next-generation sequencing (NGS) of the microRNAome [17]. The NGS also screened for potential epigenetically repressed miRs using NSCLC cell lines treated with vehicle or the DNA methylation inhibitor 5-aza-2′-deoxycytidine (DAC). The specificity of methylation to lung cancer and its association with tumor histology, stage, patients’ demography and/or smoking history was evaluated using qualitative and quantitative methylation assays. The results were confirmed using various qualitative and quantitative methylation and expression assays and independently validated using publicly available methylation data from The Cancer Genome Atlas (TCGA) database. Finally, the role of epigenetic silencing of ANK1, its effects one genome-wide transcription of genes, and the pathways it impacts were studied using siRNA-mediated knockdown of the gene in NSCLC cell lines.
2. Materials and Methods
2.1. Tissue samples and cell lines
Lung tumors from 262 NSCLC patients were obtained from frozen tumor banks at the University of New Mexico (UNM), Johns Hopkins, and the Mayo Clinic. Distant normal lung tissue (DNLT) obtained from the resected lobe was available for a subset of these cases. Normal human bronchial epithelial cells (NHBEC) collected through diagnostic bronchoscopy [31] and peripheral blood mononuclear cells (PBMC) from cancer-free smokers (n=10) were used as normal control. All samples were obtained with written informed consent from patients, and the study was approved by the institute’s Ethics Committee. Five human bronchial epithelial cell lines (HBEC1, 2, 3, 13, and 14) immortalized as described [32] were obtained from Drs. Shay and Minna, Southwestern Medical Center, Dallas, TX. Twenty-five NSCLC cell lines (Table S1) obtained from and authenticated by the American Type Culture Collection (Manassas, VA) were also used. Experiments were conducted in cell lines passed for a maximum of 6 months post-resuscitation.
2.2. DNA methylation
DNA extraction, modification, and methylation analysis using Combined Bisulfite Modification and Restriction Analysis (CoBRA) and Methylation-Specific PCR (MSP) were performed as described [33] using primers and amplification conditions in Tables S2. Quantitative methylation data for ANK1 including its promoter CpG islands and the miR-486 containing last intron was obtained from our recent HumanMethylation450 beadchip (HM450K) analysis of lung tumor-normal pairs and cell lines [34,35]. The publicly available HM450K data for 809 NSCLC cases from TCGA was used for independent validation of results. Treatment of NSCLC cell lines with the DNA methylation inhibitor 5-aza-2′-deoxycytidine (DAC) was performed as described [33].
2.3. Gene, miRNA, and protein expression
For gene expression, total RNA was isolated and reverse transcribed as described [36]. For miRNA expression, 10ng total RNA was reverse transcribed using the TaqMan® MicroRNA Reverse Transcription Kit from Applied Biosystems (Foster City, CA) according to the manufacturer’s protocol. TaqMan assays from Applied Biosystems were used to quantify ANK1 and miR-486-5p expression using ACTB (β-actin) and RNU6B as endogenous controls, respectively. Samples were run at least twice in duplicate and expression of each target gene relative to the endogenous control (ΔCT) and the reference control samples (Δ ΔCT) was calculated in fold-change as described [37]. For protein expression, cells were lysed in RIPA buffer and 60μg total protein was used to detect endogenous ANK1 and Beta-actin with anti-ANK1 (NBP1-71805) and anti-B-actin (A2228) antibodies from Novus Biologicals (Littleton, CO, USA) and Sigma-Aldrich, (Carlsbad, CA, USA), respectively.
2.4. Transfection
Cells were transfected with ANK1-specific siRNA (siANK1), miR-486-5p mimic, or scrambled controls from Applied Biosystems using Lipofectamine 2000 (Invitrogen, Santa Clara, CA) as described [38]. Four different siRNAs targeting different sequences of ANK1 (Fig. S1), siANK1#1 (s223810), siANK1#2 (s1364), siANK1#3 (s1362), and siANK1#4 (s1363) were used. ANK1 and miR-486-5p expression following transfections were determined as described above. Cell survival and migration were compared between siControl and siANK1 transfected cells using MTT and wound closure assays, respectively as described [36]. Genome-wide gene expression changes between siControl and siANK1 cells were evaluated 48h post-transfection using Illumina Whole-Genome Gene Expression Beadchip (Illumina, San Diego, CA) as described by the manufacturer and the pathways regulated by the most significantly altered genes were identified using Ingenuity pathway analysis software.
2.5. Statistical analysis
Gene methylation and patient characteristics including age, gender, smoking status, and tumor histology were summarized with mean and standard deviation for continuous variables and proportions for categorical variables. The association between methylation and patient characteristics was assessed by Fisher’s exact test. ANK1 and miR-486-5p expression in siControl vs. siANK1 transfected cells was compared using one-way analysis of variance (ANOVA). All analyses were conducted in SAS 9.2.
