Abstract
Cisplatin (CDDP) resistance is a major clinical problem associated with poor prognosis in gastric cancer (GC) patients. In this study, we performed integrated analysis of TCGA data from microRNAs (miRNAs) expression matrix of GC patients who received CDDP-based chemotherapy with GEO dataset which contains differential miRNAs expression profiles in CDDP-resistant and -sensitive cell lines. We identified miR-148a-3p downregulation as a key step involved in CDDP resistance. Using a cohort consisting 105 GC patients who received CDDP-based therapy, we found that miR-148a-3p downregulation was associated with a decrease in patients’ disease-free survival (DFS, P=0.0077). A series of experiment data demonstrated that: 1) miR-148a-3p was downregulated in CDDP-resistant GC cell lines; 2) miR-148a-3p reconstitution sensitized CDDP-resistant cells to CDDP treatment through promoting mitochondrial fission and decreasing AKAP1 expression level; 3) AKAP1 played a novel role in CDDP resistance by inhibiting P53-mediated DRP1 dephosphorylation; 4) miR-148a-3p reconstitution in CDDP-resistant cells inhibits the cyto-protective autophagy by suppressing RAB12 expression and mTOR1 activation. Taken together, our study demonstrates that miR-148a-3p could be a promising prognostic marker or therapeutic candidate for overcoming CDDP resistance in GC.
Keywords: miR-148a-3p, AKAP1, RAB12, Cisplatin sensitivity, Gastric cancer
1. Introduction
Gastric cancer (GC) is one of the most common malignancies world-wide with high prevalence in East Asia. In fact, GC is the second leading cause of cancer-related death worldwide[1]. Cisplatin (CDDP) or Fluorouracil-based chemotherapy is the first-line regimen used to treat patients with locally advanced or metastatic GC [2]. However, more than 50% of patients exhibit intrinsic or acquired drug resistance with 5-year survival rates approximately 20% [3]. CDDP drug resistance could be determined by several factors, such as reduction in drug influx, improvement in drug efflux or metabolism, increase in DNA repair, activation of pro-survival signaling, and inhibition of pro-apoptotic pathways [4]. The molecular mechanisms underlying these phenomena remain to be explored, and new therapeutic strategies for treating GC are urgently needed.
Uncontrolled proliferative capacity and oncogenic features of cancer cells require a sustained high energy source that meets the cancer cell energy demands. Genes or non-coding RNA associated with energy synthesis and metabolism play an important role in carcinogenesis and tumor progression [5–7]. Mitochondria are an essential energy source in cells where changes of mitochondria metabolic homeostasis directly determine the fate of cancer cells [8]. It has been shown that mitochondrial fusion and fission, collectively termed mitochondrial dynamics, are determinants of mitochondrial stability [9]. Both excessive fusion and fission can cause mitochondrial dysfunction in cancer cells [10]. Furthermore, mitochondria-dependent apoptosis is recognized as one of the most important intrinsic pathways leading to cancer cell death in response to anti-cancer therapies [11]. Recent studies have shown that mitochondrial fission is a critical initiating step of mitochondria-mediated apoptosis [12]. Dynamin-Related Protein 1 (DRP1) is an upstream regulator of mitochondrial fission [13]. Several studies demonstrated that targeting DRP1 and mitochondrial fission may be an effective approach in cancer treatment [14]. Additionally, Fission, Mitochondrial 1 (FIS1), which serves as a receptor to recruit DRP1 to mitochondria, has also been uncovered to promote CDDP sensitivity in tongue squamous cell carcinoma [15]. This information suggests a close relationship between DRP1-mediated mitochondrial fission and CDDP sensitivity in cancer.
Autophagy is a stress response intracellular degradation system that utilizes organelles and proteins to produce energy for promoting cancer cells survival [16]. Autophagy inhibitors could enhance apoptosis (type I cell death) in many types of cancer [17–19]. Therefore, cancer cells may have the ability to use mitochondria and autophagy as two major sources for energy production that is required for multiple oncogenic cellular functions including resistance to cell death, proliferation, and invasion. However, excessive autophagic process can also lead to autophagic cell death (ACD), also known as type II cell death [20], this might be due to either autophagic digestion of cytoplasmic substances reaching a lethal threshold or cellular selective degradation of cargos needed for survival. Thus, autophagic process is a “double-edged sword” in cancer cells and needs to be carefully studied.
MicroRNAs (miRNAs) function as the central controllers of gene expression, which regulate mRNAs by cleaving them or preventing their translation [21]. Previous studies have shown that miRNAs regulate key cellular functions and are deregulated in several cancer types [22, 23], including GC [24]. miR-148a-3p is a member of the miR-148/152 family, which has been established with tumor suppressor functions [25–27]. However, molecular and mechanistic data regarding the relationship between miR-148a and CDDP sensitivity or resistance is lacking. Using bioinformatics analyses combining TCGA with GEO datasets, we demonstrate that miR-148a-3p plays an important role in CDDP cytotoxicity of GC cells. Further experiments elucidate that miR-148a-3p modulates CDDP sensitivity by simultaneous regulating Ras-related protein Rab-12, a member of the RAS Oncogene Family, (RAB12)-mediated autophagy and A-Kinase Anchoring Protein 1 (AKAP1)-mediated mitochondrial fission. These findings suggest that the miR-148a-3p-RAB12 and -AKAP1 axes can serve as a potential therapeutic target in the treatment of GC as well as a biomarker to predict responsiveness to CDDP in management of GC patients.
2. Materials and Methods
All the materials and methods, and abbreviations are included in Supplementary Materials and Methods.
3. Results
3.1 miR-148a-3p expression level is decreased in CDDP-resistant GC tissues and associated with poor clinical outcome
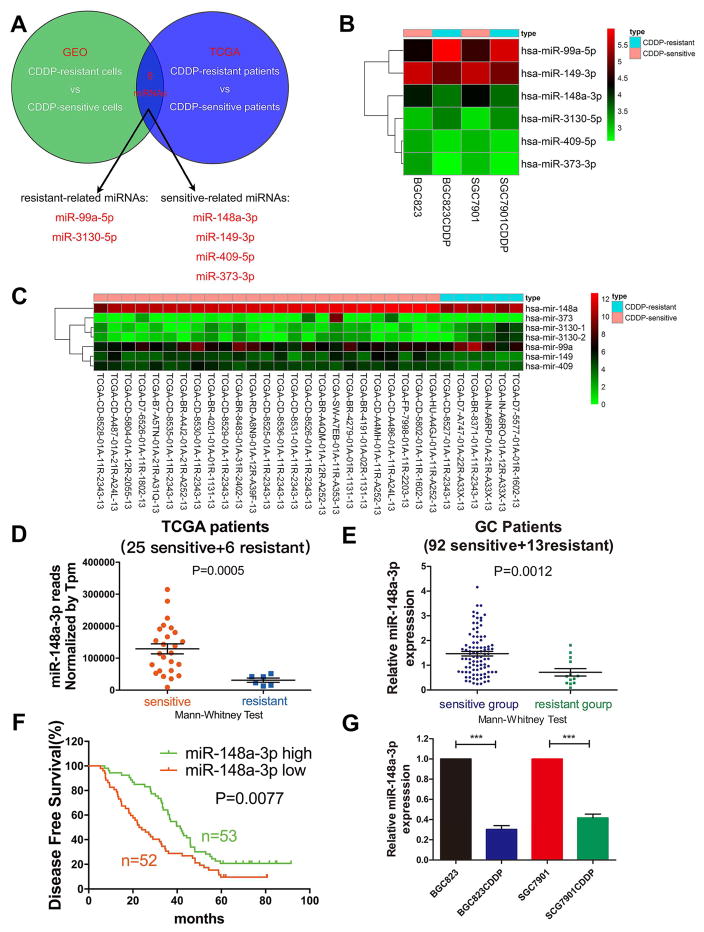
To find out the miRNAs associated with CDDP sensitivity and resistance in GC, miRNA expression levels were analysed by combining TCGA database, including 31 GC patients who received CDDP-based chemotherapy after surgery, with a GEO database containing microarray results pertaining to CDDP-resistant or CDDP-sensitive GC cells (GSE86195). The integrated analysis identified two upregulated CDDP resistant-related miRNAs (miR-99a-5p and miR-3130-5p) and four downregulated CDDP sensitive-related miRNAs (miR-148a-3p, miR-149-3p, miR-373-3p, and miR-409-5p) (Fig. 1A and Table S1). Expression of miR-148a-3p was decreased both in CDDP-resistant BGC823 and SGC7901 cells compared with their parental CDDP-sensitive cells, based on the GEO dataset (Fig. 1B). Reduced expression of miR-148a-3p was also observed in six CDDP-resistant patients, as compared to twenty-five CDDP-sensitive subjects based on TCGA database (Fig. 1C). Additionally, miR-148a-3p was one of the miRNAs that had the highest average expression level (32775 reads) among patients on TCGA (Fig. 1D). These findings suggested that miR-148a-3p may be involved in CDDP sensitivity. We next investigated the expression level of miR-148a-3p in human GC tissue samples. miR-148a-3p expression level was significantly downregulated in CDDP-resistant GC tissues (n=13), as compared with CDDP-sensitive GC tissues (n=92) (Fig. 1E). In patients who received CDDP-based therapy, high expression levels of miR-148a-3p in GC tissues were associated with significantly increased five-year disease free survival, as compared with low expression group (Fig. 1F). Similar result was acquired when we used cut-off value to group survival data (Supplementary Fig. S1A–C). Clinicopathological characteristics analysis in the same patient groups showed that miR-148a-3p expression level was negatively correlated with tumor size, clinical stage, T classification, and CDDP resistance (Table 1). Besides, miR-148a-3p expression levels were also significantly decreased in CDDP-resistant BGC823 and SGC7901 cells (i.e. BGC823CDDP and SGC7901CDDP), as compared with parental-sensitive cells (i.e. BGC823 and SGC7901) (Fig. 1G). The resistance to CDDP was validated in both BGC823CDDP and SGC7901CDDP cells using CCK-8 assay (Supplementary Fig. S1D–F).
Figure 1. miR-148a-3p downregulation was associated with CDDP resistance in GC indicating poor prognosis.
(A) The schematic diagram of screening strategy of miRNAs associated with CDDP resistance in GC. (B) miRNA profiling data from GEO database was analyzed by Perl programming language and R-language software (limma package) with fold changes ≥1.5 and P<0.05. Heat map of six miRNAs (upregulated or downregulated in both TCGA and GEO databases) was generated using R-language software (pheatmap package) (C) 31 GC patients were selected from TCGA database who received CDDP-based chemotherapy after surgery. Differential expression miRNAs (fold change ≥2 and P<0.05) were analyzed using R-language software (edgeR package) and Tpm correction. Heat map was generated as mentioned in panel b. (D) miR-148a-3p normalized reads (Tpm correction) between CDDP-resistant and -sensitive patients in TCGA database. (E) miR-148a-3p expression level in 105 GC patient tissue samples (CDDP resistant and sensitive groups). (F) Kaplan-Meier analysis of 5-year disease-free survival of 105 GC patients who received CDDP-based chemotherapy with high or low miR-148a-3p expression. (G) RT-PCR analysis of miR-148a-3p expression level in CDDP-resistant BGC823CDDP/SGC7901CDDP cell lines and parental CDDP-sensitive BGC823/SGC7901 cell lines. Graph represents mean ± SEM; *P<0.05, **P<0.01, ***P<0.001.
Table 1.
Correlation of relative miR-148a-3p expression with the clinicopathological characteristics of 105 patients who accepted cisplatin-based chemotherapy with gastric cancer.
| Characteristics | Number | No. of patients
|
Pvalue | ||
|---|---|---|---|---|---|
| miR-148a-3phigh | miR-148a-3plow | ||||
| Age(y) | ≥ 60 | 43 | 23 | 20 | 0.607 |
| < 60 | 62 | 30 | 32 | ||
| Gender | Male | 80 | 43 | 37 | 0.230 |
| Female | 25 | 10 | 15 | ||
| Tumor size(cm) | ≥ 3.5 | 49 | 19 | 30 | 0.025* |
| < 3.5 | 56 | 34 | 22 | ||
| Histological grade Well-moderately | 33 | 18 | 15 | 0.572 | |
| Poorly-sig net | 72 | 35 | 37 | ||
| Clinical stage | II | 39 | 26 | 13 | 0.011* |
| III | 66 | 27 | 39 | ||
| T classification | T1-T2 | 31 | 21 | 10 | 0.022* |
| T3-T4 | 74 | 32 | 42 | ||
| N classification | N0 | 14 | 9 | 5 | 0.267 |
| N1-N3 | 91 | 44 | 47 | ||
| Cisplatin chemosensitivity | Sensitive | 92 | 51 | 41 | 0.007** |
| Resistant | 13 | 2 | 11 | ||
P<0.05,
P<0.01,
P<0.001.
3.2 miR-148a-3p enhances CDDP cytotoxicity of GC cells in vitro
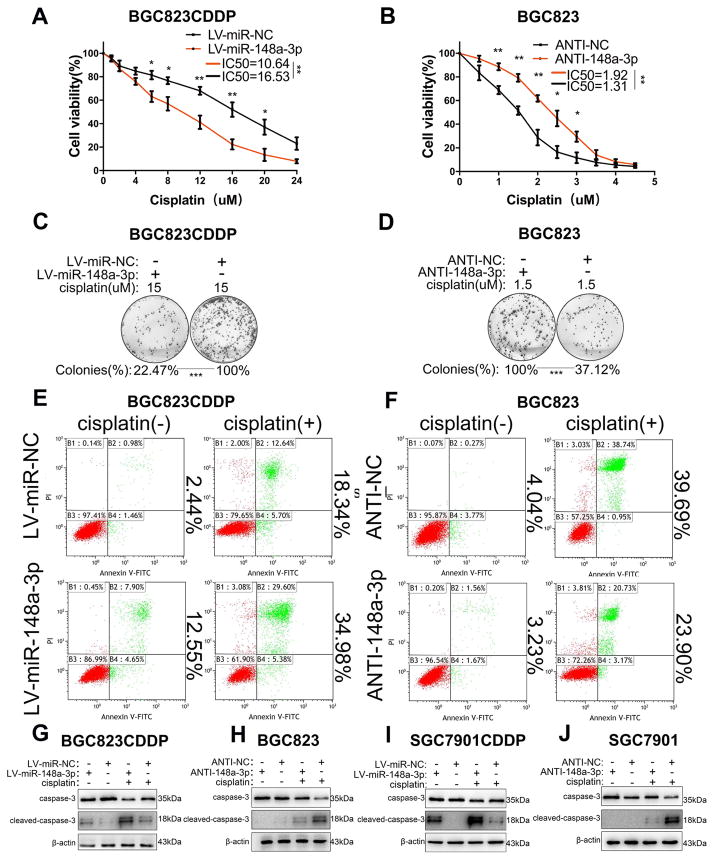
Stable miR-148a-3p reconstitution in BGC823CDDP and SGC7901CDDP cells decreased the CDDP IC50s in contrast to control cells (Fig. 2A and Supplementary Fig. S2A). On the other hand, miR-148a-3p inhibition significantly increased the IC50s of BGC823 and SGC7901 cells (Fig. 2B and Supplementary Fig. S2B). Consistently, reconstitution of miR-148a-3p also decreased the numbers of cell colony and promoted apoptosis in colony formation assay (Fig. 2C and Supplementary Fig. S2C) and flow cytometry analysis (Fig. 2E and Supplementary Fig. S2E, G, I). Whereas miR-148a-3p stable suppression could observe the opposite results (Fig. 2D, F and Supplementary Fig. S2D, F, H, J). Western blot was utilized to investigate the possible mechanism underlying. miR-148a-3p reconstitution in BGC823CDDP or SGC7901CDDP cells increased cleaved caspase-3 protein level, but decreased the inactivated form of caspase-3 protein level, with or without CDDP treatment, as compared with controls (Fig. 2G and I). Whereas, we detected the opposite findings in miR-148a-3p inhibited BGC823 and SGC7901 cells (Fig. 2H and J). Our findings imply that miR-148a-3p could play an important role in sensitizing GC cells to CDDP through enhancing CDDP-induced cell apoptosis.
Figure 2. miR-148a-3p sensitized GC cells to CDDP treatment in vitro.
(A, B) Cell viability was determined using CCK-8 in cells treated with indicated concentrations of CDDP for 48h. Average IC50s of three independent experiments were calculated. (C, D) The average colony numbers (%) of three independent experiments were calculated. (E, F) Flow cytometry analysis of apoptosis rates in BGC823CDDP (with or without miR-148a-3p reconstitution) or BGC823 cells (with or without miR-148a-3p knockdown) after CDDP treatment. Quantification data was in Supplementary Fig. S2E–F. (G–J) Caspase-3 and cleaved caspase-3 protein levels were analyzed by Western blot. β-actin was used as internal reference. CDDP concentrations: 15 μM in BGC823CDDP, 8 μM in SGC7901CDDP, 1.5 μM in BGC823 and SGC7901. Graph represents mean ± SEM; *P<0.05, **P<0.01, ***P<0.001.
3.3 miR-148a-3p sensitizes GC cell to CDDP by increasing mitochondrial fission-induced apoptosis
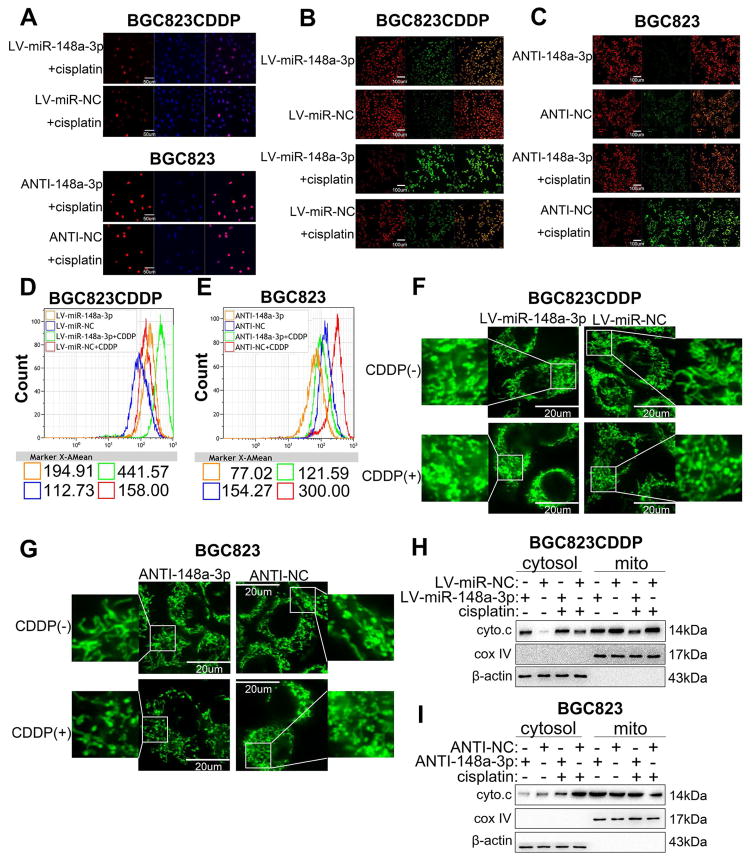
Immunostaining of phospho-H2AX (yH2AX) and ELISA assay for DNA-CDDP adducts detection [28] were used to test whether miR-148a-3p facilitated CDDP sensitivity in GC cells through inhibiting the DNA damage repair process. Under CDDP treatment conditions, we did not detect significant differences in yH2AX following reconstitution or inhibition of miR-148-3p in resistant and sensitive cells, respectively (Fig. 3A and Supplementary Fig. S3A–D). There were also no significant changes in the DNA-CDDP adduct amounts in the same cells (Supplementary Fig. S3E–H). Subsequently, mitochondrial membrane potential (MMP) and reactive oxygen species (ROS) levels were tested to explore the potential mechanism of caspase-3 activation. Of note, we detected a reduction in MMP levels (Fig. 3B and Supplementary Fig. S3I, K, M) with an elevation in ROS levels (Fig. 3D and Supplementary Fig. S4A, C, D) following reconstitution of miR-148a-3p with or without CDDP treatment. Converse results were determined in miR-148a-3p inhibited BGC823 and SGC7901 cells (Fig. 3C and Supplementary Fig. S3J, L, N) (Fig. 3E and Supplementary Fig. S4B, E, F). Collectively, these results suggested that miR-148a-3p promoted mitochondrial dysfunction induced by CDDP in GC cells. We next used the Mito-Tracker Green probes to detect mitochondrial fission in GC cells. Our data demonstrated that numbers of mitochondrial fragment were significantly increased following miR-148a-3p reconstitution in BGC823CDDP and SGC7901CDDP cells with or without CDDP treatment (Fig. 3F and Supplementary Fig. S4G, I, J), but decreased in miR-148a-3p suppressed BGC823 and SGC7901 cells (Fig. 3G and Supplementary Fig. S4H, K, L). Moreover, Western blot indicated that protein levels of cytochrome C were increased in cytosol of miR-148a-3p reconstituted cells and diluted in mitochondria (Fig. 3H and Supplementary Fig. S4M). However, an inverse results were found after miR-148a-3p inhibition in BGC823 and SGC7901 cells (Fig. 3I and Supplementary Fig. S4N). Together, these results suggest that miR-148a-3p increases CDDP-induced apoptosis through enhancing mitochondrial fission and dysfunction in GC cells.