3. Results
3.1. MiR-486-5p is epigenetically repressed in lung cancer
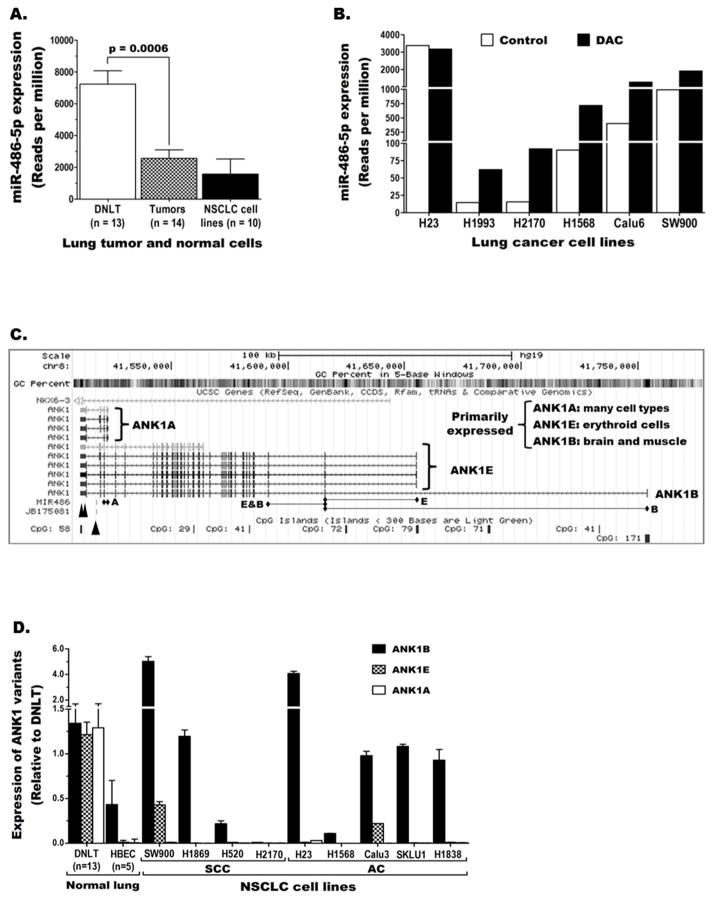
We have recently screened the microRNAome of NSCLC cell lines (n=10) and lung tumor-normal pairs (n=14) from lung adenocarcinoma patients using NGS [17]. MiR-486-5p was one of the miRs with markedly reduced expression in NSCLC cell lines and lung tumors compared to normal lung. The level of miR-486-5p in each tumor was lower than the corresponding DNLT (supplementary Fig. S2A) and pairwise comparison revealed that the overall expression in the tumors was significantly lower (p=0.0006) than the normal lung (Fig. 1A). Although miR-486-5p down-regulation and its effects in promoting lung carcinogenesis have been previously reported [3,8–10], the mechanism of repression is unknown. MicroRNAome analysis of NSCLC cell lines treated with vehicle or DAC as described [33] showed that DAC treatment increased miR-486-5p expression in 5/10 cell lines with lowest expression by 2 to 8-fold (Fig. 1B). These results, which were verified with miR-specific TaqMan expression assays (Fig. S2B), suggest DNA methylation is likely involved in miR-486-5p repression. However, our HM450K-based quantitative methylation data [34,35] revealed that the ~5kb region containing miR-486 is strongly methylated in all samples including DNLT (Fig. S2C). Evaluation of the region in normal lung epithelial (NHBEC) and blood (PBMC) cells using CoBRA also showed strong methylation (Table S3) indicating this methylation is a normal epigenetic modification that does not explain the tumor-specific repression of miR-486-5p in lung cancer.
Figure 1. Expression of miR-486-5p and its host gene ANK1 in lung cancer.
Next generation sequencing reveals miR-486-5p expression is (A) reduced in lung cancer and (B) could be induced by the DNA methylation inhibitor 5-aza-2′-deoxycytidine (DAC). C) Schematic representation depicting genomic structure of ANK1, its various transcript variants, CpG islands, and miR-486 located in the last intron. Diamond headed lines indicate exons targeted by the transcript specific TaqMan assays. Double and single arrow heads indicate the binding sites for the different siRNAs used in the study and the location of miR486, respectively. D) Expression of the three ANK1 transcript variants in normal and malignant lung tissue/cells.
3.2. MiR-486-5p and its host gene show similar expression pattern in lung cancer
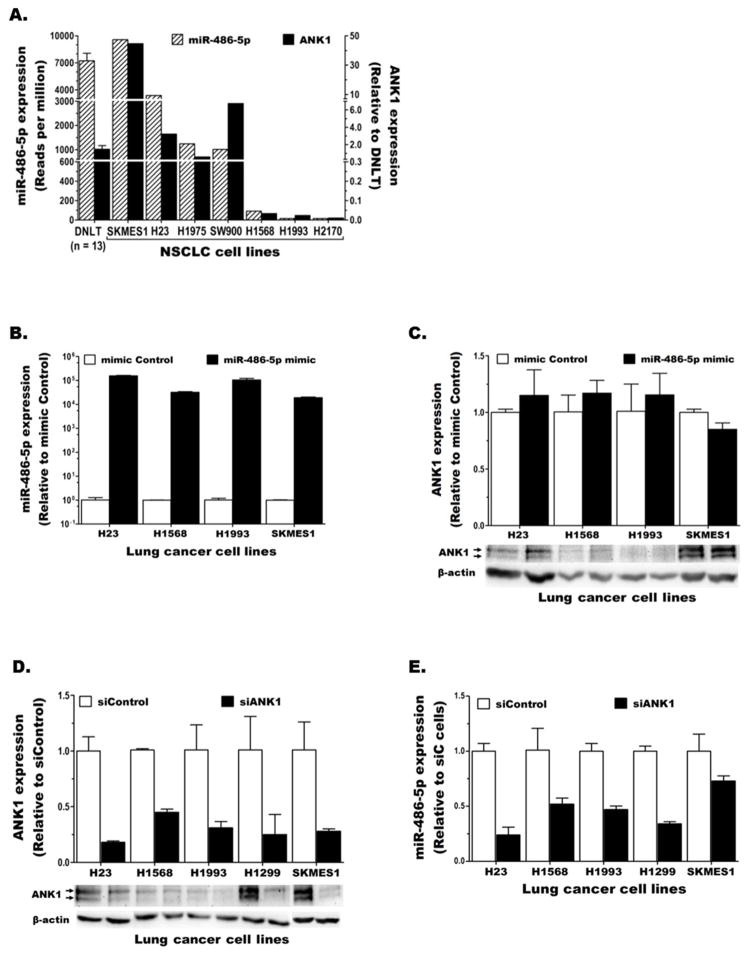
Many intragenic miRs are co-transcribed (thus co-regulated) with their host gene [20–22,39]. MiR-486 is located within the last intron of ANK1 (Fig. 1C), a large gene with three distinct promoter regions that regulate tissue-specific expression of at least three groups of transcript variants, ANK1B, ANK1E, and ANK1A [24–26]. We quantified the expression of these variants in lung cancer using variant-specific TaqMan gene expression assays (illustrated in Fig. 1C). Although all three ANK1 variants were detected in DNLT that contains various cell types, ANK1B was the main/only transcript expressed in lung epithelial cells (the precursor cells for NSCLC) and lung cancer cells (Fig. 1D). Also a low level of ANK1E (but no ANK1A) expression was detected in a few NSCLC cell lines (Fig. 1D). Hence, total ANK1 expression in lung cancer was measured using a TaqMan assay that detects both ANK1B and ANK1E (E&B line in Fig. 1C). Comparison of ANK1 (total) and miR-486-5p levels in lung cancer revealed that the miR and its host follow similar expression pattern (Fig. 2A).