Figure 3. miR-148a-3p increased CDDP-induced cell death through promoting mitochondrial fission-mediated apoptosis.
(A) Immunofluorescence staining of phosphorylated H2AX (yH2AX) using confocal microscopy in BGC823CDDP and BGC823 cells treated with CDDP (48h). Scale bar = 50 μm. (B, C) Mitochondrial membrane potential was detected in BGC823CDDP and BGC823 cells by immunofluorescence JC-1 staining. Scale bar = 100 μm. (D, E) Cellular ROS levels were measured by DCFH-DA staining in cells with CDDP 48h treatment. (F, G) BGC823CDDP and BGC823 cells transfected with Mito-Tracker Green probe. Mitochondrial morphology in individual cells was dyed green color by the probe, and identified by confocal microscopy. Fragmented mitochondria were shortened, punctate, and sometimes rounded, whereas filamentous mitochondria showed a thread-like tubular structure. Quantitation of mitochondria were evaluated as mentioned in Materials and Methods. Mitochondrial morphology was observed in BGC823CDDP and BGC823 cells using Mito-Tracker Green staining after CDDP or PBS exposure for 48 h. Scale bar = 20 μm. Mitochondrial fission positive cell percentages were calculated, as mentioned in Supplementary Fig. S4G–H. (H, I) Western blot analysis of cytochrome C (cyto.c) protein levels in cytosol or mitochondrial (mito) of BGC823CDDP (with or without miR-148a-3p reconstitution) and BGC823 cells (with or without miR-148a-3p inhibition) with or without 48h CDDP treatment. β-actin: internal control in cytosol. Cox IV: internal control in mitochondrial fragments. CDDP concentrations: same as in Figure 2. Graph represents mean ± SEM; *P<0.05, **P<0.01, ***P<0.001.
3.4 miR-148a-3p promotes CDDP-induced cell death by inhibiting autophagosome formation in GC cells
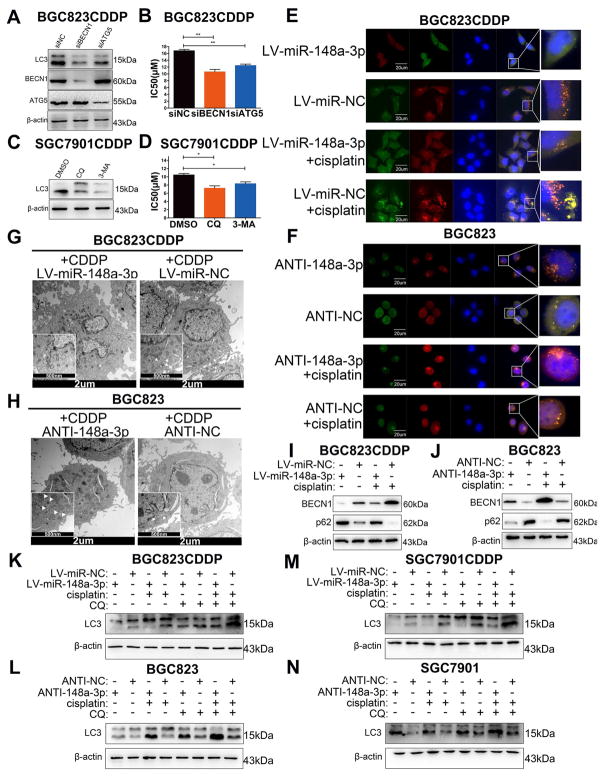
Recent studies have uncovered that miRNAs could affect CDDP sensitivity in cancer cells by regulating autophagy [29, 30]. Therefore, we explored whether miR-148a-3p could also promotes CDDP-induced cell death through regulating gastric cancer cells’ intracellular autophagy. MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3), which is known as Atg8 in yeast, is a commonly used marker to monitor autophagy. During autophagy process, LC3-I, the lipid-insoluble form of LC3, binds to phosphatidylethanolamine (PE), and then converted into lipid-soluble LC3-II, eventually participates in autophagosome formation[31]. LC3-II can be quickly degraded by lysosome, thus we can measure autophagy flux by the level changes of LC3-II with or without lysosome inhibitor chloroquine (CQ)[32]. The protein levels of type II LC3 (LC3-II) was increased in BGC823CDDP and SGC7901CDDP cells contrasted with their parental CDDP-sensitive cells with or without CDDP or autophagy inhibitor CQ treatment (Supplementary Fig. S5A). Of note, CDDP and CQ combination treatment also induced more significant differences of protein level of LC3-II between CDDP-resistant cells and –sensitive cells compared with CDDP or CQ treatment alone (Supplementary Fig. S5A). Additionally, transient knockdown of BECN1 or ATG5 in BGC823CDDP cells significantly decreased the protein level of LC3-II and CDDP IC50 as compared with controls (Fig. 4A and B), and similar results were discovered in SGC7901CDDP cells with autophagy inhibitor CQ or 3-Methyladenine (3-MA) treatment (Fig. 4C and D). These results suggested that CDDP resistance was partially attributed to activation of cyto-protective autophagy. After GFP-mRFP-LC3 plasmid transfection, we found that the numbers of both autophagosome and autolysosome were significantly decreased in miR-148a-3p-reconstituted BGC823CDDP and SGC7901CDDP cells (Fig. 4E and Supplementary Fig. S5C, D, G, I, J), but elevated after inhibition of miR-148a-3p in BGC823 and SGC7901 cells (Fig. 4F and Supplementary Fig. S5 Supplementary Fig. S5E, F, H, K, L). Similar findings were obtained using transmission electron microscopy (TEM) (Fig. 4G, H and Supplementary Fig. S6A, B). Western blot analysis demonstrated that miR-148a-3p-reconstituted CDDP-resistant cells showed increased protein expression level of p62 but declined protein level of BECN1 after CDDP stimulation (Fig. 4I and Supplementary Fig. S6C). However, decreased p62 and increased BECN1 expression were observed in miR-148a-3p-inhibited BGC823 and SGC7901 cells (Fig. 4J and Supplementary Fig. S6D). Moreover, miR-148a-3p reconstitution also dramatically decreased, whereas stable suppression of miR-148a-3p significantly increased LC3-II protein levels with or without CDDP or CQ treatment (Fig. 4K–N). The descent of CDDP-induced apoptosis by miR-148a-3p suppression in BGC823 cells was restored by transient knockdown of BECN1 or ATG5 using siRNA (siBECN1/siATG5) (Supplementary Fig. S6E and F) or in SGC7901 cells with CQ/3-MA (Supplementary Fig. S6G and H). Taken together, our results show that miR-148a-3p promotes CDDP-induced apoptosis through decreasing GC cell autophagosome formation.
Figure 4. miR-148a-3p enhanced CDDP cytotoxicity by inhibiting cyto-protective autophagy.
(A) Western blot analysis of LC3-II, BECN1, and ATG5 protein levels in cells with BECN1 or ATG5 siRNA knockdown (C) or with CQ or 3-MA treatment. (B, D) CDDP IC50s of cells in Panel A&C. (E, F) Immunofluorescence analysis using GFP-mRFP-LC3 staining. Scale bar = 20 μm. The numbers of LC3 puncta (yellow puncta for autophagosome, red puncta for autolysosome) were quantified in Supplementary Fig. S5C–F. (G, H) Transmission Electron Microscope images of endogenic autophagic microstructure in BGC823CDDP and BGC823 cells. The white arrows refer to cellular autophagosome or autolysosome that has a double layer structure. Scale bar = 2 μm or 500 nm. (I, J) Western blot analysis of BECN1 and p62 protein levels in cells with or without CDDP treatment. (K–N) LC3-II protein levels were detected in cells with CDDP (48h) and/or CQ (1h) treatment. CQ, Chloroquine. DMSO, Dimethyl Sulfoxide. 3-MA, 3-Methyladenine. β-actin: internal control. CDDP concentrations: same as in Figure 2. Graph represents mean ± SEM; * = P<0.05, ** = P<0.01, *** = P<0.001.
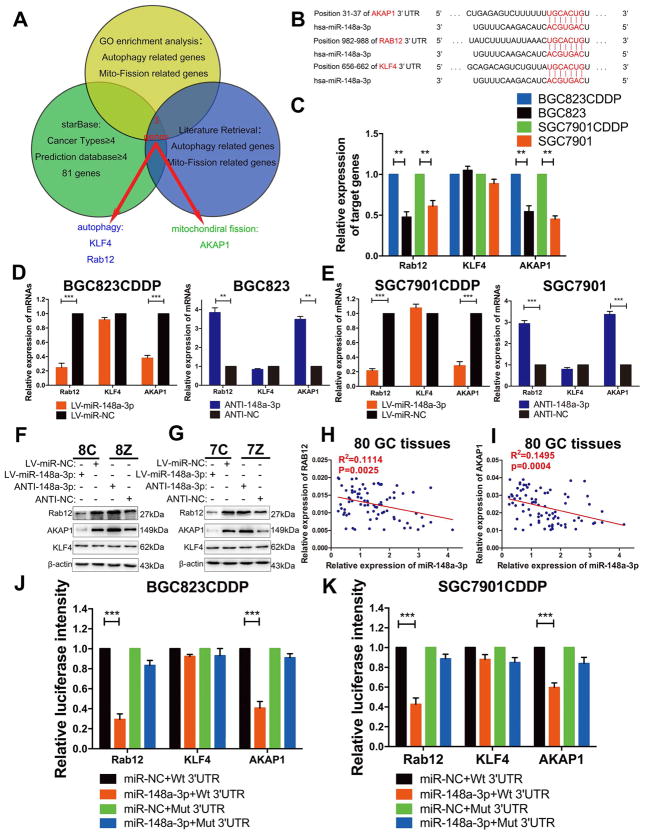
3.5 AKAP1 and RAB12 are direct downstream targets of miR-148a-3p
To investigate the mechanisms underlying the regulation of autophagy and mitochondrial fission by miR-148a-3p, we screened genes by literature retrieval and GO enrichment analysis to find genes that may be involved in the autophagic process or mitochondrial fission. These candidate genes were further screened using starBase v2.0 (http://starbase.sysu.edu.cn/), to fulfil the following criteria: 1) ≥4 types of cancer with CLIP-seq data showing that the gene could directly interact with miR-148a-3p, and 2) ≥4 online predictive programs implying that the gene may be one of the potential targets of miR-148a-3p (Fig. 5A). Based on the results, we identified two potential target genes (RAB12 and KLF4) of miR-148a-3p that had been reported to be involved in autophagic processes and one gene (AKAP1) that may regulate mitochondrial fission (Fig. 5B) [33–35]. RAB12 and AKAP1 mRNA levels were significantly upregulated, whereas KLF4 mRNA levels were unchanged in CDDP-resistant cells compared with CDDP-sensitive cells (Fig. 5C). Furthermore, RAB12 and AKAP1 mRNA (Fig. 5D and E) and protein expression levels (Fig. 5F and G) were dramatically descended in miR-148a-3p-reconstituted BGC823CDDP and SGC7901CDDP cells, but ascended when miR-148a-3p was suppressed in BGC823 and SGC7901 cells; no changes in KLF4 levels were detected. At the meantime, using 80 GC patients’ tissue samples, who received CDDP-based chemotherapy after gastrectomy, miR-148a-3p expression level was significantly negatively correlated with RAB12 or AKAP1 mRNA level (Fig. 5H and I). Luciferase reporter plasmids containing AKAP1, RAB12 or KLF4 wildtype (contain miR-148a-3p binding sites) or mutant 3′UTRs (lack of miR-148a-3p binding sites) were transfected into BGC823CDDP and SGC7901CDDP cells with or without miR-148a-3p reconstitution. Fluorescence intensities of AKAP1 and RAB12 plasmids with wild-types 3′UTR were significantly decreased in miR-148a-3p reconstituted cells, whereas fluorescence intensities of RAB12 and AKAP1 plasmids with mutant types 3′UTR had no significant difference. However, the fluorescence intensity of KLF4 plasmid with both wild-type and mutant type 3′ UTR were unchanged after miR-148a-3p reconstitution (Fig. 5J and K). These results illustrate that RAB12 and AKAP1 are direct targets of miR-148a-3p. It is worth mentioning that when we searched potential miRNAs which may regulate both RAB12 and AKAP1, we found that miR-92b-3p also meets our screening criteria except for miR-148a-3p (Table S2). However, in our validation results, both miR-92b-3p upregulation or downregulation had no influence on AKAP1 and RAB12 protein levels (Supplementary Fig. S7A–F). Besides, there was no statistical difference of miR-92b-3p expression in CDDP resistant patients compared with CDDP sensitive patients based on TCGA database (Supplementary Fig. S7G). Therefore, we concluded that miR-92b-3p cannot regulate AKAP1 and RAB12, at least in CDDP resistance of GC.
Figure 5. miR-148a-3p directly bound to the 3′UTRs of AKAP1 and RAB12.
(A) Bioinformatics analysis process showed the potential targets of miR-148a-3p. (B) Specific miR-148a-3p binding sites in the 3′ UTRs of AKAP1, RAB12 and KLF4 genes. (C) RT-PCR analysis of mRNA expression levels of AKAP1, Rab12, and KLF4 in BGC823CDDP, SGC7901CDDP and their parental CDDP-sensitive cells (BGC823 and SGC7901). (D, E) AKAP1, RAB12, KLF4 mRNA (RT-PCR) and (F, G) protein expression levels (Western blot) with or without miR-148a-3p reconstitution in CDDP-resistant cells or miR-148a-3p inhibition in CDDP-sensitive cells. 8C: BGC823CDDP; 8Z: BGC823; 7C: SGC7901CDDP; 7Z: SGC7901. (H, I) Correlations between miR-148a-3p and RAB12 or AKAP1 mRNA levels in 80 GC patients tissue samples measured by Pearson correlation coefficient respectively. (J, K) Luciferase reporter plasmids containing AKAP1, RAB12 or KLF4 wildtype (Wt, contain miR-148a-3p binding sites) or mutant 3′UTRs (Mut, lack of miR-148a-3p binding sites) were transfected into BGC823CDDP and SGC7901CDDP cells with or without miR-148a-3p reconstitution. β-actin: internal control. CDDP concentrations: same as in Figure 2. Graph represents mean ± SEM; *P<0.05, **P<0.01, ***P<0.001.
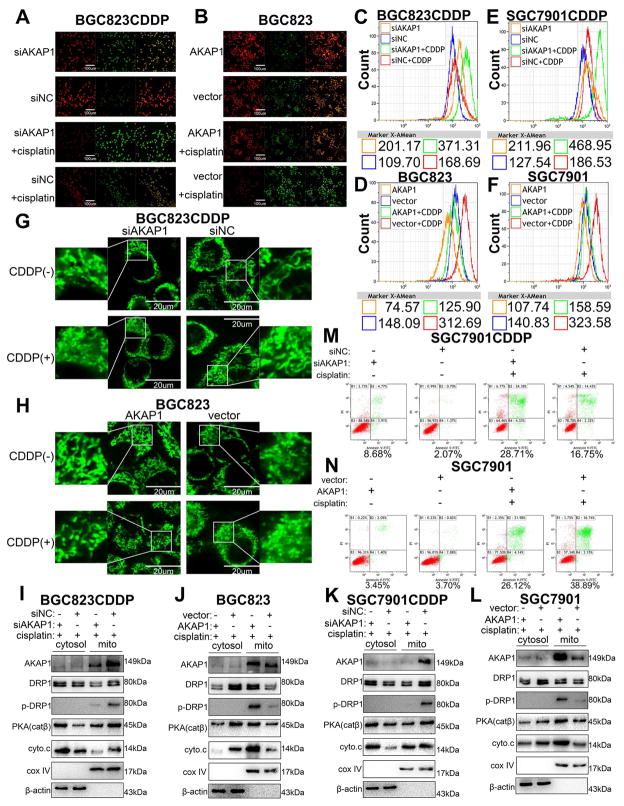
3.6 AKAP1 facilitates GC cell CDDP resistance by promoting PKA-mediated phosphorylation of DRP1
JC-1 staining results demonstrated that knockdown of AKAP1 in BGC823CDDP or SGC7901CDDP cells decreased the mitochondrial membrane potential (MMP) (Fig. 6A and Supplementary Fig. S8C, E, G), and these results were further validated in AKAP1 overexpressed BGC823 and SGC7901 cells (Fig. 6B and Supplementary Fig. S8D, F, H). However, cellular ROS levels were significantly increased in AKAP1-downregulated cells, but decreased in AKAP1-upregulated cells with or without CDDP exposure (Fig. 6C–F and Supplementary Fig. S8I–L). In addition, AKAP1 knockdown in BGC823CDDP or SGC7901CDDP cells significantly accelerated CDDP-induced mitochondrial fission (Fig. 6G and Supplementary Fig. S9A, C, D); and the opposite results were observed in AKAP1-reconstituted cells (Fig. 6H and Supplementary Fig. S9B, E, F). Western blot analysis revealed that PKA (catalytic subunit β) and cytochrome C protein expression levels were decreased in mitochondria segment but increased in cytosol; phosphorylated DRP1 (p-DRP1, Ser637) level was decreased in the mitochondria after AKAP1 knockdown compared with control cells (Fig. 6I and K). Alternatively, opposite findings were detected in cells with AKAP1 reconstitution (Fig. 6J and L). Flow cytometry results manifested that CDDP-induced apoptosis was significantly promoted in SGC7901CDDP cells with AKAP1 knockdown (Fig. 6M and Supplementary Fig. S9G), but suppressed in AKAP1 overexpressed SGC7901 cells (Fig. 6N and Supplementary Fig. S9H). These findings indicate that the CDDP resistance-inducing effect of AKAP1 is mediated by DRP1 phosphorylation and inactivation.