Figure 2. Comparison of miR-486-5p and ANK1 expression in lung cancer.
A) Expression patterns of ANK1 and miR-486-5p in normal lung and NSCLC cell lines. B–E) NSCLC cell lines were transiently transfected with control or miR-486-5p mimic (B and C) and control or ANK1-specific siRNA, siANK1#4, (D and E) and the level of miR-486-5p (B and E) and ANK1 (C and D) mRNA (top) and protein (bottom) expression were determined.
3.3. MiR-486-5p is co-expressed with ANK1 but does not repress its host
Approximately 20% of intragenic miRs are predicted to target their host gene [23]. Using the mirSVR miRNA target prediction method from miRanda we identified two miR-486-5p binding sites within the ANK1 3′UTR. However, both sites have low mirSVR scores (−0.2046 and −0.1013) for potentially down-regulating ANK1 expression. These predictions were experimentally tested by comparing miR-486-5p and ANK1 expression in multiple NSCLC cell lines following transfection with miR-486-5p-mimic or scrambled control. The results showed that increasing miR-486-5p in NSCLC cells (Fig. 2B) did not significantly alter ANK1 mRNA or protein expression (Fig. 2C). Conversely, the effect of epigenetic repression of ANK1 on miR-486-5p expression was investigated using siRNA-mediated knockdown of ANK1 expression in NSCLC cell lines (Fig. S3A–B). The results revealed that ANK1 knockdown in lung cancer (Fig. 2D) also reduced miR-486-5p expression (Fig. 2E). Together, these results indicate that miR-486-5p is co-expressed with its host ANK1 but does not target it for repression.
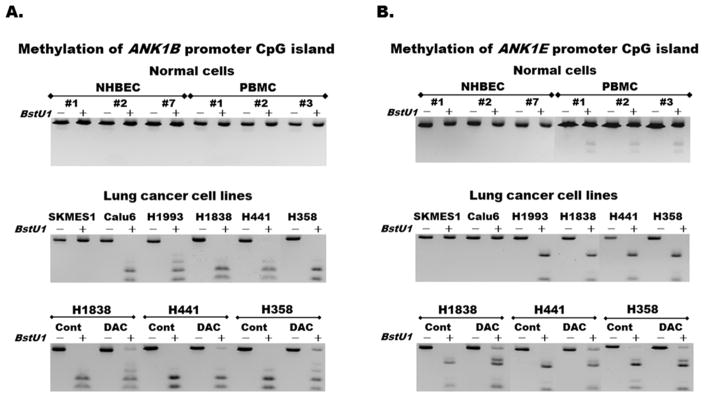
3.4. ANK1 promoter CpG islands are aberrantly methylated in lung cancer
ANK1 has 8 CpG islands scattered across its ~230kb DNA (Fig. 1C). Methylation of two of these CpG islands that cover the promoter regions of ANK1B (CpG:171) and ANK1E (CpG:79) were screened in normal and NSCLC cells using the semi-quantitative methylation assay (CoBRA) and the relationship with ANK1 and miR-486-5p expression was compared. Both CpG islands were unmethylated in NHBEC but completely methylated in some lung cancer cell lines (Fig. 3A and 3B). ANK1B promoter was unmethylated in NSCLC cell lines with higher ANK1 and miR-486-5p expression such as SKMES1, H23, and H1299, but completely methylated in cell lines with lower expression of the gene and the miR such as Calu6, H1568, and H1993 (Fig. 3A and 2A). Although ANK1E promoter was also completely methylated in some lung cancer cell lines (Fig. 3B middle panel), its weak methylation in PBMC from cancer-free donors (Fig. 3B top panel) indicate that its methylation is not as cleanly tumor-specific as ANK1B promoter. The reversibility of ANK1B and ANK1E promoter methylation (Fig. 3A–B bottom panels) and expression (Fig. S4A–C) are shown in cells treated with growth media (control) or the DNA methyltransferases inhibitor (DAC) for 96 hours as described [33].
Figure 3. Aberrant methylation of ANK1 promoter CpG islands in lung cancer.
Methylation of (A) ANK1B and (B) ANK1E promoter CpG islands in normal (top panels) and lung cancer (middle and bottom panels) was evaluated using the semi-quantitative Combined Bisulfite Modification and Restriction Analysis (CoBRA). Partial or complete digestion of PCR products into smaller fragments following the addition (+) of the BstU1 restriction enzyme indicates partial or complete methylation. Partial demethylation of the two promoter CpG islands (bottom panels) following treatment with the methylation inhibitor DAC compared to control shown by the re-appearance of some undigested bands, indicates reversibility of these epigenetic changes.
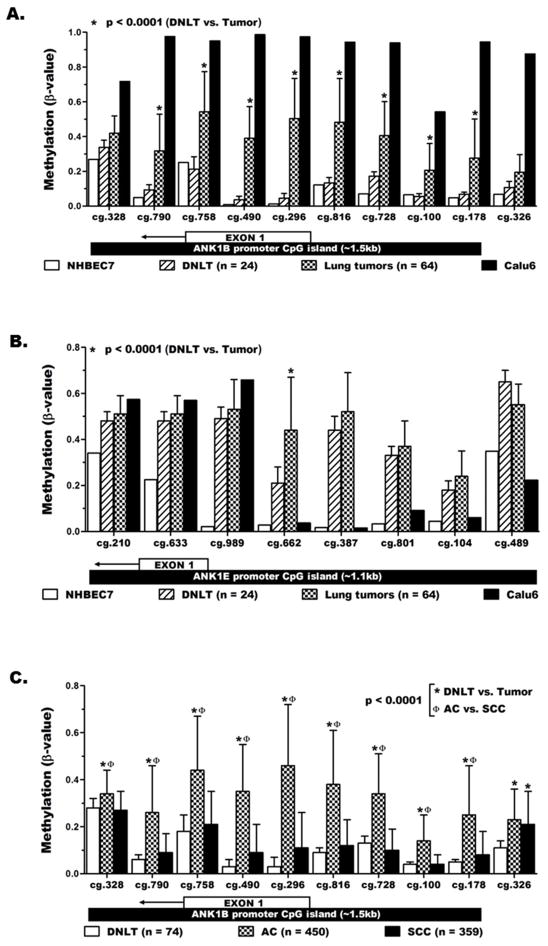
The methylation levels of ANK1B and ANK1E promoters in lung cancer were also quantified using our previous HM450K data [34,35]. A total of 18 HM450K probes interrogated the methylation levels of CpGs across the ANK1B and ANK1E promoters in NHBEC, DNLT (n=24), lung tumors (n=64), and Calu6. In agreement with the CoBRA results (Fig. 3A), each of the 10 ANK1B promoter probes displayed the lowest methylation levels (β-values) in NHBEC7 and the highest levels in Calu6 (Fig. 4A). The average methylation level of each ANK1B probe in DNLT was comparable to the levels in NHBEC7 (unmethylated control) but was significantly lower than the methylation levels in the primary tumors. Details of the methylation data including the average and range of methylation levels for each probe in the lung tumor-normal pairs are shown in Table S4. The prevalence for aberrant methylation of each probe was also calculated using β ≥ 0.20 as a threshold to define aberrant methylation (Table S4). In contrast to ANK1B, the 8 probes across the ANK1E promoter showed higher methylation levels in DNLT compared to NHBEC7 (DNLT β-values are closer to tumors than NHBEC7, Fig. 4B). The weak methylation of ANK1E promoter seen in all PBMC by CoBRA (Fig. 3B) along with the presence of PBMC in the DNLT (but not in NHBEC7) likely contributed to the higher methylation in DNLT.