Figure 6. AKAP1 restrained mitochondrial fission and reinforced CDDP resistance in GC cells.
(A) Immunofluorescence analysis of JC-1 in BGC823CDDP cells (with or without AKAP1 knockdown) and (B) BGC823 cells (with or without AKAP1 overexpression) with CDDP 48h treatment. Scale bar = 100 μm (C–F) ROS levels were determined in BGC823CDDP, SGC7901CDDP (with or without AKAP1 inhibition), BGC823 and SGC7901 cells (with or without AKAP1 reconstitution) after 48h CDDP exposure. (G, H) Mitochondrial morphology in individual cells was dyed green color by the probe, and identified by confocal microscopy. Mitochondrial fragments were detected in AKAP1-knockdown BGC823CDDP cells, AKAP1-reconstituted BGC823 and control cells using Mito-Tracker Green probes after 48h CDDP treatment. Scale bar = 20 μm. (I–L) Western blot analysis of AKAP1, DRP1, p-DRP1(Ser637), PKA (cat β), and cytochrome c (cyto.c) protein levels in CDDP resistant cells (with or without knockdown of AKAP1) or CDDP sensitive cells (with or without AKAP1 overexpression) with 48h CDDP treatment. (M, N) Flow cytometry analysis of cell apoptotic rates was measured in SGC7901CDDP cells (with or without AKAP1 knockdown) and SGC7901 cells (with or without AKAP1 overexpression). β-actin: internal control for cytosol. Cox IV: internal control for mitochondrial fragments. CDDP concentrations: same as in Figure 2. Graph represents mean ± SEM; *P<0.05, **P<0.01, ***P<0.001.
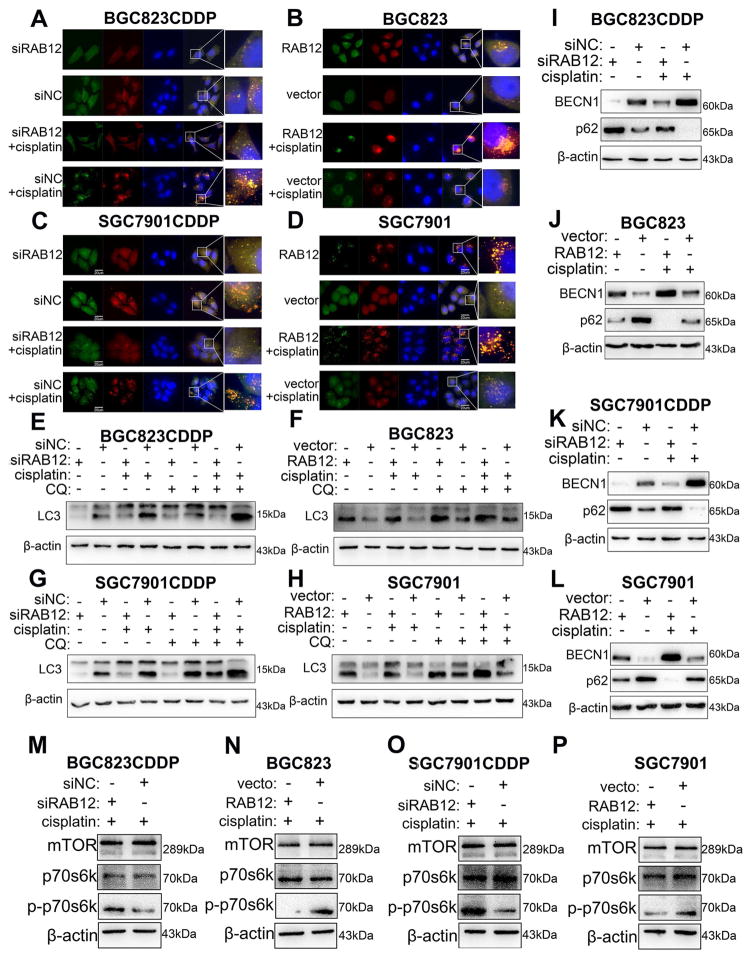
3.7 RAB12 inhibits CDDP-induced cell death by promoting autophagy via suppressing mTORC1 activity
Immunofluorescence analysis after GFP-mRFP-LC3 transfection indicated that RAB12 knockdown in BGC823CDDP and SGC7901CDDP cells reduced, whereas reconstitution of RAB12 in BGC823 and SGC7901 cells augmented both the amounts of autophagosome and autolysosome with or without CDDP treatment (Fig. 7A–D and Supplementary Fig. S10A–D). Western blot results illustrated that RAB12 knockdown not only decreased LC3-II protein level with or without CDDP, CQ alone or combination treatment (Fig. 7E and G), but also decreased BECN1 and increased p62 protein levels in the same cells (Fig. 7I and K). Alternatively, RAB12 overexpression showed the opposite findings (Fig. 7F, H, J and L). Additionally, we investigated the role of the mammalian target of rapamycin (mTOR) signaling pathway in RAB12-mediated autophagy[36]. As expected, the levels of p-p70s6k, readout of mTORC1 activity, was remarkably enhanced in BGC823CDDP and SGC7901CDDP cells following knockdown of RAB12 with CDDP treatment (Fig. 7M and O). The weak activity of mTOC1 (p-p70s6k) was observed in BGC823 and SGC7901 cells, in which RAB12 was ectopic expressed (Fig. 7N and P). Flow cytometry analysis determined that RAB12 knockdown promoted apoptosis caused by CDDP in BGC823CDDP cells (Supplementary Fig. S10G and H), whereas RAB12 reconstitution in BGC823 cells decreased CDDP-induced apoptosis compared with controls (Supplementary Fig. S10I and G). Therefore, our results suggest that RAB12 can reinforce GC cell CDDP resistance by inhibiting mTORC1 activity.
Figure 7. RAB12 promoted autophagy and decreased CDDP-induced cell apoptosis.
CDDP resistant cells (BGC823CDDP, SGC7901CDDP) with RAB12 transiently knockdown, and CDDP sensitive cells (BGC823, SGC7901) with RAB12 overexpression were utilized to investigate the role of RAB12 in autophagy and apoptosis. (A–D) Immunofluorescence staining of GFP-mRFP-LC3 in cells treated with CDDP for 48h. Scale bar = 20 μm. Western blot analysis of LC3-II (E–H), BECN1 and p62 (I–L) protein levels in cells with or without CDDP (48 h) or CQ (1h) treatment. (M–P) Pan-mTORC1 (total mTORC1), p-p70s6k (Thr389) and p70s6k protein levels were detected by Western blot in cells with 48h CDDP treatment. β-actin: internal control. CDDP concentrations: same as in Figure 2. Graph represents mean ± SEM; *P<0.05, **P<0.01, ***P<0.001.
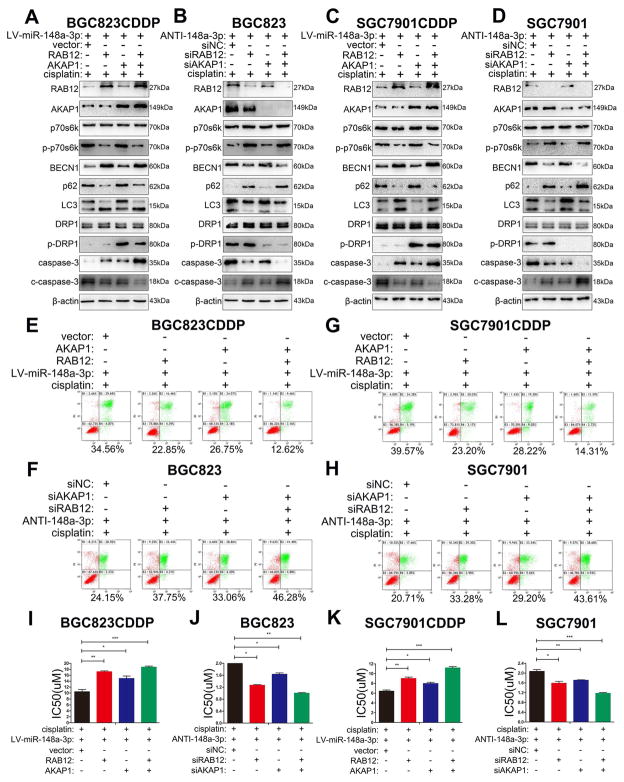
3.8 AKAP1 and RAB12, the two direct targets of miR-148-3p, are essential for the resistance to CDDP in gastric cancer
Whether targeting AKAP1 and/or RAB12 could sensitize the effect of CDDP and mimic miR-148a-3p functions remains to be explored. Western blot results demonstrated an increase in LC3-II, BECN1 protein levels and a decrease of cleaved caspase-3 and p62 following RAB12 overexpression in BGC823CDDP or SGC7901CDDP cells with miR-148-3p suppression (Fig. 8A and C). At the meantime, there was an increase in p-DRP1 (Ser637) protein level and a decrease in cleaved-caspase-3 after AKAP1 overexpression (Fig. 8A and C). Of note, the combined RAB12 and AKAP1 overexpression showed lower levels of cleaved caspase-3 protein, as compared with a single protein overexpression (Fig. 8A and C). These findings were further validated following single or combined knockdown of AKAP1 or RAB12 in BGC823 or SGC7901 cells with miR-148-3p suppression (Fig. 8B and D). Flow cytometry (Fig. 8E, G and Supplementary Fig. S11A, C) and cell viability (Fig. 8I and K) analyses indicated that AKAP1 or RAB12 overexpression relieved CDDP induced the cell death showing an increase of CDDP IC50s in miR-148-3p-reconstituted CDDP resistant cells. These results confirmed that suppression of AKAP1 and RAB12 is an essential step in sensitizing to CDDP. Furthermore, our results from AKAP1 or RAB12 knockdown in BGC823 or SGC7901 parental cells with miR-148a-3p inhibition and CDDP treatment further validated our findings (Fig. 8F, H, J, L and Supplementary Fig. S11B, D). Next, we explored whether there has potential regulation relationship between these two proteins. Inhibiting AKAP1 expression by two interference sequences not only had no significant influence on RAB12 expression in CDDP-resistant cells after CDDP stimulation (Supplementary Fig. S12A, B), but also could not change cellular autophagic level (Supplementary Fig. S12C–F). Consistently, neither RAB12 suppression had effect on AKAP1 expression or mitochondrial fission (Supplementary Fig. S13A–F). Thus, we concluded that there is no crosslinking between AKAP1 and RAB12.