Figure 4. Quantitative validation of ANK1 methylation in lung cancer.
The methylation level (mean ± SD) of (A) ANK1B and (B) ANK1E promoter CpG islands was quantitatively determined using whole genome methylation data from the HumanMethylation450 beadchip (HM450K). The location of each probe with respect to the promoter CpG island region and first exon of the specific transcript variant is depicted below the x-axis labels. C) HM450K data for large lung tumor and normal samples from the publicly available TCGA database validated the tumor-specific (not present in normal lung) methylation of ANK1B promoter in lung cancer and revealed its strong association with lung adenocarcinoma (AC) than squamous cell carcinoma SCC.
3.5. ANK1B methylation discriminates lung tumors by histology and smoking history
Screening aberrant methylation in patient samples using simple, highly sensitive, qualitative assays such as MSP requires a clean tumor-specific methylation. Thus, due to ANK1E promoter methylation in PBMC and DNLT, MSP was used to screen only the ANK1B promoter in lung tumor-normal pairs from 262 NSCLC cases. Aberrant methylation of ANK1B promoter was found in 45% of lung tumors while none of the DNLT (0/25), NHBEC (0/5), or PBMC (0/5) were methylated (Table S3). Comparison of ANK1B methylation with clinical characteristics of the patients (Table 1) revealed that methylation occur from early stage (Stage-I) in AC (46%, 56/123) and SCC (29%, 10/35) and remained at similarly high frequency in the advanced (Stages II – IV) AC (59%, 44/75) and SCC (26%, 7/27). After adjustment for smoking, the prevalence for ANK1B methylation was significantly higher in lung adenocarcinoma (AC=101/200, 51%) compared to squamous cell carcinoma (SCC=17/62, 27%), p<0.001; HR=3.513 (CI: 1.818–6.788) (Table 2). Within the same cancer histology, the prevalence for ANK1B methylation was similar between current and former smokers in AC and SCC. In contrast, ANK1B methylation was significantly more prevalent in AC from smokers, current [27/47 (57%), p=0.047], former [46/81 (57%), p=0.027], or all smokers combined [73/128 (57%), p=0.014] than from never smokers [28/72 (39%)]. Targeted sequencing data for the clinically relevant EGFR activating mutation hotspots was available for the tumors from never smokers. Interestingly, ANK1 methylation was found in 21/44 (48%) EGFR wild-type tumors compared to 7/28 (25%) EGFR mutant tumors indicating a borderline (p = 0.054) inverse relationship between the two abnormalities (Table S5).
Table 1.
ANK1B methylation by demographic and clinical characteristics of NSCLC patients.
| Characteristics | ANK1B methylation vs. patients’ characteristics | |||
|---|---|---|---|---|
|
| ||||
| Lung AC (n = 200), n (%) * | Lung SCC (n = 62), n (%) * | |||
|
| ||||
| Unmethylated | Methylated | Unmethylated | Methylated | |
| 99 (49.5%) | 101 (50.5%) | 45 (72.6%) | 17 (27.4%) | |
|
| ||||
| Age (years): | ||||
| median ± SD | 66 ± 12 | 67 ± 11 | 69 ± 7 | 71 ± 7 |
|
| ||||
| Sex: | ||||
| Female | 61 (62%) | 64 (64%) | 19 (42%) | 8 (47%) |
| Male | 38 (38%) | 37 (37%) | 26 (58%) | 9 (53%) |
|
| ||||
| Stage: ** | ||||
| Stage I | 67 (68%) | 56 (56%) | 25 (56%) | 10 (59%) |
| Stage II | 10 (10%) | 17 (17%) | 6 (13%) | 6 (35%) |
| Stage III | 15 (15%) | 19 (19%) | 7 (16%) | - |
| Stage IV | 6 (6%) | 8 (8%) | 7 (16%) | 1 (6%) |
| Unknown | 1 (1%) | 1 (1%) | - | - |
|
| ||||
| Smoking status: | ||||
| Current smokers | 20 (20%) | 27 (27%) | 12 (27%) | 3 (18%) |
| Former smokers | 35 (35%) | 46 (46%) | 33 (71%) | 14 (820%) |
| Never smokers | 44 (44%) | 28 (28%) | - | - |
|
| ||||
| Survival (months) | ||||
| Median ± SD | 34 ± 24 | 36 ± 28 | 36 ± 46 | 38 ± 57 |
Percentiles are calculated for each methylation group within the AC or SCC tumor histology.
Despite a slight increase in AC, the prevalence for ANK1 methylation in early stage (Stage I) and advanced (Stage II – IV) tumors was comparable.
Table 2.