Figure 8. miR-148a-3p enhanced CDDP cytotoxicity by simultaneously inhibiting AKAP1 and RAB12 expression in GC cells.
miR-148a-3p reconstituted CDDP resistant cells (BGC823CDDP, SGC7901CDDP) with RAB12, AKAP1 single or combined overexpression or miR-148a-3p inhibited CDDP sensitive cells (BGC823, SGC7901) with RAB12, AKAP1 single or combined knockdown were tested with 48h CDDP treatment. (A–D) Western blot analysis of RAB12, AKAP1, p70s6k, p-p70s6k(Thr389), BECN1, p62, LC3, DRP1, p-DRP1(Ser637), caspase-3, and cleaved caspase-3 protein levels. (E–H) Flow cytometry analysis of cell apoptosis. (I–L) CDDP IC50s calculated using CCK-8. CDDP concentrations: same as in Figure 2. β-actin: internal control. Graph represents mean ± SEM; *P<0.05, **P<0.01, ***P<0.001.
3.9 miR-148a-3p sensitization of GC cells to CDDP in vivo includes suppression of AKAP1 and RAB12 expression levels
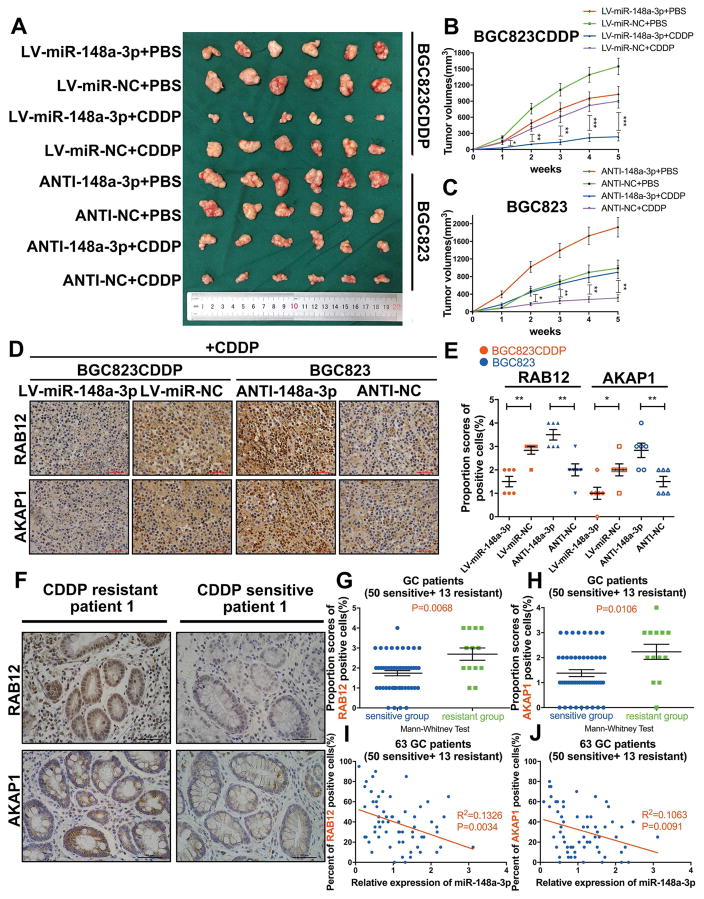
We generated xenograft tumor models to validate our previous findings. Tumor xenograft data indicated that miR-148a-3p reconstitution in CDDP-resistant cells can significantly decrease, whereas miR-148a-3p inhibition in CDDP-sensitive cells promoted xenograft tumor growth following CDDP treatment (Fig. 9A, B and C). RT-PCR results showed that miR-148a-3p expression was negatively correlated with AKAP1 or RAB12 mRNA level in xenograft tumors (Supplementary Fig. S14A–C). Consistently, immunohistochemistry staining of tumor xenograft samples indicated the similar results. (Fig. 9D and E). Analysis of human tissue samples showed significant higher protein expression levels of RAB12 and AKAP1 in tissue samples from GC patients with CDDP resistance, as compared to CDDP-sensitive GC patients (Fig. 9F and Supplementary Fig. S15). Similar as our previous findings, IHC scores data showed that RAB12 or AKAP1 protein expression was significantly increased in CDDP-resistant GC tissues (n=13), as compared to CDDP-sensitive GC tissues (n=50) (Fig. 9G and H). Furthermore, a negative correlation was identified between miR-148a-3p expression level and RAB12 or AKAP1 protein level in these 63 GC tissue samples (Fig. 9I and J). Univariate analysis of clinicopathological characteristics indicated that expression level of AKAP1 (Supplementary Table S3) or RAB12 (Supplementary Table S4) in GC cancer samples was associated with CDDP resistance in GC patients (P<0.05). Based on these data, we conclude that miR-148a-3p sensitizes GC cells to CDDP by targeting RAB12 and AKAP1.
Figure 9. miR-148a-3p sensitized GC cells to CDDP through decreasing AKAP1 and RAB12 expression levels in vivo.
Reconstitution of miR-148a-3p in BGC823CDDP cells or inhibition of miR-148a-3p in BGC823 cells were performed to validate our findings on the role of miR-148a-3p in promoting sensitivity of gastric cancer cells to CDDP. (A) Representative xenograft tumors of sacrificed mice at end of experiment with or without CDDP treatment (5 mg/kg, three times a week). (B, C) Growth curves of subcutaneous xenograft tumors. (D) AKAP1 and RAB12 expression levels were shown in representative xenograft tumors by Immunohistochemistry (IHC) (400x magnification, scale bars = 50 μm). (E) IHC scores quantification of RAB12 and AKAP1 expression levels. (F) IHC staining of AKAP1 or RAB12 in a CDDP-resistant patient and a CDDP-sensitive GC patient (400x magnification, scale bars = 50 um, more samples are shown in Supplementary Fig. S11). (G, H) IHC scores of AKAP1 and RAB12 were calculated in 13 CDDP-resistant and 50 CDDP-sensitive patient tissues. (I, J) The correlations between miR-148a-3p expression and AKAP1 or RAB12 positive cells percentages were calculated by Pearson correlation analysis. Graph represents mean ± SEM; *P<0.05, **P<0.01, ***P<0.001.
4. Discussion
CDDP resistance in cancer patients is a common clinical challenge worldwide. In this study, we investigated miRNAs that were associated with CDDP sensitivity or resistance in GC using TCGA and GEO databases. We found that miR-148a-3p was significantly downregulated in CDDP-resistant GC patient tissue samples and cell lines. Moreover, miR-148a-3p was expressed at significantly higher levels than other miRNAs in TCGA database (32775 reads/per sample), which suggested that it may have a greater effect than other miRNAs.
Based on this discovery, we designed a series of experiments and showed that miR-148a-3p enhanced CDDP-induced activation of caspase-3 and apoptosis. Inhibition of DNA damage repair and promotion of mitochondria-mediated apoptotic pathways are two major factors that are known to facilitate caspase-3-dependent apoptosis following the exposure of CDDP. However, miR-148a-3p had no effect on DNA repair in GC cells, implying that miR-148a-3p may decrease CDDP resistance through other mechanisms in GC cells.
P53 is one of the most important tumor-suppressors in cancer. CDDP induces p53 phosphorylation and translocation to mitochondria, binding to Bcl-2, inducing mitochondrial outer membrane permeabilization (MOMP) and apoptosis [37]. Of note, it has been reported that in conditions of CDDP treatment, the activated P53 could also translocate to mitochondria to induce DRP1 dephosphorylation and activation, thereby promoting mitochondrial fission and cell death in gynecological cancer [38]. These findings indicated that CDDP cytotoxicity involves DRP1 activation. AKAP1, acting as a scaffold, presents multiple macromolecules such as PKA to downstream targets at the outer mitochondrial membrane (OMM) controlling the phosphorylation state of these targets [33]. It has been shown that AKAP1-PKA axis can protect cells from hypoxia-induced cell death in myocardial infarction. This protective effect is restrained by ubiquitin ligase Siah2-mediated degradation of AKAP1 [39]. However, the role of AKAP1 is not well understood in cancer. Our results indicated that following miR-148a-3p downregulation, AKAP1 is upregulated in CDDP-resistant GC tissues and cells, antagonizing CDDP-induced mitochondrial fission by phosphorylation and inactivation of DRP1. These results highlighted the function of AKAP1 as a natural executor to protect cells from DRP1 activation (dephosphorylation) caused by CDDP, without affecting DRP1 expression. Therapeutic regimens targeting AKAP1 may restore the anti-cancer effects of CDDP in CDDP resistant cases. Notably, in addition to PKA, AKAP1 also recruits various other macromolecules to the OMM, such as protein tyrosine phosphatase D1 (PTPD1) and the non-receptor tyrosine kinase Src [40, 41]. Thus, additional mechanisms may be involved in GC cell apoptosis apart from AKAP1-PKA-mediated DRP1 phosphorylation. However, this hypothesis needs to be investigated by additional studies.
Autophagy can play a cellular-protective role to promote GC cell survival and CDDP resistance [42]. However, a prolonged and excessive self-eating process could lead to cancer cell death [43]. Herein, we discovered that autophagy played a protective role in GC cells when treated with CDDP. RAB12 is a member of the Ras oncogene family that influences autophagic processes [44]. In fact, RAB12 is a potent inducer of autophagy by inhibiting mTORC1 activity [36] or accelerating autolysosome maturation [34]. Our findings indicated that mTORC1-depressing effects of RAB12 facilitate early autophagosome formation to protect GC cells from CDDP-induced cell death. Of note, it has been shown that RAB12 could be a potential oncogene involved in colorectal cancer, although the mechanism remains unclear [45]. Our finding demonstrated that miR-148a-3p can significantly reduce autophagic flux and autophagosome formation by regulating RAB12 supporting the observed complex roles of miRNAs in cancer cells.
It’s worth noting that although we have proved that miR-148a-3p function as a crucial executor of both AKAP1 and RAB12, there may also have some other regulatory mechanisms, such as post-translational regulation. Thirstrup K et al. found that phosphorylation of RAB12 in Ser106 is associated with sporadic Parkinson’s disease[46]; Carlucci A et al. showed that AKAP121 levels are regulated by the ubiquitin/proteasome pathway[47]. These findings leading us to speculate that whether there is the possible cross-regulatory mechanism between RAB12 and AKAP1 involving protein phosphorylation or ubiquitylaytion. Moreover, Ranganathan G et al. demonstrated that AKAP1 includes an RNA-binding K homology (KH) domain, which could bind to 3′-UTR of Lipoprotein Lipase (LPL) mRNA, and participate in lipolysis[48]. Similarly, Small GTPase protein family, such as RAB7, has also been revealed that plays a significant role in lipolysis[49]. This potential connection indicates that AKAP1 may directly or indirectly regulate RAB12 by its RNA-binding capacity. All these problems that we have not solved in this study need to be further investigated in the future.
Several clinical trials are currently using microRNAs to reverse drug resistance in cancer [50]. The advantage of using miRNAs compared to existing strategies relies on miRNAs ability to suppress several oncogenic cellular pathways by targeting multiple genes in unrelated signaling networks. Based on our findings, we suggest that approaches utilizing miR-148a-3p reconstitution in therapy could reinforce CDDP sensitivity in GC. However, there is still need of a lot research work for applying miR-148a-3p plus CDDP as a clinical treatment for advanced GC, which is just theoretically feasible. Furthermore, at present, miRNA-based therapeutics remain face the challenges. For example, how to overexpress miR-148a-3p expression at local organ or tumor, and which concentration of miR-148a-3p is the most effective for the patients who are accepting CDDP treatment, also has the minimal side effects. These problems are what we cannot solve at present.
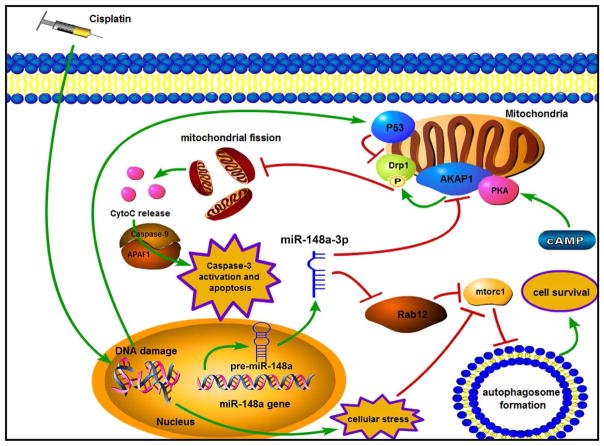
In conclusion, we demonstrated that miR-148a-3p reconstitution sensitizes GC cells to CDDP treatment by simultaneously targeting RAB12 and AKAP1. We found that following CDDP treatment miR-148a-3p reconstitution activates mitochondrial fission and apoptosis in GC cells by targeting AKAP1 and promoting activation of P53 and DRP1. In addition, miR-148a-3p expression also targets RAB12 to suppress autophagosome formation in GC in response to CDDP treatment (Fig. 10). These data suggested that the miR-148a-3p-RAB12 and -AKAP1 axes may serve as a novel prognostic and therapeutic target to overcome CDDP resistance for locally advanced or metastatic GC.
Figure 10.
Schematic model of the regulatory process of miR-148a-3p in CDDP sensitivity of gastric cancer.
Supplementary Material
Highlights.
miR-148a-3p is significant downregulated in CDDP-resistant GC tissues and cells.
miR-148a-3p influences prognosis in patients who received CDDP-based chemotherapy.
miR-148a-3p enhances CDDP sensitivity by targeting RAB12 and AKAP1 in GC.
AKAP1 phosphorylates DRP1 to abrogate CDDP-induced mitochondrial fission.
RAB12 promotes CDDP-induced protective autophagy by suppressing mTORC1 activation.
Acknowledgments
This work was supported by the National Natural Science Foundation Project of International Cooperation (NSFC-NIH, 81361120398); a Research Career Scientist award (1IK6BX003787) from the United States Department of Veterans affairs (W. El-Rifai); grants from the United States National Institutes of Health (R01CA177372 and R01CA93999, W. El-Rifai); the National Natural Science Foundation of China (81572362); the Program for Development of Innovative Research Team in the First Affiliated Hospital of NJMU; the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, JX10231801); 333 Project of Jiangsu Province (BRA2015474); Jiangsu Key Medical Discipline (General Surgery); and Jiangsu Key Lab of Cancer Biomarkers, Prevention and Treatment, Collaborative Innovation Center for Cancer Personalized Medicine, Nanjing Medical University.
Footnotes
Conflict of interest statement
The authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016 doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 3.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 4.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 5.Wu H, Ng R, Chen X, Steer CJ, Song G. MicroRNA-21 is a potential link between non-alcoholic fatty liver disease and hepatocellular carcinoma via modulation of the HBP1-p53-Srebp1c pathway. Gut. 2015 doi: 10.1136/gutjnl-2014-308430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keckesova Z, Donaher JL, De Cock J, Freinkman E, Lingrell S, Bachovchin DA, Bierie B, Tischler V, Noske A, Okondo MC, Reinhardt F, Thiru P, Golub TR, Vance JE, Weinberg RA. LACTB is a tumour suppressor that modulates lipid metabolism and cell state. Nature. 2017;543:681–686. doi: 10.1038/nature21408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sumita K, Lo YH, Takeuchi K, Senda M, Kofuji S, Ikeda Y, Terakawa J, Sasaki M, Yoshino H, Majd N, Zheng Y, Kahoud ER, Yokota T, Emerling BM, Asara JM, Ishida T, Locasale JW, Daikoku T, Anastasiou D, Senda T, Sasaki AT. The Lipid Kinase PI5P4Kbeta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis. Mol Cell. 2016;61:187–198. doi: 10.1016/j.molcel.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarosiek KA, Fraser C, Muthalagu N, Bhola PD, Chang W, McBrayer SK, Cantlon A, Fisch S, Golomb-Mello G, Ryan JA, Deng J, Jian B, Corbett C, Goldenberg M, Madsen JR, Liao R, Walsh D, Sedivy J, Murphy DJ, Carrasco DR, Robinson S, Moslehi J, Letai A. Developmental Regulation of Mitochondrial Apoptosis by c-Myc Governs Age- and Tissue-Specific Sensitivity to Cancer Therapeutics. Cancer Cell. 2017;31:142–156. doi: 10.1016/j.ccell.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasahara A, Scorrano L. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014;24:761–770. doi: 10.1016/j.tcb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Suliman HB, Piantadosi CA. Mitochondrial Quality Control as a Therapeutic Target. Pharmacol Rev. 2016;68:20–48. doi: 10.1124/pr.115.011502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ni Chonghaile T, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, del Moore VG, Deng J, Anderson KC, Richardson P, Tai YT, Mitsiades CS, Matulonis UA, Drapkin R, Stone R, Deangelo DJ, McConkey DJ, Sallan SE, Silverman L, Hirsch MS, Carrasco DR, Letai A. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science. 2011;334:1129–1133. doi: 10.1126/science.1206727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerveny KL, Tamura Y, Zhang Z, Jensen RE, Sesaki H. Regulation of mitochondrial fusion and division. Trends Cell Biol. 2007;17:563–569. doi: 10.1016/j.tcb.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Fell SM, Surova O, Smedler E, Wallis K, Chen ZX, Hellman U, Johnsen JI, Martinsson T, Kenchappa RS, Uhlen P, Kogner P, Schlisio S. The 1p36 Tumor Suppressor KIF 1Bbeta Is Required for Calcineurin Activation, Controlling Mitochondrial Fission and Apoptosis. Dev Cell. 2016;36:164–178. doi: 10.1016/j.devcel.2015.12.029. [DOI] [PubMed] [Google Scholar]
- 15.Fan S, Chen WX, Lv XB, Tang QL, Sun LJ, Liu BD, Zhong JL, Lin ZY, Wang YY, Li QX, Yu X, Zhang HQ, Li YL, Wen B, Zhang Z, Chen WL, Li JS. miR-483-5p determines mitochondrial fission and cisplatin sensitivity in tongue squamous cell carcinoma by targeting FIS1. Cancer Lett. 2015;362:183–191. doi: 10.1016/j.canlet.2015.03.045. [DOI] [PubMed] [Google Scholar]