Aberrant methylation of ANK1 promoter CpG island discriminates lung cancer by patients’ smoking history and tumor histology.
| Smoking status | Methylation n (%) | Logistic regression | ||
|---|---|---|---|---|
|
| ||||
| Lung AC | SCC | Odds ratio (95% Confidence interval) | p-value | |
|
| ||||
| Smokers (S) † | 73/128 (57%) | 17/62 (27%) | 3.513 (1.818 – 6.788) | < 0.001 |
| Current (CS) | 27/47 (57%) | 3/15 (20%) | 5.400 (1.344 – 21.703) | 0.009 |
| Former (FS) | 46/81 (57%) | 14/47 (30%) | 3.098 (1.443 – 6.652) | 0.003 |
| Never smokers (NS) | 28/72 (39%) | - | - | - |
|
| ||||
| Total | 101/200 (51%) | 17/62 (27%) | 2.701 (1.448 – 5.035) | 0.001 |
|
| ||||
| Comparisons by smoking status with in a specific histology | CS vs. FS | - | 1.027 (0.497 – 2.124) | 0.942 |
| CS vs. NS | - | 2.121 (1.005 – 4.480) | 0.047 | |
| FS vs. NS | - | 2.065 (1.082 – 3.942) | 0.027 | |
| S vs. NS | - | 2.086 (1.157 – 3.759) | 0.014 | |
| - | CS vs. FS | 0.589 (0.144 – 2.417) | 0.450 | |
Smokers (S) are current and former smokers combined.
3.6. The histologically distinct ANK1B methylation in lung cancer is independently validated
Our findings were independently validated using HM450K data for 809 NSCLC patients from the publicly available TCGA database. The large normal lung (n=74), AC (n=450) and SCC (n=359) samples valuated by TCGA also allowed quantitative comparison of ANK1B methylation between these two most common lung cancer subtypes. Methylation of the 10 ANK1B promoter probes in the TCGA (Fig. 4C) and our DNLT samples (Fig. 4A) was comparable. Similarly, methylation of these probes in TCGA samples was significantly higher in AC than DNLT, p<0.0001 (Fig. 4C). In contrast, with the exception of probe cg.326 (located outside of the ANK1B CpG island), methylation of all probes in the SCC samples was similar to DNLT. Furthermore, the significantly higher methylation of ANK1B promoter probes found in TCGA AC than SCC samples (p<0.0001) also confirmed our qualitative MSP results (Table 2). Details of ANK1B methylation for the TCGA samples, including comparisons of the prevalence for aberrant methylation between DNLT vs. AC (Table S6), DNLT vs. SCC (Table S7), and AC vs. SCC (Table S8) are shown as described above and the data across ANK1 is shown in Fig. S5.
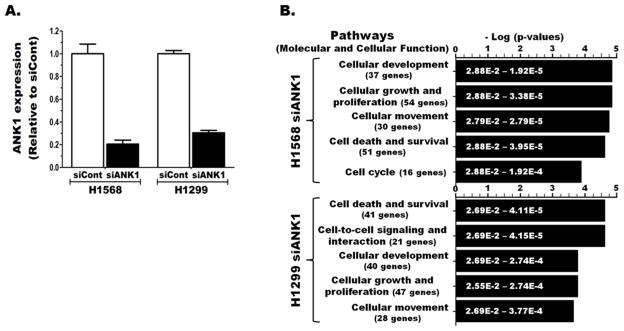
3.7. ANK1 knockdown in NSCLC alters important cancer related pathways
The impact of epigenetic silencing of ANK1 (and its co-expressed miR) in lung cancer was investigated in vitro using siRNA (another epigenetic mechanism) to repress its expression. ANK1 expression in siANK1 transfected H1568 and H1299 was reduced >70% compared to the corresponding siCont transfected cells (Fig. 5A). Comparison of the genome-wide gene expression between the siCont and siANK1 transfected cells revealed that ANK1 knockdown in these cell lines led to ≥ 1.5-fold changes in the expression of 255 and 209 genes, respectively. The most significantly affected cellular and molecular pathways in both cell lines were important regulators of cancer development and progression (Fig. 5B). Although the two cell lines share 4 of the top 5 significantly altered molecular pathways, ANK1 knockdown in neither cell line caused discernible difference in cell death, survival, or migration (data not shown).
Figure 5. ANK1 knockdown in lung cancer impacts cancer related pathways.
A) ANK1 expression in two NSCLC cell lines 48 h after transfection with control (siCon) or ANK1-specific (siANK1#4) siRNA. B) The genome-wide impact of these knockdowns was evaluated using whole transcriptome array and the top pathways affected by significantly altered genes were identified using pathway analysis.
4. Discussion
This study demonstrated that miR-486-5p and its host gene ANK1 are co-expressed, epigenetically repressed in lung cancer through aberrant hypermethylation of ANK1 promoter, and both could be re-expressed by inhibiting DNA methylation with 5-aza-2′-deoxycytidine (DAC). The last intron of ANK1 that contains miR-486 and its surrounding region is completely methylated in all normal and lung cancer samples evaluated indicating that DNA methylation in this region is normal, non-cancer-related, modification. In contrast, the promoter CpG island of ANK1B, the only/major ANK1 transcript variant in lung epithelial cells, was strongly methylated in lung cancer, but not in normal lung or blood cells. This aberrant ANK1B promoter methylation was significantly associated with lung adenocarcinoma and tumors from smokers. Epigenetic silencing of ANK1 in lung cancer either through promoter hypermethylation in vivo or siRNA-mediated knockdown in vitro reduced the expression of both ANK1 and miR-486-5p, and significantly altered cancer-related pathways. However, unlike some intragenic miRs that target their host gene for degradation in a negative-feedback loop regulation [40–42], increasing miR-486-5p in NSCLC cells using its mimic did not affect ANK1 expression. These findings were verified using various qualitative and quantitative assays and independently validated using quantitative methylation data for 809 lung tumors from TCGA. Together, our findings for the first time demonstrate that aberrant methylation of ANK1 promoter is a highly prevalent and tumor-specific abnormality in NSCLC and contributes to lung carcinogenesis by epigenetic repression of the gene and the tumor-suppressor miR-486-5p it hosts.