- 16.Endo S, Nakata K, Ohuchida K, Takesue S, Nakayama H, Abe T, Koikawa K, Okumura T, Sada M, Horioka K, Zheng B, Mizuuchi Y, Iwamoto C, Murata M, Moriyama T, Miyasaka Y, Ohtsuka T, Mizumoto K, Oda Y, Hashizume M, Nakamura M. Autophagy is Required for Activation of Pancreatic Stellate Cells, Associated With Pancreatic Cancer Progression and Promotes Growth of Pancreatic Tumors in Mice. Gastroenterology. 2017 doi: 10.1053/j.gastro.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Chang Y, Yan W, He X, Zhang L, Li C, Huang H, Nace G, Geller DA, Lin J, Tsung A. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143:177–187 e178. doi: 10.1053/j.gastro.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 18.Dolma S, Selvadurai HJ, Lan X, Lee L, Kushida M, Voisin V, Whetstone H, So M, Aviv T, Park N, Zhu X, Xu C, Head R, Rowland KJ, Bernstein M, Clarke ID, Bader G, Harrington L, Brumell JH, Tyers M, Dirks PB. Inhibition of Dopamine Receptor D4 Impedes Autophagic Flux, Proliferation, and Survival of Glioblastoma Stem Cells. Cancer Cell. 2016;29:859–873. doi: 10.1016/j.ccell.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu FQ, Fang T, Yu LX, Lv GS, Lv HW, Liang D, Li T, Wang CZ, Tan YX, Ding J, Chen Y, Tang L, Guo LN, Tang SH, Yang W, Wang HY. ADRB2 signaling promotes HCC progression and sorafenib resistance by inhibiting autophagic degradation of HIF1alpha. J Hepatol. 2016;65:314–324. doi: 10.1016/j.jhep.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 20.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gougelet A, Sartor C, Bachelot L, Godard C, Marchiol C, Renault G, Tores F, Nitschke P, Cavard C, Terris B, Perret C, Colnot S. Antitumor activity of an inhibitor of miR-34a in liver cancer with beta-catenin-mutations. Gut. 2016;65:1024–1034. doi: 10.1136/gutjnl-2014-308969. [DOI] [PubMed] [Google Scholar]
- 23.Zhao M, Luo R, Liu Y, Gao L, Fu Z, Fu Q, Luo X, Chen Y, Deng X, Liang Z, Li X, Cheng C, Liu Z, Fang W. miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR-p-PI3K/AKT-c-JUN. Nat Commun. 2016;7:11309. doi: 10.1038/ncomms11309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han TS, Hur K, Xu G, Choi B, Okugawa Y, Toiyama Y, Oshima H, Oshima M, Lee HJ, Kim VN, Chang AN, Goel A, Yang HK. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut. 2015;64:203–214. doi: 10.1136/gutjnl-2013-306640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo SL, Peng Z, Yang X, Fan KJ, Ye H, Li ZH, Wang Y, Xu XL, Li J, Wang YL, Teng Y, Yang X. miR-148a promoted cell proliferation by targeting p27 in gastric cancer cells. Int J Biol Sci. 2011;7:567–574. doi: 10.7150/ijbs.7.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang SH, Li X, Zhou LS, Cao ZW, Shi C, Zhou CZ, Wen YG, Shen Y, Li JK. microRNA-148a suppresses human gastric cancer cell metastasis by reversing epithelial-to-mesenchymal transition. Tumour Biol. 2013;34:3705–3712. doi: 10.1007/s13277-013-0954-1. [DOI] [PubMed] [Google Scholar]
- 27.Zheng B, Liang L, Wang C, Huang S, Cao X, Zha R, Liu L, Jia D, Tian Q, Wu J, Ye Y, Wang Q, Long Z, Zhou Y, Du C, He X, Shi Y. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574–7583. doi: 10.1158/1078-0432.CCR-11-1714. [DOI] [PubMed] [Google Scholar]
- 28.Srivastava AK, Han C, Zhao R, Cui T, Dai Y, Mao C, Zhao W, Zhang X, Yu J, Wang QE. Enhanced expression of DNA polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells. Proc Natl Acad Sci U S A. 2015;112:4411–4416. doi: 10.1073/pnas.1421365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujiwara N, Inoue J, Kawano T, Tanimoto K, Kozaki K, Inazawa J. miR-634 Activates the Mitochondrial Apoptosis Pathway and Enhances Chemotherapy-Induced Cytotoxicity. Cancer Res. 2015;75:3890–3901. doi: 10.1158/0008-5472.CAN-15-0257. [DOI] [PubMed] [Google Scholar]
- 30.He J, Yu JJ, Xu Q, Wang L, Zheng JZ, Liu LZ, Jiang BH. Downregulation of ATG14 by EGR1-MIR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy. Autophagy. 2015;11:373–384. doi: 10.1080/15548627.2015.1009781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merrill RA, Strack S. Mitochondria: a kinase anchoring protein 1, a signaling platform for mitochondrial form and function. Int J Biochem Cell Biol. 2014;48:92–96. doi: 10.1016/j.biocel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sirohi K, Chalasani ML, Sudhakar C, Kumari A, Radha V, Swarup G. M98K-OPTN induces transferrin receptor degradation and RAB12-mediated autophagic death in retinal ganglion cells. Autophagy. 2013;9:510–527. doi: 10.4161/auto.23458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riz I, Hawley TS, Hawley RG. KLF4-SQSTM1/p62-associated prosurvival autophagy contributes to carfilzomib resistance in multiple myeloma models. Oncotarget. 2015;6:14814–14831. doi: 10.18632/oncotarget.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsui T, Fukuda M. Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino-acid transporter PAT4. EMBO Rep. 2013;14:450–457. doi: 10.1038/embor.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X, Fraser M, Moll UM, Basak A, Tsang BK. Akt-mediated cisplatin resistance in ovarian cancer: modulation of p53 action on caspase-dependent mitochondrial death pathway. Cancer Res. 2006;66:3126–3136. doi: 10.1158/0008-5472.CAN-05-0425. [DOI] [PubMed] [Google Scholar]
- 38.Kong B, Tsuyoshi H, Orisaka M, Shieh DB, Yoshida Y, Tsang BK. Mitochondrial dynamics regulating chemoresistance in gynecological cancers. Ann N Y Acad Sci. 2015;1350:1–16. doi: 10.1111/nyas.12883. [DOI] [PubMed] [Google Scholar]
- 39.Kim H, Scimia MC, Wilkinson D, Trelles RD, Wood MR, Bowtell D, Dillin A, Mercola M, Ronai ZA. Fine-tuning of Drp1/Fis1 availability by AKAP121/Siah2 regulates mitochondrial adaptation to hypoxia. Mol Cell. 2011;44:532–544. doi: 10.1016/j.molcel.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cardone L, Carlucci A, Affaitati A, Livigni A, DeCristofaro T, Garbi C, Varrone S, Ullrich A, Gottesman ME, Avvedimento EV, Feliciello A. Mitochondrial AKAP121 binds and targets protein tyrosine phosphatase D1, a novel positive regulator of src signaling. Mol Cell Biol. 2004;24:4613–4626. doi: 10.1128/MCB.24.11.4613-4626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livigni A, Scorziello A, Agnese S, Adornetto A, Carlucci A, Garbi C, Castaldo I, Annunziato L, Avvedimento EV, Feliciello A. Mitochondrial AKAP121 links cAMP and src signaling to oxidative metabolism. Mol Biol Cell. 2006;17:263–271. doi: 10.1091/mbc.E05-09-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu L, Gu C, Zhong D, Shi L, Kong Y, Zhou Z, Liu S. Induction of autophagy counteracts the anticancer effect of cisplatin in human esophageal cancer cells with acquired drug resistance. Cancer Lett. 2014;355:34–45. doi: 10.1016/j.canlet.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 43.Huang Y, Zhou J, Luo S, Wang Y, He J, Luo P, Chen Z, Liu T, Tan X, Ou J, Miao H, Liang H, Shi C. Identification of a fluorescent small-molecule enhancer for therapeutic autophagy in colorectal cancer by targeting mitochondrial protein translocase TIM44. Gut. 2016 doi: 10.1136/gutjnl-2016-311909. [DOI] [PubMed] [Google Scholar]
- 44.Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21:348–358. doi: 10.1038/cdd.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida T, Kobayashi T, Itoda M, Muto T, Miyaguchi K, Mogushi K, Shoji S, Shimokawa K, Slida, Uetake H, Ishikawa T, Sugihara K, Mizushima H, Tanaka H. Clinical omics analysis of colorectal cancer incorporating copy number aberrations and gene expression data. Cancer Inform. 2010;9:147–161. doi: 10.4137/cin.s3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thirstrup K, Dachsel JC, Oppermann FS, Williamson DS, Smith GP, Fog K, Christensen KV. Selective LRRK2 kinase inhibition reduces phosphorylation of endogenous Rab10 and Rab12 in human peripheral mononuclear blood cells. Sci Rep. 2017;7:10300. doi: 10.1038/s41598-017-10501-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlucci A, Adornetto A, Scorziello A, Viggiano D, Foca M, Cuomo O, Annunziato L, Gottesman M, Feliciello A. Proteolysis of AKAP121 regulates mitochondrial activity during cellular hypoxia and brain ischaemia. EMBO J. 2008;27:1073–1084. doi: 10.1038/emboj.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ranganathan G, Phan D, Pokrovskaya ID, McEwen JE, Li C, Kern PA. The translational regulation of lipoprotein lipase by epinephrine involves an RNA binding complex including the catalytic subunit of protein kinase A. J Biol Chem. 2002;277:43281–43287. doi: 10.1074/jbc.M202560200. [DOI] [PubMed] [Google Scholar]
- 49.Schroeder B, Schulze RJ, Weller SG, Sletten AC, Casey CA, McNiven MA. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology. 2015;61:1896–1907. doi: 10.1002/hep.27667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.