MiR-486-5p suppresses the transformation, growth, and migration of cancer cells by directly targeting and mediating the degradation of oncogenes. Thus, abnormal repression of this miR in cancer provides a growth advantage by enhancing the tumor-promoting properties of its targets. Among miR-486-5p target oncogenes that get activated following its downregulation in lung cancer include: CDK4, which promote cell cycle progression [3], ARHGAP5, which regulates cellular adhesion, motility, and polarity to promote cell migration and invasion [10], the growth promoting IGF signaling genes IGF1, IGF1R, and PIK3R1 [9], and the proto-oncogene serine/threonine kinase PIM-1 [8]. The tumor-suppressor role of miR-486-5p is also demonstrated in other cancers where it targets PIM-1 in breast cancer [43], SNAI1 in prostate cancer [44], and CLDN10, CITRON, and PIK3R1 in HCC [45,46] [45]. Although downregulation of this important tumor-suppressor miR in various cancer types is well-known, the mechanism of repression had not been delineated. Our study now fills this knowledge gap by demonstrating that the tumor-specific and highly prevalent promoter methylation of its host gene ANK1 simultaneously suppresses expression of the gene and miR-486-5p in lung cancer.
Co-expression of miR-486-5p and ANK1 may not be unique among intragenic miRs. About one-third human miRs are located within introns of annotated genes and are transcribed by RNA polymerase II as part of the host transcription unit [21,22,39]. Expression of these miRs and their host genes largely coincides indicating that both may be generated from a common precursor transcript [22]. Splicing of the primary transcript produces a mature mRNA while the miR containing intron is cleaved by Drosha to release a pre-miR (~70 nt) and subsequently processed by Dicer to a mature miR (~22 nt). A synchronized interplay between the spliceosome and microprocessor (miR biogenesis) machineries ensures efficient production of a mature mRNA and miR(s) from a single primary transcript [21,47]. The coordinated repression of miR486-5p and ANK1 we showed in NSCLC is also supported by direct and/or indirect evidence for ANK1/miR-486 co-regulation in multiple cell types. Hall et al., demonstrated that ANK1 and miR-486-5p are upregulated following DNA damage and regulate complementary pathways [48]. In hematopoietic progenitor cells, MYB binds to ANK1 promoter and activates ANK1 and miR-486-3p expression to shift commitment from megakaryocyte to erythroid lineage [28]. Shaham et al., showed that GATA1 binds to ANK1 promoter and regulates ANK1 and miR-486-5p expression in erythroid cells [27]. In muscle cells, MLK binding to ANK1 promoter induces dose dependent ANK1 and miR-486 expression [29], whereas myostatin binding suppresses expression of both [30]. Down regulation of miR-486-5p and miR-486-3p following ANK1 knockdown has also been recently reported [49]. The emerging role of siRNA as the gene specific guide of RNA-induced initiation of transcriptional gene silencing (RITS) complex [50,51] that suppresses transcription or causes degradation of the primary transcript prior to the micro-processing of the intronic miR could explain these findings.
We showed that DAC treatment increased ANK1 and miR-486-5p expression in NSCLC, especially in cell lines with the lowest expression. The tumor-specific ANK1B promoter methylation that was detected in lung cancer but not normal lung and blood cells was also associated with reduced ANK1 and miR-486-5p expression. Likewise, downregulation of miR-486-5p in NSCLC cells following siRNA-mediated ANK1 knockdown supports that ANK1 promoter methylation could similarly co-repress both in vivo. Although ANK1 has three major transcript variants [24-26], ANK1B is the only or predominant variant expressed in lung epithelial cells and lung cancer cells, hence the major source of miR-486 co-expression in these cells. However, the lack of significant phenotypic changes following in vitro knockdown of ANK1 and miR-486-5p suggest that these abnormalities are likely contributors rather than drivers of lung cancer development. Taken together, the independently validated, highly prevent, aberrant ANK1B promoter methylation shown in this study likely explains the frequently reported repression of miR-486-5p in lung cancer. The fact that this abnormality is more common in lung AC than SCC from smokers and AC from never-smokers indicates that epigenetic repression of ANK1 and miR-486-5p may play more important role in smoking-induced development and progression of lung AC.
Supplementary Material
Highlights.
MiR-486-5p is commonly repressed in NSCLC but the mechanism is unclear.
MiR-486-5p is located within ANK1 intron and is co-expressed with its host gene.
Aberrant methylation of ANK1 promoter represses both ANK1 and miR-486-5p in NSCLC.
ANK1 is primarily methylated in lung adenocarcinoma from smokers.
ANK1/miR-486-5p co-repression contributes to smoking induced lung adenocarcinoma.
Acknowledgments
Data generated by The Cancer Genome Atlas (TCGA) pilot project established by the NCI and NHGRI was used to validate part of our findings. The dbGaP accession number for TCGA data is phs000178.v8.p7. Information about TCGA and the investigators and institutions that constitute the TCGA research network can be found at http://cancergenome.nih.gov/.
Funding:
This study was supported by grants from the National Institutes of Health (R21CA161561, R01CA183296, and in part P30CA11800 and R01CA193532).
Appendix A. Supplementary data
Supplementary data related to this article can be found online.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
This article has supplemental data available online.
Disclosure: This study was supported by grants from the National Institutes of Health (R21CA161561, R01CA183296, and in part P30CA11800 and R01CA193532).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PloS one. 2009;4:e8003. doi: 10.1371/journal.pone.0008003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shao Y, Shen YQ, Li YL, Liang C, Zhang BJ, Lu SD, He YY, Wang P, Sun QL, Jin YX, Ma ZL. Direct repression of the oncogene CDK4 by the tumor suppressor miR-486-5p in non-small cell lung cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan X, Qin W, Zhang L, Hang J, Li B, Zhang C, Wan J, Zhou F, Shao K, Sun Y, Wu J, Zhang X, Qiu B, Li N, Shi S, Feng X, Zhao S, Wang Z, Zhao X, Chen Z, Mitchelson K, Cheng J, Guo Y, He J. A 5-microRNA signature for lung squamous cell carcinoma diagnosis and hsa-miR-31 for prognosis. Clin Cancer Res. 2011;17:6802–6811. doi: 10.1158/1078-0432.CCR-11-0419. [DOI] [PubMed] [Google Scholar]
- 5.Oh HK, Tan AL, Das K, Ooi CH, Deng NT, Tan IB, Beillard E, Lee J, Ramnarayanan K, Rha SY, Palanisamy N, Voorhoeve PM, Tan P. Genomic loss of miR-486 regulates tumor progression and the OLFM4 antiapoptotic factor in gastric cancer. Clin Cancer Res. 2011;17:2657–2667. doi: 10.1158/1078-0432.CCR-10-3152. [DOI] [PubMed] [Google Scholar]
- 6.Ali S, Saleh H, Sethi S, Sarkar FH, Philip PA. MicroRNA profiling of diagnostic needle aspirates from patients with pancreatic cancer. British journal of cancer. 2012;107:1354–1360. doi: 10.1038/bjc.2012.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Namlos HM, Meza-Zepeda LA, Baroy T, Ostensen IH, Kresse SH, Kuijjer ML, Serra M, Burger H, Cleton-Jansen AM, Myklebost O. Modulation of the osteosarcoma expression phenotype by microRNAs. PloS one. 2012;7:e48086. doi: 10.1371/journal.pone.0048086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang W, Tian X, Bai F, Han R, Wang J, Shen H, Zhang X, Liu Y, Yan X, Jiang F, Xing L. Pim-1 kinase is a target of miR-486-5p and eukaryotic translation initiation factor 4E, and plays a critical role in lung cancer. Molecular cancer. 2014;13:240. doi: 10.1186/1476-4598-13-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng Y, Dai Y, Hitchcock C, Yang X, Kassis ES, Liu L, Luo Z, Sun HL, Cui R, Wei H, Kim T, Lee TJ, Jeon YJ, Nuovo GJ, Volinia S, He Q, Yu J, Nana-Sinkam P, Croce CM. Insulin growth factor signaling is regulated by microRNA-486, an underexpressed microRNA in lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15043–15048. doi: 10.1073/pnas.1307107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Tian X, Han R, Zhang X, Wang X, Shen H, Xue L, Liu Y, Yan X, Shen J, Mannoor K, Deepak J, Donahue JM, Stass SA, Xing L, Jiang F. Downregulation of miR-486-5p contributes to tumor progression and metastasis by targeting protumorigenic ARHGAP5 in lung cancer. Oncogene. 2014;33:1181–1189. doi: 10.1038/onc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 12.Shen L, Kantarjian H, Guo Y, Lin E, Shan J, Huang X, Berry D, Ahmed S, Zhu W, Pierce S, Kondo Y, Oki Y, Jelinek J, Saba H, Estey E, Issa JP. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol. 2010;28:605–613. doi: 10.1200/JCO.2009.23.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sozzi G, Boeri M, Rossi M, Verri C, Suatoni P, Bravi F, Roz L, Conte D, Grassi M, Sverzellati N, Marchiano A, Negri E, La Vecchia C, Pastorino U. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol. 2014;32:768–773. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xing L, Su J, Guarnera MA, Zhang H, Cai L, Zhou R, Stass SA, Jiang F. Sputum microRNA biomarkers for identifying lung cancer in indeterminate solitary pulmonary nodules. Clin Cancer Res. 2015;21:484–489. doi: 10.1158/1078-0432.CCR-14-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu L, Todd NW, Xing L, Xie Y, Zhang H, Liu Z, Fang H, Zhang J, Katz RL, Jiang F. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. International journal of cancer. 2010;127:2870–2878. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tellez CS, Juri DE, Do K, Bernauer AM, Thomas CL, Damiani LA, Tessema M, Leng S, Belinsky SA. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer research. 2011;71:3087–3097. doi: 10.1158/0008-5472.CAN-10-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tellez CS, Juri DE, Do K, Picchi MA, Wang T, Liu G, Spira A, Belinsky SA. miR-196b Is Epigenetically Silenced during the Premalignant Stage of Lung Carcinogenesis. Cancer research. 2016;76:4741–4751. doi: 10.1158/0008-5472.CAN-15-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu W, Dong Z, Liang J, Guo X, Guo Y, Shen S, Kuang G, Guo W. Downregulation of Potential Tumor Suppressor miR-203a by Promoter Methylation Contributes to the Invasiveness of Gastric Cardia Adenocarcinoma. Cancer investigation. 2016;34:506–516. doi: 10.1080/07357907.2016.1242010. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Dong Z, Liang J, Guo Y, Guo X, Shen S, Kuang G, Guo W. Methylation-mediated repression of potential tumor suppressor miR-203a and miR-203b contributes to esophageal squamous cell carcinoma development. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37:5621–5632. doi: 10.1007/s13277-015-4432-9. [DOI] [PubMed] [Google Scholar]
- 20.Hinske LC, Franca GS, Torres HA, Ohara DT, Lopes-Ramos CM, Heyn J, Reis LF, Ohno-Machado L, Kreth S, Galante PA. miRIAD-integrating microRNA inter- and intragenic data. Database : the journal of biological databases and curation. 2014;2014 doi: 10.1093/database/bau099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim YK, Kim VN. Processing of intronic microRNAs. The EMBO journal. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. Rna. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinske LC, Galante PA, Kuo WP, Ohno-Machado L. A potential role for intragenic miRNAs on their hosts’ interactome. BMC genomics. 2010;11:533. doi: 10.1186/1471-2164-11-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallagher PG, Forget BG. An alternate promoter directs expression of a truncated, muscle-specific isoform of the human ankyrin 1 gene. The Journal of biological chemistry. 1998;273:1339–1348. doi: 10.1074/jbc.273.3.1339. [DOI] [PubMed] [Google Scholar]
- 25.Gallagher PG, Romana M, Tse WT, Lux SE, Forget BG. The human ankyrin-1 gene is selectively transcribed in erythroid cell lines despite the presence of a housekeeping-like promoter. Blood. 2000;96:1136–1143. [PubMed] [Google Scholar]
- 26.Yocum AO, Steiner LA, Seidel NE, Cline AP, Rout ED, Lin JY, Wong C, Garrett LJ, Gallagher PG, Bodine DM. A tissue-specific chromatin loop activates the erythroid ankyrin-1 promoter. Blood. 2012;120:3586–3593. doi: 10.1182/blood-2012-08-450262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaham L, Vendramini E, Ge Y, Goren Y, Birger Y, Tijssen MR, McNulty M, Geron I, Schwartzman O, Goldberg L, Chou ST, Pitman H, Weiss MJ, Michaeli S, Sredni B, Gottgens B, Crispino JD, Taub JW, Izraeli S. MicroRNA-486-5p is an erythroid oncomiR of the myeloid leukemias of Down syndrome. Blood. 2015;125:1292–1301. doi: 10.1182/blood-2014-06-581892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bianchi E, Bulgarelli J, Ruberti S, Rontauroli S, Sacchi G, Norfo R, Pennucci V, Zini R, Salati S, Prudente Z, Ferrari S, Manfredini R. MYB controls erythroid versus megakaryocyte lineage fate decision through the miR-486-3p-mediated downregulation of MAF. Cell death and differentiation. 2015;22:1906–1921. doi: 10.1038/cdd.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Small EM, O’Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, Richardson JA, Olson EN. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4218–4223. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hitachi K, Nakatani M, Tsuchida K. Myostatin signaling regulates Akt activity via the regulation of miR-486 expression. The international journal of biochemistry & cell biology. 2014;47:93–103. doi: 10.1016/j.biocel.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Belinsky SA, Palmisano WA, Gilliland FD, Crooks LA, Divine KK, Winters SA, Grimes MJ, Harms HJ, Tellez CS, Smith TM, Moots PP, Lechner JF, Stidley CA, Crowell RE. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer research. 2002;62:2370–2377. [PubMed] [Google Scholar]
- 32.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, Milchgrub S, Smith AL, Souza RF, Gilbey L, Zhang X, Gandia K, Vaughan MB, Wright WE, Gazdar AF, Shay JW, Minna JD. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer research. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 33.Tessema M, Willink R, Do K, Yu YY, Yu W, Machida EO, Brock M, Van Neste L, Stidley CA, Baylin SB, Belinsky SA. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer research. 2008;68:1707–1714. doi: 10.1158/0008-5472.CAN-07-6325. [DOI] [PubMed] [Google Scholar]
- 34.Tessema M, Yingling CM, Picchi MA, Wu G, Liu Y, Weissfeld JL, Siegfried JM, Tesfaigzi Y, Belinsky SA. Epigenetic Repression of CCDC37 and MAP1B Links Chronic Obstructive Pulmonary Disease to Lung Cancer. J Thorac Oncol. 2015;10:1181–1188. doi: 10.1097/JTO.0000000000000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tessema M, Yingling CM, Snider AM, Do K, Juri DE, Picchi MA, Zhang X, Liu Y, Leng S, Tellez CS, Belinsky SA. GATA2 is epigenetically repressed in human and mouse lung tumors and is not requisite for survival of KRAS mutant lung cancer. J Thorac Oncol. 2014;9:784–793. doi: 10.1097/JTO.0000000000000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tessema M, Yingling CM, Thomas CL, Klinge DM, Bernauer AM, Liu Y, Dacic S, Siegfried JM, Dahlberg SE, Schiller JH, Belinsky SA. SULF2 methylation is prognostic for lung cancer survival and increases sensitivity to topoisomerase-I inhibitors via induction of ISG15. Oncogene. 2012;31:4107–4106. doi: 10.1038/onc.2011.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Tessema M, Yingling CM, Grimes MJ, Thomas CL, Liu Y, Leng S, Joste N, Belinsky SA. Differential epigenetic regulation of TOX subfamily high mobility group box genes in lung and breast cancers. PloS one. 2012;7:e34850. doi: 10.1371/journal.pone.0034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome research. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paraboschi EM, Cardamone G, Rimoldi V, Duga S, Solda G, Asselta R. miR-634 is a Pol III-dependent intronic microRNA regulating alternative-polyadenylated isoforms of its host gene PRKCA. Biochimica et biophysica acta. 2017;1861:1046–1056. doi: 10.1016/j.bbagen.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 41.Qian J, Tu R, Yuan L, Xie W. Intronic miR-932 targets the coding region of its host gene, Drosophila neuroligin2. Experimental cell research. 2016;344:183–193. doi: 10.1016/j.yexcr.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 42.Yuva-Aydemir Y, Xu XL, Aydemir O, Gascon E, Sayin S, Zhou W, Hong Y, Gao FB. Downregulation of the Host Gene jigr1 by miR-92 Is Essential for Neuroblast Self-Renewal in Drosophila. PLoS genetics. 2015;11:e1005264. doi: 10.1371/journal.pgen.1005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang G, Liu Z, Cui G, Wang X, Yang Z. MicroRNA-486-5p targeting PIM-1 suppresses cell proliferation in breast cancer cells. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:11137–11145. doi: 10.1007/s13277-014-2412-0. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Zhang T, Yang K, Zhang M, Wang K. miR-486-5p suppresses prostate cancer metastasis by targeting Snail and regulating epithelial-mesenchymal transition. OncoTargets and therapy. 2016;9:6909–6914. doi: 10.2147/OTT.S117338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang XP, Hou J, Shen XY, Huang CY, Zhang XH, Xie YA, Luo XL. MicroRNA-486-5p, which is downregulated in hepatocellular carcinoma, suppresses tumor growth by targeting PIK3R1. The FEBS journal. 2015;282:579–594. doi: 10.1111/febs.13167. [DOI] [PubMed] [Google Scholar]
- 46.Sun H, Cui C, Xiao F, Wang H, Xu J, Shi X, Yang Y, Zhang Q, Zheng X, Yang X, Wu C, Wang L. miR-486 regulates metastasis and chemosensitivity in hepatocellular carcinoma by targeting CLDN10 and CITRON. Hepatology research : the official journal of the Japan Society of Hepatology. 2015;45:1312–1322. doi: 10.1111/hepr.12500. [DOI] [PubMed] [Google Scholar]
- 47.Agranat-Tamir L, Shomron N, Sperling J, Sperling R. Interplay between pre-mRNA splicing and microRNA biogenesis within the supraspliceosome. Nucleic acids research. 2014;42:4640–4651. doi: 10.1093/nar/gkt1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hall AE, Lu WT, Godfrey JD, Antonov AV, Paicu C, Moxon S, Dalmay T, Wilczynska A, Muller PA, Bushell M. The cytoskeleton adaptor protein ankyrin-1 is upregulated by p53 following DNA damage and alters cell migration. Cell death & disease. 2016;7:e2184. doi: 10.1038/cddis.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Omura N, Mizuma M, MacGregor A, Hong SM, Ayars M, Almario JA, Borges M, Kanda M, Li A, Vincent A, Maitra A, Goggins M. Overexpression of ankyrin1 promotes pancreatic cancer cell growth. Oncotarget. 2016;7:34977–34987. doi: 10.18632/oncotarget.9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nature reviews. Genetics. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.